Abstract
Coffee breeding and improvement efforts can be greatly facilitated by availability of a large repository of simple sequence repeats (SSRs) based microsatellite markers, which provides efficiency and high-resolution in genetic analyses. This study was aimed to improve SSR availability in coffee by developing new genic−/genomic-SSR markers using in-silico bioinformatics and streptavidin-biotin based enrichment approach, respectively. The expressed sequence tag (EST) based genic microsatellite markers (EST-SSRs) were developed using the publicly available dataset of 13,175 unigene ESTs, which showed a distribution of 1 SSR/3.4 kb of coffee transcriptome. Genomic SSRs, on the other hand, were developed from an SSR-enriched small-insert partial genomic library of robusta coffee. In total, 69 new SSRs (44 EST-SSRs and 25 genomic SSRs) were developed and validated as suitable genetic markers. Diversity analysis of selected coffee genotypes revealed these to be highly informative in terms of allelic diversity and PIC values, and eighteen of these markers (∼27%) could be mapped on a robusta linkage map. Notably, the markers described here also revealed a very high cross-species transferability. In addition to the validated markers, we have also designed primer pairs for 270 putative EST-SSRs, which are expected to provide another ca. 200 useful genetic markers considering the high success rate (88%) of marker conversion of similar pairs tested/validated in this study.
Introduction
Coffee tree belongs to the genus Coffea, comprising two main cultivated species C. arabica L. (2n = 4x = 44) and C. canephora Pierre ex A. Froehner (diploid, 2n = 2x = 22), yielding arabica and robusta type of coffees, respectively. Arabica coffee is known for excellent cup quality but suffers from a narrow genetic base due to its domestication history and susceptibility to diseases and pests. In contrast, robusta coffee though poor in quality has better adaptability to various stresses. To keep pace with the environment and also of the sensibilities of market, there is a continuous need for genetic improvement of coffee, which unfortunately is severely constrained owing to inherently slow pace of tree breeding using conventional methods, and variety of other reasons [1], [2]. The situation demands development of new, easy, practical technologies that can provide acceleration, reliability and directionality to the breeding efforts, as well as characterization of cultivated/secondary gene pool for proper utilization of the available germplasm in coffee genetic improvement programs. In this context, DNA polymorphism based genetic markers becomes important that have proven to be of immense value in characterization and genetic improvement of plant germplasm resources.
Among different types of DNA markers, microsatellites or SSR markers are the most ideal for studying genetic diversity, population structure, phylogenetic relationships, construction of frame-work linkage maps, QTL interval mapping, marker-assisted selection (MAS), etc., thereby aiding in genetic improvement of crop plants [1]. In the last few years a number of efforts have lead to development of a few hundred SSR markers in coffee [2]–[12], but these are insufficient to realize the full potential of markers for mapping/linkage studies in coffee, more so in arabicas which have an extremely narrow genetic base. Moreover, most of the described markers are poorly validated, especially for their utility in cultivated genepool comprising arabicas and robusta coffee. The situation thus calls for newer efforts to generate additional validated markers for them being of any gainful utility in marker-based genetic studies/coffee breeding.
With advancements in genomic studies, there has been an huge burst in the EST sequences in the public domain that provide an easy and economic/cost-effective opportunity to identify and develop EST based SSR markers, which have the additional advantage of assessing the functionally effective genetic diversity [13], [14], and also have very high cross-species transferability [8], [11]. In this study, we have used the coffee EST database containing 13,175 unigene [15] to identify SSRs in the expressed part of coffee genome, and use the same to develop novel coffee-specific EST-SSRs for use as efficient genetic markers. Thus we describe here 44 new validated genic-SSRs, and another set of 270 putative similar markers that need further validation. In addition, we also describe 25 new genomic SSR markers that were developed using an affinity capture approach based SSR-enriched partial, small-fragment genomic library.
Materials and Methods
Plant material and DNA extraction
The plant material used for the validation of SSR markers comprised a set of 16 elite coffee genotypes belonging to C. arabica (tetraploid arabicas) and C. canephora (diploid robustas) and 14 related wild species belonging to Coffea and Psilanthus [2] that were available in the Coffee Germplasm Bank maintained at Central Coffee Research Institute, Balehonnur, Karnataka, India. The fresh leaf samples collectedfrom each genotype were used for DNA isolation as described by Aggarwal et al. [16]. The DNA isolated from robusta variety CxR was used for constructing SSR enriched small-insert genomic library.
Microsatellite screening of coffee transcriptome, identification of SSRs and marker development
An EST database of robusta coffee comprising 13,175 unigene ESTs [15] was downloaded from ftp site (ftp://ftp.sgn.cornell.edu/coffee/) maintained by Sol Genomics Network (SGN, http://www.sgn.cornell.edu/coffee.pl). The database was used for: (i) identification and localization of SSRs using microsatellite search module MISA (MIcroSAtellite, http://www.pgrc.ipk-gatersleben.de/misa), and the criteria being- a minimum repeat core of 12 bp, considering the base complementarities and a minimum distance of 50 bp between two SSRs; (ii) selecting the ‘usable/candidate SSRs’ for marker development, being those that carried a minimum of 18-bp long repeat core (nine repeat units of DNRs, six of TNRs, five of TtNRs, four of PNRs, or three of HNRs) (iii) designing of primer pairs for the selected usable SSR sequences using PRIMER 3 tool embedded in MISA and/or GENETOOL Lite version 1.0 (http://www.biotools.com/downloads/brochures/GeneTool2.pdf); and (iv) standardizing PCR conditions followed by validation of working primer pairs for genetic studies as described earlier [2].
Construction of an SSR-enriched small-insert genomic library/development of genomic SSRs
A partial genomic DNA library enriched for microsatellite repeats was constructed using the methods described earlier [17]. Briefly, the method involved: one-step restriction digestion of genomic DNA with Hae III enzyme (NEB) and ligation of resulting fragments with ds Mlu-I adaptor (Mlu-F: CTC TTG CTT ACG CGT GGA CTA and Mlu-R: pTAG TCC ACG CGT AAG CAA GAG CAC) [18]; amplification of the restricted-ligated DNA pool using Mlu-F primer; SSR enrichment of the amplified DNA pool using liquid phase hybridisation (in 6X SSC) with streptavidin coated paramagnetic beads (Dynal) attached with biotinylated equimolar pool of four oligos (CA)15, (GA)15, (GAA)15 and (CAA)15. This was followed by amplification of the hybridized/trapped genomic DNA fragments by PCR and construction of partial genomic library in TA vector (Invitrogen) as per the manufacturer’s instructions. A number of positive (white) recombinant clones were randomly picked up from the library, amplified and sequenced for both the strands using M13 universal primers on ABI 3730 DNA Analyzer (Applied Biosystems, USA). The sequences were aligned and edited using Autoassembler (Applied Biosystems, USA). The SSR-positive sequences were identified and used for development of new genomic SSRs as described earlier [2].
The amplified PCR products generated using all the new SSRs were resolved using capillary-based ABI 3730 DNA Analyzer and were precisely sized for major, comparable and conspicuous peaks using GeneMapper 3.7 (Applied Biosystems), using default parameters.
Statistical, genetic and diversity analysis
The data for EST-SSRs and genomic SSRs were analyzed separately for various genetic parameters, viz., mean, standard deviation, expected heterozygosity (He), Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium, (LD), using Arlequin ver 3.1 [19], polymorphism information content (PIC) and private alleles (Pas) using Convert ver. 1.3.1 [20]. Cross-taxa transferability (Tmark) was calculated over 15 species (except C. canephora) as proportion of primers showing successful amplification vis-à-vis all the tested primers whereas primer conservance (Ctaxa) was calculated as proportion of the species displaying successful amplification vis-à-vis all the tested markers.
Genetic diversity analysis to infer generic relatedness/affinities was performed over informative Pms (polymorphic markers) for cultivated genotypes/related species using MicroSatellite Analyzer [21] with Nei’s genetic distance [22]. The genetic distance matrices were used to construct Neighbour Joining (NJ) consensus tree using Phylip ver 3.6 [23], which was viewed using Treeview ver 1.6.6 [24].
We also attempted mapping of the new markers on a robusta linkage map using JoinMap ver 4.0 [25] as described earlier [2], [26] using group LOD score of 5.0.
Results
In this study, we undertook in-silico analysis of a robusta coffee transcriptome to identify and develope coffee-specific EST-SSR markers. Simultaneously, we also attempted development of genomic-SSRs by constructing a small-insert SSR enriched partial genomic DNA library. The new markers (44 EST-SSRs and 25 genomic-SSRs) were validated for their utility in genetic studies using panels of elite coffee genotypes, and related taxa of coffee for cross-species transferability. The details of these new markers viz., locus designation, primer sequences, repeat motifs, amplification temperature, amplicon size, and SGN ID or Genbank accession numbers are given in Tables 1 and 2. Details of additional primer pairs for 270 putative EST-SSRs, which were designed but need to be validated have been provided in Table 3.
Table 1. Details of the new EST-SSR markers developed using EST database and validated using diverse coffee genotypes in the present study.
| Sl. No. | Marker Id | Primer sequence | Repeat motif | Allele size (bp)* | Source EST ID** | Linkage group |
| 1 | CCESSR01 | F: TGGTAGCACTGTCGGAAGCATAT | (AGC)7 | 239 | 119516 | – |
| R: GACCCATCTAACTTGCTGCATTTT | ||||||
| 2 | CCESSR02 | F: AAGATATGTTTTAGCCCAAGTAGTGAC | (AT)11 | 168 | 119534 | – |
| R: ATTGGTTGGTACTGTTTAGCTGTTCAT | ||||||
| 3 | CCESSR03 | F: CAGCCGTATCAGCACCAGCAT | (TC)10 | 217 | 119559 | CLG02 |
| R: TTCCCAACCCGTCAAAGTCCT | ||||||
| 4 | CCESSR04 | F: GTCGACAACAGCGCTCAAGATC | (TCT)6 | 259 | 119613 | – |
| R: CAAAAAAGACTGGAAAGAGGGTATTAG | ||||||
| 5 | CCESSR05 | F: AGGGGCTGGTTATTTTTTGGG | (GCA)6 | 141 | 119723 | – |
| R: GGGGGTAAATACGGGAAAGCAGA | ||||||
| 6 | CCESSR06 | F: AACCCTCCCTCCTCCACCTTTTC | (AC)23 | 224 | 119781 | CLG09 |
| R: GGAGGTGGTGGTGGTGTAAAAAAAG | ||||||
| 7 | CCESSR07 | F: CCCCCTTCCCATTTTTTCCCCT | (AGC)6 | 194 | 119864 | – |
| R: CCGGGTGGTAGAGAGATTGCTGCA | ||||||
| 8 | CCESSR08 | F: AAACCACAACATCCCCCAAGAGT | (GCA)8 | 149 | 120008 | – |
| R: GGCAGTAGAATTGGAGCGGTGAAA | ||||||
| 9 | CCESSR09 | F: CCCCCACCCACTTCTCTTTG | (TTC)8 | 184 | 120113 | – |
| R: ACAACAAACGAACGCTCTCTGATAA | ||||||
| 10 | CCESSR10 | F: GCAGAAGAAGCACCAGTAGCAGAAGAAG | (GCT)7 | 207 | 120120 | – |
| R: TGCCTTCTACTCTTCACTCTTCTCCACT | ||||||
| 11 | CCESSR11 | F: AGGGAAAAAGAAGAGTGAGAAGAATATT | (GAA)6 | 140 | 120217 | – |
| R: GGGACACTCACATATACTGCAAACTTAG | ||||||
| 12 | CCESSR12 | F: CCGCCATCCCTTTTGCCTTTC | (CCA)6 | 200 | 120227 | CLG02 |
| R: ACGGCAGCAGAAGTGGAGGTGTT | ||||||
| 13 | CCESSR13 | F: GCGGGGTAGTTTTGGGAATATGG | (TTA)6-tt-(TTC)6 | 124 | 120252 | CLG02 |
| R: TTTGGGGTCCTTTTTCTTTCACACAT | ||||||
| 14 | CCESSR14 | F: CTTGCCCCCTTCCCTCCCACTC | (CT)10 | 232 | 120329 | – |
| R: TTCGGCTCCTTGTGTTTGGGTA | ||||||
| 15 | CCESSR16 | F: AGAGCAATGAGAAACAAACGAAACT | (CAT)6 | 173 | 120439 | – |
| R: AGGTGCCCAACTATCCCAGAAT | ||||||
| 16 | CCESSR17 | F: CTCCACACCAACAAAATCCCACTT | (CAG)6 | 169 | 120475 | – |
| R: CCCACATCCTGAGTCTGCTGCTAA | ||||||
| 17 | CCESSR18 | F: GGGGAGGATGCTTATGATATGAGG | (CAC)6 | 152 | 120498 | – |
| R: TCCGGTTCACCTGCTTTTCCTT | ||||||
| 18 | CCESSR19 | F: CATCGTATCTCGCCCTCTCTCTTTC | (CA)13 | 219 | 120514 | – |
| R: CCACAACAAGTACAACCAACCGAAAC | ||||||
| 19 | CCESSR20 | F: TTCTGGCCGATTGATTGTGAT | (AAG)7 | 143 | 120538 | – |
| R: GCGACAAGGCTGACAAACTACTAC | ||||||
| 20 | CCESSR21 | F: CGAGCTAGTGCAGACAGATTGAGAT | (AG)17 | 164 | 120568 | CLG13 |
| R: GTCCTTGGCGAAATCCCTCAG | ||||||
| 21 | CCESSR22 | F: CCCTCAATCTCGTCCCCCTCT | (TC)9 | 153 | 120823 | – |
| R: CCCTCCATAAATCTTCTTCACGTACTC | ||||||
| 22 | CCESSR23 | F: GGCCTCTCTTTAATTTTCTTGTCTTTTTTC | (TTC)8 | 160 | 120860 | – |
| R: ATGGAGGGTAGGGTTTCGAGAGTGA | ||||||
| 23 | CCESSR26 | F: AACCGGCCTTCTTGTATGATTCTCTA | (CAT)8 | 206 | 121392 | CLG07 |
| R: TTGGCTAACCCTCACTCTCTCCCTACTA | ||||||
| 24 | CCESSR27 | F: GCCCACTCCATTCGTACTTGTTTCC | (CT)12 | 120 | 121464 | – |
| R: GCGGTGCTGCTCAATGCTCAT | ||||||
| 25 | CCESSR28 | F: AAAATGAGTGACGATGGGAAAGACA | (CCA)7 | 193 | 121482 | – |
| R: GAGGGAAGCCGATCACTGGTTG | ||||||
| 26 | CCESSR29 | F: GGCGCTAGAGTTGGTTGTTTGC | (CTTCT)5 | 95 | 121548 | – |
| R: CAGGCATTGGAACCAGCGAAC | ||||||
| 27 | CCESSR31 | F: AGAAGAGTACTGAAGGCCTGGAAGA | (GTG)6 | 220 | 121671 | – |
| R: AGCATCTGCAGCCTCCATAGC | ||||||
| 28 | CCESSR32 | F: CTTGGCGTTTAGCGTTCTCACATT | (TA)9 | 163 | 121811 | – |
| R: GCTCAACCAAACCAATACATACCTCTT | ||||||
| 29 | CCESSR33 | F: GCCCGCATGGACGACTTGGA | (AGG)7 | 227 | 121876 | CLG13 |
| R: CGCTTGACGTATCCTTTGGCCTCT | ||||||
| 30 | CCESSR34 | F: GCATTGCTCCCCCCACTTCA | (CTC)6 | 168 | 121905 | – |
| R: GAGCATGGGGACGAGGAGGA | ||||||
| 31 | CCESSR35 | F: CTGCTAATGCTGCTGAAAAAGAGATACC | (AG)10 | 107 | 121994 | CLG13 |
| R: GGCTGTGAATTCTTGTGACTTGTGACT | ||||||
| 32 | CCESSR36 | F: AGCCTTCTGCAATTCCCTCGTACA | (AG)9 | 100 | 122089 | – |
| R: GGCGTCGTAGAGGGCATTCAGA | ||||||
| 33 | CCESSR38 | F: GCCCGAGGGTTAGATTGATCA | (AG)12 | 162 | 122114 | – |
| R: CTTGTCTTCTGTTTGATTTTGTGTTCTA | ||||||
| 34 | CCESSR39 | F: GCGACCGGACGACCAAAAATAAT | (GAA)7 | 131 | 122147 | – |
| R: CGCCGTCGTCAGAGTCATAATAATCA | ||||||
| 35 | CCESSR40 | F: CGTGGGGGTTTGTTTTTCTCG | (AGG)7 | 205 | 122194 | – |
| R: GTCCCCCCTCAGCCGTTTTTG | ||||||
| 36 | CCESSR41 | F: GGGCTGCAGGCTTGTCACCAC | (GA)10 | 205 | 122322 | – |
| R: AATCGGTTTAGTTTTTTGTTTCCTCAC | ||||||
| 37 | CCESSR42 | F: CGGGCGGAAACGGTCAGATC | (GCA)7 | 117 | 122295 | – |
| R: TGCCGTTGTTGTTGTCCAGGTG | ||||||
| 38 | CCESSR43 | F: CCCAGCAAGAACTCAACCCCATCA | (TTC)8 | 172 | 122653 | CLG02 |
| R: TGGCCTAATGAAGATGACGTTGCTGATG | ||||||
| 39 | CCESSR44 | F: AGGAATAATGGAGGAGACGTTGTTG | (CTT)7 | 236 | 122680 | CLG02 |
| R: GCACAAATCCCAGTACTTCCTCATAGA | ||||||
| 40 | CCESSR45 | F: AAATGGCCGAGATAGAGAAGGAGAAG | (AGG)7 | 135 | 122764 | – |
| R: CCCACTCCTCCGCGGTACTGATC | ||||||
| 41 | CCESSR47 | F: GCAGCAACAATCACTTCCACAGC | (GCA)6 | 198 | 122922 | – |
| R: TGCTGTTGTAACTGCGGGATTTG | ||||||
| 42 | CCESSR48 | F: GCAACCTTATCTAGATTCAACTTCAACTT | (AAATCA)5 | 188 | 122975 | – |
| R: CGGGAAGAAATGGCAGCCTATAC | ||||||
| 43 | CCESSR49 | F: GCGGCCATCCTTGTCTTCG | (ATC)8 | 186 | 122978 | CLG02 |
| R: TAGCCGCTGACGTAATCTTCCTT | ||||||
| 44 | CCESSR50 | F: GGGATGATGTGGATTCTATGGTCTACTA | (CAG)7 | 108 | 123181 | CLG10 |
| R: ATGCCATTTTAACACTTCCTCCTCA |
CCESSR: CCMB Coffee EST SSR marker;
F: forward primer;
R: reverse primer;
–: Unmapped;
CLG: Combined Linkage Group [1];
*: Predicted amplicon size based on source EST sequence;
