Abstract
The purpose of this study was to probe active site structure and dynamics of human cytochrome P4502E1 and P4502A6 using a series of related short chain fatty aldehydes. Binding efficiency of the aldehydes was monitored via their ability to inhibit the binding and activation of the probe substrates p-nitrophenol (2E1) and coumarin (2A6). Oxidation of the aldehydes was observed in reactions with individually expressed 2E1, but not 2A6, suggesting alternate binding modes. For saturated aldehydes the optimum chain length for inhibition of 2E1 was 9 carbons (KI=7.8 ±0.3 μM), whereas for 2A6 heptanal was most potent (KI=15.8 ±1.1 μM). A double bond in the 2-position of the aldehyde significantly decreased the observed KI relative to the corresponding saturated compound in most cases. A clear difference in the effect of the double bond was observed between the two isoforms. With 2E1, the double bond appeared to remove steric constraints on aldehyde binding with KI values for the 5–12 carbon compounds ranging between 2.6 ± 0.1 μM and 12.8± 0.5 μM, whereas steric effects remained the dominant factor in the binding of the unsaturated aldehydes to 2A6 (observed KI values between 7.0± 0.5 μM and >1000 μM). The aldehyde function was essential for effective inhibition, as the corresponding carboxylic acids had very little effect on enzyme activity over the same range of concentrations, and branching at the 3-position of the aldehydes increased the corresponding KI value in all cases examined. The results suggest that a conjugated π-system may be a key structural determinant in the binding of these compounds to both enzymes, and may also be an important feature for the expansion of the active site volume in 2E1.
Keywords: Cytochrome P450, Aldehydes, Inhibition, 2E1, 2A6
1. Introduction
The function of microsomal cytochrome P450 enzymes is generally a protective one, where foreign chemicals entering the body can be oxidized in a way that makes them more readily excreted. However, the human microsomal cytochrome P450 isoforms 2E1 and 2A6 have been implicated in the deleterious effects of a variety of drugs and environmental agents, including acetaminophen [1], nitrosamines [2,3], and carbon tetrachloride [4], to name a few. There is also evidence to suggest that the inhibition of these isoforms during times of exposure to these agents can provide a level of protection against the harmful effects [5–7]. Consequently, identification of selective inhibitors of these isoforms and understanding the basis for selectivity has been the focus of a number of recent studies. It has been established through crystallographic analysis and binding studies that the active sites of these two isoforms are relatively small compared with other microsomal P450s, with the 2A6 isoform being the more sterically restricted [8–10]. Comparisons between the two enzymes suggest that 2A6 is more structurally rigid, whereas the 2E1 active site may have more flexibility, and can change in response to different substrates [11,12]. Indeed, active site volumes ranging from 190 Å3 to 470 Å3 have been measured for 2E1 with different substrates bound [13]. This is consistent with spectroscopic studies indicating that the active site of this isoform displays a significant level of compressibility under high pressure, whereas the 2A6 active site is considerably more unyielding [14,15]. Crystallographic and computational studies suggest that π-stacking interactions in the active site may facilitate the binding of planar low molecular weight molecules to both isoforms, and in 2E1, these interactions may be the basis for active site reorganization associated with observed substrate inhibition [16,17].
The goal of the current study was to explore structure-activity relationships mediating the binding of a series of related aldehydes to human Cytochrome P4502E1 and P4502A6 in order to gain additional insight into the structural features contributing to selectivity of substrates and inhibitors toward these two isoforms, along with features that facilitate active site reorganization and expansion. In particular, factors such as chain length, branching in the 3-position and unsaturation in the 2-position were evaluated for their influence on KI. Aldehydes were selected on the basis of prior studies involving rabbit liver cytochrome P450s identifying aldehydes as a class of P450 inhibitors, and the availability of a wide range of structurally related compounds belonging to this class [18]. In addition, aldehydes of the type described in this study can be generated physiologically via lipid peroxidation, thus insight gained through this study may guide future studies related to toxicology and cellular oxidative stress.
2. MATERIALS AND METHODS
2.1 Chemicals
The aldehyde compounds used in this study were purchased from Acros Organics. Coumarin, 7-hydroxycoumarin (umbelliferone), NADPH, p-nitrophenol and 4-nitrocatechol were from Sigma Chemical Co. Human liver S9 fractions were purchased from Moltox Inc, Ashville, NC.
