Abstract
A precise representation of the spatial distribution of hydrophobicity, hydrophilicity and charges on the molecular surface of proteins is critical for the understanding of the interaction with small molecules and larger systems. The representation of hydrophobicity is rarely done at atom-level, as this property is generally assigned to residues. A new methodology for the derivation of atomic hydrophobicity from any amino acid-based hydrophobicity scale was used to derive 8 sets of atomic hydrophobicities, one of which was used to generate the molecular surfaces for 35 proteins with convex structures, 5 of which, i.e., lysozyme, ribonuclease, hemoglobin, albumin and IgG, have been analyzed in more detail. Sets of the molecular surfaces of the model proteins have been constructed using spherical probes with increasingly large radii, from 1.4 to 20 Å, followed by the quantification of (i) the surface hydrophobicity; (ii) their respective molecular surface areas, i.e., total, hydrophilic and hydrophobic area; and (iii) their relative densities, i.e., divided by the total molecular area; or specific densities, i.e., divided by property-specific area. Compared with the amino acid-based formalism, the atom-level description reveals molecular surfaces which (i) present an approximately two times more hydrophilic areas; with (ii) less extended, but between 2 to 5 times more intense hydrophilic patches; and (iii) 3 to 20 times more extended hydrophobic areas. The hydrophobic areas are also approximately 2 times more hydrophobicity-intense. This, more pronounced “leopard skin”-like, design of the protein molecular surface has been confirmed by comparing the results for a restricted set of homologous proteins, i.e., hemoglobins diverging by only one residue (Trp37). These results suggest that the representation of hydrophobicity on the protein molecular surfaces at atom-level resolution, coupled with the probing of the molecular surface at different geometric resolutions, can capture processes that are otherwise obscured to the amino acid-based formalism.
Introduction
The shape of, and the physico-chemical properties on the protein molecular surfaces govern the specific molecular interactions in protein-ligand complexes [1]. Therefore, studies as diverse as those on protein folding [2], protein conformational stability [3], inter- and intra- protein interactions [4], molecular recognition [5] and docking [6]; as well as applications-orientated ones, such as drug design [7], [8], protein and peptide solubility [9], crystal packing [10], and enzyme catalysis [11], benefit from an accurate and precise representation of the molecular surfaces. Furthermore, for large, intricate protein complexes, such as ion-channels [12], mechano-sensitive channels [13], or molecular chaperones [14], where the biomolecular functionality occurs on the inner molecular surface of the complex, makes the precision of the representation of molecular surfaces even more imperative.
A relatively under-studied aspect of the construction of molecular surfaces is the resolution at which the hydrophobicity is represented. Because the biomolecular recognition is a geometrically-localized and charge- and hydrophobicity-specific event, its accurate description requires the representation of molecular surfaces with the finest resolution possible. However, while the charges are atom-localized and therefore their representation at high spatial resolution is immediate, the assignment of hydrophobicity based on residues inherently translates into its representation at a much lower resolution than that for electrical properties. Several studies [15]–[20] developed “atomic hydrophobicities” proposing different sets of atom types, but a sensitivity analysis regarding the number of atom types, as well as study comparing the protein molecular surfaces obtained using atom- or amino acid-level hydrophobicity is lacking.
Separate from the physical resolution of hydrophobicity, i.e., at atom- or amino acid-level, the impact of using different geometrical resolutions for the construction of the molecular surface has been also relatively under-studied. Indeed, the representation of the molecular surface, which relies on procedures [21]–[27] that use the protein structure deposited in databases, such as Protein Database, PDB [28], usually uses a geometrical resolution between 1.4 to 5 Å, which represents the size of the small molecular species the proteins interact with. However, as discussed before [29], there are many situations that justify the use of larger probes because the protein interacts with larger objects, e.g., membrane lipid rafts [30], cytoskeleton proteins [31], amyloid plaques [32], biomaterials surface [33], biomedical micro-devices [34], [35] and chromatographic media [36]. Also, from the methodology point of view, the probing of the molecular surfaces with at different geometrical resolutions, i.e., using different probe radii, can reveal structural features of the proteins, e.g., shielding of the hydrophobic core [29].
To this end, the present study proposes a methodology for the derivation of atomic hydrophobicity from any hydrophobicity scale, runs a sensitivity analysis to assess the suitability of alternative atom types, and compares the results obtained with atom- and amino acid-level representation of hydrophobicity on molecular surfaces.
Methods
Terminology and definitions
Usually, hydrophobicity defines the property of a physico-chemical unit, i.e., a material, a surface, a molecule, or a chemical group, which reflects a particular density and geometrical distribution of water molecules around that unit. When this property, measured by various methods, reflects the repelling of water molecules, this value, usually negative, is also denominated as hydrophobic. Conversely, when the property reflects an increased density of water molecules around the unit, the measured property, with values usually positive, is denominated as hydrophilicity. A physico-chemical unit, in particular a molecule or a chemical group, could contain various sub-units, e.g., chemical groups, or atoms, respectively, which have distinct and different hydrophobicities and/or hydrophilicities. If at least two, non-contiguous units present a hydrophobic and a hydrophilic character, respectively, the unit is deemed amphiphilic. To avoid confusions resulting from the overlap of terms for different parameters, and for the purposes of the analysis of the characterization of protein molecular surfaces, the following terminology will be used:
hydrophobicity is the measured hydrophobicity of a unit, i.e., atom, or amino-acid, which is hydrophobic and which does not have an amphiphilic character, i.e., an atom, or which is assumed, or assigned not to have an amphiphilic character, i.e., amino-acids;
total hydrophobicity is the sum of the hydrophobicities of the units, i.e., atoms, or amino-acids, which are exposed on the protein molecular surface, weighted with their respective exposed areas;
hydrophilicity is the measured hydrophobicity of a unit, e.g., amino-acid or atom, which is hydrophilic and which does not have an amphiphilic character, i.e., an atom, or which is assumed, or assigned not to have an amphiphilic character, i.e., amino-acids;
total hydrophilicity is the sum of the hydrophilicities of the units, i.e., atoms, or amino-acids, which are exposed on the protein molecular surface, weighted with their respective exposed areas;
overall hydrophobicity is the hydrophobicity of the amphiphilic protein (previously [29] denominated as amphiphilicity), calculated as the algebraic sum of the total hydrophobicity and total hydrophilicity of the units exposed on the molecular surface, calculated by either using amino-acid, or atom-based methodologies.
Proteins
A set of 35 proteins (Table 1) selected from the Protein Bata Bank [28], comprising several representative types, i.e., lactalbumins, lysozymes, ribonucleases, hemoglobins and related proteins, albumins and antibodies, have been selected for the comparison of amino acid- and atom-level representation of amphiphilicity. For the purposes of this contribution the chosen proteins need to have a convex shape. Indeed, the probing of proteins that exhibit concave shapes, most notably channel proteins, by probes with increasing radii will produce unreliable results, because much of their interior molecular surface will be inaccessible to larger probes. Finally, to ensure a representative comparison between the atomic- and amino acid-based hydrophobicity, the selected set of proteins is identical with the one used in a previous contribution [29], which reports on the probing of protein molecular surfaces with probes of different sizes.
Table 1. Proteins used for the analysis of molecular surfaces.
| Cluster | Protein no. | Protein name | PDB code | Atoms | Residues | Chains |
| 1 | 1 | α lactalbumin | 1A4V | 1092 | 123 | 1 |
| 2 | 2 | porcine β-lactoglobulin | 1EXS | 1248 | 160 | 1 |
| 3 | bovine β-lactoglobulin | 1BEB | 2473 | 324 | 2 | |
| 3 | 4 | chicken egg-white lysozyme | 1LYZ | 1001 | 129 | 1 |
| 5 | turkey egg-white lysozyme | 135L | 994 | 129 | 1 | |
| 6 | hen egg-white lysozyme | 2LYM | 1001 | 129 | 1 | |
| 7 | triciclic lysozyme | 2LZT | 1001 | 129 | 1 | |
| 8 | mutant phage T4 lysozyme | 1L35 | 1305 | 164 | 1 | |
| 9 | T4 lysozyme | 1LYD | 1309 | 164 | 1 | |
| 4 | 10 | ribonuclease-A | 8RAT | 951 | 124 | 1 |
| 11 | ribonuclease-A | 1RBX | 956 | 124 | 1 | |
| 12 | bovine ribonuclease-A | 3RN3 | 957 | 124 | 1 | |
| 13 | ribonuclease-A | 1AFU | 1894 | 248 | 2 | |
| 5 | 14 | human oxyhemoglobin | 1HHO | 2192 | 287 | 2 |
| 15 | human carbonmonoxy hemoglobin | 2HCO | 2192 | 287 | 2 | |
| 16 | horse hemoglobin | 2DHB | 2201 | 287 | 2 | |
| 17 | human hemoglobin A | 1BUW | 4342 | 574 | 4 | |
| 18 | human hemoglobin | 1Y4F | 4368 | 574 | 4 | |
| 19 | hemoglobin mutant | 1A01 | 4368 | 574 | 4 | |
| 20 | human hemoglobin | 1Y4P | 4376 | 574 | 4 | |
| 21 | hemoglobin mutant | 1A00 | 4382 | 574 | 4 | |
| 22 | human hemoglobin | 1Y46 | 4382 | 574 | 4 | |
| 23 | human deoxyhemoglobin | 2HHB | 4384 | 574 | 4 | |
| 24 | human hemoglobin | 1Y4G | 4366 | 574 | 4 | |
| 25 | hemoglobin mutant | 1A0U | 4386 | 574 | 4 | |
| 26 | hemoglobin mutant | 1A0Z | 4386 | 574 | 4 | |
| 27 | recombinant hemoglobin | 1C7D | 4396 | 576 | 3 | |
| 6 | 28 | human serum albumin complex with octadecanoic acid | 1E7I | 4496 | 585 | 1 |
| 29 | recombinant human serum albumin | 1UOR | 4617 | 585 | 1 | |
| 30 | serum albumin | 1E_78 | 4302 | 585 | 1 | |
| 31 | human serum albumin | 1AO6 | 4600 | 585 | 1 | |
| 32 | human serum albumin | 1BM0 | 4600 | 585 | 1 | |
| 7 | 33 | immunoglobulin | 1IGY | 10002 | 1294 | 4 |
| 34 | immunoglobulin | 1IGT | 10196 | 1316 | 4 | |
| 35 | intact human IgG B12 | 1HZH | 10355 | 1344 | 4 |
Note: The proteins marked in bold are model proteins and those in italics have been also used for the analysis of statistical strength.