**: Source EST ID as per the downloaded SGN database (ftp://ftp.sgn.cornell.edu/coffee/).
Table 2. Details of the new genomic SSR markers developed using streptavidin-avidin affinity capture SSR-enriched library in the present study.
| Sl. No. | Primer Id | Primer sequence | Repeat motif | Ta (°C) | Allele size (bp)* | GeneBank acc. No. | Linkage group |
| 1 | CCRM02 | F: AATGGTGGCAGTCCTGAAAGATC | (GA)12 | 57 | 268 | KM874369 | – |
| R: AACATCAACTTTCCTGGTCTTC | |||||||
| 2 | CCRM06 | F: TTCTTATCACCTTGGGCTACCTTTCTTC | (AG)8 | 57 | 146 | KM874370 | – |
| R: AAGCGGTTTAGTTTTTTGTTTCCTCAC | |||||||
| 3 | CCRM07 | F: TAAAGGATGGTATATGTGGCTGGAGTA | (AT)8 | 57 | 126 | KM874371 | CLG01 |
| R: CCACAGCCTCGGCATTTACTATATAT | |||||||
| 4 | CCRM10 | F: AAAAAGACAAGATTCAACCTGCAGTAGT | (GT)9 | 57 | 104 | KM874372 | – |
| R: TTCCCACCCCCCAAAAAAAA | |||||||
| 5 | CCRM14 | F: ATTTGATTTCTTCTTTCTCTGTTGTC | (CT)22 | 55 | 130 | KM874373 | – |
| R: ACAAAAGCCCTGAAAATAATAGATCTA | |||||||
| 6 | CCRM15 | F: CGAAATTGACGAAGCTCTTGTT | (CTT)6 | 57 | 243 | KM874374 | – |
| R: TTGCTAGTTTCGAAATCGTGTAAGGAC | |||||||
| 7 | CCRM16 | F: TCCTATAGCAGAAACACAAAATGACACAG | (TC)26 | 55 | 223 | KM874375 | – |
| R: GGTTTTTGGGTTCTTTTTAGCATATACA | |||||||
| 8 | CCRM17 | F: TAAGCGTTGGAATTCCTCACTCTATCT | (CT)17 | 55 | 228 | KM874376 | CLG01 |
| R: ACAGCTAAAGAAACAATGAACCAGT | |||||||
| 9 | CCRM19 | F: GTTTTTTTTTTCTTTTTTCTTTTTGAGCT | (GA)26 | 57 | 252 | KM874377 | CLG06 |
| R: AAGGCAATGTTGGTCAGCAGTGG | |||||||
| 10 | CCRM21 | F: CACCCCTCCCATCCGTTGAAACAT | (GA)16 | 57 | 258 | KM874378 | CLG06 |
| R: AATGATGCTCCCAGTGTTTGATGA | |||||||
| 11 | CCRM22 | F: CTTGCAGTTTACTTCCCTTTGGTTG | (GA)29 | 57 | 241 | KM874379 | – |
| R: TTTTCTTCTGTATATTGTTGGAGTTCTTC | |||||||
| 12 | CCRM23 | F: CGGCAGTGTGGTCCCCTTTGAAT | (GTT)6 | 57 | 141 | KM874380 | – |
| R: AAAAAAAAACTCACACTCTATCAAAACTAAGG | |||||||
| 13 | CCRM24 | F: GAGTGTGAGTTTTTTTTTGTGACCTTAA | (GA)9.(GA)9 | 57 | 213 | KM874381 | – |
| R: ACCCCACATTCCTCTCATCCATTC | |||||||
| 14 | CCRM28 | F: GGGGCAACAAGTGGTAGGATATGAAGAC | (CA)10 | 57 | 209 | KM874382 | – |
| R: CGCCTTCACTATGGTTTTGCCTTCTAA | |||||||
| 15 | CCRM31 | F: CTTTTATGTCTATCTGTCTCTGCTTTTC | (CA)10 | 57 | 114 | KM874383 | – |
| R: CCTGCAGTAGTTTCACCCTTTATCC | |||||||
| 16 | CCRM33 | F: ACAGCCGTTGAACTTATGGGATTACA | (CT)12 | 57 | 118 | KM874384 | – |
| R: ACAAAGGGATGGAGAGGATGGAATATAC | |||||||
| 17 | CCRM34 | F: CCCCAGAACGAAAGGCAATCAT | (GA)10 | 57 | 165 | KM874385 | CLG05 |
| R: TTGGGACTATTTATACTGGGGAAGAA | |||||||
| 18 | CCRM35 | F: GGGGTTAAATCAGGGGAAAAGTGG | (TG)12 | 57 | 144 | KM874386 | – |
| R: AAGCGAGGGAGAGAGCAGCAGATC | |||||||
| 19 | CCRM36 | F: CCATGGGGCAAAAGGCAAATTCTAT | (CT)18 | 57 | 171 | KM874387 | – |
| R: TCCAGACCGCCGTTCACGAAGTATA | |||||||
| 20 | CCRM37 | F: TGCTTCCCTTCTCATTCTGGTACTTT | (GT)10 | 57 | 146 | KM874388 | – |
| R: AATCCATCAACAACTTCAGCATACCA | |||||||
| 21 | CCRM38 | F: TGAGAATTAAAGCAGCAGGGGTATG | (CA)10 | 57 | 204 | KM874389 | CLG05 |
| R: GCAAAAAAAGGCAAAAGCATTACATC | |||||||
| 22 | CCRM40 | F: ATTCACGCTTTCATTACTTTTCTC | (CT)29 | 57 | 176 | KM874390 | CLG05 |
| R: TTTGTATTTCCTTTCCATTTCTTTTGTA | |||||||
| 23 | CCRM41 | F: AGCAGAAACACAAAATGACACAGAGCA | (CT)24 | 57 | 161 | KM874391 | – |
| R: AATGGTCCAAGGAAAATGAAAAATGTT | |||||||
| 24 | CCRM42 | F: CGGAGAAGAGCAATATACAAGCAAGG | (GA)13 | 55 | 143 | KM874392 | CLG11 |
| R: GCCACCCCAGAACTTTTGCAA | |||||||
| 25 | CCRM45 | F: CTTCAAGCAAAATTTTCAACAGCACAG | (GA)10 | 57 | 187 | KM874393 | – |
| R: GGCCCTTTTTTAGTCTCACCACATT |
CCRM: CCMB CXR Microsatellite marker;
Ta: annealing temperature;
F: forward primer;
R: reverse primer;
–: Unmapped;
CLG: Combined Linkage Group [1];
*: Expected amplicon size in the robusta variety CxR.
Table 3. Details of primer pairs for additional new EST-SSR markers designed in the present study.
| Sl No. | Marker ID | Unigene ID* | SSR repeat unit | Forward Primer | Predicted Tm (°C)forward primer | Reverse Primer | Predicted Tm (°C)reverse primer | Predictedamplicon size (bp) |
| 1 | CCESSR51 | 119463 | (AGAGCA)5 | AGCAGAGATCGAGACAGAGAGA | 59 | ACATATCTGAAACCCTCGGC | 59 | 477 |
| 2 | CCESSR52 | 119606 | (CT)9 | AATCGGAGGATTTGTGCTTC | 59 | GCGCCTAAATCACCCATATT | 59 | 390 |
| 3 | CCESSR53 | 119736 | (GA)11 | CCGCGGTCAGTCTTACTACA | 59 | ACACAAATCAACACCCATCC | 58 | 266 |
| 4 | CCESSR54 | 119787 | (GGA)6 | CGCAATCTTGAATGAGGAGA | 59 | TGGGAGGTTGATCATCTGAA | 59 | 341 |
| 5 | CCESSR55 | 119820 | (CT)11 | AGGACAGGAGTGTGATCCCT | 59 | GCCATGTCCTCCTTTCGTAT | 59 | 376 |
| 6 | CCESSR56 | 119897 | (TC)11 | AGGTTTTTGTGTCCCTTTGG | 59 | CATCGATGAAAAGCAGCAGT | 59 | 230 |
| 7 | CCESSR57 | 119919 | (AT)15 | GAATTTTGTCAGCCAGAGCA | 59 | GACGGAAAGATTCTGGCTTT | 58 | 184 |
| 8 | CCESSR58 | 119987 | (TCC)7 | AGCTACGCTAGGCAATTGGT | 59 | GACAACAACAACAGCCAACA | 58 | 296 |
| 9 | CCESSR59 | 120008 | (TC)12 | GGAACAAGACTCTCTTGCCA | 58 | TCATCACACAAGGAGGCAAT | 59 | 418 |
| 10 | CCESSR60 | 120008 | (TG)9 | ATTGCCTCCTTGTGTGATGA | 59 | CCGGTCGATCAACAATCTTA | 59 | 198 |
| 11 | CCESSR61 | 120045 | (TCT)6 | GGTGAAGGGTCCTTACCTGA | 59 | GAGATGTGCTACTGGCTACTGC | 59 | 232 |
| 12 | CCESSR62 | 120064 | (GATT)5 | CTCTTGTTTCCAACCCAACC | 59 | CGGACACTGTGAGGAGAGAA | 59 | 320 |
| 13 | CCESSR63 | 120107 | (TCT)6 | AGTCCAGTCCAGTCCTGTCC | 59 | CCGATATGATTTTGGTGCTG | 59 | 432 |
| 14 | CCESSR64 | 120121 | (GCT)7 | CGAAGTTGTGCAGGATGAAC | 59 | GGAGCTGCTTGCTCTTCTTT | 59 | 241 |
| 15 | CCESSR65 | 120179 | (AT)10 | GTGATGCTCGGTGTATCTGG | 59 | ACTAGAGGCCGAGAATTGGA | 59 | 203 |
| 16 | CCESSR66 | 120206 | (AGA)10 | GATGAGCTCCAAAACAAGCA | 59 | AAAACTTCCCAGGCTTCAGA | 59 | 447 |
| 17 | CCESSR67 | 120260 | (CTCC)5 | GTCGTCTGTTCCTCCTCGAC | 60 | CCACTAATCCCGAGCAAAAT | 59 | 273 |
| 18 | CCESSR68 | 120291 | (AAG)8 | CCACGCGAATAATCATCAAC | 59 | AAGCACCTTATCCCCAACAG | 59 | 469 |
| 19 | CCESSR69 | 120316 | (TCA)6 | TTGGAAAACCATAGAAGGGC | 59 | ACCCTCATCAATCTCTTGCC | 59 | 408 |
| 20 | CCESSR70 | 120320 | (GCCACC)4 | TTAAATCAGCCCTCAAGCCT | 59 | CTTGAAATCTTGCGCACTGT | 59 | 343 |
| 21 | CCESSR71 | 120322 | (TTC)6 | TTAGAAAAGCTGCGAGACGA | 59 | TTGACCATTTCCCCTTCTTC | 59 | 342 |
| 22 | CCESSR72 | 120543 | (AT)9 | GATTGCTCTCTTTCTTCGGG | 59 | TCCGCCCCAGTTAGATTTAT | 59 | 382 |
| 23 | CCESSR73 | 120545 | (AT)9 | GTTTTCCGGCTAGCTTGTTC | 59 | GTGCATGAGGTGAAAATGGA | 60 | 214 |
| 24 | CCESSR74 | 120579 | (CCT)7 | TGTAATATTGCTTCGCTCGG | 59 | AAGGGATGATGCCTAGTTGG | 59 | 485 |
| 25 | CCESSR75 | 120596 | (TCC)7 | GGCTCAATCGATAAGCAACA | 59 | CATGAACATATCCTGGAGCG | 59 | 429 |
| 26 | CCESSR76 | 120656 | (AAC)6 | AAGCGAAGATGGAGAGCAAT | 59 | CCGCCTTTTGTAGAAGAACC | 59 | 316 |
| 27 | CCESSR77 | 120656 | (TCA)6 | TTTTAGGCCAATCCTTCACC | 59 | GTATGTGAGGGCGTAACGTG | 59 | 454 |
| 28 | CCESSR78 | 120720 | (GAA)6 | GAGCCCTCTTTTTCCTCCTT | 59 | CCGTAATGATACAAACCCCC | 59 | 326 |
| 29 | CCESSR79 | 120764 | (CATATA)4–13 bp-(AT)9 | AGCAGCACCAGACTAGCTCA | 59 | GTCCCAGAAACGAAGATGGT | 59 | 399 |
| 30 | CCESSR80 | 120821 | (ACAGG)4 | CAGAATCCCATATTCCCCAC | 59 | TTGCTGTCTGATTTCCAAGC | 59 | 324 |
| 31 | CCESSR81 | 120883 | (TCAAT)4 | CAGATTCCGGTGTCACAAAG | 59 | TTTGCTGCAGTGTCACTTGT | 58 | 107 |
| 32 | CCESSR82 | 120965 | (TCGAAA)4 | GACTATGGATGGCTTTGCCT | 59 | GGCGGATTGGTTGATAGAAT | 59 | 397 |
| 33 | CCESSR83 | 121010 | (CCTGCA)4 | TGAATCAACTCCTGCTCCTG | 59 | GGTAGTTCCAGCCACCAGAT | 59 | 115 |
| 34 | CCESSR84 | 121086 | (TC)11 | GGATCAAACGTGGCTAAGGT | 59 | AAGAAAAGGGCTACAACGGA | 59 | 203 |
| 35 | CCESSR85 | 121131 | (AAG)6 | CCCCGGGCTGCAGAAACAAGT | 60 | AGCTCCGGTAAGCCTCAATA | 59 | 389 |
| 36 | CCESSR86 | 121240 | (GCA)6 | AACCCTCCACTCCATTTCAG | 59 | ACTATTGTTGCTGCTGCTGG | 59 | 347 |
| 37 | CCESSR87 | 121289 | (TTC)6 | GAAGCAACCCCTCAACTGAT | 59 | AGCACCCTCTGCTTCAATCT | 59 | 345 |
| 38 | CCESSR88 | 121377 | (GT)9 | CACGCGGATAATACACCG | 59 | AGTTGCTCCTGCTTCTCCAT | 59 | 191 |
| 39 | CCESSR89 | 121392 | (GCT)6 | GCACTGTTTTGAATGGTTGG | 59 | TCCGCACTACAAGTACCGTC | 59 | 159 |
| 40 | CCESSR90 | 121439 | (GATTA)4 | TACTTGAGCGATCAGAACCG | 59 | TAATCCTGCGTGCTATTTGG | 59 | 455 |
| 41 | CCESSR91 | 121548 | (CTTCT)5 | AGGCGCTAGAGTTGGTTGTT | 59 | CAAAGTAAGCAGCAGGCATT | 58 | 108 |
| 42 | CCESSR92 | 121580 | (TC)11 | CCTACATCCCACGACCTTCT | 59 | GTGGTAGTGCTTTGGTGGTG | 59 | 361 |