2.2 Enzymatic assays: P4502E1
All assays for 2E1 (p-nitrophenol) were carried out according to published protocols (19) with slight modifications. S9 fractions (0.5 mg) were incubated in 100 mM phosphate buffer (pH 7.4) in the presence of 20–100 μM p-nitrophenol and 1.0 mM NADPH for a total of 30 min at 37 °C in a 0.5 mL total reaction volume. Reactions were terminated by addition of ice-cold 6% perchloric acid and incubation on ice for an additional 10 min, followed by centrifugation to remove precipitated protein. Cleared supernatant (50 μL) was then analyzed by RP-HPLC on a 150 × 4.6 C18 column with a mobile phase consisting of 35% acetonitrile, 64.5% H2O, 0.5% acetic acid at a flow rate of 1.0 mL/min. Absorbance detection at 340 nm was used to quantify the 4-nitrocatechol product. A Shimadzu LC 20A Series HPLC system consisting of an SPD-20A UV/Vis detector, LC 20AT solvent delivery, and a Sil 20A autosampler was used for analysis of samples.
2.3 Inhibition of P4502E1
All inhibitors, including the aldehydes and acids, were prepared as aqueous stocks by diluting 4.0 μg inhibitor into 100 mL DI H2O immediately prior to their use. Initially, a dose-response curve was generated for each of the aldehydes with regard to their relative inhibitory effects on P4502E1. Reactions contained 0.5 mg S9 fractions, 50 μM p-nitrophenol, 1.0 mM NADPH and aldehyde concentrations ranging from 5–370 μM in a 0.50 mL volume of 0.1 M phosphate buffer (pH 7.4). Samples were quenched and processed for HPLC analysis as described earlier. To obtain more detailed kinetic parameters, Michaelis-Menten kinetic experiments were then performed to probe the mode and potency of inhibition using inhibitor concentrations of either 30 μM or 60 μM. These concentrations were chosen based on the results of the screening experiments in which it was determined that this was the lowest concentration to produce measurable inhibition for all of the compounds being examined except for dodecyl aldehyde. The substrate concentrations were varied from 20.0–100 μM in a total reaction volume of 0.50 mL for 30 min, and reactions were processed as described above. A minimum of 4 independent trials were carried out for each experiment and the standard deviations are reported.
2.4 Enzymatic assays: P4502A6
The assay that was used to measure inhibition of 2A6 is one that utilizes the conversion of coumarin into 7-hydroxycoumarin. The procedure is modified from that of Waxman and Chang [20]. Liver S9 fractions (0.50 mg) were incubated in a reaction mixture containing 3.0 μM coumarin, 100 mM potassium phosphate buffer (pH 7.4) and 1.0 mM NADPH in a total volume of 0.50 mL. Following a 30 min incubation at 37 °C the protein was precipitated with 100 μL of 6% perchloric acid and placed on ice for 10 minutes. Samples were centrifuged at 13500 rpm for 10 minutes and 40 μL of the cleared supernatant was injected onto a RP-C18 HPLC column with a mobile phase consisting of 59% DI water, 40% methanol, and 1% acetic acid, at a flow rate of 1.0 mL/min.
2.5 Inhibition of P4502A6
Inhibitors were prepared as aqueous stock solutions by diluting 4.0 μg inhibitor into 100 mL DI H2O immediately prior to their use. Dose-response curve was generated for each of the aldehydes with regard to their relative inhibitory effects on P4502A6. Reactions contained 0.5 mg S9 protein, 3.0 μM coumarin, 1.0 mM NADPH and aldehyde concentrations ranging from 5–370 μM in a 0.50 mL volume of 0.1 M phosphate buffer (pH 7.4). Samples were quenched and processed for HPLC analysis as described in the previous sections. Michaelis-Menten kinetic experiments were then performed using inhibitor concentrations of either 30 μM or 60 μM. Again, these inhibitor concentrations were selected based on the results of the screening experiments, as with the 2E1 reactions. The substrate concentrations were varied from 0.5–6.0 μM in a total reaction volume of 0.50 mL for 30 min, and reactions were processed as described above. A minimum of 4 independent trials were carried out for each experiment and the standard deviations are reported.