The selected proteins have various molecular weights (14 to 148 [kDa]), residues (123 to 1344), isoelectric points (4.5 to 11) and shapes (globular, Y-shaped). Five representative proteins, i.e., lysozyme, ribonuclease, hemoglobin, albumin and IgG (Table 1, in bold) have been selected for an in-depth comparison of the atom-level and amino acid-level representation of hydrophobicities. The full results are presented in the Supporting Information section.
A subset of the hemoglobin class has been selected to test the fine differences between the hydrophobicity represented at atom- and amino acid-level resolution. Briefly, the subset comprises eight mutant structures of the deoxy forms of the protein, with the same number of residues (574), but with (i) the Trp37 residue, i.e., 1A0U and 1A0Z, for the crystal form 1 and 2, respectively; and with residues replacing the Trp37 residue by (ii) Tyr37, i.e., structures 1Y46 and 1A00, for crystal 1 and 2, respectively; (iii) Ala37, i.e., 1Y4F and 1A01, for crystal form 1 and 3, respectively; (iv) Glu37, i.e., 1Y4P, for crystal form 1; and (v) Gly37, i.e., 1Y4G for crystal form 1. A full description of these single residue mutations has been reported elsewhere [37].
Derivation of atomic hydrophobicities
The atomic amphiphilicities have been calculated as independent variables of the following system of linear equations:
for j = 1 to 20; and for each jth amino acid AAj:
| (1) |
where j = amino acid index; i = atom type index; AAj = the jth amino acid; hypho_ati – atomic hydrophobicity for atom type i; nij – number of atoms of type i in amino acid j; hypho_aaj – hydrophobicity of the amino acid j. This system of equations has been solved using several sets of atom types, proposed according to their chemical nature and charge. The number of atom types tested, m, was from 8 to 13. The proposed atom type matrices, [nij], are presented in File S1. The system of equations (Eq. 1) has multiple solutions because the number of equations is not equal with the number of variables. For each of these number of atom types the solution retained was the one that presented the best fit, i.e., the smallest standard deviation between the target values (amino acid hydrophobicities) and estimated ones (atomic hydrophobicities multiplied with the nij respective to amino acid j). The solution that represents the best fit, and consequently the one that has been retained for further calculations contains 12 atom types. The respective matrix, [ni12], is presented in Table 2.
Table 2. Atom types and relative atomic hydrophobicity (small correlation matrix, see text).
| No. | Name | Types | Atomic hydrophobicity [kcal mol−1] | |
| DGwif | DGoct | |||
| 1 | Cl | Aliphatic C | 0.2169 | −0.2485 |
| 2 | Cr | Aromatic C | −0.2607 | −0.3332 |
| 3 | Cx | C bonded to a heteroatom, less O | −0.1217 | −0.0217 |
| 4 | Cox | C bonded to O | −0.2645 | 0.2613 |
| 5 | Clp | Aliphatic C – positively charged | 1.5299 | 0.5564 |
| 6 | Cln | Aliphatic C – negatively charged | −0.9227 | −0.9126 |
| 7 | N | N in amide backbone | −0.0062 | −0.0763 |
| 8 | Np | N – positively charged (amino) | 0.3544 | 0.3748 |
| 9 | Nl | N in lysine | −0.1231 | −0.7263 |
| 10 | O | O in amide backbone | 0.4881 | 0.9277 |
| 11 | On | O negatively charged in COOH and OH | 0.7653 | 1.6749 |
| 12 | S | S in Cys and Met | 0.4989 | 3.029 |
Note: DGwif and DGoct are the free energies of transfer of AcEL-X-LL peptides from water to bilayer, or octanol interface, respectively [38].
Atom-based hydrophobicities
The initial test of fitness versus the number of atom types, from 8 to 13 atom types, used two hydrophobicity scales, i.e., the hydrophobicity of an amino acid embedded in a penta-peptide [38] as a measure of the enthalpy for its transfer (i) through a lipid membrane (DGwif); and (ii) from water to octanol (DGoct). The results of these calculations are presented in File S2. For the best fit of the atom types (m = 12), additional sets of atomic hydrophobicities have been calculated from other hydrophobicity scales, namely (i) Kyte-Doolittle, KD [39]; (ii) Hopp-Woods, HW [40]; (iii) logP [41] (cf. its implementation in HyperChem); (iv) two “estimated hydrophobic effects”, for “residue burial”, RB; and for “side chain burial”, SCB, [42]; (v) two measurements of HPLC retention, i.e., retn21 and retn74, [43]; (vi) position-specific apparent free energy of membrane insertion, ΔGapp(i) app, at position 0, DGapp_0, [44]; (vii) water-to-bilayer transfer free energy scale, ΔGsc wbi [45]; and (viii) unified hydrophobicity scale (UHS) for the water-membrane transfer free energy [46]. The results of these calculations are presented in File S3.
Molecular surfaces
The molecular surfaces of the selected proteins have been constructed using Connolly’s algorithm [22], [23], which records the position of the points of contact (or at a distance equivalent to the van der Waals radius of the respective atoms) between a virtual rolling probing ball with a set radius and the atoms on the surface of the protein. For amino acid-based overall hydrophobicity, total hydrophobicity and hydrophilicity, their spatial distribution was determined through the allocation, at the point of contact, of the hydrophobicity of the amino acid, weighted by the ratio of the probed surface per the total area of the amino acid. A similar procedure was used for mapping the spatial distribution of the atom-based hydrophobicities. The procedure involved the allocation of specific atomic hydrophobicity weighted with the ratio between the probed atomic area and the total atomic area. The results of the calculations regarding the exposed area vs. probe radii are presented in File S4.
The calculations used an in-house program [29], which is an upgrade of the Connolly’s original software code [23], [47], embedded in a Windows interface. The program has been run on a personal computer with a 64-bit operating system, an Intel Core i7-3630QM CPU @2.40 GHz, and an installed memory of 8GB. The 4D points (x, y, z coordinates and molecular property) have been visualized using DS Viewer Pro. (from Accelerys Inc.). The molecular surfaces have been constructed for all 35 proteins in the dataset (Table 1), for probe radii ranging from 1.4 Å to 20 Å. Beyond probe radii of 20 Å it was found [29] that the change of the properties on the molecular surface is negligible. Consequently the calculations stopped at this threshold.
Protein properties on the molecular surface
Three types geometrical and physico-chemical properties have been calculated on the molecular surface of the selected proteins: (i) global properties (i.e., total surface; overall hydrophobicity, total hydrophobicity and hydrophilicity, for amino acid- and atom-based calculations); (ii) property relative densities (i.e., overall and total hydrophobic and hydrophilic relative density, calculated by dividing the property value to the total molecular area); and (iii) property specific densities (calculated by dividing the respective property, e.g. total hydrophobicity, to the area that property turns up, e.g. hydrophobic area). For the comparison purposes, the overall hydrophobicity, i.e., the algebraic sum of hydrophobicity expressed in negative numbers; and hydrophilicity expressed in positive numbers, has been calculated for both amino acid-based and atom-based hydrophobicity scales. This methodology, applied here to atom-based properties, was used before [29] but only for amino acid-based properties. The full results are presented in File S5.
The hemoglobins subset has been separately analyzed using the same procedures. To compare the molecular surface properties with the hydrophobicity of the single residue replacement, the values for the proteins that present two crystallographic forms, i.e., 1A0U and 1A0Z for Trp; 1Y46 and 1A00 for Tyr; and 1Y4F and 1A01 for Ala, have been averaged, but those with a single crystallographic form, i.e., 1Y4P for Glu and 1Y4G for Gly, remained unchanged. The full results regarding this subset are presented in File S6.
Results and discussion
1. Atomic hydrophobicity
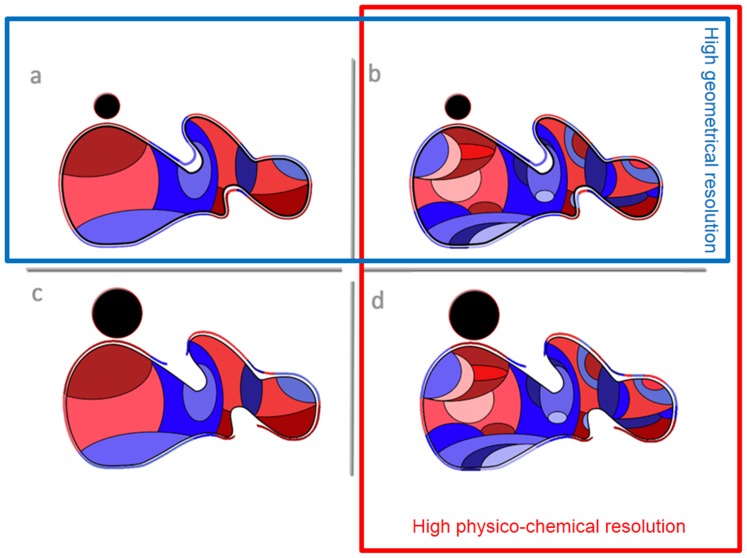
Because the charge is an atom-based property, the spatial representation of charges on the protein molecular surface can be inherently performed at atom-level resolution. In contrast, the spatial distribution of hydrophobicity cannot be usually represented at high resolution, because of two reasons. First, as the hydrophobicity is usually assigned to amino acids not to atoms, its spatial representation on the protein molecular surface is constructed at several-atoms resolution, i.e., from patches comprising several atoms belonging to a parent amino acid, which is probed by the molecular surface probing ball. Intuitively, an atom-level representation of hydrophobicity would allow a more precise quantification of the properties manifested on the molecular surface and inference of the molecular recognition between protein and small molecular species. For instance, the role arginine, which comprises chemical groups with various hydrophobicities along the molecule, plays in protein-protein interactions is difficult to be understood within the framework of an evenly distributed hydrophobicity. A schematic of the differences between a molecular surface which is represented at amino acid- and at atomic level is presented in Figure 1, a and b, respectively. Furthermore, constructing the molecular surface using larger probes, which could be relevant to the analysis of the interaction of proteins with larger objects [29], e.g., nanoparticles, flat surfaces, will result in more uncertain quantification of the hydrophobicity, as the molecular surface is represented by a collection of atoms which represent a decreasingly-smaller fraction of their parent amino acids. This situation is presented schematically in Figure 1, c and d, respectively.