| 43 | CCESSR93 | 121610 | (AT)9 | CTGCCCTATGATGAATGGTG | 59 | TCATCTCAAGCATCGTCTCC | 59 | 157 |
| 44 | CCESSR94 | 121752 | (TGCTCC)4 | CCTCTCATGCCAGCAACTAA | 59 | AAAGCACTGGGAACTGAAGC | 59 | 258 |
| 45 | CCESSR95 | 121841 | (CT)10 | TGGAGCAGAGATTGTCAAGG | 59 | CCAGCTGGAACTTCCCTG | 59 | 349 |
| 46 | CCESSR96 | 122004 | (AT)19 | ACATTCGAAAACTTGGGGAG | 59 | CAAGGTCTTGGGTTTGTCCT | 59 | 232 |
| 47 | CCESSR97 | 122006 | (AG)14 | CTCGTGCTGTAACCCTCTCA | 59 | AGTGTGATGGAACGCGAATA | 59 | 439 |
| 48 | CCESSR98 | 122012 | (AGC)6 | GACGCGCAGTCTTTCAAGTA | 59 | GCTTCACAGTCGTCTTCCAA | 59 | 373 |
| 49 | CCESSR99 | 122077 | (CGC)6 | TCGGACTGCTATAGTGGACG | 59 | TCCTTCGAGACGTTAGCCTT | 59 | 215 |
| 50 | CCESSR100 | 122089 | (AG)9 | TTCTGCAATTCCCTCGTACA | 59 | AGGGGTCAGAAGAAGGGATT | 59 | 214 |
| 51 | CCESSR101 | 122184 | (AT)10 | AGGGGTGGATTCAGTAGACG | 59 | ATGCAATTGAACCGACTCAG | 59 | 466 |
| 52 | CCESSR102 | 122195 | (AGGCTC)6 | AGTCAGCTTCTTGCTGCTCA | 59 | CCCTCCAGAGTACCCATTGT | 59 | 336 |
| 53 | CCESSR103 | 122254 | (TCGT)5 | GGAATGAGCTAAAAGCCCAG | 59 | TTACCGTGTAGGTTGGTGGA | 59 | 318 |
| 54 | CCESSR104 | 122257 | (TTCA)5 | GCTCAGTGAGGTTCCAAACA | 59 | ACCCTGTAGGTTGGTGGAGA | 59 | 310 |
| 55 | CCESSR105 | 122324 | (AGA)6 | AGGCCATAGTGGTGAACACA | 59 | ACAAAGCACAGAACCAACCA | 59 | 319 |
| 56 | CCESSR106 | 122340 | (TC)10 | AGCTGATTGAGGATGGCTCT | 59 | CATTGGCTTTCCCCAATACT | 59 | 484 |
| 57 | CCESSR107 | 122383 | (CTT)8 | CCTTCTCCCTCTTCCCTTCT | 59 | TGAGTGACAGCGTCAACAAA | 59 | 459 |
| 58 | CCESSR108 | 122387 | (CTT)7 | GTGGGAAATCTCAAATGGCT | 59 | CTCTTTCTGCTGGGTTGTGA | 59 | 143 |
| 59 | CCESSR109 | 122514 | (AGG)6 | GAAAGACCCCAAACCAAGAA | 59 | GGAGTCATCAACAATGGCAC | 59 | 340 |
| 60 | CCESSR110 | 122533 | (TA)10 | GTGCCGTTGTTTGTTTCATC | 59 | GTGCATGAGTGGATTCAAGG | 59 | 324 |
| 61 | CCESSR111 | 122550 | (TCA)6 | CTTCATGGACCATTCTCACG | 59 | TAGCCGAAACAGAGCATTTG | 59 | 290 |
| 62 | CCESSR112 | 122619 | (GAA)6 | TCCCCTGAATTGGACTCTTC | 59 | ACAAAGGCCAACGTTTTACC | 59 | 452 |
| 63 | CCESSR113 | 122646 | (AGC)6 | GCATCCTTGTATTCGGAGGT | 59 | ACCACTCGCTTCAACCTCTT | 59 | 115 |
| 64 | CCESSR114 | 122681 | (AAAG)5 | CCAGGCTAAGTGCTCATCAA | 59 | GGCAGCGCAGTAAAGTCATA | 59 | 209 |
| 65 | CCESSR115 | 122758 | (TC)13 | CTCCCATTTCCTCTTTCTCT | 55 | TAGGGTTTTGAAGGGCAATC | 59 | 274 |
| 66 | CCESSR116 | 122793 | (TC)18 | GAGAAGTTTGCCAGAACCCT | 58 | ATCAATCTTCAAAGGGCCAC | 59 | 453 |
| 67 | CCESSR117 | 122797 | (CT)12 | CTCGTCCCTCTCCCTGTACT | 58 | ACCACAACAGCGAACATCAT | 59 | 160 |
| 68 | CCESSR118 | 122811 | (AAG)7 | CTTTCCTCAGCTTCACCACA | 59 | CCCCTGTTTCTGGTCTTGTT | 59 | 371 |
| 69 | CCESSR119 | 122842 | (ATA)6 | TGCAAGTGTGATGAATGTGG | 59 | CTAAGGCGAGAAAACAAGGG | 59 | 431 |
| 70 | CCESSR120 | 122880 | (GAAGCA)4 | TAGGGGGACCAATAGCAAAG | 59 | ACACGTCTTGCGACAAAGTC | 59 | 212 |
| 71 | CCESSR121 | 122996 | (TTA)6 | CCGGACAGGAACAAGAAAAT | 59 | CATTCAATTGCCTGACCATC | 59 | 443 |
| 72 | CCESSR122 | 123010 | (TCC)6–30 bp-(TCA)7 | GTCTGCATCTGCTTGCTCAT | 59 | AAACCATCTATCCCAGCCAG | 59 | 438 |
| 73 | CCESSR123 | 123023 | (CTGCCT)4 | GACAGAGGAATCCTTGCCAT | 59 | TCCCAGAAAAATCACCTTCC | 59 | 494 |
| 74 | CCESSR124 | 123110 | (AACTA)4 | GAGAAGCGAACCACACTTGA | 59 | ACTTGAGCAGCAACCCTTTT | 59 | 183 |
| 75 | CCESSR125 | 123132 | (CTATGA)4 | GAGCGCTATGGATGAGCATA | 59 | GCTGTACAGGATTGGGAGGT | 59 | 478 |
| 76 | CCESSR126 | 123134 | (CTG)6 | ACCACCACAAGCACAACAAT | 59 | TGACCCCCAACAAAACAAAG | 61 | 457 |
| 77 | CCESSR127 | 123160 | (TA)11 | CTGATGGGCGTCATCAATAC | 59 | GGGAAAGGGAGTCTCAACAA | 59 | 394 |
| 78 | CCESSR128 | 123185 | (ACG)6 | GAGGAAATTCGGGAAGTTGA | 59 | CATCGTCATCATCATCGTCA | 59 | 280 |
| 79 | CCESSR129 | 123240 | (TTC)6 | CCAGAACCAACTAGGGTTTT | 56 | TCACTTGAATTGCAGAAGGC | 59 | 498 |
| 80 | CCESSR130 | 123248 | (GCA)6 | TCTTCTACAGCTGCCACCAC | 59 | ATTGCGAAGAATGAAGGGTC | 59 | 137 |
| 81 | CCESSR131 | 123291 | (CT)10 | CTTCTCCAAACGGACCAAAC | 60 | CTCAGCCTCCATTGCACTAA | 59 | 172 |
| 82 | CCESSR132 | 123309 | (AT)20 | CGTAAATTAGGGGCGTTGTT | 59 | TTGGCACAAACCTGATGGTA | 60 | 227 |
| 83 | CCESSR133 | 123323 | (ACAT)5 | TCTCCCCATGGTACTTCACA | 59 | CCCATGCATTTCCAGACATA | 59 | 463 |
| 84 | CCESSR134 | 123329 | (TC)13 | TCACTGCCTGAGGCTTTATG | 59 | GAGCATGGTCCCGATAAGAT | 59 | 380 |
| 85 | CCESSR135 | 123332 | (AG)10 | TTGTTCTCTCCAGGGGAGTC | 59 | CGTAGAACCAATCCAGCAGA | 59 | 210 |
| 86 | CCESSR136 | 123342 | (AC)9 | AATATGAATGGAGGCGAAGG | 59 | ACAATGTCCCGAACTGTTGA | 59 | 347 |
| 87 | CCESSR137 | 123362 | (AAT)6 | GGTCTGAAAGTGGCCAAAAT | 59 | GGGAGATACGCTCAAGGTGT | 59 | 496 |
| 88 | CCESSR138 | 123381 | (CGG)6 | AAACCTAGTGGTGGAGGTGG | 59 | CAGCTTGGTTCCAGTCTCAA | 59 | 240 |
| 89 | CCESSR139 | 123445 | (TC)9 | AATCTTGTGGAGGAAATGCC | 59 | GCTTTTTGCTTCGGTACCTC | 59 | 349 |
| 90 | CCESSR140 | 123562 | (CT)9 | CGAGGCTCTATCCTCCTCTC | 58 | CTGACGCATCTGACCTCACT | 59 | 182 |
| 91 | CCESSR141 | 123682 | (CTC)7–10 bp-(TTC)7 | AGTACCTCGACAACCCCAAC | 59 | AGGCTCTGCCAAATCAAAGT | 59 | 479 |
| 92 | CCESSR142 | 123741 | (CAGCAA)4 | GAAGTTTGAAGCCTGAAGCC | 59 | TTCCGCGTATCTGAATCTTG | 59 | 478 |
| 93 | CCESSR143 | 123742 | (TTGTA)4 | AACCGATTTGATTCAGAGCC | 59 | CTCCACTCCACTGGGTTTTT | 59 | 294 |
| 94 | CCESSR144 | 123743 | (TTGTA)4 | AACCGATTTGATTCAGAGCC | 59 | AAAAAGGTCAAACGTCAGCC | 59 | 269 |
| 95 | CCESSR145 | 123795 | (GCG)8 | AGGCTTGAAGACGAAATGCT | 59 | GCTCTTTCCAGTCGATCTCC | 59 | 309 |
| 96 | CCESSR146 | 123812 | (ATCTTT)4 | ACACTCGACCCTCAAGTTCC | 59 | AGTGATCATCCATCCATCCA | 59 | 482 |
| 97 | CCESSR147 | 123819 | (CTTC)5 | ACGAGGGCAAAACAAAGAAG | 59 | TCAAGACGAAGAAATGCAGG | 59 | 112 |
| 98 | CCESSR148 | 123822 | (GCT)6 | ATCCGCGAAGCTACTTGTTT | 59 | ACGAATTTCTGGTCCACTCC | 59 | 133 |
| 99 | CCESSR149 | 123903 | (CCG)6 | GGCTCTGGAGGCTTTGAATA | 59 | GGCGAGCATGATTAGACAGA | 59 | 256 |
| 100 | CCESSR150 | 123909 | (CT)14 | CTTCCAGGTTCCTTGTCCTC | 59 | CCATCAGCACTTCCATCATC | 59 | 389 |
| 101 | CCESSR151 | 123967 | (TTTC)5 | CTCTTCCCTACTTTGCTGCC | 59 | AGGGTTTCTGTCGCTGTTCT | 59 | 374 |
| 102 | CCESSR152 | 123993 | (TG)10 | GGCCTAGTACTCCCAATCCA | 59 | ATGATGCAGTTGATCCTCCA | 59 | 251 |
| 103 | CCESSR153 | 124161 | (CT)12 | CTCGGCTCCCAAATATTCAT | 59 | ACATGCGGTTATGCACTCAT | 59 | 453 |
| 104 | CCESSR154 | 124171 | (CCA)6 | ACCACCACCTCCAGAGAATC | 59 | CGTAGAAATTGCTATCCGCA | 59 | 421 |
| 105 | CCESSR155 | 124195 | (AGC)6 | ATCCCCATCAGAAGACCTCA | 59 | GAAGCTCGACAAACAGGACA | 59 | 296 |
| 106 | CCESSR156 | 124198 | (GAACT)4 | CCCAGAAGAAATCCCAGTGT | 59 | GACCAATCGAGGCCAATAAC | 59 | 240 |
| 107 | CCESSR157 | 124268 | (TGC)6 | TTTCTCTCACACGCCAAAAG | 59 | GCTGATGATGCGATCAAAGT | 59 | 406 |
| 108 | CCESSR158 | 124269 | (CCT)7 | AGTCGTCTTCTGGTGGCTTT | 59 | GAGGAGGAGGTGGTGGTG | 59 | 195 |
| 109 | CCESSR159 | 124309 | (AAAGG)4 | TTCGAGTTCTATGGCTCGTG | 59 | TGGTGGTATGGAAAACAGGA | 59 | 345 |
| 110 | CCESSR160 | 124355 | (CAAAA)4 | TCGCCTTTTCTCTCACTTCA | 59 | GCTCGTTTCCTCTTGACTCC | 59 | 359 |
| 111 | CCESSR161 | 124358 | (ACA)6 | ATGAGGATGAGGAGGGATTG | 59 | TCCCTCAAGCTGTCTTTCCT | 59 | 407 |
| 112 | CCESSR162 | 124358 | (AAG)6 | AGCTTCCTCCAGAACCTCAA | 59 | ATCATCATCAGGGCCTCCT | 59 | 147 |
| 113 | CCESSR163 | 124365 | (AAC)6 | TTGCTCTTCCACTTTCATCG | 59 | CTTCACGAAATGCCTCAGAA | 59 | 171 |
| 114 | CCESSR164 | 124426 | (TTTTTA)5 | GACGTGGAGGGATCAGTTTT | 59 | CAATTCCGACGTTTCATCAG | 59 | 140 |
| 115 | CCESSR165 | 124428 | (TTTTTA)4 | GACGTGGAGGGATCAGTTTT | 59 | TGAATATGACCCGCATGAGT | 59 | 239 |
| 116 | CCESSR166 | 124436 | (GAG)6 | CGAGTACCATTGGGATGATG | 59 | ACTGTCATTTGAGTGCTCCG | 59 | 304 |
| 117 | CCESSR167 | 124457 | (AC)9 | CAAAGAAAAACCGCTCTCG | 59 | AGTGGAGGATTTGGAAAACG | 59 | 367 |
| 118 | CCESSR168 | 124461 | (AATA)6 | CTCTGCCCACTTTCTCCTTC | 59 | CACTTGGGCATCTGAGAAGA | 59 | 354 |
| 119 | CCESSR169 | 124465 | (TTCA)6 | TCCCCCATTCATTTGGTAGT | 59 | GCAAGGAGACCTTCAAGAGG | 59 | 296 |
| 120 | CCESSR170 | 124468 | (TTCCTC)4–24 bp-(ATC)6 | TCTCCCATTCATTCCAGGAT | 59 | GTAAGTGCGGATGATGATGG | 59 | 342 |
| 121 | CCESSR171 | 124565 | (ATT)6 | GCTCTGTTCAGGAAGGGAAG | 59 | GTATCAGAATCCCCACCCAA | 60 | 406 |
| 122 | CCESSR172 | 124577 | (AAG)6 | TGGCTTTTCTCCGTTATCCT | 59 | TTCTTCTTGGTGCTTGATCG | 59 | 293 |
| 123 | CCESSR173 | 124612 | (CGG)6 | TCCTTTCCTAGCCTTCTGGA | 59 | AGAGAAAAAGAGCGCGAGAG | 59 | 331 |
| 124 | CCESSR174 | 124697 | (GAC)6 | GAAAATCGAGAAAGGGTGGA | 59 | GAGTGGTCCTCAAGTCCCAT | 59 | 242 |