2.6 Reversibility Studies
To determine whether the inhibition of each P450 isoform by the individual aldehydes was reversible or irreversible, microsomal samples were pre-incubated with (1) Buffer alone, (2) Buffer + Aldehyde and (3) Buffer + Aldehyde + NADPH, as described in previously by Raner et al. [18].
2.7 Aldehyde Oxidation by human P450 enzymes
Oxidation of trans-2-octenal was carried out in a total reaction volume of 200 μL, and contained expressed human P450 (2E1 or 2A6 supersomes, Gentest, 50 nM final), 100 mM potassium phosphate buffer (pH 7.4), 100 μM trans-2-octenal, and 1 mM NADPH. The reaction was quenched with 6% perchloric acid as described previously. A C18 RP HPLC column (4.6 × 250) from Phenominex was used with a mobile phase consisting of 55% acetonitrile and 45% water, both containing 0.1% TFA at a flow rate of 1.4 mL/min and detection was at 230 nm. A sample of trans-2-octenoic acid was acquired from Sigma-Aldrich and used to generate a standard curve for quantification of product. The reactions with human 2E1 supersomes were linear for 45 min at 37°C, so all additional reactions were carried out under these conditions. The 2A6 supersomes failed to produce NADPH-dependent acid product. The oxidation of trans-2-decenal, trans-2-nonenal, trans-2-octenal and trans-2-heptenal by the expressed cytochrome P4502E1 in supersomes was also carried out with each of the aldehydes present at the following concentrations: trans-2-decenal (110 μM), trans-2-nonenal (120 μM), trans-2-octenal (100 μM) and trans-2-heptenal (110 μM), along with 1.0 mM NADPH in a 100 mM phosphate buffer at pH 7.4. Reactions were carried out for 45 min at 37 °C and quenched as described above. Control reactions without NADPH were carried out in parallel, and the reaction mixtures were analyzed using HPLC under the same conditions as described for trans-2-octenoic acid alone. The effects of ethanol (0.001%, 0.01%, 0.10% and 1.0%) on the activity in 2E1 supersomes was also monitored under identical conditions.
2.8 Analysis of the kinetic data
Michaelis-Menton plots were prepared for each of the various activity data sets in the presence and absence of inhibitor. All data was fit using Slidewrite version 4.1 (Advanced Graphics Software Inc) and the non-linear regression analysis function using the Michaelis-Menton equation. Based on Michaelis-Menton plots for each aldehyde, the inhibition could be described using the competitive model, so values for KI were calculated using this model, where Vmax remains constant, α is defined as Kmapp/Km and KI = [I]/(α − 1).
2.9 Graphical images of P450 active sites
The active site images of the 2E1 and P4502A6 enzymes were generated using crystal structure coordinates deposited in the protein databank. The file PDB: 1Z10 was used for the image of P4502A6 with coumarin bound; PDB: 3T3Z was used for P4502E1 with pilocarpene bound; PDB: 3E6I was used to generate the P4502E1 image with indazole bound; and the PDB: 3LC4 coordinates were used for P4502E1 image with imidazolyl-dodecanoic acid bound. RasWin 2.7.5.2 Molecular Graphics software was used to generate each of these images based on the respective pdb files.