Figure 1. Schematics of different representations of molecular surfaces.
Top row: representation of the hydrophobicity on the molecular surface, at (a) low, amino acid-level; and (b) high, atomic-level resolutions. Bottom row, the same representation for molecular surfaces probed with larger probes. Scheme upgraded from [29], which reports the mapping at low, amino acid-based hydrophobicity (i.e., a and c).
Second, in contrast with the charges, hydrophobicities are not represented in a standardized manner, with more than 100 hydrophobicity scales being presently proposed. Although “hydrophobic potentials” have been proposed [48]–[51], including some for atomic level representations [41], [50]–[53], the non-standardized hydrophobicity, in particular at atom level, precludes their universal use.
There are several possible avenues for the derivation of atomic hydrophobicities, either independent of the Accessible Solvent Area (ASA), as used primarily in this work; or accounting for ASA when solving the system of equations (Eq. 1). Probing the molecular surface at different geometrical resolutions –a central methodological tool for assessing the structuring of the molecular surface, will result in different ASA’s for different probe radii (see File S7). Consequently, if ASA’s are used as weighting factors for the calculation of atomic hydrophobicities, then the solution of the system of equations (Eq. 1) will be dependent on the radius of the probe used for the construction of the molecular surface. Equally important, the equivalence between atomic hydrophobicities and amino acid ones from which they are derived will cease, thus making the comparison between the two methods of constructing the distribution of hydrophobicity and hydrophilicity on the protein molecular surface inoperable. Furthermore, if ASA’s are used for the calculation of atomic hydrophobicities, their equivalent formalism with atomic charges also cease to exist, making their possible use for the development of hydrophobic potential also inoperable. A full treatment of the modes of calculation of atomic hydrophobicities is presented in File S8. For all these reasons, and although we report results obtained both accounting or not ASA’s (see File S3 and S4), the further analysis will mainly use the atomic hydrophobicities obtained from Eq. 1.
2. Derivation and use of atomic hydrophobicities
While several atomic hydrophobicity scales have been proposed in the last decades, they present several limitations. For example they (i) are estimated from large QSAR databases where amino acids represent a small fraction of the archived molecules [54], [55], thus skewing the results away from the residues of interest for the analysis of proteins; or (ii) propose a small number of atom types, e.g., m = 5 [15], [16], [56], m = 6,7 [18], [20], m = 8 [17], thus potentially not being able to describe the molecular surface with sufficient atom-specificity; or (iii) use proprietary parameters [41], [57]; (iv) use “hydrophobic potentials” (the analogue to electrostatic potentials), usually embedded in proprietary software [41], [58]–[60]; or (v) result from the compilation of several different sources [61], [62]. Most importantly, none of these atom-based hydrophobicity scales are derived from amino acid-based ones, therefore making the comparison of molecular surfaces constructed using amino acid-, or atom-based hydrophobicities difficult. The methodology for the derivation of atom-based hydrophobicity proposed here attempts to address many of these limitations.
Several sets of atomic hydrophobicities are proposed, each calculated for a number of representative atom types, varying from 8 atom types, i.e., starting with the set proposed by Efremov at al. [17], to 14. The selection of the atom types was based on the chemical structure and environment of the respective amino acid. For m = 8 the atom types are: Cl – aliphatic C; Cr – aromatic carbon; Cx – carbon linked to a heteroatom; N – uncharged nitrogen; O – uncharged oxygen; S – sulphur; Np – positive charged nitrogen; and On – negatively charged oxygen. For m = 12 this set was expanded by splitting the C atoms types according to their charge, i.e., in conformity with the charges assigned by the Amber force field [63]; and creating a new atom type for the N atom in lysine. The representative atom types for m = 12 are presented in Table 2.
The criterion for the choosing the optimum number of atom types has been the overall (i.e., for all 20 amino acids) best fit of the estimated atom-based hydrophobicities compared to the actual amino acid-based ones used for calculations. Two hydrophobicity scales, i.e., the hydrophobicity of an amino acid embedded in a penta-peptide, [38] derived from the thermodynamic measurements of the enthalpy of the transfer of the respective peptide through a lipid membrane (DGwif); and from water to octanol (DGoct), respectively, have been used to calculate the best fit between atom-based and amino acid based hydrophobicities. The best fit increased moderately, but steadily, with the increase of the number of atom types, m, from 8 to 12. For m = 13 the improvement of the fit ceased and for m = 14 the system could not be solved anymore. The detailed discussion on these results is presented in File S8 and a full description of the data is presented in Files S1–S5. The evolution of the fit with the number of atom types is presented in Table 3.
Table 3. Fit for 12 atom types for different hydrophobicity scales.
| Scale | R2 | Standard deviation |
| DGwif | 0.95 | 0.86 |
| DGoct | 0.97 | 1.64 |
| DGhx | 0.97 | 8.51 |
| DGsa | 0.82 | 5.5 |
| KD | 0.89 | 2.79 |
| WD | 0.95 | 1.88 |
| Log P | 0.96 | 1.61 |
| DGaa_0 | 0.87 | 0.28 |
| DGsc_wbi | 0.97 | 0.36 |
| DGwm_UHS | 0.82 | 0.18 |
Note: DGwif, DGoct, DGhx and DGsa are the free energies of transfer of AcEL-X-LL peptides from water to bilayer, or octanol interface, respectively [38]; Kyte-Doolittle, KD [39]; Hopp-Woods, HW [68]; partition coefficient, logP [41]; position-specific apparent free energy of membrane insertion at position 0, DGapp_0 [44]; water-to-bilayer transfer free energy scale, DGsc_wbi [45]; and unified hydrophobicity scale (UHS) for the water-membrane transfer free energy, DGwm_UHS [46].
3. Protein overall hydrophobicity on the molecular surface
Once the optimum set of atom-based hydrophobicity, i.e., atom types and the values of the atomic hydrophobicities, has been established, one can quantify the protein overall hydrophobicity manifested on its molecular surface, and compare it with the one calculated with the classical amino acid-based hydrophobicity. The following discussion will focus on five representative proteins, i.e., lysozyme, ribonuclease, hemoglobin, albumin and IgG, which have vastly different molecular weights, i.e., from 129 to 1344 residues (Table 1, in bold); and shapes, i.e., globular, ellipsoidal and Y-shaped. While the following results are discussed for the DGwif-derived hydrophobicity only, similar results are obtained for all other hydrophobicity scales. The full results for all 35 model proteins are presented in File S5.
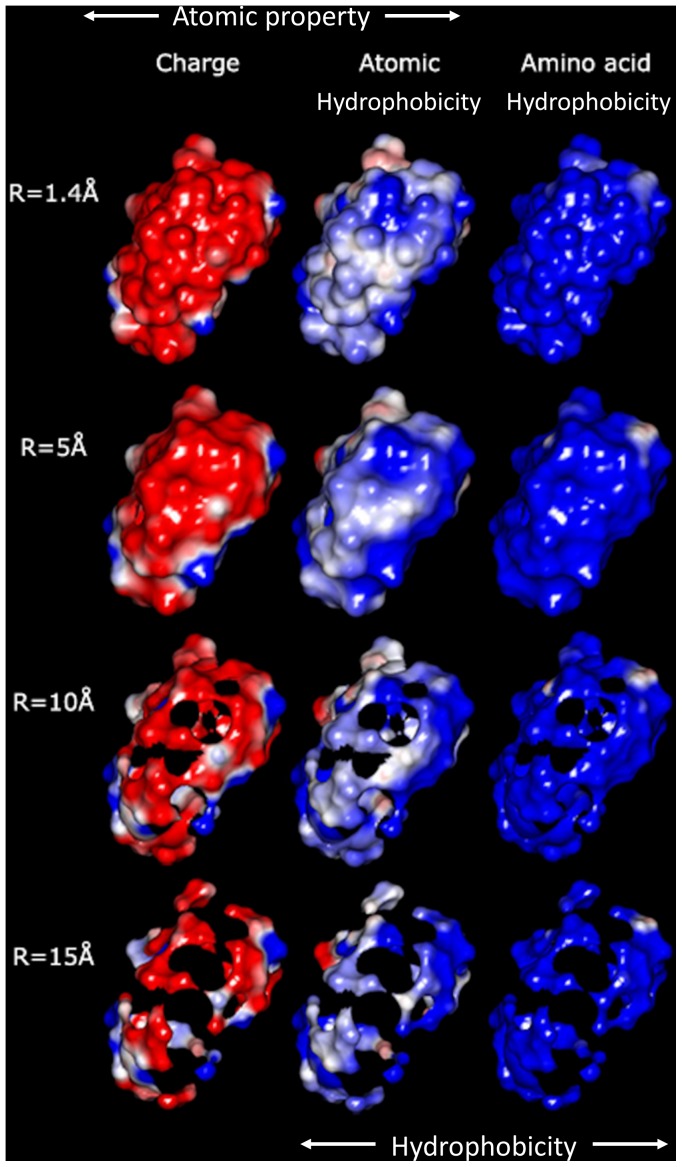
The comparison of the molecular surface (Figure 2 for ribonuclease) allows a qualitative discrimination between properties calculated at atom-level resolution, but of a different nature, i.e., charges and atomic hydrophobicity (Figure 2, left and middle columns, respectively); as well as between those of the same nature, i.e., hydrophobicity, but calculated at atom- and amino acid-level (Figure 2, middle and right columns, respectively). A preliminary inspection shows that the distribution of atomic hydrophobicity, despite being physico-chemically similar with the amino acid hydrophobicity, from which it is actually derived, resembles far more the distribution of charges on the molecular surface. Indeed, the molecular surface represented by amino acid hydrophobicity remains largely, and evenly, hydrophilic, regardless of the geometrical resolution it is probed at. Conversely, the molecular surface represented by atomic hydrophobicity offers a far more varied landscape. For instance, several hydrophobic ‘fingers’, not detected by the amino acid hydrophobicity molecular surface, but visible as near-zero charges on the charge molecular surface (Figure 3, left column), remain apparent, regardless of the probe radii. A more detailed graphical representation of the evolution of the property-molecular surface is presented in File S9.
Figure 2. Comparison between the representation of atom-based properties, i.e., charges (left column; red = negative, blue = positive), atomic hydrophobicity (middle column; red = hydrophobic and blue = hydrophilic region); with amino acid-based properties, i.e., amino acid-based hydrophobicity (right column) on the molecular surface of ribonuclease (PDB ID: 1AFU).