| 125 | CCESSR175 | 124700 | (GCT)6 | ACACCAACGACGGGTAAGAT | 59 | CAGCCAGATTCGAATACCCT | 59 | 346 |
| 126 | CCESSR176 | 124759 | (AG)11 | GGGATTGCTGCATAGGAAAT | 59 | CATCTCAATCAAATGCCCAC | 59 | 290 |
| 127 | CCESSR177 | 124765 | (CCA)7a(AAC)6 | CTGCGACTCAGACATCCACT | 59 | ACGCTGTCTCCTCCGTAACT | 59 | 444 |
| 128 | CCESSR178 | 124767 | (TCAT)5 | GCCGGCTAGTCTATACGGAG | 59 | ACCTTGAAGTTTGGTGGAGC | 59 | 369 |
| 129 | CCESSR179 | 124817 | (TC)9 | TCATTGCTGCTTCAGTCTGTC | 59 | TCTCTTCAAAGCGGATTCCT | 59 | 354 |
| 130 | CCESSR180 | 124851 | (TTC)11 | GACTTCTCTCTCCCCCTTCC | 59 | TTGGAGCGTAATGTCGTAGG | 59 | 422 |
| 131 | CCESSR181 | 124871 | (AGC)6 | ACCAATTTCAAACCTCCAGC | 59 | CGAACACCACATGCATTACA | 59 | 475 |
| 132 | CCESSR182 | 124949 | (CCT)7 | CCCCTTAAAATCGATTCCCT | 59 | CGGAGAGGAATTTTCGACAT | 59 | 328 |
| 133 | CCESSR183 | 124969 | (AATTG)4 | TCTAGCTTGAGGGGATCGTT | 59 | TAAGCCTAAGCAAAGCAGCA | 59 | 243 |
| 134 | CCESSR184 | 124995 | (GAGAAG)4 | CACCAAAGCCATGAAAGCTA | 59 | TCCCAAATCTCCTCTCCATC | 59 | 224 |
| 135 | CCESSR185 | 125025 | (TC)11 | CCTCCTTAAAACCCTAAACCG | 59 | TCTAGTATTTTGGTGGGGGC | 59 | 302 |
| 136 | CCESSR186 | 125054 | (TA)9 | TCTTGTCTTGCACATTTCCC | 59 | GGTCTCCGTTGTTGAGGATT | 59 | 283 |
| 137 | CCESSR187 | 125099 | (ACC)6 | TTCCCGCTTTACCAGATACC | 59 | TTCCTCCAACAGACAACAGC | 59 | 388 |
| 138 | CCESSR188 | 125102 | (ACC)6 | TTCCCGCTTTACCAGATACC | 59 | AAACATAAGGTTGGGGTGGA | 59 | 281 |
| 139 | CCESSR189 | 125104 | (AT)11 | CTGGCTCTGACATGCAATTT | 59 | TTCAGAAGCGCGTAGTCTGT | 59 | 387 |
| 140 | CCESSR190 | 125128 | (TC)12 | TGCCACTTTCTTCTCCTCCT | 59 | CTGGAAAAGGTGAAAAGGGA | 59 | 418 |
| 141 | CCESSR191 | 125139 | (CAGCGG)4 | AGCACATCCAACACCACAGT | 59 | AACCCAACGTTAAGGGACAC | 59 | 204 |
| 142 | CCESSR192 | 125151 | (ACA)8 | ATTCCGAATCGGGTACAGAG | 59 | AATGTACATCCCTTCCCCAC | 59 | 400 |
| 143 | CCESSR193 | 125186 | (AAG)6 | GCCTCAACCACTTGCCTATT | 59 | CGTGATCATGATGCCCTAAC | 59 | 372 |
| 144 | CCESSR194 | 125232 | (AT)13 | ATTCAGTTGCAGCTGTGGAG | 59 | ATGATCTGGGAAAGGACAGG | 59 | 314 |
| 145 | CCESSR195 | 125265 | (GT)14 | TGATGATAGCTCCGCTTTTG | 59 | AGCCAATCACCAGCAAGATT | 60 | 254 |
| 146 | CCESSR196 | 125286 | (CTC)8 | GGGTCCATGTTCGTCTTTTT | 59 | GAGACCAAGCATTGAAGCAA | 59 | 304 |
| 147 | CCESSR197 | 125289 | (ATTTT)5 | GAGGATCACTTTTGCCCCT | 59 | GAAAAAGGTCAAGACATCCC | 56 | 470 |
| 148 | CCESSR198 | 125382 | (AC)16 | CTCTCCCCATCCCTCAACT | 59 | GTGAAGGAGGTGGTGGTCTT | 59 | 365 |
| 149 | CCESSR199 | 125416 | (AT)16 | ATCATCCCCTCGATAGCAAC | 59 | TCCCGAAAGGAAAGAATGAG | 59 | 183 |
| 150 | CCESSR200 | 125444 | (TG)11 | TGGTTGTAGGTGGTCGTCAT | 59 | GCTCGGGGATATCGATCTTA | 59 | 495 |
| 151 | CCESSR201 | 125484 | (CTCCGA)4 | AGGGTTTTTCGGAGATGACA | 60 | CCGGGAAAACTAAGAGCAAG | 59 | 424 |
| 152 | CCESSR202 | 125529 | (TC)9 | GTGCTCGTGCAATTGAAACT | 59 | ATGCCGAGTGGATGTCTATG | 59 | 490 |
| 153 | CCESSR203 | 125676 | (AAACAA)4 | TGATCTTGATCCCCTCATCA | 59 | GATGCATTCACAAAACCGTC | 59 | 393 |
| 154 | CCESSR204 | 125734 | (GAG)6 | TCCTTGGCTCGAGATCTTTT | 59 | TTTGTGTCCTCTTCTCGCAC | 59 | 434 |
| 155 | CCESSR205 | 125776 | (CT)14 | AGGATTGCCTTCCGTTTGTA | 60 | GATTCAACTCGTGGGACCTT | 59 | 225 |
| 156 | CCESSR206 | 125893 | (CAT)7 | CCCCTTGTTTCTCTTTGTGG | 60 | TGCATCATCCCTTGTATCGT | 59 | 271 |
| 157 | CCESSR207 | 126139 | (GGC)6 | TCCGTTCCTCTTTCTCGTTT | 59 | CACAGCACCCATGGTAACAA | 60 | 411 |
| 158 | CCESSR208 | 126219 | (GAG)6 | TGGTCTTCAATCACCAAGGA | 59 | GACACATTGCACGTAGTCCC | 59 | 134 |
| 159 | CCESSR209 | 126243 | (CAC)8 | GGTGGACGAGGCTTTTATGT | 59 | CTGTGAGCAAACTTGACGGT | 59 | 180 |
| 160 | CCESSR210 | 126250 | (CAC)9 | GCCAAAATCCCTGTTCTCAT | 59 | TGTGGATGCACCAGATTCTT | 59 | 245 |
| 161 | CCESSR211 | 126255 | (GAA)6 | ACGAAAGGGTTGAAGGTGAC | 59 | GACAGAGACGACGAAGACCA | 59 | 412 |
| 162 | CCESSR212 | 126427 | (ATC)7 | TCCAATGGTTTACAGGAGCA | 59 | TCTCCGGTGTAAAACTGCTG | 59 | 341 |
| 163 | CCESSR213 | 126427 | (CAGGAG)6 | GGCGGCTTCGGTATTATTTA | 59 | GCACTAGTTTCTTGGACTGGG | 59 | 184 |
| 164 | CCESSR214 | 126506 | (TTTC)6 | TTCATTGTCTTTGAGCCAGG | 59 | CCCCATCACTCATTCCTTCT | 59 | 339 |
| 165 | CCESSR215 | 126540 | (AAG)9 | AAATCGAATGGCTTGGAGAC | 59 | GTGCAAGTAAATCCGAGCAA | 59 | 309 |
| 166 | CCESSR216 | 126661 | (AGT)6 | TTTGTCATTTCTCTCGCTGC | 59 | AGTTGGGGAGAATTGACCTG | 59 | 440 |
| 167 | CCESSR217 | 126701 | (TTCA)5 | AAAGGATACGGCTCACAAGG | 59 | GAATGAATGAACGAGGGGTT | 59 | 124 |
| 168 | CCESSR218 | 126701 | (TTCA)6 | AAGGAGCCAATCGTTCATTC | 59 | TTACCCTGTAGGTTGGTGGAG | 59 | 175 |
| 169 | CCESSR219 | 126730 | (CT)10 | GAGTTGGCTTGAAAGCATGA | 59 | CCATCCCCAAGGAAAGTGTA | 60 | 371 |
| 170 | CCESSR220 | 126767 | (GA)9 | GCACAAGACCTTCCCTTTGT | 59 | CTCTTGCACGTCGTTTTGTT | 59 | 428 |
| 171 | CCESSR221 | 126881 | (GCA)6 | CGGTGCTGCAGTTTCACTAC | 60 | CAAGGCAGCAAGTATACCGA | 59 | 351 |
| 172 | CCESSR222 | 126910 | (AAT)8 | CCAACAGAAAAGTTGGGGAC | 59 | TTAGCCGAATCGTTATTCCC | 59 | 163 |
| 173 | CCESSR223 | 127055 | (TTC)6 | GAAATCCCACTCGGTGAAGT | 59 | CCCACCCCGAAGATATAAAA | 59 | 401 |
| 174 | CCESSR224 | 127171 | (AAGG)5 | ATTTCAGTCTGGGTCTTCCC | 58 | TGAGGCAGACCTTGTTCTTG | 59 | 470 |
| 175 | CCESSR225 | 127376 | (GT)10 | TCCCAGAACAAAACGAGACA | 59 | CTTGTCGTAAACGGCTTCAA | 59 | 270 |
| 176 | CCESSR226 | 127478 | (CT)23 | CAGCCTCCGGTTAGCAAG | 60 | TGTCCAATACCCACTCTTCG | 59 | 179 |
| 177 | CCESSR227 | 127479 | (ACC)10 | GTACCCCCACAACACTCACA | 59 | GATGCTTCTCATCGTCTCCA | 59 | 404 |
| 178 | CCESSR228 | 127712 | (TTTA)5 | GATTTTGTGCCAAAGGGAAG | 60 | GCTTTGACCGGAGAAGTCAT | 59 | 115 |
| 179 | CCESSR229 | 127774 | (CCT)6 | CCGCCGTTAAATCAAACTCT | 59 | CAAAGAAAGCTGCTCACTGC | 59 | 120 |
| 180 | CCESSR230 | 127785 | (AT)13 | GCTGCTGCTTTGCTTGTTCT | 61 | AGCTTCAGAAATGGTGGCAT | 60 | 333 |
| 181 | CCESSR231 | 127800 | (GAA)9 | GCTACAGCGTGCAGAAAAAG | 59 | TACTCCTCAGGCCTCCATTT | 59 | 318 |
| 182 | CCESSR232 | 127828 | (TCA)6 | ACCTGGGAGGACTGATAACG | 59 | CCAGATTCCATACTTGGGCT | 59 | 177 |
| 183 | CCESSR233 | 127910 | (AAAAGG)4 | GAAGACTCTGTAAAGGAACGACC | 58 | GTGTTATGGCTCAACCATGC | 59 | 347 |
| 184 | CCESSR234 | 127938 | (CTG)8 | GCGGCTTAGTTCTATCCTCG | 59 | ATGGCTGACAGCTTCCTCTT | 59 | 442 |
| 185 | CCESSR235 | 127992 | (TAA)6 | AGTGGGAAAAGTGAAGGTGG | 59 | ATTAATCCCCTCCTCCGAGT | 59 | 225 |
| 186 | CCESSR236 | 128005 | (CTC)8 | AGTCAGGTATGCTGCCATTG | 59 | GGAAATCTTTGGGGTCTGAA | 59 | 322 |
| 187 | CCESSR237 | 128130 | (CCT)6 | CGTCAATAAATTGGTGTGGC | 59 | GGGAGAATGGGAGATGAAGA | 59 | 142 |
| 188 | CCESSR238 | 128268 | (TA)11 | CCGTCGTTACTGCAAAAGAA | 59 | ACAGAGGGGTTTCAAACAGG | 59 | 254 |
| 189 | CCESSR239 | 128301 | (AAGAAC)4 | TCCCAAGTCCCTATCTCTGC | 59 | CAGCTGATGGTTTAGGAGCA | 59 | 123 |
| 190 | CCESSR240 | 128306 | (TCT)7 | TCTGTGTCCCTTTTCAGCAG | 59 | GGATGGTCAATGCAGATGAG | 59 | 221 |
| 191 | CCESSR241 | 128309 | (CGG)8 | ATATCATGGATGGTGCTGCC | 61 | TCCAGATTGTGCTGCTTTTC | 59 | 355 |
| 192 | CCESSR242 | 128315 | (TTC)6 | GCACGAGGCTTTCTCTCTTT | 59 | CAGCAGGAAACTGGAGAAGTC | 59 | 260 |
| 193 | CCESSR243 | 128377 | (AG)12 | GGTTCTTCTGTGTGGGTGTG | 59 | GTAGAAGCCAGCTAATGCCC | 59 | 107 |
| 194 | CCESSR244 | 128432 | (TG)15 | CACACACCCAACACACAGAA | 59 | GCGAGAGGAATTTTGACCAG | 60 | 482 |
| 195 | CCESSR245 | 128504 | (GAG)6 | TCCATCCCATATTCCCATCT | 59 | GTCGGGTCTCTTCTTCTTGC | 59 | 454 |
| 196 | CCESSR246 | 128527 | (GA)10 | CACCAACAATTCCCTATCCC | 59 | TCCATCTCCAACACTACCCA | 59 | 427 |
| 197 | CCESSR247 | 128527 | (CTG)6 | CACCAACAATTCCCTATCCC | 59 | TCCATCTCCAACACTACCCA | 59 | 427 |
| 198 | CCESSR248 | 128527 | (CTG)6 | TGGGTAGTGTTGGAGATGGA | 59 | CAATGACCGCCTGTATTCTG | 59 | 260 |
| 199 | CCESSR249 | 128550 | (ATC)6 | CTAGTAGCCATGGTCACCCA | 59 | TTCTTCGTAGGCAGCTCTGA | 59 | 416 |
| 200 | CCESSR250 | 128587 | (TC)11 | GAGGCAAACCAAATGGAGAT | 59 | ATTGAAAAGCCTGCTGAACC | 59 | 358 |
| 201 | CCESSR251 | 128673 | (CT)9 | TTCTCTGCTGCTGCTCATTT | 59 | GGCACACATTTATCCACTCG | 59 | 245 |
| 202 | CCESSR252 | 128894 | (AAAAG)4 | GCCCAAAGCTGTTAGAGACC | 59 | TCCTTGTCATACGCTTGCTC | 59 | 344 |
| 203 | CCESSR253 | 129060 | (TCT)7 | CAATTTGCAGTCAGTCCGTT | 59 | ACATCTGCCACTGTCTGAGC | 59 | 216 |
| 204 | CCESSR254 | 129129 | (GAG)7 | ACAGATGCTTTATGGGAGGG | 59 | GGAAAGGTATTGCTGGGGTA | 59 | 272 |
| 205 | CCESSR255 | 129269 | (GCA)9 | TGAAGGTTCACTGATTGGGA | 59 | TGCTGTTGTAACTGCTGCTG | 59 | 477 |