3. RESULTS
3.1 Inhibition of P4502E1 by alkanals and alkenals
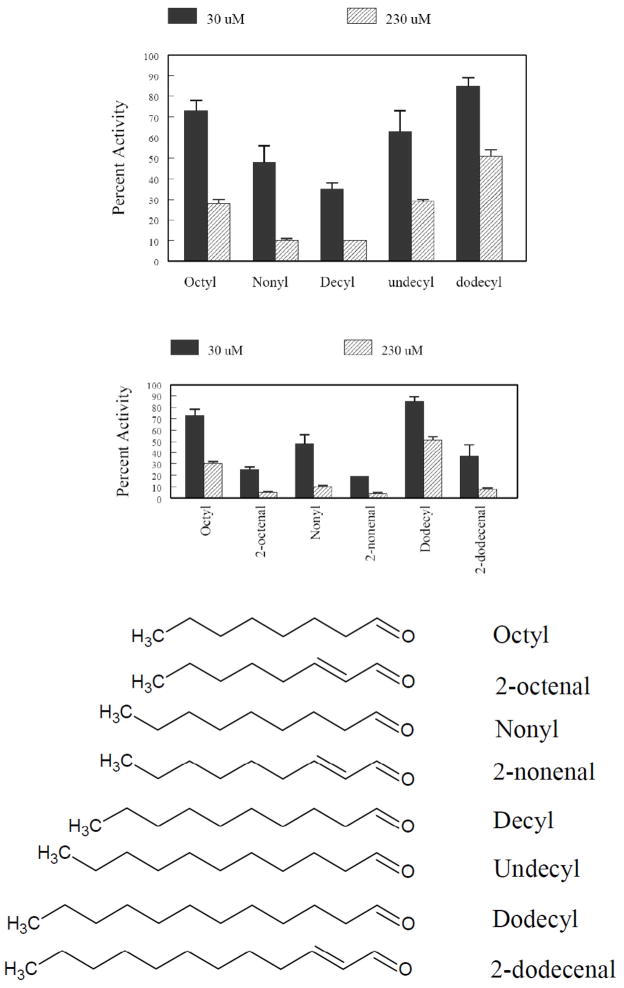
Dose-response inhibition studies using p-nitrophenol oxidation as a measure for 2E1 activity were initially carried out using saturated straight-chain aldehydes ranging from 8–12 carbon atoms and 2-unsaturated aldehydes with 8-, 9- and 12-carbon atoms (Figure 1) at fixed inhibitor concentrations of 30 μM and 230 μM. Inhibition was observed for each aldehyde as a dose-dependent reduction in activity, relative to a control in which no aldehyde was present. The selection of these concentrations was based empirically on their ability to highlight the differences in potency among this group of aldehydes. Maximum inhibition was observed for the nonyl and decyl aldehydes, and unsaturation significantly increased the level of inhibition, suggesting the extended π-system of the unsaturated aldehydes leads to more favorable binding in the active site.
Figure 1.
Effects of saturated and 2-unsaturated aldehydes on P4502E1 activity. (A) Effect of chain length on P450 inhibition by satuated aldehydes at 30 and 230 μM aldehyde concentration. (B) Comparison of the effects of saturated vs 2- unsaturated aldehydes on P4502E1 inhibition at 30 and 230 μM aldehyde concentration. (C) Structures of aldehydes used for these comparative studies.
The reversibility of inhibition was examined for each of the aldehydes represented in Figure 1 by pre-incubating the aldehydes with S9 fractions in the presence and absence of NADPH for 15 min at 37 °C. Dilution of the pre-incubation mixture resulted in complete restoration of the p-nitrophenol oxidation activity of the samples, thus all of these saturated and unsaturated aldehydes (data not shown) appeared to be reversible inhibitors of 2E1 under the conditions used. As a positive control, the experiment was carried out using 11-undecylenic aldehyde at a concentration of 25 μM. Following a 15 min pre-incubation at 37 °C in the presence of 1 mM NADPH, a loss of ~50% of the activity of 2E1 was observed (data not shown). This compound possesses a terminal olefin group, which is known to cause irreversible damage to the heme [21–23].
Detailed inhibition studies were subsequently carried out in which the inhibition constants (KI) were calculated for the aldehydes, both saturated and unsaturated, with carbon chain lengths, between 5–12, using the standard Michaelis-Menten approach (Table 1). For all the aldehydes tested, Vmax was unaltered, once again indicating competitive-reversible type inhibition, thus the competitive model of inhibition was applied for KI determination. Consistent with the preliminary screening data, the KI values indicate that the 9- and 10-carbon aldehydes, possessing a 2-double bond (nonenal and decenal) were the most potent inhibitors of the human 2E1 isoform. It is also worth noting that a strong dependence on chain length was observed for the saturated compounds, whereas less dependence on chain length was observed with unsaturated aldehydes. Furthermore, the unsaturated compounds were all much more potent than the respective saturated compounds, with KI values as much as 30-fold lower in the case of the 11- and 12-carbon aldehydes.
Table 1.