The molecular surface is probed with decreasing geometrical resolution (from top to bottom).
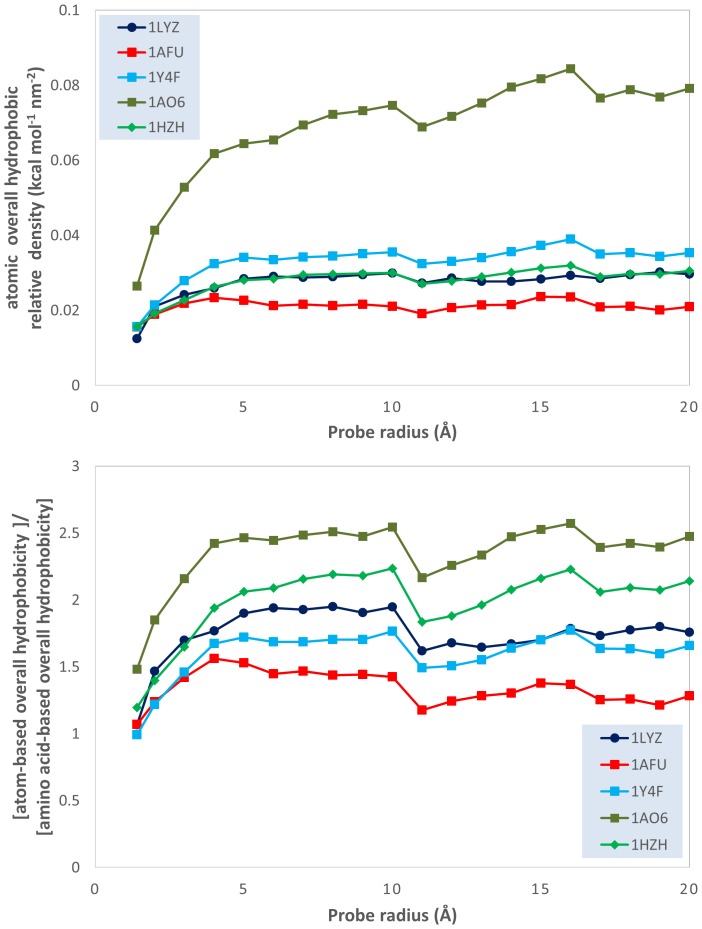
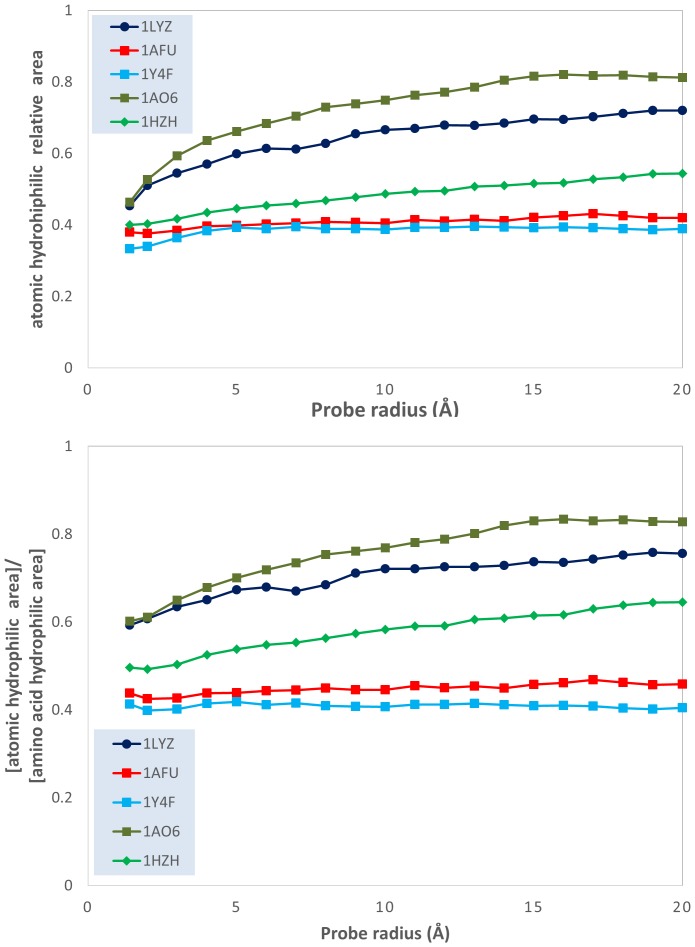
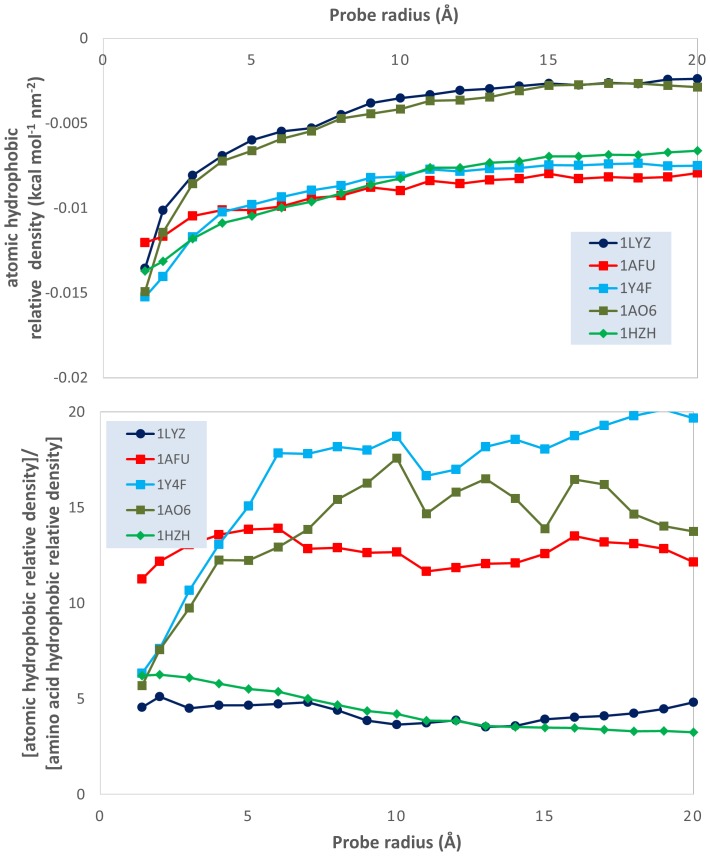
Figure 3. Evolution of the ratio between atom-based overall hydrophobicity and total molecular surface area (relative density of the atomic hydrophobicity); and of the ratio of the atomic and the amino acid overall hydrophobicities; vs. probe radii for 5 model proteins: lysozyme (1LYZ); ribonuclease-A (1AFU); human hemoglobin (1Y4F); human serum albumin (1AO6); human IgG (1HZH).
Atomic and amino acid based hydrophobicities
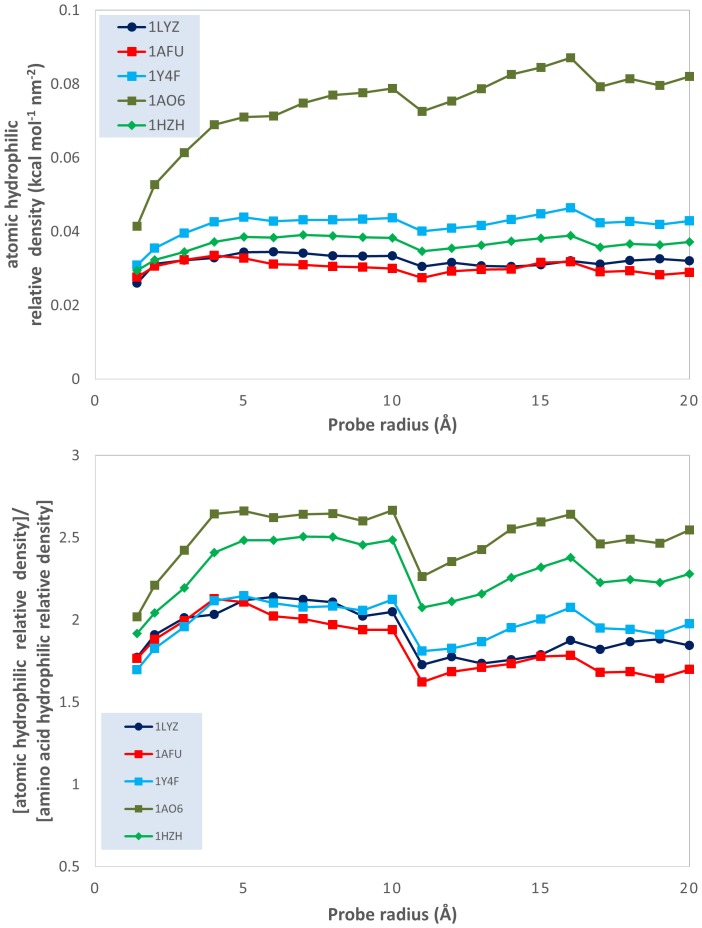
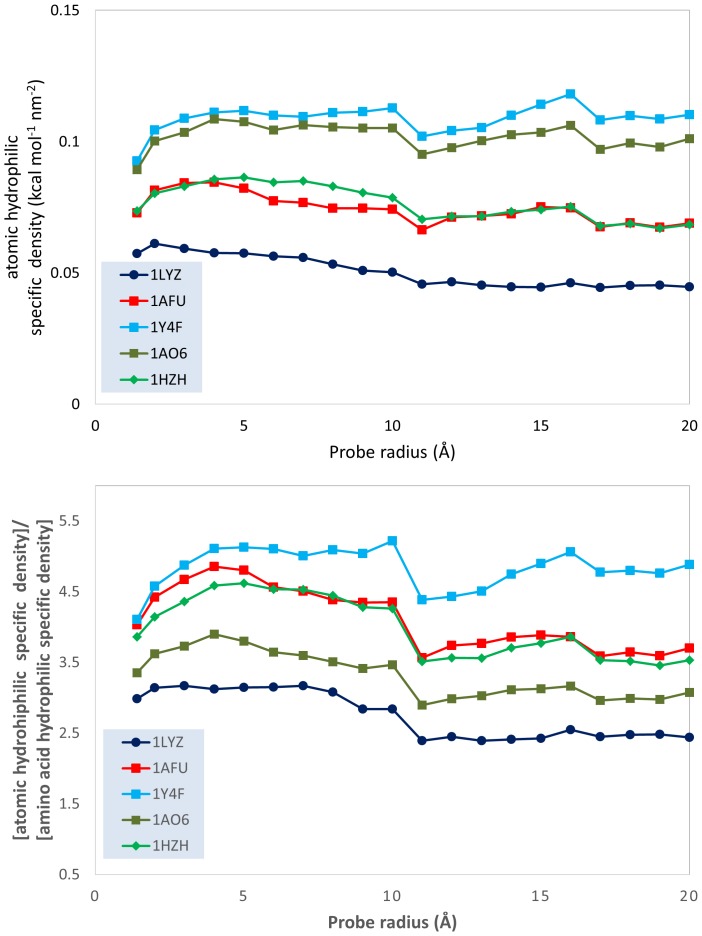
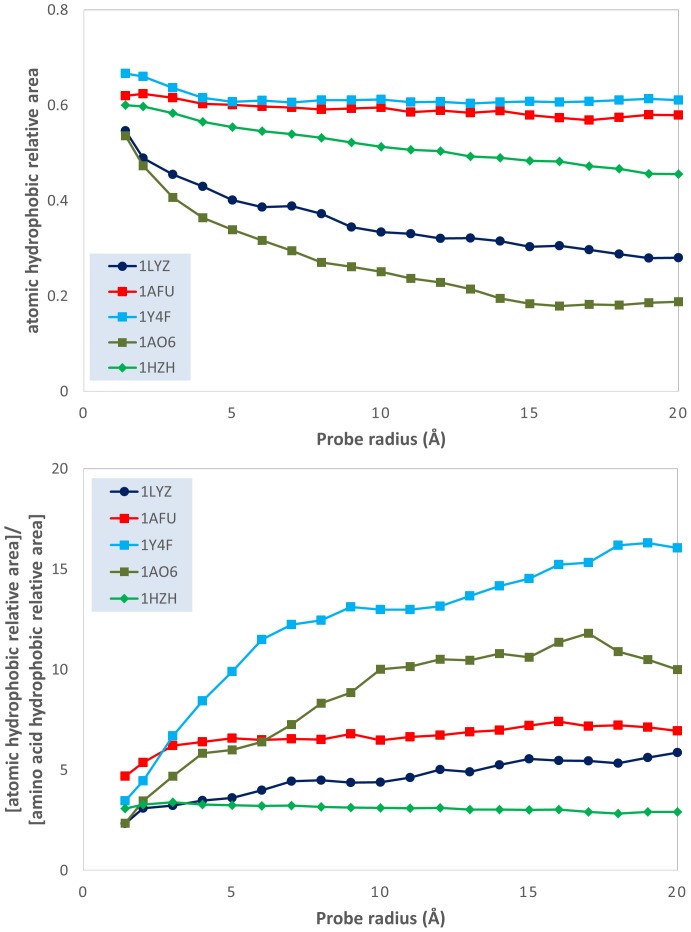
This qualitative analysis is also supported by quantitative data, which could also provide a more detailed physical insight. The variation of the atomic physico-chemical properties, i.e., overall hydrophobicity, total hydrophilicity and hydrophilicity, as well as their derived measures, i.e., relative area (hydrophilic or hydrophobic area divided by total molecular surface area), relative density (overall hydrophobicity, total hydrophobicity or hydrophilicity divided by the total molecular surface area) and specific density (hydrophilicity or hydrophobicity divided by their respective area) with the variation of the probe radius is presented in Figures 3–9 (top panels); and a synthetic overview of these parameters is presented in Table 4. Table 4 also presents the comparison between the atomic and their homologue amino acid properties (also presented in Figures 3–9, bottom panels).
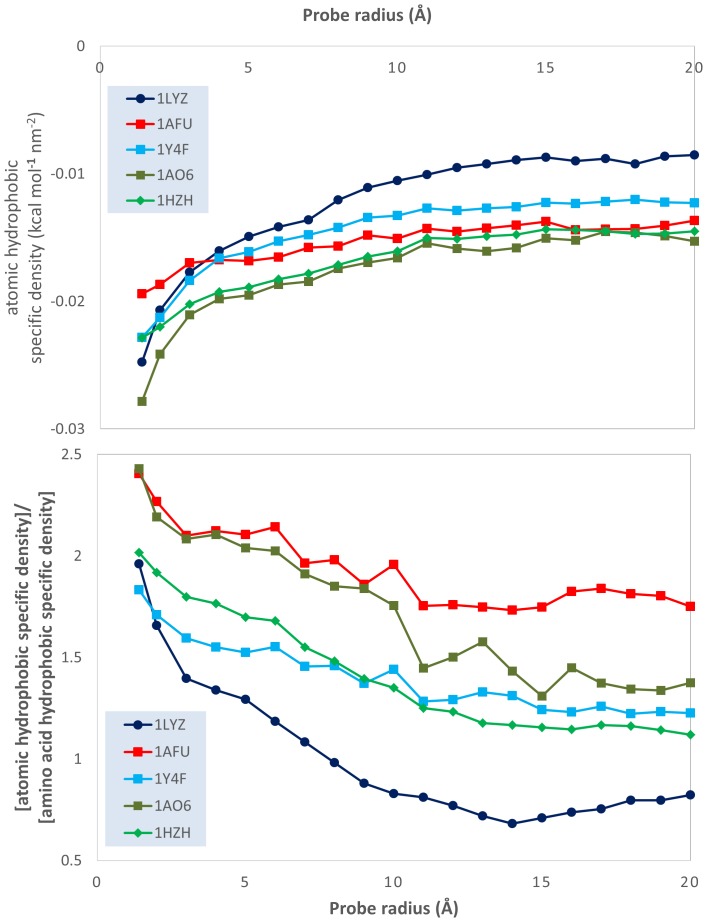
Figure 9. Evolution of the ratio between the atomic hydrophobicity and the hydrophobic area (hydrophobic specific density); and of the ratio between the atomic and the amino acid hydrophobicity specific densities; vs. probe dimensions for 5 model proteins: lysozyme (1LYZ); ribonuclease-A (1AFU); human hemoglobin (1Y4F); human serum albumin (1AO6); human IgG (1HZH).
Table 4. General comparison of the evolution of molecular surface properties with the probe radius, calculated at atom- and amino acid level.
| Property (definition) | Atomic property relationship vs. probe radius (R) increase | Atomic property ratio to amino acid homologue |
| Overall hydrophobicity | ||
| Relative density1,2: Overall hydrophobicity/Molecular surface area | Slight increase for most proteins Large increase (from ∼0.02 to ∼0.08 kcal nm−2) for 1AO63 [Figure 3 top] | Generally larger (up to 2.5x); Generally increase for R = 1 to 5 Å, then constant [Figure 3 bottom] |
| Hydrophilicity | ||
| Relative area: Hydrophilic area Total molecular area | Constant, i.e., ∼40% of the total area (1AFU, 1Y4F) or increase from 40% to 50% (1Y4F), 60% (1LYZ) and 80% (1AO6) [Figure 4 top] | Generally smaller, between 40% to 80% Constant (40%, 1Y4F, 1AFU), or increase from 50% up to 80% [Figure 4 bottom] |
| Relative density: Total hydrophilicity Molecular surface area | Slight increase for most proteins Large increase (from ∼0.02 to ∼0.08 kcal nm−2) for 1AO6 [Figure 5 top] | Generally larger –1.5 to 2.5x Generally constant with R [Figure 5 bottom] |