| 206 | CCESSR256 | 129431 | (AAG)9 | CGCCACCAGTACAGATCAAG | 59 | TCCGCATATCGAACACTCAT | 59 | 496 |
| 207 | CCESSR257 | 129439 | (CA)10 | AGTCACCACACAGATGCTGC | 60 | GGCCATTCAAGGTTTCTTGT | 59 | 296 |
| 208 | CCESSR258 | 129524 | (GAA)8 | CTAATCTTCCACGGCTCTCC | 59 | TTTTGTTCAGCAGCAAGGTC | 59 | 412 |
| 209 | CCESSR259 | 129581 | (AAAAG)4 | CCAACCTCACGTCTCACCTA | 59 | GCCAGTGACACAGACGACTT | 59 | 102 |
| 210 | CCESSR260 | 129646 | (TC)11 | CTTCAGCCACAACAGATGCT | 59 | AAAAGAGCACACAGTGCAGC | 59 | 179 |
| 211 | CCESSR261 | 129668 | (CT)12 | CATGATTATGCGTTGGCTTC | 59 | GGTTGTCATGTTGAATGAGG | 56 | 163 |
| 212 | CCESSR262 | 129745 | (CAC)10 | GCCTCCAGTCAACAAACAAA | 59 | TGGTGCTTCTCTTCCTTGTG | 59 | 252 |
| 213 | CCESSR263 | 129793 | (CA)10 | TAGCGGGGAAAATTGATAGG | 59 | AGGGGCTTTTTCTCCATCTT | 59 | 487 |
| 214 | CCESSR264 | 129797 | (TCC)6 | TCGATGATGGCTACAGCTTC | 59 | TTGCTCCTGAGAAACACTGG | 59 | 312 |
| 215 | CCESSR265 | 129906 | (TA)11 | GACCAGAGAGAGACACCTACTTTT | 57 | CTCTTCCTAGCAGCAGCAGA | 59 | 186 |
| 216 | CCESSR266 | 129913 | (CAG)6 | TTCCTTTAAAACCGTCAGGG | 59 | CTGCATGCTCTGTTGTTCCT | 59 | 264 |
| 217 | CCESSR267 | 129917 | (AAGAGC)5 | GAGATGGAGAGACCATCATCC | 58 | AACTAAGACTTTTGCCGCGT | 59 | 267 |
| 218 | CCESSR268 | 129950 | (TCCACT)4 | TTCCCCATCTAAGGCAAAGT | 59 | CTGATTCAGGGCTCTCCATT | 59 | 403 |
| 219 | CCESSR269 | 129957 | (GAGCAG)4 | AACCCACCTTCCTCTCCTTT | 59 | CATGCATATCGGACACAACA | 59 | 294 |
| 220 | CCESSR270 | 129964 | (GAT)7 | AAGGCTCGACATGTTTTTCC | 59 | CCTACGCAACATCATCAACC | 59 | 221 |
| 221 | CCESSR271 | 129972 | (AG)9 | CTTTCCGCCTTTTCGTACTC | 59 | AACACACGCACAACACACAG | 59 | 392 |
| 222 | CCESSR272 | 130030 | (AG)19 | GGCTGCAAGAACATAGAGCA | 59 | GAAATCCTTGTTGGCTGGTT | 59 | 369 |
| 223 | CCESSR273 | 130066 | (GAG)6 | AAAGAAAGAGCCCTCCACAA | 59 | GCTCCAAAACCACCATTACC | 59 | 212 |
| 224 | CCESSR274 | 130235 | (TGC)6 | GGTTTATGACGTTGTGGACG | 59 | GGCCTATGATGCGGTAGAAT | 59 | 198 |
| 225 | CCESSR275 | 130276 | (GGC)7 | GGCGGCTACTCAGAGACTTC | 59 | TGGAACTCCAATTCCTTTCC | 59 | 300 |
| 226 | CCESSR276 | 130329 | (AT)13 | AGGCTGCGTTCCATATAAGA | 57 | CCCAGAAACTCCATTTCGAT | 59 | 410 |
| 227 | CCESSR277 | 130342 | (ATC)6 | CTCTGCAGAACAACCCTTCA | 59 | CCAACAGCTAAAAGTGCCAA | 59 | 296 |
| 228 | CCESSR278 | 130353 | (CA)11 | TCTCTCTCTCTCCATTCGCA | 59 | GGTACGAGGTTGATCGGAGT | 59 | 154 |
| 229 | CCESSR279 | 130353 | (CAC)6 | ATGGCTCCATCATCTCATCA | 59 | ATTGGTGGGTCCTTCAATTC | 59 | 215 |
| 230 | CCESSR280 | 130368 | (AG)17 | GGCTGTCTTAAGGCCCTAGTT | 59 | ATCAAGTGATGCTGTCGAGC | 59 | 436 |
| 231 | CCESSR281 | 130475 | (CTC)7 | CGCCAACACCCCTTATCTAT | 59 | CATTGAGGAATTGGAACGTG | 59 | 485 |
| 232 | CCESSR282 | 130516 | (CCT)6 | ACCCTGCTCCACCTTATCAC | 59 | GACAGAAACCTTCATCCCGT | 59 | 357 |
| 233 | CCESSR283 | 130575 | (AGA)7 | GTGGTCAGTATGGTTGCTGG | 59 | AGCCACAAAAGAACCATTCC | 59 | 456 |
| 234 | CCESSR284 | 130635 | (AG)13 | AGCACCGTCAGACTCTCCAT | 60 | TAAGCCAAACGTCGTCGTAG | 59 | 283 |
| 235 | CCESSR285 | 130647 | (CAC)6 | GAGGCCGAGTTACGAGAGTC | 59 | AGCATGCCAACCCTTTATTC | 59 | 366 |
| 236 | CCESSR286 | 130785 | (TAAAA)4 | TGCTCGGACCCTCTAAGTTT | 59 | TGGAGTCGTTTAAAAAGGGG | 59 | 334 |
| 237 | CCESSR287 | 130804 | (AG)10 | CGTTACAGAATTTGCGGATG | 59 | CAACTCTAAGCCGTCGATGA | 59 | 360 |
| 238 | CCESSR288 | 130947 | (GCAGAG)4 | TTTTGAACACGTCAAGGCTC | 59 | CGAAGAGGGGACTGAGAAAG | 59 | 273 |
| 239 | CCESSR289 | 130972 | (TCA)9 | GCACAACCCATATGACGAAG | 59 | GGAGGAACAGTTGGAGAATGA | 59 | 107 |
| 240 | CCESSR290 | 130978 | (AG)16 | TTATTATTGCACAGCCCCG | 60 | AACAATCCATTCTTGCTCCC | 59 | 388 |
| 241 | CCESSR291 | 130993 | (TC)14 | CGGTGGGGTTCTGATAATTT | 59 | CGGTTTCAAGCTAACGAACA | 59 | 313 |
| 242 | CCESSR292 | 131127 | (TC)11 | ATCGTGTTTAGCAGTGACGC | 59 | GCAGATATCATATACTCCCGTC | 55 | 143 |
| 243 | CCESSR293 | 131130 | (AAG)6 | CACTCACTCAACACTTCGCA | 59 | CCATCTTCCACCACCTTTCT | 59 | 426 |
| 244 | CCESSR294 | 131306 | (TCGCA)4 | CTTTGGGGGCTGAGAGTAAG | 59 | GCTCCATATTGCAACGTCTT | 58 | 432 |
| 245 | CCESSR295 | 131334 | (CCA)7a(AAC)6 | TAATACATCCACTCCGCCAA | 59 | TACTCATCAACAACCTCGCC | 59 | 320 |
| 246 | CCESSR296 | 131479 | (TG)9 | GCAATTGTGGATGGTCTGAG | 59 | AAACACACTGGGAAAGAGCA | 58 | 424 |
| 247 | CCESSR297 | 131560 | (ACC)6 | CAATCTGTTCCCACGTATGC | 59 | ACAAGTGCTCCAACAAACCA | 59 | 328 |
| 248 | CCESSR298 | 131583 | (AT)12 | CCACACACTGCAGATTGGTT | 60 | AAGATCTCTGGGGCATCAAC | 59 | 453 |
| 249 | CCESSR299 | 131633 | (CTT)6 | AGGTTCGAGCCAGGAAGATA | 59 | TTGCCTTGGTTGATGACTTC | 59 | 141 |
| 250 | CCESSR300 | 131669 | (AG)12 | TCCGGGAAGGATAGTGGTAG | 59 | TTGCTGAAACAGTCTCCCTC | 58 | 203 |
| 251 | CCESSR301 | 131714 | (GA)22 | ATTAGTGCGGAATTCCCTTG | 59 | TGATAATCTCGAGTGACCGC | 59 | 491 |
| 252 | CCESSR302 | 131742 | (TA)18 | TAATGCACGCACAAACACAC | 59 | ATCATTGCAAATCTCCTCCC | 59 | 112 |
| 253 | CCESSR303 | 131761 | (CA)10 | CATGGATTGGAATTCTGCTG | 59 | CAGATTAGCTCCGCAATCAA | 59 | 454 |
| 254 | CCESSR304 | 131806 | (ATG)8 | GCACCTAAGAGAGGAGTGCC | 59 | ATGAGTTTGGCTGGATGTGA | 59 | 401 |
| 255 | CCESSR305 | 131831 | (GCCTGC)4 | CCGATTTCGCTCGTAGTGTA | 59 | TATCAATACGGACTGCTGGG | 59 | 462 |
| 256 | CCESSR306 | 131899 | (GAT)9 | TCCGCTGGTCACAAGAAAT | 59 | ATCAAACTTCGACACCCCTC | 59 | 184 |
| 257 | CCESSR307 | 131901 | (AAC)6 | AAACTTTGATATCCCGGCTG | 59 | GCCGCCTTTTGTAGAAGAAC | 59 | 175 |
| 258 | CCESSR308 | 131958 | (TCA)9 | GATTCCAGTATTTCGGCTCC | 59 | TTGCTCAGATTAGGCTGTCG | 59 | 211 |
| 259 | CCESSR309 | 131966 | (AC)10 | TTTGAACTTGCTGGAGCATC | 59 | TGGTGAACTTCTGTTTTCCC | 58 | 392 |
| 260 | CCESSR310 | 132036 | (AAG)6 | CTCCTCAACACTCCTGACCA | 59 | CCCTCCTGAAGCTGCTTATC | 59 | 194 |
| 261 | CCESSR311 | 132205 | (GT)10 | CCTTGATTGTCACGTGTATGC | 59 | AAGATATGCCCAGGTCATGC | 60 | 424 |
| 262 | CCESSR312 | 132207 | (TCGT)7 | AAAGGATACGGCTCACAAGG | 59 | ACCCTGTAGGTTGGTGGAGA | 59 | 198 |
| 263 | CCESSR313 | 132325 | (AAG)8 | TGAGCACTCAAGGGAAATTG | 59 | TTCGTCCAGGGAGTAGCTTT | 59 | 462 |
| 264 | CCESSR314 | 132326 | (AAG)6 | GCTTTTGTCCAAGGGATGAT | 59 | GGAGCTCCCAGGAGGTAGTA | 58 | 336 |
| 265 | CCESSR315 | 132398 | (TA)11 | ACGTTTATTACGGCGGATTC | 59 | ATCGCCGTCCCATAACTATT | 58 | 174 |
| 266 | CCESSR316 | 132466 | (AGA)6 | TCTGTCTGGGCAACACTATAC | 55 | AGAAAGGGAGGTGGAGGAAT | 59 | 338 |
| 267 | CCESSR317 | 132484 | (CT)9 | TTTGCAGTTATGACTTGGGC | 59 | TCCTCACTGGAAAGGGATTC | 59 | 449 |
| 268 | CCESSR318 | 132502 | (GTCTT)4 | GGGGATCAGCGTAAGAATGT | 59 | TTTCCACCCACGTAATAGCA | 59 | 346 |
| 269 | CCESSR319 | 132516 | (TCA)6 | CCGTCCACGTTCACATCTAC | 59 | AGTAGCGCGTAGTGACCTGA | 59 | 130 |
| 270 | CCESSR320 | 132606 | (AGC)6 | GACCAGAAGAACAGCGATGA | 59 | CTCCTTCTTCTTCTCGGCAG | 59 | 285 |
*: Unigene ID as per downloaded from the SGN ftp site ftp://ftp.sgn.cornell.edu/coffee/.
Types, Frequency and Distribution of SSRs in the coffee transcriptome
The coffee EST unigene database analyzed here, comprised 13,175 unigenes having a total length of 8923 kb and an average lengthof ca. 677 bases/unigene [15]. These ESTs were found to contain a total of 2,589 SSRs (having a minimum numbers of repeats as: six for DNRs, four for TNRs and three for all other HO-NRs) located in 2,028 unigenes. The identified 2,589 SSRs comprised- 502 DNRs, 1285 TNRs, 503 TtNRs, 144 PNRs and 155 HNRs, which differed significantly in their relative distribution and abundance accross the unigene ESTs (Tables S1 & S2). The mean length of repeat iterations (RI) for all the SSRs was 5.2, whereas average length of DNRs was maximum (9.6 RI) followed by TNRs (4.6 RI). Among the individual SSRs, AC had the maximum average repeat length of 10.5 RI, followed by AG (8.6 RI), AT (8.3 RI), CG (6.3 RI); all the TNRs had RI in the range of 4.3 to 4.8, whereas all other larger SSRs had an average RI of less the three. The identified SSRs having a repeat core of 18 bp or more were selected as candidate ‘usable’ SSRs for further primer designing/marker conversion. Overall, in-silico analysis of the EST unigenes revealed one EST-SSR (having a minimum repeat core of 12 bp) per 3.4 kb and one usable SSR (having a minimum repeat core of 18 bp) per 15.9 kb of robusta transcriptome (Table S2). Among the individual SSRs, the most abundant EST-SSR motif was AG, followed by AAG.
