Abstract
The purpose of this study was to identify the major molecular components in the secretory and maturation stages of amelogenesis through transcriptome analyses. Ameloblasts (40 sections per age group) were laser micro-dissected from Day 5 (secretory stage) and Days 11–12 (maturation stage) first molars. PolyA+ RNA was isolated from the lysed cells, converted to cDNA, and amplified to generate a cDNA library. DNA sequences were obtained using next generation sequencing and analyzed to identify genes whose expression had increased or decreased at least 1.5-fold in maturation stage relative to secretory stage ameloblasts. Among the 9198 genes that surpassed the quality threshold, 373 showed higher expression in secretory stage, while 614 genes increased in maturation stage ameloblasts. The results were crosschecked against a previously published transcriptome generated from tissues overlying secretory and maturation stage mouse incisor enamel and 34 increasing and 26 decreasing expressers common to the two studies were identified. Expression of F2r, which encodes protease activated receptor 1 (PAR1) that showed 10-fold higher expression during the secretory stage in our transcriptome analysis, was characterized in mouse incisors by immunohistochemistry. PAR1 was detected in secretory, but not maturation stage ameloblasts. We conclude that transcriptome analyses are a good starting point for identifying genes/proteins that are critical for proper dental enamel formation and that PAR1 is specifically expressed by secretory stage ameloblasts.
Keywords: Amelogenesis, enamel, F2R, ORAI1, PAR1, STIM1
Introduction
Amelogenesis imperfecta (AI) by the strict definition is a collection of inherited disorders manifested by isolated enamel malformations. Enamel malformations also occur in syndromes. Genes known to be involved in the etiology of isolated AI include AMELX, ENAM, FAM83H, KLK4, MMP20, WDR72, C4orf26, SLC24A4 (1), LAMB3 and COL17A1. However, defects in these genes cannot be identified in about half of all probands when AI is the predominant phenotype. Genes known to cause syndromic AI include COL17A1, LAMA3, LAMB3, LAMC2, ITGA6, ITGB4, GJA1, CNNM4, ORAI1 (2), FAM20A, STIM1 (3), and ROGDI. Isolated and syndromic forms of AI can be impossible to distinguish clinically, so genetic testing that identifies the genetic defect causing the disorder would provide important diagnostic information and inform whether or not other tissues besides the teeth are likely to become affected.
Occasionally mutations result in gain-of-function or dominant negative effects that cause pathology beyond what occurs due to a simple loss of function. Such is the case when autosomal dominant enamel defects are caused by FAM83H truncation mutations (4) and autosomal dominant dentin defects are caused by mutations in DSPP (5). Despite the possibilities of these kinds of pathological mechanisms, it can generally be inferred from the finding of inherited enamel defects that the mutated gene performs some necessary, non-redundant function in ameloblasts, the cells that make dental enamel. If the enamel defects are isolated, the defective gene is likely to be specialized for enamel formation. If syndromic, the gene serves necessary functions in ameloblasts as well as in other cells. The fact that there are so many unidentified genes that cause AI suggests that we are unaware of many genes/proteins that are critical for dental enamel formation.
Many of the genes essential for amelogenesis were discovered through investigations of normal enamel formation. Examples include AMELX, ENAM, KLK4, and MMP20. Other necessary genes were discovered through genetic studies, for example FAM83H, WDR72, CNNM4, STIM1, ORAI1, and FAM20A. Recently SLC24A4 mutations were shown to cause AI (1), after expression of this gene was shown to be dramatically increased during the maturation stage of amelogenesis (6). Basic research into the mechanisms of tooth development and studies investigating the genetic etiologies of AI are mutually supportive. Identifying the genes/proteins that function during normal amelogenesis discovers new AI candidate genes, while discovering the genes that cause inherited enamel defects identifies proteins with critical functions during amelogenesis.
In this study, we perform transcriptome analyses specifically on secretory and maturation stage ameloblasts. The findings corroborate the results of a previous transcriptome analysis of tissues overlying secretory and maturation stage mouse incisor enamel (6). We demonstrate that protease activated receptor 1 (PAR1), encoded by F2r, is specifically expressed by secretory stage ameloblasts.