| Specific density: Total hydrophilicity Hydrophilic area | Rather constant or a slight decrease (1LYZ) [Figure 6 top] | Much larger, i.e., 2.5–5.5x; Slight decrease with R [Figure 6 bottom] |
| Hydrophobicity | ||
| Relative area: Hydrophobic area Total molecular area | Constant, i.e., ∼40% of the total area (1AFU, 1Y4F) or decrease from 60% to 50% (1Y4F), 40% (1LYZ) and 20% (1AO6) [Figure 7 top] | Much larger, between 2.5x to 17x Constant (1HZH) or increase with R [Figure 7 bottom] |
| Relative density: Total hydrophobicity Molecular surface area | Slight decrease4 from −0.015 to −0.01 kcal nm−2 (1HZH, 1AFU, 1Y4F). Large decrease, from ∼0.015 to ∼0.005 kcal nm−2 (1AO6, 1LYZ) [Figure 8 top] | Much larger 5 to 20x Generally constant with R [Figure 8 bottom] |
| Specific density: Total hydrophobicity Hydrophobic area | Decrease from −0.03 and 0.02 to −0.02 and −0.01 kcal nm−2 [Figure 9 top] | Generally larger, i.e., 1–2.5x;Decrease with R [Figure 9 bottom] |
Notes:
1. Overall hydrophobicity is the algebraic sum of hydrophilicity (positive sign) and hydrophobicity (negative sign). Consequently, the increase of the overall hydrophobicity means that it is more hydrophilic.
2. The relative density of the overall hydrophobicity is equal to its specific density.
3. PDB codes for model proteins: lysozyme (1LYZ); ribonuclease-A (1AFU); human hemoglobin (1Y4F); human serum albumin (1AO6); human IgG (1HZH).
4. Hydrophobicity is expressed in negative numbers. Consequently, a decrease in hydrophobicity will be represented by a move towards 0.
The qualitative (Figure 2) and quantitative data (Figures 3–9 and Table 4) allows for the construction of the following framework regarding the structuring of the protein molecular surfaces:
Overall hydrophobicity. The slight, or –for albumin- considerable, increase of the density of overall hydrophobicity with the probe radius (Figure 3, top) indicates that protein molecular surfaces are more hydrophilic towards their outer edges, which is consistent with the “hydrophobic core” model. Moreover, the considerable (approximately two times) higher values obtained for atom-level density of overall hydrophobicity compared with amino acid ones (Figure 3, bottom) suggest that amino acid-based formalism underestimates the “hydrophobic core” structuring of the molecular surface.
Total hydrophilicity. The slight increase of the atomic hydrophilic relative area with the probe radius (Figure 4, top) and the slight-to-considerable increase of the atomic hydrophilic relative density with the probe radius (Figure 5, top) also supports the “hydrophobic core” model. However, this observation needs to be qualified: the atom-based calculations reveal lower hydrophilic areas (Figure 4, bottom) and higher hydrophilic relative densities (Figure 5, bottom) than the homologue values obtained by amino acid-based calculations. This apparent contradiction can be reconciled if we assume that the hydrophilic areas are more “hydrophilicity intense” than predicted by amino acid calculations. The much higher atomic hydrophilic specific density than its amino acid counterpart (Figure 6, bottom) also supports this interpretation.