Experimentally determined inhibition constants (KI) for the inhibition of Human cytochrome P4502A6 by straight chain saturated and unsaturated aldehydes ranging in size from 5–12 carbons. KI values were calculated using a competitive model for inhibition and a minimum of 3 trials were carried out for each aldehyde.
| P4502A6 | P4502E1 | |||
|---|---|---|---|---|
| Aldehyde | KI (μM) Saturated | KI (μM) Unsaturated | KI (μM) Saturated | KI (μM) Unsaturated |
| Pentyl | 126 ± 16 | 1270 ± 150 | 155 ± 13 | 13 ± 1 |
| Hexyl | 57 ± 7 | 33 ± 3 | 26 ± 2 | 6.7 ± 0.2 |
| Heptyl | 16 ±1 | 9 ± 1 | 32 ± 1 | 5.6 ± 0.1 |
| Octyl | 25 ± 2 | 7 ± 1 | 13 ± 1 | 4.1 ± 0.1 |
| Nonyl | 55 ± 6 | 34 ± 3 | 8.0 ±1 | 2.9 ± 0.2 |
| Decyl | 106 ± 29 | 59 ± 6 | 17 ± 1 | 3.3 ± 0.1 |
| Undecy | 68 ± 7 | 103 ± 22 | 117 ± 15 | 3.4 ± 0.1 |
| Dodecyl | 48 ± 9 | 128 ± 35 | 85 ± 8 | 2.6 ± 0.1 |
3.2 Effects of branching on inhibition of 2E1
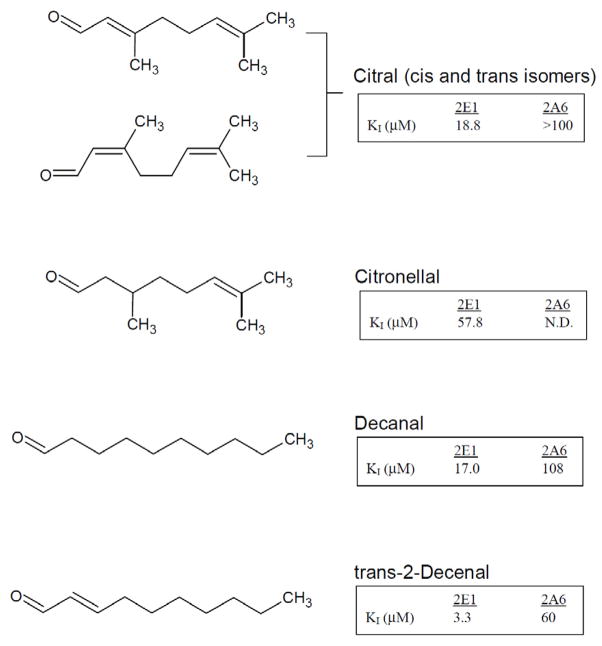
Additional aldehydes (citral and citronellal) with branching at the 3-position were evaluated for inhibitory potency against 2E1. Citral (3,7-dimethyl 2,6-octadienal) as shown in Figure 2, is a naturally occurring mixture of 2,3 -unsaturated aldehyde isomers with branching methyl groups at carbon 3 and 7. Citronellal (3,7-dimethyl 6-octenal ), shown in figure 3, also has a 3-methyl branch but does not have a double bond in the 2-position. Both aldehydes showed reversible competitive inhibition profiles when evaluated using the Michaelis-Menten model. KI values were measured for both of these compounds and were compared with those of the linear 10-carbon saturated and unsaturated aldehydes. By comparing citral to citronellal it is clear that the double bond at the 2-position dramatically impacts inhibition; KI values of 18.8 μM vs 57.8 μM, respectively. As for the effects of branching, both decanal and citronellal are saturated at the 2-position and possess 10 carbon atoms, like decanal. The KI for Decanal, however, is about 8-fold lower than the branched citronellal (7.0 μM vs 57.8 μM). Likewise, a comparison between decenal and citral showed a 5-fold increase in KI (3.3 μM to 15.7 μM) with the inclusion of a branching methyl at the 3-position of the aldehyde.
Figure 2.
Structure and measured inhibition constants for branched saturated and unsaturated aldehydes, citral, citronellal, and the corresponding linear, saturated and unsaturated 10 carbon aldehydes, decanal and trans-2-decenal.