Materials and methods
All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Michigan. Detailed methods describing tissue preparation and sectioning, laser micro-dissection (LMD) and library construction, identification of differentially expressed genes, PAR1 immunohistochemistry are provided in the Supplementary Figure S1.
Results and discussion
A total of 987 genes were identified with q-values ≤0.05 (based upon false discovery rate analyses) and a minimum fold change ≥1.5 as being significantly increased or decreased in maturation relative to secretory stage ameloblasts. Of these, 614 showed higher expressions in the maturation stage (Figure S2) and 373 had higher expression in secretory stage ameloblasts (Figure S3). There were 48 ion transporter genes with stage-specific expression, including Slc31a2, Slc6a8, Slc24a4, Slc23a2, Slc4a11, Slc26a7, Slc34a2, Cnnm2 and Cnnm4 and Cftr, and 32 protease-associated genes, including Bmp1, Adamts1, Adamts2, Mmp13, Mmp15, and Mmp23. Other stage-specific genes are associated with endocytosis (Sorl1, Tfrc, and Ldlr) and signaling (Shh, Fxyd4, Atp6v1a, Batf3). As transcriptome analyses contain false positives and negatives, additional experimental verification is needed before concluding that any specific gene is expressed by ameloblasts and in a stage-specific pattern.
As a first step, we compared our ameloblast transcriptome to the one made by dissecting tissues overlying secretory and maturation stage mouse incisor enamel (6). Commonalities between the staged incisor transcriptome and our staged molar ameloblast transcriptome included 26 genes that decreased during the maturation stage and 34 genes that increased during maturation (Figure 1). Among the genes more highly expressed during the secretory stage were Enam and Mmp20, both already known to be more highly expressed by secretory stage ameloblasts. Another gene that stood out was F2r, which encodes protease activated receptor 1 (PAR1) and is also known as the coagulation factor II receptor or thrombin receptor. PAR1 is the prototypical member of a family of four G-protein-coupled receptors that are activated by proteolytic cleavage of its N-terminal exodomain, inducing a conformation change that stimulates signal transduction. PARs are internallized and sorted to lysosomes after a single activation. Kallikrein 4 (KLK4) can activate PAR1 (7), but KLK4 is specifically expressed by maturation-not secretory stage ameloblasts. We therefore performed additional studies to better characterize the expressions of protease activated receptors during amelogenesis. RT-PCR of RNA isolated from the enamel organ epithelia of day 7 mandibular first molars (which contains ameloblasts in both the secretory and maturation stages of development) detected the expression of PAR1 and PAR4, but not PAR2 or PAR3 (Figure 2a). Immunohistochemistry of mouse mandibular incisors detected PAR1 (F2R) in secretory, but not maturation, stage ameloblasts, which is consistent with the results of the transcriptome analyses (Figure 2b). The F2r null mouse phenotype was originally reported in 1996 (8), but no mention has been made then or since of an enamel phenotype.
Figure 1.
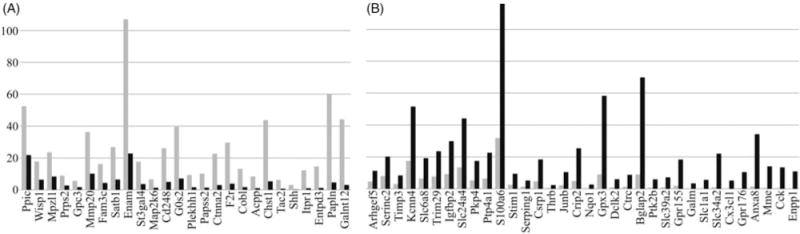
Expression levels of stage-specific genes. The bar graphs include genes that show stage specific expression in the mouse molar ameloblast transcriptome as well as in the published rat incisor transcriptome (6). Expression levels are plotted in fragments per kilobase of transcript per million mapped reads (FPKM). (A) 26 genes with elevated expression during the secretory stage. (B) 34 genes with elevated expression during the maturation stage.
Figure 2.