Total hydrophobicity. The above conclusion is also supported by hydrophobicity calculations. Indeed, the slight-to-considerable decrease of the hydrophobic relative area with the probe radius (Figure 7, top) and the considerable decrease of the hydrophobic relative density (Figure 8, top) support the “hydrophobic core” model. However, the much larger prediction of the hydrophobic areas by atomic based calculations compared with amino acid ones (approximately 5 times even for the smallest radius considered, but above 10–15 times for some proteins (Figure 7, bottom) suggests a much larger extent of the hydrophobic molecular surface predicted by atom-based calculations than amino acid ones. Apparently, the “hydrophobic intensity” of these extended hydrophobic areas is also considerably higher (Figure 8, bottom) than those calculated from amino acid properties. The higher, but decreasing with the probe radius, ratio between the atomic hydrophobic specific density and its amino acid counterpart (Figure 9, bottom) results from the coupling of the decrease of the former (Figure 9, top) and the constant values for the latter [29].
Atom-based description of protein molecular surfaces. The observation that the atom-based representation of the molecular surfaces has considerably higher resolution, coupled with the fact that the respective atomic hydrophobicities have been derived directly from a chosen amino acid hydrophobicity scale, leads to the description of the protein molecular surface with better accuracy and precision than that using amino acid hydrophobicities. While both atom- and amino acid-based calculations describe the protein molecular surface as hydrophilic, and more so with the increase of the probe radius, the atom-level description reveals a “leopard skin” design, with more intense hydrophobic and hydrophilic patches than the rather uniform-hydrophilic surface predicted by the amino acid calculations. Moreover, considering the specific hydrophobic density, the validity of the hydrophobic core concept appears not to be fully supported by amino acid-based calculations, especially for large proteins (where it should be the most apparent, [64], [65]), but it is valid if atom-based hydrophobicity is used. These observations lead to the conclusion that atom-based hydrophobicities offer a better representation of the protein molecular surface, as demonstrated by the general agreement with the “hydrophobic core” concept. The molecular surfaces depicted in Figure 2 support these conclusions.
Figure 4. Evolution of the ratio between the atomic hydrophilic area and the total molecular surface area; and of the ratio between the atomic and the amino acid hydrophilic areas; vs. probe dimensions for 5 model proteins: lysozyme (1LYZ); ribonuclease-A (1AFU); human hemoglobin (1Y4F); human serum albumin (1AO6); human IgG (1HZH).
Figure 5. Evolution of the ratio between the atomic hydrophilicity and the total molecular surface area (hydrophilic relative density); and of the ratio between the atomic and the amino acid hydrophilicity relative densities; vs. probe dimensions for 5 model proteins: lysozyme (1LYZ); ribonuclease-A (1AFU); human hemoglobin (1Y4F); human serum albumin (1AO6); human IgG (1HZH).
Figure 6. Evolution of the ratio between the atomic hydrophilicity and the hydrophilic area (hydrophilic specific density); and of the ratio between the atomic and the amino acid hydrophilicity specific densities; vs. probe dimensions for 5 model proteins: lysozyme (1LYZ); ribonuclease-A (1AFU); human hemoglobin (1Y4F); human serum albumin (1AO6); human IgG (1HZH).
Figure 7. Evolution of the ratio between the atomic hydrophobic area and the total molecular surface area; and of the ratio between the atomic and the amino acid hydrophobic relative areas; vs. probe dimensions for 5 model proteins: lysozyme (1LYZ); ribonuclease-A (1AFU); human hemoglobin (1Y4F); human serum albumin (1AO6); human IgG (1HZH).
Figure 8. Evolution of the ratio between the atomic hydrophobicity and the total molecular surface area (hydrophobic relative density); and of the ratio between the atomic and the amino acid hydrophobicity relative densities; vs. probe dimensions for 5 model proteins: lysozyme (1LYZ); ribonuclease-A (1AFU); human hemoglobin (1Y4F); human serum albumin (1AO6); human IgG (1HZH).
4. Analysis of a homologous set of proteins
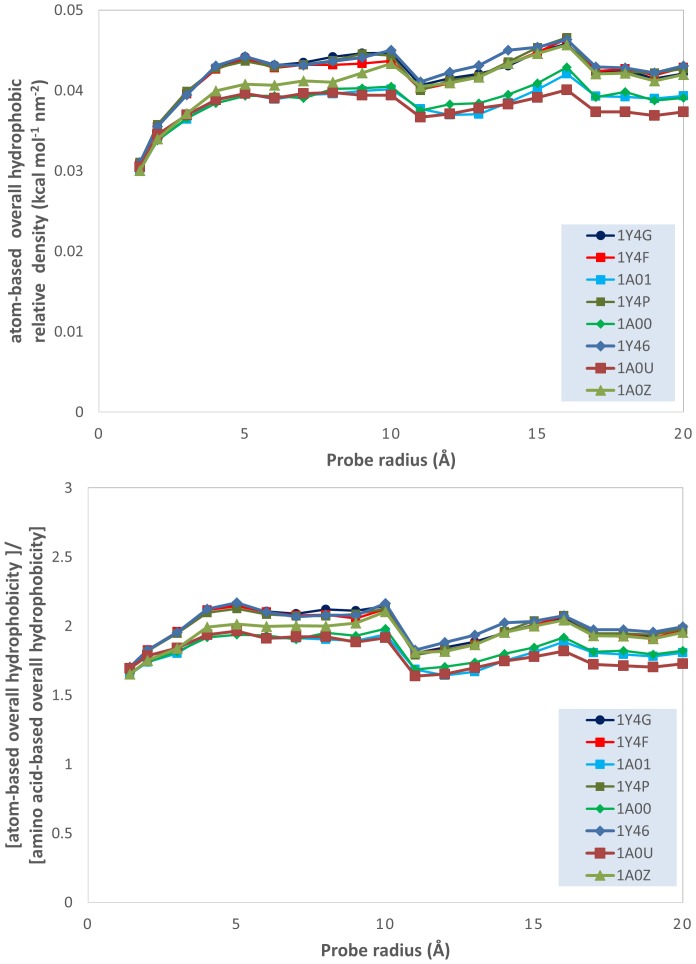
A more precise comparison of the differences between the atom- and amino acid-based hydrophobicity quantified on the protein molecular surfaces is occasioned by the analysis of a sub-set of hemoglobin single-residue mutants. Because the proteins in this sub-dataset are much more similar between themselves than the rest of the proteins in the overall, larger data set, as only one residue (Trp37) is different, and because this replacement, with Ala, Gly, Glu and Tyr, did not lead to substantial changes in the tertiary structure of the hemoglobins [37], the evolution of the molecular surface parameters with the probe radius is expected to be much closer than that for very different proteins. While this assumption is qualifiedly true, all conclusions drawn from the analysis of very different proteins, as described in the above section, are validated by the analysis on the hemoglobin dataset (see File S6). For example, the evolution of the density of the overall hydrophobicity with the radius of the probe (Figure 10), reveals an increasingly hydrophilic surface with the decrease of the probing resolution; and a higher hydrophilicity (approximately two times) of the molecular surface than that predicted by the amino acid calculations.
Figure 10. Evolution of the ratio between atom-based overall hydrophobicity and total molecular surface area (relative density of the atomic overall hydrophobicity); and of the ratio of the atomic and the amino acid overall hydrophobicities; vs. probe dimensions for hemoglobin subset.
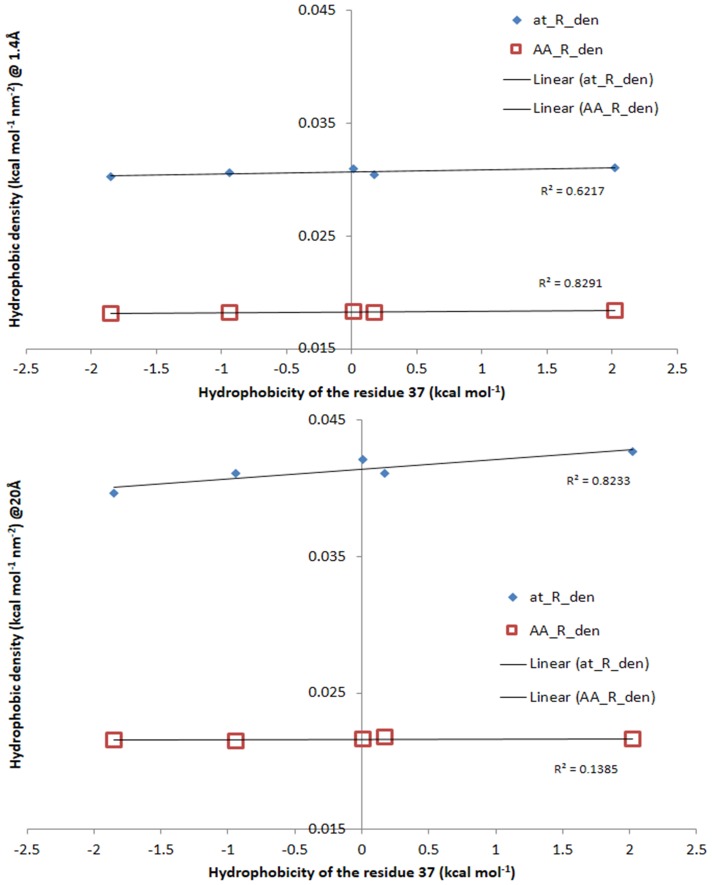
Working with very similar set of proteins could lead to important conclusions following the removal of the “noise” caused by too large variations. For instance, the amino acid-based overall hydrophobicity density is essentially identical for all hemoglobins, for both the finest and the coarsest probe, i.e., 1.4 Å and 20 Å, respectively (Figure 11, top and bottom, respectively). However, while the density of the atomic hydrophobicity for the finest probing resolution is also identical for all hemoglobins (albeit larger than amino acid homologue), the calculations for the coarsest probing shows an overall hydrophobicity density that seems to be protein-specific and correlated with the hydrophobicity of the amino acid that replaced the Trp37 in the natural hemoglobin structure.