Development of microsatellite markers from usable EST-SSRs
Only 483 (18.7%) of the total 2589 identified SSRs had a repeat core>18 bp, which were used for marker conversion. Primer pairs could be designed for 320 of these SSRs, of which randomly chosen 50 pairs were further tested for validation studies. These included SSRs with DNRs (30%), TNRs (64%), PNRs, HNRs (2% each) and complex SSRs (see Table 1 for marker ID, primer sequences, repeat motifs, amplicon size, sequence ID and functional identity). Of the selected 50 primer pairs, 44 could be successfully amplified as single locus SSR marker, indicating 88% primer to marker conversion ratio. Considering this high conversion ratio, another ca. 200 useful genetic markers are expected from the remaining 270 putative EST-SSRs (Primer IDs: CCESSR51 to CCESSR320) that are lsited in Table 3.
Identification and development of genomic SSRs using affinity capture
Sequencing of randomly chosen 66 recombinant clones from the small-insert SSR-enriched robusta genomic library (prepared in this study), revealed 81 potential SSRs distributed in 62 sequences. A redundancy analysis of these sequences indicated presence of a total of 60 non-redundant sequences of which 56 were SSR +ive (93.3% of non-redundant sequences) containing 72 non-redundant SSRs. Non-redundant dataset contained 10 sequences with more than one SSR either in compound formation or separated by>50 bp distance. The non-redundant SSRs contained 56.9% AG, 33.3% AC, 2.8% AT, 2.8% AAC, 1.4% AAG and 2.7% A/T repeat motifs (Table S3).
From the 56 SSR+ive sequences, a total of 41 primer pairs could be designed successfully (with five pairs containing two SSRs each). Of these, 25 pairs (encompassing 28 SSRs) resulted in robust PCR amplifications (Table 2), and all of them could further be validated as single locus markers indicating ∼61% primer to marker conversion ratio.
Validation of EST- SSRs for use in genetic studies
All the new 44 EST-SSRs resulted in good amplicons exhibiting low to medium allelic diversity when tested on a panel of 16 elite robusta and arabica genotypes (Figure 1). Overall, a maximum of six and seven alleles (NA) with an average of 2.1 and 3 alleles/SSR were obtained for the tested markers of which 65.9% and 81.8% were polymorphic/informative for tetraploids and robusta genotypes, respectively (Table 4). Fifteen markers in the case of tetraploids and eight for robustas were found to be monomorphic. Moreover, 14 markers resulted in double alleles (i.e. consistent presence of two allelic amplicons across the tested samples) indicative of duplicated loci in case of all the tested tetraploid arabicas. In general, no private alleles were evident except in one robusta genotype (Sln274) for marker CCESSR14.
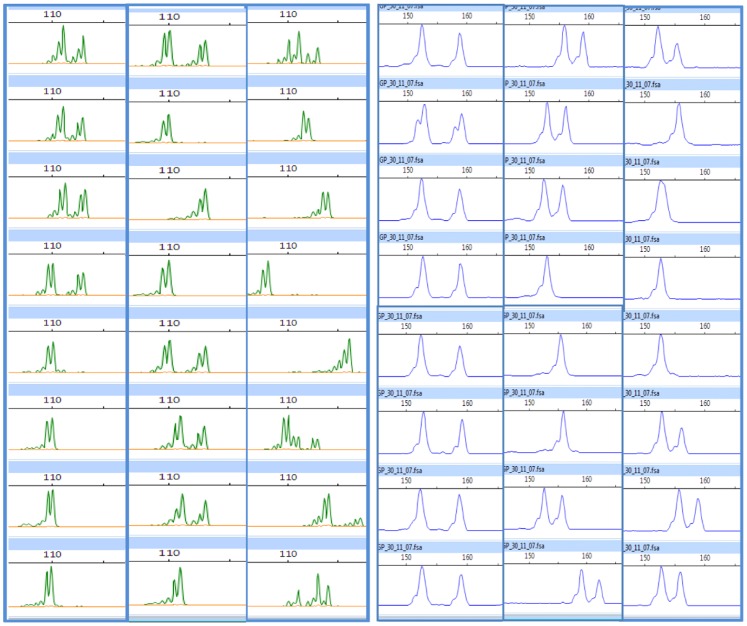
Figure 1. Representated Gene Scan profiles showing the SSR alleles obtained using the new SSR markers for some of the coffee genotypes tested in the study for marker validation.
The right side set of 3 panels is for the genomic-SSR CCRM-33, and the similar set on the left is for genic-SSR CCESSR05. The 3 panels in each set represent 8 genotypes each of arabica and robusta coffee, and 8 of the related coffee species, respectively, from right to left end.
Table 4. Allelic diversity attributes of the newly developed 44 EST-SSRs when tested over cultivated and wild related coffee genera.
| Species: | C. arabica (n = 8) | C. canephora (n = 8) | Coffea spp. (n = 12) | Psilanthus spp. (n = 2) | ||||||||||||||
| Primer ID | NA | PA | Allele range (in bp) | Ho | He | PIC | NA | PA | Allele range (in bp) | Ho | He | PIC | NA | PA | Allele range (in bp) | NA | PA | Allele range (in bp) |
| CCESSR01 | 2 | 0 | 251–254 | 0.00 | 0.50** | 0.36 | 2 | 0 | 251–254 | 0.50 | 0.40 | 0.30 | 7 | 3 | 246–260 | 4 | 0 | 246–281 |
| CCESSR02 | 5 | 0 | 181–209 | 0.33 | 0.67* | 0.57 | 5 | 0 | 183–207 | 0.38 | 0.71* | 0.62 | 10 | 0 | 178–209 | 1 | 0 | 183 |
| CCESSR03 | 6 | 0 | 236–260 | DL | 5 | 0 | 232–244 | 1.00 | 0.78 | 0.68 | 12 | 1 | 232–260 | 3 | 0 | 226–242 | ||
| CCESSR04 | 2 | 0 | 252–255 | 0.00 | 0.50** | 0.36 | 2 | 0 | 252–255 | 0.50 | 0.40 | 0.30 | 6 | 2 | 247–261 | 2 | 0 | 247–252 |
| CCESSR05 | 2 | 0 | 153–159 | DL | 4 | 0 | 153–162 | 0.63 | 0.68 | 0.61 | 4 | 0 | 153–162 | 2 | 0 | 154–159 | ||
| CCESSR06 | 1 | 0 | 223 | MM | 6 | 0 | 225–242 | 0.88 | 0.72 | 0.63 | 17 | 5 | 105–243 | 2 | 0 | 225–227 | ||
| CCESSR07 | 2 | 0 | 211–133 | 0.38 | 0.33 | 0.26 | 2 | 0 | 211–233 | 0.50 | 0.40 | 0.30 | 8 | 4 | 211–239 | 4 | 4 | 220–240 |
| CCESSR08 | 2 | 0 | 151–167 | DL | 2 | 0 | 164–167 | 0.50 | 0.40 | 0.30 | 7 | 1 | 151–176 | 2 | 0 | 161–167 | ||
| CCESSR09 | 3 | 0 | 138–202 | 0.00 | 0.62** | 0.50 | 5 | 0 | 129–212 | 0.50 | 0.76* | 0.67 | 10 | 0 | 129–212 | 1 | 0 | 188 |
| CCESSR10 | 3 | 0 | 158–235 | 0.83 | 0.68** | 0.55 | 2 | 0 | 221–138 | 1.00 | 0.53* | 0.38 | 13 | 3 | 158–247 | 3 | 0 | 212–227 |
| CCESSR11 | 3 | 0 | 147–155 | DL | 3 | 0 | 147–155 | 0.38 | 0.54 | 0.43 | 4 | 0 | 147–160 | 1 | 0 | 147 | ||
| CCESSR12 | 1 | 0 | 218 | MM | 1 | 0 | 218 | MM | 4 | 1 | 205–221 | 1 | 0 | 208 | ||||
| CCESSR13 | 4 | 0 | 122–140 | 0.75 | 0.59 | 0.51 | 4 | 0 | 131–141 | 0.50 | 0.73 | 0.62 | 11 | 3 | 115–144 | 3 | 0 | 122–130 |
| CCESSR14 | 5 | 0 | 243–257 | 0.50 | 0.61 | 0.54 | 6 | 1 | 231–256 | 0.43 | 0.60 | 0.54 | 14 | 0 | 145–257 | 2 | 0 | 231–136 |
| CCESSR16 | 1 | 0 | 190 | MM | 1 | 0 | 190 | MM | 1 | 0 | 190 | 1 | 0 | 181 | ||||
| CCESSR17 | 2 | 0 | 182–191 | 0.88 | 0.53 | 0.37 | 3 | 0 | 179–191 | 0.50 | 0.58 | 0.48 | 5 | 2 | 179–191 | 2 | 0 | 182–185 |
| CCESSR18 | 1 | 0 | 169 | MM | 1 | 0 | 169 | MM | 1 | 0 | 169 | 2 | 1 | 169–172 | ||||
| CCESSR19 | 3 | 0 | 230–236 | DL | 4 | 0 | 224–234 | 0.75 | 0.70 | 0.56 | 9 | 1 | 224–239 | 4 | 0 | 224–232 | ||
| CCESSR20 | 2 | 0 | 153–162 | DL | 4 | 0 | 153–166 | 0.88 | 0.74 | 0.64 | 8 | 2 | 153–189 | 2 | 1 | 166–177 | ||
| CCESSR21 | 2 | 0 | 170–175 | 0.00 | 0.23 | 0.19 | 2 | 0 | 170–175 | 0.00 | 0.40* | 0.30 | 9 | 4 | 168–211 | 4 | 1 | 168–176 |
| CCESSR22 | 1 | 0 | 158 | MM | 7 | 0 | 159–180 | 1.00 | 0.85 | 0.77 | 12 | 2 | 158–180 | 2 | 0 | 151–153 | ||
| CCESSR23 | 2 | 0 | 168–174 | 0.88 | 0.53 | 0.37 | 2 | 0 | 174–177 | 0.00 | 0.23 | 0.19 | 7 | 1 | 166–178 | 3 | 1 | 167–172 |
| CCESSR26 | 4 | 0 | 214–227 | 0.86 | 0.78 | 0.67 | 3 | 0 | 214–227 | 0.57 | 0.47 | 0.39 | 9 | 3 | 212–236 | 1 | 0 | 218 |
| CCESSR27 | 1 | 0 | 132 | MM | 5 | 0 | 122–139 | 0.50 | 0.65 | 0.56 | 13 | 5 | 115–195 | 3 | 0 | 122–195 | ||
| CCESSR28 | 1 | 0 | 197 | MM | 1 | 0 | 189 | MM | 5 | 1 | 186–213 | 2 | 2 | 194–207 | ||||
| CCESSR29 | 3 | 0 | 96–106 | DL | 5 | 0 | 96–111 | 1.00 | 0.75* | 0.66 | 5 | 0 | 96–111 | 3 | 0 | 96–106 | ||
| CCESSR31 | 1 | 0 | 150 | MM | 4 | 0 | 150–182 | 0.63 | 0.74 | 0.64 | 7 | 2 | 140–182 | 2 | 2 | 167–182 | ||
| CCESSR32 | 1 | 0 | 181 | MM | 4 | 0 | 179–185 | 0.38 | 0.66* | 0.57 | 12 | 5 | 177–204 | 2 | 1 | 187–189 | ||
| CCESSR33 | 2 | 0 | 236–250 | DL | 2 | 0 | 236–241 | 0.88 | 0.53 | 0.37 | 6 | 0 | 230–256 | 2 | 0 | 223–236 | ||
| CCESSR34 | 2 | 0 | 185–191 | 0.13 | 0.53* | 0.37 | 2 | 0 | 174–185 | 0.13 | 0.13 | 0.11 | 10 | 3 | 135–224 | 2 | 2 | 193–222 |
| CCESSR35 | 1 | 0 | 115 | MM | 4 | 0 | 122–128 | 0.75 | 0.73 | 0.62 | 9 | 1 | 115–130 | 2 | 0 | 111–120 | ||
| CCESSR36 | 1 | 0 | 112 | MM | 2 | 0 | 112–114 | 0.25 | 0.23 | 0.19 | 4 | 1 | 102–124 | 2 | 0 | 102–114 | ||
| CCESSR38 | 2 | 0 | 177–179 | 0.63 | 0.46 | 0.34 | 3 | 0 | 177–181 | 0.25 | 0.24 | 0.21 | 11 | 4 | 170–185 | 2 | 0 | 177–179 |
| CCESSR39 | 1 | 0 | 150 | MM | 1 | 0 | 147 | MM | 2 | 0 | 147–150 | 2 | 0 | 143–150 | ||||
| CCESSR40 | 2 | 0 | 220–222 | DL | 3 | 0 | 220–223 | 0.25 | 0.24 | 0.21 | 7 | 3 | 217–115 | 1 | 0 | 222 | ||
| CCESSR41 | 3 | 0 | 212–219 | DL | 4 | 0 | 214–223 | 0.71 | 0.76 | 0.65 | 9 | 0 | 212–226 | 1 | 0 | 139 | ||
| CCESSR42 | 1 | 0 | 129 | MM | 1 | 0 | 129 | MM | 5 | 1 | 120–141 | 1 | 0 | 132 | ||||
| CCESSR43 | 2 | 0 | 184–190 | DL | 3 | 0 | 187–199 | 0.33 | 0.32 | 0.27 | 5 | 0 | 178–199 | 3 | 0 | 174–187 | ||
| CCESSR44 | 3 | 0 | 250–259 | 0.88 | 0.58 | 0.45 | 3 | 0 | 238–259 | 1.00 | 0.59* | 0.46 | 8 | 1 | 236–259 | 3 | 0 | 247–159 |
| CCESSR45 | 2 | 0 | 144–151 | DL | 1 | 0 | 151 | MM | 8 | 3 | 137–166 | 2 | 0 | 125–151 | ||||
| CCESSR47 | 1 | 0 | 211 | MM | 1 | 0 | 211 | MM | 2 | 0 | 211–213 | 1 | 0 | 207 | ||||
| CCESSR48 | 2 | 0 | 199–211 | DL | 2 | 0 | 192–205 | 0.13 | 0.13 | 0.11 | 4 | 0 | 192–211 | 1 | 0 | 182 | ||
| CCESSR49 | 2 | 0 | 192–203 | DL | 3 | 0 | 192–203 | 0.50 | 0.43 | 0.35 | 7 | 1 | 188–203 | 2 | 0 | 185–197 | ||
| CCESSR50 | 1 | 0 | 122 | MM | 2 | 0 | 116–122 | 0.38 | 0.33 | 0.26 | 4 | 0 | 116–128 | 2 | 0 | 122–128 | ||
| Range | 1–6 | 0 | 0.00–0.88 | 0.23–0.78 | 0.19–0.67 | 1–7 | 0–1 | 0.00–1.00 | 0.13–0.85 | 0.11–0.77 | 1–17 | 0–5 | 1–4 | 0–4 | ||||
| Average | 2.14 | 0.00 | 0.47 | 0.54 | 0.43 | 3.00 | 0.02 | 0.54 | 0.53 | 0.44 | 7.52 | 1.57 | 2.11 | 0.34 | ||||
| SD (±) | 1.21 | 0.00 | 0.36 | 0.13 | 0.16 | 1.57 | 0.15 | 0.26 | 0.22 | 0.19 | 3.62 | 1.59 | 0.89 | 0.81 | ||||
| SE (±) | 0.18 | 0.00 | 0.10 | 0.03 | 0.03 | 0.24 | 0.02 | 0.04 | 0.04 | 0.03 | 0.55 | 0.24 | 0.14 | 0.12 | ||||
Note: NA: Number of amplified alleles; PA: Number of Private Alleles; Ho: Observed heterozygosity; He: Expected heterozygosity; PIC: Polymorphism Information Content; PI: Probability of Identity; NA: Not amplified; *: Significant HW dis-equilibrium at P<0.05; **: Highly significant HW dis-equilibrium at P<0.01; The putative DL (duplicated loci) markers were not considered for calculation of various estimates as these appear to be fixed exhibiting no segregation.