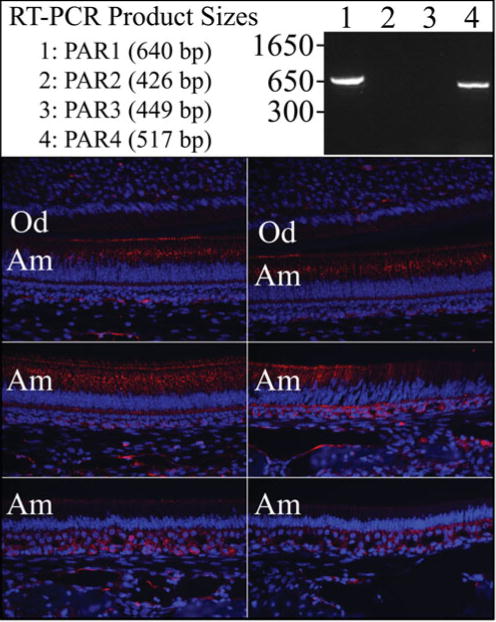
F2r analyses. Top: RT-PCR of PAR1–PAR4 transcripts from enamel organ epithelia (EOE) of Day 7 mouse molars. PCR conditions were: annealing 58 °C for 30 s; extention at 72 °C for 1 min; 25 cycles. The primers used were: PAR1, F: ctcctcaaggagcagaccac, R: tgcagggactaatgggattc; PAR2, F: tgtgattggtttgcccagta, R: tcgtgacaggtggtgatgtt; PAR3, F: ttctgccagtcactgtttgc, R: ctcgccaaatacccagttgt; PAR4, F: gctggtgctgcactattcaa, R: cacatagcccagcctagctc. Bottom: PAR1 Immunohistochemistry on D14 mandibular incisor. Key: Am, ameloblasts; Od, odontoblasts.
Among the 34 genes in common with the previous transcriptome that increased during the maturation stage (6), two are already associated with inherited enamel defects: Stim1 (3) and Slc24a4 (1). STIM1 is a transmembrane protein with an N-terminal Ca2+ binding domain in the endoplasmic reticulum (ER) that senses depletion of ER Ca2+ stores (9). Activated STIM1 migrates to ER-plasma membrane junctions where it turns on the Ca2+ channel ORAI1. Orai1 showed a 2.15-fold increase in expression during the maturation stage with a q-value of 0.07, so Orai1 just missed being listed as a gene with increased maturation stage expression in our transcriptome. Like STIM1 and SLC24A4, ORAI1 mutations are associated with inherited enamel defects (2). Acting together STIM1 and ORAI1 mediate store-operated calcium entry (SOCE), in which ORAI1 Ca2+ channels on one side of a cell open following depletion of Ca2+ stores in the ER. SLC24A4 is a potassium-dependent Na+/Ca2+ exchanger (also known as NCKX4) that transports Ca2+ out of the cell.
Based upon these observations, we propose a working hypothesis that satisfies the basic requirement for movement of Ca2+ from the blood supply, across the enamel organ epithelia, and into the mineralizing enamel matrix and posits that the directional transcellular transport of Ca2+ through ameloblasts is driven by the consumption of Ca2+ by mineral deposition. The fluid surrounding developing enamel is appears to be low in Ca2+ and nearly at equilibrium with the enamel mineral (10). We suggest that low extracellular Ca2+ activity helps draw Ca2+ out of the cell through transporters (via SLC24A4 assisted by a Na+ gradient) on the distal side of maturation ameloblasts, which depletes ER Ca2+ stores and stimulates (via STIM1) an infusion of Ca2+ through ORAI1 channels on the proximal side of maturation ameloblast nearest the blood supply.
In this study, we provide the results of transcriptome analyses of secretory and maturation stage ameloblasts laser dissected from Day 5 and Days 11/12 mouse first molars, which includes a list of 987 genes with significantly increased or decreased expression in maturation relative to secretory stage ameloblasts. We demonstrate that F2r is specifically expressed by secretory stage ameloblasts. Based upon the finding of elevated maturation stage expression in genes implicated in the etiologies of inherited enamel malformations and also with Ca2+ homeostasis, we propose a hypothesis on the transcellular transport of Ca2+ in maturation stage ameloblasts.
Supplementary Material
Supplementary Figure S1–S3: Genes with increased or decreased expression in maturation relative to secretory stage ameloblasts.