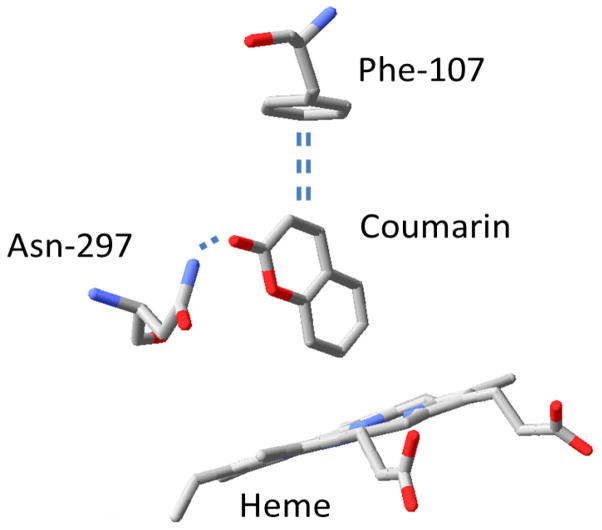
Figure 3.
Key residues in the active site structure of human cytochrome P4502A6 with the substrate coumarin bound. Hydrogen bond interactions between the carbonyl oxygen on the substrate and the amide group from Asn-297 and π-π interactions between Phe-107 and the aromatic ring of coumarin are shown ( generated using RasWin 2.7.5.2 and coordinates from PDB: 1Z10)
3.3 Effects of aldehyde function
The importance of the aldehyde group for inhibition was probed by monitoring 2E1 inhibition using the corresponding 8- to 12-carbon saturated carboxylic acids. Over a range of 1.0–100 μM, only lauric acid inhibited 2E1 significantly (data not shown). At a concentration of 50 μM, lauric acid reduced the activity of 2E1 by 50%, which is still modest inhibition relative to the aldehydes. Due to the lack of observed inhibition at high concentrations of carboxylic acid, KI values were not determined.
3.4 Inhibition of P4502A6 by alkanals and alkenals
As with the 2E1 isoform, all of the aldehydes tested, both saturated and unsaturated, inhibited coumarin hydroxylation via a reversible competitive mode. Values for KI were determined for each of the saturated and unsaturated aldehydes used with 2E1, and these values are presented in Table 1. A trend similar to that observed with 2E1 was also observed for 2A6. However, the optimal chain length appeared to be slightly shorter for 2A6, with a carbon chain length of 7–8 being most potent. Comparison of the saturated vs unsaturated aldehydes again suggested that the double bond in the 2-position improved the interaction with the active site, as indicated by a 2- to 3-fold decrease in KI for the unsaturated aldehydes over nearly the entire range. It is worth pointing out that with 2A6 inhibition, dodecanal and undecanal appeared to be outliers in the general trends observed, as both of these saturated aldehydes inhibited more potently than the smaller decanal, and more potently than the corresponding unsaturated counterparts. Another clear distinction between the 2E1 and 2A6 data can be seen in the behavior of the unsaturated aldehydes. Where the 2E1 isoform appeared to be almost insensitive to chain length, the inhibitors of the 2A6 isoform had an optimal chain length of 7 or 8, and above or below this value the potency of inhibition decreased in a continuous manner, with the 2 notable exceptions previously pointed out.
3.5 other structural determinants for P4502A6 inhibition by aldehydes
As with P4502E1, saturated carboxylic acids were ineffective at inhibiting the 2A6 isoform at concentrations as high as 100 μM, thus KI values were not determined. Interestingly, branching in the aldehyde also resulted in a dramatic loss in potency as demonstrated by the use of citral and citronellal as inhibitors of 2A6. Only citral inhibited 2A6, and here the KI was still >100 μM (data not shown).