The PIC values were comparable (0.19–0.67 and 0.11–0.77), and no significant differences were seen in the observed/expected heterozygosity (Ho/He: t-value = 0.70; P = 0.49; and t-value = 0.68; P = 0.40) for the new markers across the tested tetraploids and diploid robustas, respectively. However, significant differences were observed in the total number of amplified alleles (NA: t = 3.74, P<0.005), as well as, the behaviour of the polymorphic markers (Pms) when tested for HWE and LD in the tested tetraploids and the robusta genotypes (Table 4). In general, more markers were in HWE and only a relatively small proportion of markers exhibited LD and heterozygote excess and/or deficiency in case of robustas, in comparision to tetraploid arabicas.
Validation of genomic SSRs for use in genetic studies
A total of 25 putative genomic SSRs were also validated as genetic markers (Table 5). When tested on the panel of 16 elite robusta and arabica (tetraploid) genotypes, five of these markers in arabicas and one in robustas were found to be monomorphic. Twelve ofthe polymorphic markers in arabicas resulted in double alleles (putative duplicated loci). In total, a maximum of seven and eight alleles (NA) with an average of 2.7 and 4.3 alleles/marker were obtained for the tested polymorphic markers of which 32% and 96% were informative in arabicas and robustas, respectively (Figure 1). The PIC values varied considerably, with mean PIC value being 0.47 (range 0.12–0.78) for tetraploids, which was significantly less than 0.60 (0.12–0.85) observed for robusta (Table 5). Further, the Student’s t-test revealed significant differences in NA (t = 4.09, P = 0.00) but non-significant differences in PIC estimates (t = 1.26, P = 0.13), as well as, for the observed/expected heterozygosity estimates (Ho/He) for the comparable markers of arabica and robusta genotypes.
Table 5. Allelic diversity attributes of the newly developed 25 genomic SSRs when tested over cultivated and wild related coffee genera.
| Species: | C. arabica (n = 8) | C. canephora (n = 8) | Coffea spp. (n = 12) | Psilanthus spp. (n = 2) | ||||||||||||||
| Primer ID | NA | PA | Allele range (in bp) | Ho | He | PIC | NA | PA | Allele range (in bp) | Ho | He | PIC | NA | PA | Allele range (in bp) | NA | PA | Allele range (in bp) |
| CCRM02 | 3 | 0 | 252 −262 | DL | 3 | 0 | 256– 268 | 0.71 | 0.69 | 0.67 | 7 | 2 | 252– 278 | 2 | 1 | 162 −272 | ||
| CCRM06 | 2 | 1 | 143 −156 | 0.14 | 0.14 | 0.12 | 1 | 0 | 143 | MM | 1 | 0 | 143 | 1 | 0 | 143 | ||
| CCRM07 | 5 | 0 | 124– 140 | 0.71 | 0.82 | 0.78 | 5 | 1 | 124– 146 | 1.00 | 0.78 | 0.74 | 6 | 3 | 115– 136 | 2 | 0 | 124 −128 |
| CCRM10 | 3 | 0 | 96– 106 | 0.29 | 0.38 | 0.32 | 2 | 0 | 105– 106 | 0.00 | 0.23 | 0.23 | 6 | 2 | 98– 100 | NA | – | – |
| CCRM14 | 3 | 0 | 109– 188 | DL | 7 | 2 | 110–132 | 0.63 | 0.88** | 0.83 | 8 | 5 | 109–155 | 1 | 0 | 123 | ||
| CCRM15 | 1 | 0 | 298 | MM | 3 | 0 | 243– 249 | 0.17 | 0.62** | 0.61 | 7 | 2 | 243– 361 | NA | – | – | ||
| CCRM16 | 3 | 0 | 198– 202 | 0.00 | 0.70** | 0.67 | 8 | 0 | 190– 240 | 0.88 | 0.89 | 0.84 | 11 | 3 | 180– 226 | 3 | 0 | 180 −192 |
| CCRM17 | 1 | 0 | 230 | MM | 7 | 2 | 226– 248 | 1.00 | 0.86 | 0.81 | 10 | 4 | 216– 250 | 2 | 1 | 210 −238 | ||
| CCRM19 | 4 | 0 | 217– 234 | 0.43 | 0.69 | 0.62 | 7 | 1 | 196– 252 | 0.83 | 0.92 | 0.85 | 13 | 4 | 188– 246 | 3 | 0 | 210 −226 |
| CCRM21 | 7 | 1 | 256– 290 | DL | 7 | 0 | 256– 288 | 0.63 | 0.90 | 0.85 | 16 | 6 | 234– 320 | 2 | 0 | 258 −320 | ||
| CCRM22 | 3 | 0 | 194– 203 | DL | 5 | 1 | 194– 254 | 0.25 | 0.67** | 0.63 | 10 | 4 | 194– 254 | 2 | 1 | 201 −226 | ||
| CCRM23 | 2 | 0 | 140– 162 | DL | 3 | 0 | 140– 151 | 0.38 | 0.49 | 0.46 | 7 | 1 | 140– 162 | 2 | 0 | 140– 162 | ||
| CCRM24 | 3 | 0 | 202– 213 | DL | 3 | 0 | 211– 215 | 0.63 | 0.57 | 0.56 | 8 | 1 | 202– 217 | 2 | 0 | 205– 213 | ||
| CCRM28 | 2 | 0 | 203– 206 | 0.00 | 0.26 | 0.23 | 2 | 0 | 202– 206 | 0.13 | 0.13 | 0.12 | 6 | 0 | 202– 210 | 3 | 0 | 202– 210 |
| CCRM31 | 1 | 0 | 111 | MM | 4 | 0 | 101– 110 | 0.38 | 0.44 | 0.42 | 7 | 1 | 101– 115 | 2 | 2 | 113– 118 | ||
| CCRM33 | 2 | 0 | 110– 116 | 0.14 | 0.14 | 0.33 | 3 | 0 | 110– 116 | 0.88 | 0.64 | 0.64 | 8 | 1 | 106– 122 | 2 | 0 | 112– 116 |
| CCRM34 | 2 | 0 | 130–146 | DL | 4 | 0 | 144– 163 | 0.75 | 0.75 | 0.72 | 9 | 1 | 130– 146 | 3 | 1 | 146– 163 | ||
| CCRM35 | 1 | 0 | 156 | MM | 3 | 2 | 152– 156 | 0.00 | 0.43** | 0.42 | 1 | 0 | 156 | 1 | 0 | 156 | ||
| CCRM36 | 2 | 0 | 139–146 | DL | 7 | 1 | 139– 175 | 1.00 | 0.90 | 0.85 | 11 | 3 | 139– 183 | 1 | 0 | 139 | ||
| CCRM37 | 2 | 0 | 140– 150 | DL | 3 | 0 | 140– 152 | 1.00 | 0.63 | 0.63 | 4 | 1 | 140– 150 | 2 | 1 | 140– 142 | ||
| CCRM38 | 4 | 1 | 175–217 | DL | 3 | 1 | 175– 206 | 0.38 | 0.34 | 0.33 | 12 | 5 | 163– 214 | 2 | 0 | 175– 200 | ||
| CCRM40 | 2 | 0 | 144–151 | DL | 5 | 0 | 159– 177 | 0.63 | 0.72 | 0.68 | 8 | 0 | 151– 177 | 2 | 0 | 151– 153 | ||
| CCRM41 | 1 | 0 | 101 | MM | 2 | 0 | 95– 101 | 0.14 | 0.14 | 0.14 | 2 | 0 | 95–110 | 1 | 0 | 101 | ||
| CCRM42 | 5 | 0 | 130– 161 | DL | 5 | 1 | 138– 157 | 0.75 | 0.75 | 0.72 | 13 | 2 | 121– 165 | 4 | 0 | 134– 155 | ||
| CCRM45 | 3 | 0 | 183– 195 | 0.86 | 0.69 | 0.64 | 5 | 0 | 187– 212 | 0.50 | 0.76 | 0.72 | 7 | 1 | 183– 201 | 2 | 0 | 187– 189 |
| Range | 1 −7 | 0 −1 | – | 0 −0.71 | 0.14 −0.81 | 0.12 −0.78 | 1 −8 | 0 −2 | – | 0.00 −1.00 | 0.13 −0.92 | 0.12 −0.85 | 1 −16 | 0 −6 | – | 0 −4 | 0 −2 | – |
| Mean | 2.68 | 0.12 | – | 0.32 | 0.45 | 0.47 | 4.28 | 0.48 | – | 0.57 | 0.63 | 0.60 | 7.92 | 2.08 | – | 2.04 | 0.30 | – |
| SD (±) | 1.46 | 0.33 | – | 0.32 | 0.28 | 0.24 | 1.97 | 0.71 | – | 0.33 | 0.25 | 0.22 | 3.67 | 1.78 | – | 0.77 | 0.56 | – |
| SE (±) | 0.30 | 0.07 | – | 0.07 | 0.06 | 0.05 | 0.40 | 0.15 | – | 0.07 | 0.05 | 0.05 | 0.75 | 0.36 | – | 0.16 | 0.11 | – |
Note: NA: Number of amplified alleles; PA: Number of Private Alleles; Ho: Observed heterozygosity; He: Expected heterozygosity; PIC: Polymorphism Information Content; PI: Probability of Identity; NA: Not amplified;
**: Highly significant HW dis-equilibrium at P<0.01; The putative DL (duplicated loci) markers were not considered for calculation of various estimates as these appear to be fixed exhibiting no segregation.
Further, it was notable that while>83% of the Pms were in HWE and only few markers showedsignificant heterozygote deficiency to varying extent in both arabicas and robustas, the number of marker-pairs that exhibited LD was significantly more in arabicas (28.0%; 8 of 28 pairs) that that seen in robustas (14.2%; 36 of 254 marker pairs).
Mapping of new EST- and genomic SSRs
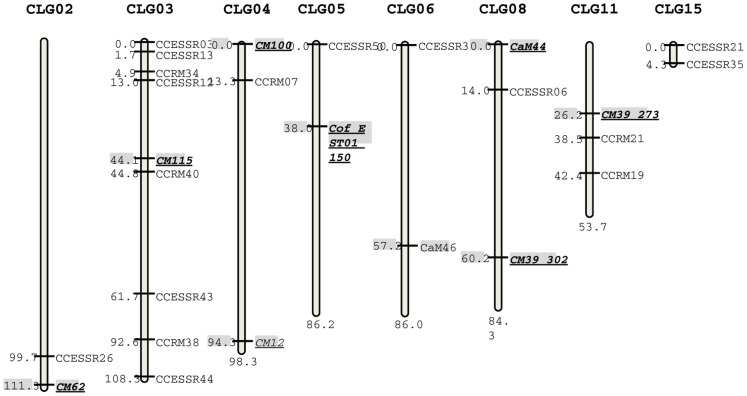
The 69 new SSR markers were also tested for their suitability in linkage mapping. In total, 11 of the 44 EST-SSRs (∼39%) and seven of the 25 genomic SSRs (28%) could be mapped onto an existing first-generation framework linkage map of robusta coffee [2], [26]. This map comprised a total of 374 mapped markers (71 SSRs, 185 RAPDs and 118 AFLPs) on 11 major and 5 minor linkage groups. The new markers developed in the present study were mapped using the existing SSRs on the map as anchor markers. The 18 new markers that could be mapped, occupied positions on eight distinct linkage groups, with eight markers on CLG03; two markers each on CLG06, CLG11, CLG15; one marker on CLG02, CLG04, CLG05, CLG08 (Tables 1 & 2). The position of these 18 markers on robusta linkage groups alongwith positions of SSRs used as anchores (CM62, CM115, CM12, CM100, Cof_EST01_150, CaM46, CaM44, CM39_302, CM39_273) is shown in figure 2.
Figure 2. Map positions of 18 new SSR markers developed in this study (11 CCESSR and 7 CCRM markers) on robusta linkage map; mapping population was derived from a cross between CxR (a commercial robusta hybrid) and a local selection Kagganahalla [26].
The SSR markers of the existing map used as anchor markers are shown in italic and bold face.
Cross-species/−genera transferability and marker conservation
New SSR-markers when tested on 13 related Coffea and two Psilanthus species, exhibited robust cross-species amplifications with alleles of comparable sizes in the tested taxa (Figure 1, Tables 4 & 5, Table S4). The EST-SSRs showed 100% transferability accross the tested Coffea and Psilanthus spp., whereas the genomic-SSRs indicated 96% amplification and transferability for Coffea spp. and 92% for the related Psilanthus spp. The analysis also indicated some private alleles (PAs), which possibly could be species-specific (Tables 4 & 5).
Generic affinities within/between cultivated and wild coffee germplasm by new SSRs
The SSR allelic data were examined for their utility in ascertaining the genetic diversity and generic inter-relationships between the cultivated, as well as, the wild coffee genepool. The average genetic distance values calculated using the EST-SSR allelic data were in general, significantly less but comparable to that obtained using the genomic SSRs for the tested arabicas, robustas, as well as, for different Coffea and Psilanthus species.
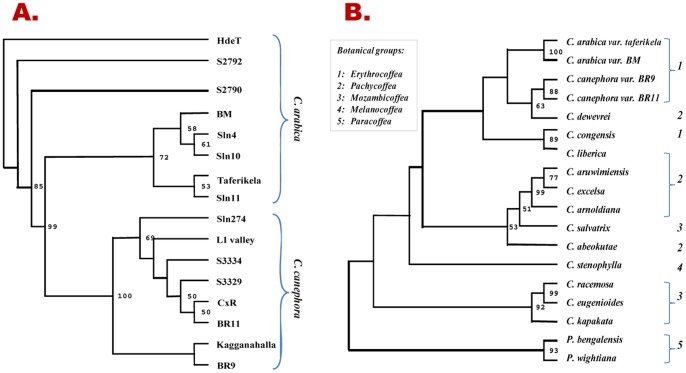
The NJ phenetic tree generated using the genetic distance estimates of EST-SSRs allelic data clearly resolved the tested germplasm in two distinct clusters, one representing all the tetraploid arabicas, while the other comprised all the diploid robusta genotypes (Figure 3a) with significant branch support. The selections formed a single cluster within the tetraploids cluster, while pure arabicas and hybrid-selections appeared in distict sub-clusters. Similarly, in clustering analysis of 14 related species (12 Coffea and two Psilanthus spp.; Figure 3b) along with two genotypes each from C. arabica and C. canephora, tetraploid Erythrocoffeas (C. arabica) and diploid Erythrocoffea (C. canephora) formed coherent clusters. Moreover, the grouping of the related taxa, in general, was as per their botanical type with few changes. Though all the entries from Erythrocoffeas fell into one cluster, it contained two entries from Pachycoffeas (C. dewevrei with C. canephora and C. liberica with C. congensis). The remaining four of the Pachycoffeas (C. excelsa, C. arnoldiana, C. aruwemiensis, C. abeokutae) grouped with each other with good bootstrap support. The C. salvatrix a Mozambicoffea was also grouped with these Pachycoffeas, while the other three Mozambicoffeas (C. racemosa, C. eugenioides, C. kapakata) and two Paracoffeas (Psilanthus spp.) appeared as independent strong groups. Single Melanocoffea species (tested in this study), C. stenophylla was not grouped with any of the above species cluster but was found close to the Coffea species than the Psilanthus spp.
Figure 3. Unrooted phenetic trees based on the allelic diversity across the tested 44 EST-SSRs showing generic affinities between the: a) C. arabica and C. canephora genotypes, and b) 14 Coffea and two Psilanthus taxa; (only>50 bootstrap values are shown).
Similar results were obtained using the data from genomic SSRs (CCRMs, data not shown).