Acknowledgments
We thank the University of Michigan DNA Sequencing Core (Robert H. Lyons, Director), Center for Computational Medicine and Bioinformatics (Dr. James Cavalcoli) and Microcopy and Image-Analysis Laboratory Biomedical Research Core Facilities (Dr. Dotty Sorenson) for their expert consultations.
This study was supported by USPHS research grants DE015846 & DE019775 from the National Institute of Dental and Craniofacial Research, National Institutes of Health.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Supplementary material available online
References
- 1.Parry DA, Poulter JA, Logan CV, Brookes SJ, Jafri H, Ferguson CH, Anwari BM, Rashid Y, Zhao H, Johnson CA, Inglehearn CF, Mighell AJ. Identification of mutations in SLC24A4, encoding a potassium-dependent sodium/calcium exchanger, as a cause of amelogenesis imperfecta. Am J Hum Genet. 2013;92:307–12. doi: 10.1016/j.ajhg.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarl CA, Picard C, Khalil S, Kawasaki T, Röther J, Papolos A, Kutok J, Hivroz C, Ledeist F, Plogmann K, Ehl S, Notheis G, Albert MH, Belohradsky BH, Kirschner J, Rao A, Fischer A, Feske S. ORAI1 deficiency and lack of store-operated Ca2+ entry cause immunodeficiency, myopathy, and ectodermal dysplasia. J Allergy Clin Immunol. 2009;124:1311–18. e7. doi: 10.1016/j.jaci.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs S, Rensing-Ehl A, Speckmann C, Bengsch B, Schmitt-Graeff A, Bondzio I, Maul-Pavicic A, Bass T, Vraetz T, Strahm B, Ankermann T, Benson M, Caliebe A, Fölster-Holst R, Kaiser P, Thimme R, Schamel WW, Schwarz K, Feske S, Ehl S. Antiviral and regulatory T cell immunity in a patient with stromal interaction molecule 1 deficiency. J Immunol. 2012;188:1523–33. doi: 10.4049/jimmunol.1102507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kweon YS, Lee KE, Ko J, Hu JC, Simmer JP, Kim JW. Effects of Fam83h overexpression on enamel and dentine formation. Arch Oral Biol. 2013;58:1148–54. doi: 10.1016/j.archoralbio.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang SK, Chan HC, Rajderkar S, Milkovich RN, Uston KA, Kim JW, Simmer JP, Hu JC. Enamel malformations associated with a defined dentin sialophosphoprotein mutation in two families. Eur J Oral Sci. 2011;119:158–67. doi: 10.1111/j.1600-0722.2011.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacruz RS, Smith CE, Bringas P, Jr, Chen YB, Smith SM, Snead ML, Kurtz I, Hacia JG, Hubbard MJ, Paine ML. Identification of novel candidate genes involved in mineralization of dental enamel by genome-wide transcript profiling. J Cell Physiol. 2012;227:2264–75. doi: 10.1002/jcp.22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gratio V, Beaufort N, Seiz L, Maier J, Virca GD, Debela M, Grebenchtchikov N, Magdolen V, Darmoul D. Kallikrein-related peptidase 4: a new activator of the aberrantly expressed protease-activated receptor 1 in colon cancer cells. Am J Pathol. 2010;176:1452–61. doi: 10.2353/ajpath.2010.090523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly AJ, Ishihara H, Kahn ML, Farese RV, Jr, Coughlin SR. Role of the thrombin receptor in development and evidence for a second receptor. Nature. 1996;381:516–19. doi: 10.1038/381516a0. [DOI] [PubMed] [Google Scholar]
- 9.Feske S. ORAI1 and STIM1 deficiency in human and mice: roles of store-operated Ca2+ entry in the immune system and beyond. Immunol Rev. 2009;231:189–209. doi: 10.1111/j.1600-065X.2009.00818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoba T, Moreno EC. The enamel fluid in the early secretory stage of porcine amelogenesis: chemical composition and saturation with respect to enamel mineral. Calcif Tissue Int. 1987;41:86–94. doi: 10.1007/BF02555250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–S3: Genes with increased or decreased expression in maturation relative to secretory stage ameloblasts.