3.6 Oxidation of aldehydes by human S9 fractions and expressed cytochrome P4502E1 and P4502A6
To determine that the aldehyde function of each inhibitor was inside the active site, reactions involving 4 of the unsaturated aldehydes, trans-2-heptenal, trans-2-octenal, trans-2-nonenal and trans-2-decenal were evaluated for the formation of the corresponding carboxylic acid. The substrate trans-2-octenal was examined initially to establish linearity over time, and the effects of ascorbate and glutathione on the reaction. This aldehyde was selected on the basis of its ability to inhibit both 2A6 and 2E1 isoforms effectively. The reaction with 2E1 supersomes was linear for 45 min at 37 °C, and beyond 45 min a marked decrease in activity was observed. In addition, inclusion of either 1.0 mM ascorbate or 1.0 mM glutathione had no effect on the rate of acid formation. Furthermore, for the 2E1-dependent reaction, increasing concentrations of ethanol (a P4502E1 inhibitor), between 0.001 and 1.0% caused a dose-dependent decrease in acid formation, with >80% inhibition at 1% and 40% inhibition at 0.1% ethanol. In contrast, 2A6 supersomes did not catalyze oxidation of trans-2-octenal. An additional experiment was carried out in which the four aldehydes; trans-2-heptenal, trans-2-octenal, trans-2-nonenal and trans-2-decenal were incubated with either expressed 2E1 or expressed 2A6, and the NADPH-dependent oxidation of each aldehyde was monitored. Supersomes containing expressed 2E1 were effective in the oxidation of all 4 aldehydes, while 2A6 supersomes again yielded no acid products.
4. DISCUSSION
Prior studies involving rabbit cytochrome P4502B4 indicated that mammalian cytochrome P450s were susceptible to aldehyde-mediated irreversible mechanism-based inhibition [18] Mechanistically, the inactivation was the result of heme destruction and was correlated to the aldehyde deformylation reaction mechanism [24,25]. No prior studies have been reported involving interactions between aldehydes and the human isoforms, including 2E1 and 2A6, both of which are of considerable toxicological importance. In the current study, no evidence for irreversible inhibition was observed with any of the aldehydes or either cytochrome P450, rather, reversible competitive inhibition was observed. However, oxidation of the aldehyde function by the expressed 2E1 enzyme was observed indicating an “aldehyde in” binding orientation in the active site for these compounds. With 2A6, a different binding orientation must be envisioned based on the inability to oxidize the aldehyde function. It should be noted that in the rabbit liver study, aldehyde concentrations used to calculate inactivation rates were in the mM range, whereas in the current study, the highest aldehyde concentrations used were mostly below 100 μM. It is conceivable that inactivation of the human P450s could occur at higher aldehyde concentrations.
All of the saturated aldehydes inhibited both P450 isoforms, with varying potency, as determined by measured KI values. Several interesting trends were observed in this data. For both 2E1 and 2A6, there appeared to be an optimal carbon chain length as it related to inhibition potency. For 2E1, the nonyl and decyl aldehyde had the lowest measured KI, whereas heptyl and octyl aldehydes were most effective against 2A6, and neither isoform was inhibited by corresponding carboxylic acids. Several recent comparative studies have been carried out concerning structural and activity relationships between the 2E1 and the 2A6 human P450 isoforms that support these experimental observations [11,12,14]. Both enzymes have relatively small active sites, and therefore binding of larger molecules may be disfavored via steric interactions, consistent with the observed pattern of inhibition. The 2A6 active site has a slightly smaller volume than 2E1 based on crystal structures with the small molecule pilocarpine bound (280 Å3 vs 337 Å3) [13], which is consistent with the smaller optimum chain length for inhibition of 2A6 (7–8 carbons for 2A6 vs 9–10 for 2E1). In addition, DeVore et al. [12], using both experimental and computational approaches showed that the 2E1 active site was slightly less polar than 2A6, however both contained a cluster of phenylalanine groups that appeared to define the active site boundaries. The reduced interaction of both isoforms with carboxylic acids relative to aldehydes is most easily justified by the less polar character of the aldehyde group. The crystal structure of 2A6 reveals an Asn side chain (Asn 297) that may help to stabilize the carbonyl group of the aldehydes via hydrogen bonding. Indeed, the substrates pilocarpene and coumarin both have carbonyl oxygens that form H-bond interactions with Asn 297 (Figure 3 shows the structure of the 2A6 active site with coumarin bound).