Discussion
SSR motifs in coffee transcriptome, and development of new EST-SSR markers
In the present study, 15.4% of the coffee ESTs were found to contain SSRs, which is comparable with our earlier study [11], but much higher than 2.7–10.8% that was reported for 18 representative dicotyledonous species [27], and 7 −10% reported for monocot species [28]. Notwithstanding this apparent enrichment/higher abundance, the SSRs in coffee transcriptome were very comparable to other plant species in observations like: 1. Abundance of TNRs than DNRs; 2. Abundance of AG among the DNRs followed by AT; 3. CG as the least abundant among the DNRs; 4. Abundance of AAG among TNRs (among the dicots); 5. Predominance of GC-rich TNRs (but not CCG/GGC) than the non-GC-rich TNRs.
A total of 18.7% of the detected EST-SSRs were found to be suitable candidates for primer design, a comparatively lower proportion (∼50%) then we reported earlier [11]. The main attributes that rendered majority of the identified SSRs unsuitable for marker development were: a shorter repeat core (<18 bp) and/or flanking sequences of low complexity (AT/GC-rich and/or regions prone to secondary structure formation) or shorter lengths seriously constraining designing of optimal primer pairs. However, in this study, primer-to-marker conversion ratio (ca. 88%), was higher than many earlier similar studies in other crops. Such differences in marker conversion ratios are expected due to differences- in the quality of primers designed), GC content of the genome, the genome complexity, and/or genome size [13].
SSR enrichment and development of genomic SSRs
The genomic DNA library constructed in this study, resulted in very high proportion of SSR+ive sequences, with very low degree of redundancy (9.1%; 6 out of 66 identified SSRs positive sequences). This was notable, as in earlier similar studies the apparent high success rates were generally confounded by high degree of redundancy [29]. Similarly, the proportion of SSR positive sequences found suitable for primer designing was also higher (87%) in our study than the average of 54±3% recorded in other species [30]. These obsrevationssuggest that the enrichment approach used in this study may be a desirable strategy for efficiently entrapping and targeting the SSRs even in genome(s) like coffee that are relatively poor in SSR motifs [2].
Utility of new EST- and genomic SSRs as genetic markers
The SSRs provide desirable markers for studying genetic diversity, germplasm characterization, constructing reference panels/bar codes, for individualization of genotypes, linkage mapping, population biology, and taxonomic relationships of related taxa [2]. Therefore, it becomes desirable to validate the new markers for their utility in genetic studies, which unfortunately has been lacking in majority of published studies describing development of coffee-specific SSR markers.
Various genetic parameters viz., allelic diversity, PIC, Ho, He, HWE, LD calculated for all the new EST and genomic SSRs and mapability on linkage map, amply suggested their possible utility as genetic markers (see Table 4 & 5). In general, the extent and pattern of allelic/genetic diversity revealed by the new markers conform to that reported earlier for the coffee genomic SSRs [5], [6], [31], and the EST-SSRs [8], [11].
Different genetic parameters/tests such as Ho, He, LD, HWE are important indicators of origin, evolution and distribution of diversity in the available genepool. The heterozygosity measures (Ho, He) for the new SSR markers indicated heterozygote decay (deficiency) in the tested germplasm. The HWE and LD analysis of the polymorphic markers were in general agreement with our earlier observations with genomic as well as EST-SSRs [5], [8], [11]. Overall, these studies indicated that the tested robusta germplasm comprised allogamous, relatively unrelated genotypes, while autogamous tetraploids comprised mostly of hybrid varieties/selections with overlapping/shared pedigrees. The results thus suggest the suitability of the new markers for reliably ascertaining genetic diversity in the coffee gene pool.
Cross-species/−generic transferability
All the new EST- and genomic SSR markers revealed very high and robust cross species/−generic amplifications with alleles of comparable sizes when tested on 12 other Coffea and two Psilanthus taxa. The data revealed that the markers described here show much higher taxa transferability than earlier published genomic−/EST-SSR markers [2], [5], [7], [11]. This is significant as successful cross-species amplification is generally restricted to related species within a genus and reduces when tested for different genera [32]. Further, it was interesting to note that the new SSRs that were monomorphic/uninformative for the tested arabica/robusta germplasm, exhibited considerable polymorphism across the tested related taxa (the only exceptions were the marker CCESSR16 and 18 that showed a very low conservation even across the Coffea spp.). Thus the new SSR markers described here strengthen the possibility of their use as Conserved Orthologous Sets (COS) for genetic characterization of different related wild coffee taxa, and also for coffee taxonomic/synteny studies.
Diversity analysis and genetic relatedness within/between Coffea and Psilanthus species
The EST−/genomic-SSRs described in this study were able to group all the 16 genotypes (representing the cultivated genepool) in phenetic clustering that was indicative of their species status and known pedigrees (Figure 1a). Similarly, the analysis 14 Coffea and two Psilanthus species, revealed generic affinities that were largely in agreement with their known taxonomic relationships (Figure 1b), based on their geographical distribution as well as Chevalier’s botanical classification [33]. Importantly, the analysis distinctly separated the two Paracoffea species (P. bengalensis and P. wightiana) from all the other Coffea spp. These results are similar to the earlier published studies undertaken to ascertain species relationships using SSRs [2], [7], [8], [11], as well as other marker approaches [34]–[36]. These results, thus, amply demonstrate that the new SSR markers developed in the present study can be considerably informative in exploring the taxonomic relationship of coffee species complex.
Conclusions
The present study describes a total of 69 new validated SSRs; 44 EST-SSRs developed from coffee transcriptome using in-silico methodology, and 25 genomic SSRs developed using SSR enrichment approach. In addition, it provides primer pairs for additional 270 putative EST-SSRs. Analysis of the identified SSR-positive ESTs also provided insights into the relative abundance and distribution pattern of different SSR motifs in the coffee transcriptome, which was found to be relatively rich in its SSR abundance. Among the identified EST-SSRs, TNRs followed by DNRs were more abundant than other SSRs, and among different types of SSR motifs, AG was the most abundant. All the 69 markers were found to be polymorphic in the tested coffee/related germplasm and their utility as efficient genetic markers could be demonstrated for diversity analysis, germplasm individualization, linkage mapping, cross-species transferability and taxonomic studies. As many of these SSRs showed a very high cross-species transferability, they can aid in conservation, management and resolving taxonomic relationships, as Conserved Orthologous Sets (COS) for Coffea and Psilanthus species and more importantly as efficient, and informative genetic landmarks on molecular linkage maps.
Supporting Information
Summary statistics of screening of the coffee unigene ESTs for SSRs.
(PDF)
Summary statistics of distribution and abundance of detected SSRs in the unigene ESTs and SSR frequency estimates for coffee transcriptome.
(PDF)
Characteristics and distribution of the detected SSR motifs (without MNRs) across the non-redundant 56 SSR+ive sequences generated using SSR enrichment approach.
(PDF)
Inter-species and inter-generic transferability of the new EST-SSRs and genomic SSR markers.
(PDF)
Acknowledgments
RKA thanks the Director, CCMB, Hyderabad for the facilities to undertake the study; Director Research, Coffee Board, Bangalore for coffee materials, and R. Phanindranath for providing lab help as and when required. PSH acknowledges CSIRfor junior and senior research fellowship during his doctoral research and DBT for the Postdoctoral fellowship.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. New genomic sequences have been submitted to GenBank/NCBI, and accession numbers are provided within the paper.
Funding Statement
This work was supported by: the FYP project, PLOMICS_BSC0301, to RKA, from the Council of Scientific & Industrial Research (Govt of India); and Project grant “BT/PR/6292/Agr/16/575/2005, dated 4/12/2006,” to RKA from the Department of Biotechnology (Govt of India), under which some of the DNA samples/sequences used in the present study were collected/carried out. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hendre PS, Aggarwal RK (2007) DNA markers: development and application for genetic improvement of coffee, In: Varshney RK, Tuberosa R (eds.), Genomic Assisted Crop Improvement: Genomics Applications in Crops Volume 2, Springer-Verlag, Germany, pp. 399–434.
- 2. Hendre PS, Phanindranath R, Annapurna V, Lalremeruata A, Aggarwal RK. (2008) Development of new genomic microsatellite markers from robusta coffee (Coffea canehora Pierre ex A. Froehner) showing broad cross-species transferability and utility in genetic studies. BMC Pl Biol. 8:51 doi: 10.1186/1471-2229-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Combes M-C, Andrzejewski S, Anthony F, Bertrand B, Rovelli P, et al. (2000) Characterization of microsatellite loci in Coffea arabica and related coffee species. Mol Ecol. 9:1178–1180. [DOI] [PubMed] [Google Scholar]
- 4.Rovelli P, Mettulio R, Anthony F Anzueto F, Lashermer P, et al.. (2000) Microsatellites in Coffea arábica L. in Coffee Biotechnology and Quality. Kluwer, Dordrecht, pp. 123–133.
- 5. Baruah A, Naik V, Hendre PS, Rajkumar R, Rajendrakumar P, et al. (2003) Isolation and characterization of nine microsatellite markers from Coffea arabica L., showing wide cross-species amplifications. Mol Ecol Notes. 3:647–650. [Google Scholar]
- 6. Moncada P, McCouch S. (2004) Simple sequence repeat diversity in diploid and tetraploid Coffea species. Genome. 47:501–509. [DOI] [PubMed] [Google Scholar]
- 7. Poncet V, Hamon P, Minier J, Carasco C, Hamon S, et al. (2004) SSR cross-amplification and variation within coffee trees (Coffea spp.). Genome. 47:1071–1081. [DOI] [PubMed] [Google Scholar]
- 8. Bhat PR, Krishnakumar V, Hendre PS, Rajendrakumar P, Varshney RK, et al. (2005) Identification and characterization of expressed sequence tags-derived simple sequence repeats markers from robusta coffee variety ‘CXR’ (an interspecific hybrid of Coffea canephora and Coffea congensis). Mol Ecol Notes. 5:80–83. [Google Scholar]
- 9. Poncet V, Dufour M, Hamon P, Hamon S, de Kocho A, et al. (2007) Development of genomic microsatellite markers in Coffea canephora and their transferability to other coffee species. Genome. 50:1156–1161. [DOI] [PubMed] [Google Scholar]
- 10. Poncet V, Rondeam M, Tranchant C, Cayrel A, Hamon S, et al. (2006) SSR mining in coffee tree EST databases: potential use of EST-SSR as markers for the Coffea genus. Mol Gen Genomics. 276:436–449. [DOI] [PubMed] [Google Scholar]
- 11. Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, et al. (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet. 114:359–372. [DOI] [PubMed] [Google Scholar]
- 12. Plechakova O, Tranchant-Dubreuil C, Benedet F, Couderc M, Tinaut A, et al. (2009) MoccaDB - an integrative database for functional, comparative and diversity studies in the Rubiaceae family. BMC Pl Biol. 9:123 doi: 10.1186/1471-2229-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Varshney RK, Graner A, Sorrells ME. (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 23:48–55. [DOI] [PubMed] [Google Scholar]
- 14.Riar DS, Rustgi S, Burke IC, Gill KS, Yenish JP (2010) EST-SSR development from 5 Lactuca species and their use in studying genetic diversity among L. serriola biotypes. J Heredity doi: 10.1093/jhered/esq103.. [DOI] [PubMed]
- 15. Lin C, Mueller LA, McCarthy J, Crouzillat D, Pe'tiard V, et al. (2005) Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts. Theor Appl Genet. 112:114–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aggarwal RK, Shenoy VV, Ramadevi J, Rajkumar R, Singh L. (2002) Molecular characterization of some Indian Basmati and other elite rice genotypes using fluorescent-AFLP. Theor Appl Genet. 105:680–690. [DOI] [PubMed] [Google Scholar]
- 17.Hendre PS, Aggarwal RK (2004) High throughput isolation of microsatellites using affinity capture, In: Aggarwal RK (ed), Course Manual, Hands-on Training course: DNA markers: Development and Applications. CCMB, Hyderabad, India, pp. 2.1–2.24.
- 18. Edwards KJ, Barker JH, Daly A, Jones C, Karp A. (1996) Microsatellite libraries enriched for several microsatellite sequences in plants. BioTechniques. 20:758–760. [DOI] [PubMed] [Google Scholar]
- 19. Excoffier L, Laval G, Schneider S. (2005) Arlequin ver 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 20. Glaubitz JC. (2004) Convert: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes. 4:309–310. [Google Scholar]
- 21. Dieringer D, Schlotterer C. (2003) MicroSatellite Analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol. 3:167–169. [Google Scholar]
- 22. Nei M. (1972) Genetic distance between populations. Am Naturalist. 106:238–292. [Google Scholar]
- 23.Felsenstein J (2004) PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle.
- 24.Page RDM (2001) TreeView Software. Available: http://taxonomy.zoology.gla.ac.uk/rod/rod.html.
- 25.van Ooijen JW (2006) JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands.
- 26.Hendre PS (2007) Development of coffee specific microsatellite markers and construction of a framework molecular linkage map of robusta coffee, Coffea canephora. PhD thesis, School of Life Sciences, Jawaharlal Nehru University, New Delhi, India.
- 27.Kumpatla SP (2004) Computational mining and survey of simple sequence repeats (SSRs) in expressed sequence tags (ESTs) of dicotyledonous plants. M.S. thesis submitted to the School of Informatics, Indiana University-Purdue University, Indianapolis, USA.
- 28. Varshney RK, Thiel T, Stein N, Langridge P, Graner A. (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett. 7:537–546. [PubMed] [Google Scholar]
- 29. Squirrel J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, et al. (2003) How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol. 12:1339–1348. [DOI] [PubMed] [Google Scholar]
- 30. Moriguchi Y, Iwata H, Ujino-IharaT, Yoshimura K, Taira H, Tsumura Y. (2003) Development and characterization of microsatellite markers for Cryptomeria japonica D. Don. Theor Appl Genet. 106:751–758. [DOI] [PubMed] [Google Scholar]
- 31.Aggarwal RK, Rajkumar R, Rajendrakumar P, Hendre PS, Baruah A, et al. (2004) Fingerprinting of Indian coffee selections and development of reference DNA polymorphism panels for creating molecular IDs for variety identification. In: Proceedings of 20th international conference on coffee science (ASIC) Bangalore, India, pp. 751–755.
- 32. Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A. (1998) Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: implications for the transferability of SSRs in plants. Mol Biol Evol. 15:1275–1287. [DOI] [PubMed] [Google Scholar]
- 33.Chevalier A (1947) Les cafeiers du globe. III. Systematique des caféiers at Faux cafeiers. Maladieset insect nuisible, in P. Lechevalier (Ed.) Encyclopedie de biologique Paris, France, pp. 356.
- 34. Orozco-Castillo C, Chalmers KJ, Powell W, Waugh R. (1996) RAPD and organelle specific PCR re-affirms taxonomic relationships within the genus Coffea . Pl Cell Rep. 15:337–341. [DOI] [PubMed] [Google Scholar]
- 35. Lashermes P, Combes M-C, Trouslot P, Charrier A. (1997) Phylogenetic relationship of coffee-tree species (Coffea L.) as inferred from ITS sequences of nuclear ribosomal DNA. Theor Appl Genet. 94:947–955. [Google Scholar]
- 36. Ruas PM, Ruas CF, Rampim L, Carvaljo VP, Ruas EA, et al. (2003) Genetic relationship in Coffea species and parentage determination of interspecific hybrids using ISSR (inter-simple sequence repeat) markers. Genet Mol Biol. 26:319–327. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary statistics of screening of the coffee unigene ESTs for SSRs.
(PDF)
Summary statistics of distribution and abundance of detected SSRs in the unigene ESTs and SSR frequency estimates for coffee transcriptome.
(PDF)
Characteristics and distribution of the detected SSR motifs (without MNRs) across the non-redundant 56 SSR+ive sequences generated using SSR enrichment approach.
(PDF)
Inter-species and inter-generic transferability of the new EST-SSRs and genomic SSR markers.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. New genomic sequences have been submitted to GenBank/NCBI, and accession numbers are provided within the paper.