Figure 11. Density of the overall hydrophobicity on the molecular surface of hemoglobin subset, at small (top) and large (bottom) probe radius vs. the hydrophobicity of the residue 37.
The hydrophobicities of the residue 37 are, from left to right, Gly, Ala, Glu, Tyr and Trp [38].
5. Computing time
For the computing system used in this study, the run time ranges from 2 sec for a small protein (lysozyme, 1LYZ, 1001 atoms) for the smallest probe radius (1.4 Å); to nearly 5000 sec for a large protein (IgG, 1HZH, 10196 atoms) for the largest probe radius considered (20 Å), as presented in Table 5. No difference has been noted between the calculations using amino acid hydrophobicities and those using atomic ones.
Table 5. Computing time (sec) for the construction of protein molecular surfaces on a personal computer.
| Protein (PDB Id) | 1LYZ | 1AFU | 1Y4F | 1AO6 | 1HZH |
| No. atoms→ Probe radius↓ | 1001 | 1894 | 4368 | 4600 | 10355 |
| 1.4 | 2 | 4 | 6 | 8 | 22 |
| 5 | 12 | 30 | 96 | 110 | 224 |
| 10 | 27 | 81 | 777 | 853 | 1447 |
| 15 | 33 | 105 | 1470 | 1678 | 3170 |
| 20 | 48 | 120 | 1722 | 1865 | 4939 |
6. Perspectives and future directions of research
The present study has demonstrated the benefits of using finer scale, atom-level description of hydrophobicity. These benefits could be further amplified pursuing several possible future directions of research:
Molecular surface databases. A recent comprehensive review of the present understanding of hydrophobicity [66], suggested that it would be beneficial to archive the data regarding the distribution of hydrophobicity and hydrophilicity on the molecular surface of the proteins, in particular those that have the structures deposited in the PDB. It was also suggested that this desideratum can be achieved through molecular simulations from which the fluctuations of the density of water molecules can be calculated. While this research avenue is certainly desirable, the calculations could be expensive and time consuming, even with the emergence of more powerful supercomputers. An interim solution could be the mapping of protein surfaces using atomic hydrophobicities, either the ones reported here, or others calculated using similar methodologies. Furthermore, once the atomic hydrophobicities of interest are derived, one can attempt to cluster the molecular surfaces of whole or parts of proteins through the comparison of atomic neighborhoods, as proposed recently [67].
Universality of atomic hydrophobicities. The present study described how atomic hydrophobicities can be derived from amino acid ones. While different niche applications would find a particular hydrophobicity scale more relevant than another, e.g., chromatography vs. lipid membranes, a standardization of atomic hydrophobicity would greatly help the transfer of knowledge from one application to another. This desideratum can be achieved via two approaches. First, one approach could consist in assigning atom types in accordance to wide-spread used force field, e.g., AMBER. This approach would have the benefit of creating ‘hydrophobic charges,’ which can then be easily used in molecular surface representations, including the calculation of ‘hydrophobic potentials’, such as those previously proposed [41]. Second, a more thorough, albeit computational intensive, approach would be to derive the atomic hydrophobicities from molecular dynamics simulations, e.g., distribution of water molecules around particular atoms, quantification of the fluctuations of water molecules distribution, as alluded above, etc. Aside from the large effort required, this approach would have the benefit of creating truly universal atomic hydrophobicities, as the procedure could be applied to any molecules, e.g., DNA, ligands, glycopeptides, etc. thus opening new avenues for fundamental studies in molecular biology or for applied research, such as drug discovery.
Conclusion
The mapping and quantification of the physico-chemical properties on the molecular surfaces of proteins using atomic hydrophobicities derived from the corresponding amino acid hydrophobicities scales, offers insights into the structuring of the protein molecular surfaces. The demonstration of the finer representation of protein molecular surfaces at atom level justifies the derivation of sets of these hydrophobicities for any chosen hydrophobicity scale that is appropriate for a specific application, thus opening the opportunity for the engineering of optimum protein-small ligand interactions, as well as protein-solid surfaces interactions. Furthermore, the results are expected to benefit both fundamental studies of protein function and drug discovery by providing a pathway for high resolution mapping of hydrophobicities on the molecular surface.
Supporting Information
Construction of various sets of atom types, from M = 8 to M = 13.
(XLSX)
Selection of the best atom types set by the regression of various sets of atom types (M = 8 to 12) for the hydrophobicity scale proposed by Wimley & White [38] .
(XLSX)
Calculation of the best atomic hydrophobicity sets for M = 12 and for various hydrophobicity scales when ASA is considered (Part 1) and when it is not considered (Part 2).
(XLSX)
Calculation of the Accessible Solvent Areas (ASA) for each atom in each amino acid as a function of the probe radius.
(XLSX)
Complete set of data regarding the calculation of physico-chemical properties on the molecular surface of the proteins in the total set ( Table 1 ), for atomic, amino acid and charges, the latter two from [29] .
(XLSX)
Complete set of data regarding the calculation of physico-chemical properties on the molecular surface of the proteins in the selected set of hemoglobins, for atomic, amino acid and charges.
(XLSX)
Example of molecular surface obtained by probing the protein with a small and a large probe.
(TIF)
Comprehensive discussion regarding the possibilities of calculation of atomic hydrophobicities.
(DOC)
Molecular surfaces of ribonuclease presented as a function of the probing resolution, from the finest (top) to the coarsest (bottom). The molecular surfaces are represented for charges (left column); amino acid-based hydrophobicity (right column); and atom-based hydrophobicity (middle columns). The atom-based molecular surfaces are presented using values directly derived from (Eq.1) – left middle column; and normalized to fit the range of the amino acid hydrophilicities – right middle column.
(TIF)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work has been supported by the Defense Advanced Research Projects Agency (DARPA, www.darpa.mil), under SymBioSys Program, Grant Contract No. F30602-00-2-0614); and by the European Union Seventh Framework Programme (FP7/2007–2013, http://ec.europa.eu/research/fp7/index_en.cfm) under grant agreement no. 214538 project Bio-Inspired Self-assembled Nano-Enabled Surfaces (BISNES). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, et al. (1992) Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proceedings of the National Academy of Sciences of the United States of America 89:2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brockwell DJ, Smith DA, Radford SE (2000) Protein folding mechanisms: new methods and emerging ideas. Current Opinion in Structural Biology 10:16–25. [DOI] [PubMed] [Google Scholar]
- 3. Takano K, Yamagata Y, Yutani K (2001) Contribution of polar groups in the interior of a protein to the conformational stability. Biochemistry 40:4853–4858. [DOI] [PubMed] [Google Scholar]
- 4. Jones S, Thornton JM (1996) Principles of protein-protein interactions. Proceedings of the National Academy of Sciences of the United States of America 93:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Janin J, Chothia C (1990) The structure of protein-protein recognition sites. Journal of Biological Chemistry 265:16027–16030. [PubMed] [Google Scholar]
- 6. Bonvin AMJJ (2006) Flexible protein-protein docking. Current Opinion in Structural Biology 16:194–200. [DOI] [PubMed] [Google Scholar]
- 7. Gordon EM, Barrett RW, Dower WJ, Fodor SPA, Gallop MA (1994) Applications of combinatorial technologies to drug discovery. II: Combinatorial organic synthesis, libray screening strategies, and future directions. Journal of Medicinal Chemistry 37:1385–1401. [DOI] [PubMed] [Google Scholar]
- 8. Eyrisch S, Helms V (2009) What induces pocket openins on protein surface patches involved in protein-protein interactions? J Comput Aided Mol Des 23:73–86. [DOI] [PubMed] [Google Scholar]
- 9. Sharp K, Nicholls A, Fine R, Honig B (1990) Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science 252:106–109. [DOI] [PubMed] [Google Scholar]
- 10. Richards FM (1977) Areas, Volumes, Packing, and Protein Structure. Annual Review of Biophysics and Bioengineering 6:151–176. [DOI] [PubMed] [Google Scholar]
- 11.Fersht A (1985) Enzyme Structure and Mechanism.: W.H. Freeman and Company.
- 12. Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, et al. (1998) The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science 280:69–77. [DOI] [PubMed] [Google Scholar]
- 13.Steinbacher S, Bass R, Strop P, Rees DC (2007) Structures of the Prokaryotic Mechanosensitive Channels MscL and MscS. Current Topics in Membranes. pp. 1–24.
- 14. Braig K, Otwinowskl Z, Hegde R, Boisvert DC, Joachimiak A, et al. (1994) The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature 371:578–586. [DOI] [PubMed] [Google Scholar]
- 15. Eisenberg D, McLachlan AD (1986) Solvation energy in protein folding and binding. Nature 319:199–203. [DOI] [PubMed] [Google Scholar]
- 16. Wesson L, Eisenberg D (1992) Atomic solvation parameters applied to molecular dynamics of proteins in solution. Protein Science 1:227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Efremov RG, Nolde DE, Vergoten G, Arseniev AS (1999) A Solvent Model for Simulations of Peptides in Bilayers. I. Membrane-Promoting α-Helix Formation. Biophysical Journal 76:2448–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vila J, Williams RL, Vásquez M, Scheraga HA (1991) Empirical solvation models can be used to differentiate native from near-native conformations of bovine pancreatic trypsin inhibitor. PROTEINS: Structure, Function, and Genetics 10:199–218. [DOI] [PubMed] [Google Scholar]
- 19.Schiffer CA, Caldwell JW, Kollman PA, Stroud RM (1993) Protein-structure prediction with a combined solvation free energy-molecular mechanics force field. Molecular Simulation 10: 121–&.
- 20. Ooi T, Oobatake M, Nemethy G, Scheraga HA (1987) Accessible surface-areas as a measure of the thermodynamic parameters of hydrations of peptides. Proceedings of the National Academy of Sciences of the United States of America 84:3086–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Can T, Chen C-I, Wang Y-F (2006) Efficient molecular surface generation using level-set methods. Journal of Molecular Graphics and Modelling 25:442–454. [DOI] [PubMed] [Google Scholar]
- 22. Connolly ML (1983) Solvent-accessible surfaces of proteins and nucleic acids. Ann Rev BiophysBioeng 221:709–713. [DOI] [PubMed] [Google Scholar]
- 23. Connolly ML (1983) Analytical molecular surface calculation. Journal of Applied Crystallography 16:548–558. [Google Scholar]
- 24. Sanner MF, Olson AJ, Spehner J-C (1996) Reduced Surface: An Efficient Way to Compute Molecular Surfaces. Biopolymers 38:305–320. [DOI] [PubMed] [Google Scholar]
- 25. Edelsbrunner H, Mücke EP (1994) Three-dimensional alpha shapes. ACM Transactions on Graphics 13:43–72. [Google Scholar]
- 26. Eisenhaber F, Lijnzaad P, Argos P, Sander C, Scharf M (1995) The double cubic lattice method: Efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. Journal of Computational Chemistry 16:273–284. [Google Scholar]
- 27. Zauhar RJ, Morgan RS (1990) Computing the electric potential of biomolecules: Application of a new method of molecular surface triangulation. Journal of Computational Chemistry 11:603–622. [Google Scholar]
- 28. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. (2000) The Protein Data Bank. Nucleic Acids Research 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nicolau DV, Paszek E, Fulga F, Nicolau DV Jr. (2013) Protein Molecular Surface Mapped at Different Geometrical Resolutions. PLoS ONE 8:e58896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicolau DV Jr, Burrage K, Parton RG, Hancock JF (2006) Identifying Optimal Lipid Raft Characteristics Required To Promote Nanoscale Protein-Protein Interactions on the Plasma Membrane. Mol Cell Biol 26:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bretschneider T, Anderson K, Ecke M, Müller-Taubenberger A, Schroth-Diez B, et al. (2009) The Three-Dimensional Dynamics of Actin Waves, a Model of Cytoskeletal Self-Organization. Biophysical Journal 96:2888–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawabata S, Higgins GA, Gordon JW (1991) Amyloid plaques, neurofibrillary tangles and neuronal loss in brains of transgenic mice overexpressing a C-terminal fragment of human amyloid precursor protein. Nature 354:476–478. [DOI] [PubMed] [Google Scholar]
- 33. Langer R, Peppas NA (2003) Advances in biomaterials, drug delivery, and bionanotechnology. AIChE Journal 49:2990–3006. [Google Scholar]
- 34. Mukhopadhyay R (2006) Devices to drool for. Analytical Chemistry 78:7379–7382. [DOI] [PubMed] [Google Scholar]
- 35. Hawkins KR, Steedman MR, Baldwin RR, E. Fu SG, Yager P (2007) A method for characterizing adsorption of flowing solutes to microfluidic device surfaces. Lab on a Chip - Miniaturisation for Chemistry and Biology 7:281–285. [DOI] [PubMed] [Google Scholar]
- 36. Nagase K, Kobayashi J, Okano T (2009) Temperature-responsive intelligent interfaces for biomolecular separation and cell sheet engineering. J R Soc Interface 6:293–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kavanaugh JS, Weydert JA, Rogers PH, Arnone A (1998) High-Resolution Crystal Structures of Human Hemoglobin with Mutations at Tryptophan 37β: Structural Basis for a High-Affinity T-State†,‡. Biochemistry 37:4358–4373. [DOI] [PubMed] [Google Scholar]
- 38. Wimley WC, White SH (1996) Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nature Structural Biology 3:842–848. [DOI] [PubMed] [Google Scholar]
- 39. Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. [DOI] [PubMed] [Google Scholar]
- 40. Hopp TP, Woods KR (1981) Prediction of protein antigenic determinants from amino acid sequences. Proceedings of the National Academy of Sciences of the United States of America 78:3824–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kellogg GE, Abraham DJ (2000) Hydrophobicity: is LogP(o/w) more than the sum of its parts? European Journal of Medicinal Chemistry 35:651–661. [DOI] [PubMed] [Google Scholar]
- 42. Karplus PA (1997) Hydrophobicity regained. Protein Science 6:1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meek JL (1980) Prediction of peptide retention times in high-pressure liquid chromatography on the basis of amino acid composition. Proc Natl Acad Sci USA 77:1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, et al. (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030. [DOI] [PubMed] [Google Scholar]
- 45. Moon CP, Fleming KG (2011) Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proceedings of the National Academy of Sciences of the United States of America 108:10174–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koehler J, Woetzel N, Staritzbichler R, Sanders CR, Meiler J (2009) A unified hydrophobicity scale for multispan membrane proteins. Proteins: Structure, Function and Bioinformatics 76:13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Connolly ML (1985) Molecular Surface Triangulation. Journal of Applied Crystallography 18:499–505. [Google Scholar]
- 48. Casari G, Sippl MJ (1992) Structure-derived hydrophobic potential: Hydrophobic potential derived from x-ray structures of globular proteins is able to identify native folds. Journal of Molecular Biology 224:725–732. [DOI] [PubMed] [Google Scholar]
- 49. Fauchére JL, Quarendon P, Kaetterer L (1988) Estimating and representing hydrophobicity potential. Journal of Molecular Graphics 6:203–206. [Google Scholar]
- 50. Lawrence CE, Bryant SH (1991) Hydrophobic potentials from statistical analysis of protein structures. Methods in Enzymology 202:20–31. [DOI] [PubMed] [Google Scholar]
- 51. Yamaotsu N, Oda A, Hirono S (2008) Determination of ligand-binding sites on proteins using long-range hydrophobic potential. Biological and Pharmaceutical Bulletin 31:1552–1558. [DOI] [PubMed] [Google Scholar]
- 52. Efremov RG, Gulyaev DI, Modyanov NN (1992) Application of three-dimensional molecular hydrophobicity potential to the analysis of spatial organization of membrane protein domains. II. Optimization of hydrophobic contacts in transmembrane hairpin structures of Na+, K+-ATPase. Journal of Protein Chemistry 11:699–708. [DOI] [PubMed] [Google Scholar]
- 53. Efremov RG, Gulyaev DI, Vergoten G, Modyanov NN (1992) Application of three-dimensional molecular hydrophobicity potential to the analysis of spatial organization of membrane domains in proteins: I. Hydrophobic properties of transmembrane segments of Na+, K+-ATPase. Journal of Protein Chemistry 11:665–676. [DOI] [PubMed] [Google Scholar]
- 54. Abraham DJ, Leo AJ (1987) Extension of the fragment method to calculate amino acid zwitterion and side chain partition coefficients. PROTEINS: Structure, Function, and Genetics 2:130–152. [DOI] [PubMed] [Google Scholar]
- 55. Ghose AK, Viswanadhan VN, Wendoloski JJ (1998) Prediction of Hydrophobic (Lipophilic) Properties of Small Organic Molecules Using Fragmental Methods: An Analysis of ALOGP and CLOGP Methods. The Journal of Physical Chemistry A 102:3762–3772. [Google Scholar]
- 56. Lesser GJ, Rose GD (1990) Hydrophobicity of amino acid subgroups in proteins. PROTEINS: Structure, Function, and Genetics 8:6–13. [DOI] [PubMed] [Google Scholar]
- 57. Gabdoulline RR, Wade RC, Walther D (2003) MolSurfer: a macromolecular interface navigator. Nucleic Acids Research 31:3349–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cozzini P, Fornabaio M, Marabotti A, Abraham DJ, Kellogg GE, et al. (2004) Free energy of ligand binding to protein: evaluation of the contribution of water molecules by computational methods. Current Medicinal Chemistry 11:3093–3118. [DOI] [PubMed] [Google Scholar]
- 59. Caron G, Ermondi G (2003) A comparison of calculated and experimental parameters as sources of structural information: the case of lipophilicity-related descriptors. Mini Reviews In Medicinal Chemistry 3:821–830. [DOI] [PubMed] [Google Scholar]
- 60. Kellogg GE, Semus SF, Abraham DJ (1991) HINT: A new method of empirical hydrophobic field calculation for CoMFA. Journal of Computer-Aided Molecular Design 5:545–552. [DOI] [PubMed] [Google Scholar]
- 61. Wolfenden R, Andersson L, Cullis PM, Southgate CCB (1981) Affinities of amino acid side chains for solvent water. Biochemistry 20:849–855. [DOI] [PubMed] [Google Scholar]
- 62. Radzicka A, Wolfenden R (1988) Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. Biochemistry 27:1664–1670. [Google Scholar]
- 63. Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, et al. (1995) A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. Journal of the American Chemical Society 117:5179–5197. [Google Scholar]
- 64. Banerji A, Ghosh I (2009) Revisiting the myths of protein interior: Studying proteins with mass-fractal hydrophobicity-fractal and polarizability-fractal dimensions. PLoS ONE 4:e7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sandelin E (2004) On Hydrophobicity and Conformational Specificity in Proteins. Biophysical Journal 86:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jamadagni SN, Godawat R, Garde S (2011) Hydrophobicity of proteins and interfaces: Insights from density fluctuations. Annual Review of Chemical and Biomolecular Engineering 2:147–171. [DOI] [PubMed] [Google Scholar]
- 67.Cristea PD, Arsene O, Tuduce R, Nicolau DV (2012) Protein surface atom neighbourhoods classification; pp. 147–150.
- 68. Hooper NM, Karran EH, Turner AJ (1997) Membrane protein secretases. Biochem J 321:265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction of various sets of atom types, from M = 8 to M = 13.
(XLSX)
Selection of the best atom types set by the regression of various sets of atom types (M = 8 to 12) for the hydrophobicity scale proposed by Wimley & White [38] .
(XLSX)
Calculation of the best atomic hydrophobicity sets for M = 12 and for various hydrophobicity scales when ASA is considered (Part 1) and when it is not considered (Part 2).
(XLSX)
Calculation of the Accessible Solvent Areas (ASA) for each atom in each amino acid as a function of the probe radius.
(XLSX)
Complete set of data regarding the calculation of physico-chemical properties on the molecular surface of the proteins in the total set ( Table 1 ), for atomic, amino acid and charges, the latter two from [29] .
(XLSX)
Complete set of data regarding the calculation of physico-chemical properties on the molecular surface of the proteins in the selected set of hemoglobins, for atomic, amino acid and charges.
(XLSX)
Example of molecular surface obtained by probing the protein with a small and a large probe.
(TIF)
Comprehensive discussion regarding the possibilities of calculation of atomic hydrophobicities.
(DOC)
Molecular surfaces of ribonuclease presented as a function of the probing resolution, from the finest (top) to the coarsest (bottom). The molecular surfaces are represented for charges (left column); amino acid-based hydrophobicity (right column); and atom-based hydrophobicity (middle columns). The atom-based molecular surfaces are presented using values directly derived from (Eq.1) – left middle column; and normalized to fit the range of the amino acid hydrophilicities – right middle column.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.