The 2-unsaturated aldehydes were generally more potent inhibitors of both 2E1 and 2A6, with some KI values nearly 10-fold lower than the corresponding saturated compounds. Computational studies by Xu et al. [26] highlight the importance of π-π interactions in the binding of 4-(methylnitrosamino)- 1-(3-pyridyl)-1-butanone (NNK) in the active site of 2A6, thus similar interactions with the planar π-system of the 2-unsaturated aldehydes could be envisioned as a basis for more efficient binding. Furthermore, based on the crystal structure of the human 2A6 isoform with coumarin bound, the nature of this π-π interaction involves the perpendicular arrangement of the π-orbitals on the aromatic ring systems of coumarin with those on Phe107 ring. [27], It is reasonable to suggest that the π-system of the 2-unsaturated aldehyde interacts favorably with Phe-107 of 2A6 in an analogous fashion, which serves to orient the substrate and restrict its mobility. Thus, increasing size would create steric interference in the rigid 2A6 active site, if these binding interactions were to be maintained [12]. This binding mode can also be used to rationalize the lack of aldehyde oxidation, as the carbonyl group of the aldehyde would be hydrogen bonded to Asn-297, which is removed from the activated oxygen in the catalytic site. The increase in potency toward 2A6 for the undecanal and dodecanal, relative to the corresponding unsaturated compounds was noted, however, it is not clear what the molecular basis for this observation is. It is tempting to rationalize this based on the greater flexibility of the large saturated aldehydes in the rigid active site. This does not account for the increased potency of these larger aldehydes relative to the smaller decanal, which should have less steric restrictions on binding. A more likely explanation is that given the hydrophobic nature of these compounds, disruption of the P4502A6/P450-reductase complex may result in the more potent observed inhibitory action.
Interestingly, for the unsaturated aldehydes, the dependence on chain length was almost completely abolished in 2E1, with the aldehydes containing 7–12 carbon atoms all inhibiting with KI in the 3–5 μM range. This is in stark contrast to the behavior of the un-saturated aldehydes with 2A6. Although the π-interactions appear to be strongly driving the binding of these aldehydes to both enzymes, the 2E1 active site appears to have the ability to accommodate larger molecules in response to the presence of the π-system of the unsaturated aldehydes. First, 2E1 lacks the H-bond donor to the carbonyl that is present in 2A6, as Asn-297 is replaced in 2E1 with Asp-295, which is excluded from the active site. According to DeVore et al. [12] the substrate pilocarpine appears to adapt to the 2A6 active site upon binding, whereas the 2E1 active site may change topology in response to substrate binding. In support of this flexibility model, the 2E1 active site volume can expand to 420–470 Å3 in response to the binding of the large substrate imidazolyl-dodecanoic acid [10].
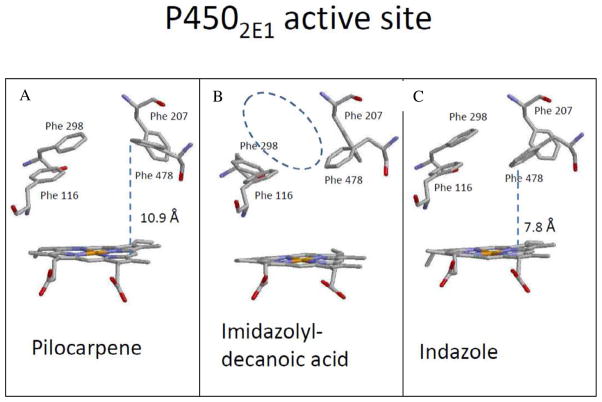
Figure 4 shows a comparison of the active site structure of P4502E1 with (A) pilocarpene (B) imidazolyl-dodecanoic acid and (C) indazole bound (substrate molecules are omitted from the structure). Although each of these substrates is structurally distinct from the aldehydes used in the current study, what is apparent is that there are residues in the active site that can rearrange in response to substrate and/or inhibitor binding. Four Phe side chains (Phe 116, Phe 207, Phe 298 and Phe 478) that form the upper surface of the active site are shown in this figure. The most significant differences on substrate binding occur in the position of Phe 298 and Phe 478 with respect to the heme. For example, in panel A, Phe 298 and Phe 478 have moved up relative to the heme, creating a larger volume directly above the heme when the bulky pilocarpene molecule binds. With indazole bound (panel C) Phe 478 is angled down toward the heme, reducing the overall volume of this cavity. Panel B shows the additional movement of Phe 298 in response to binding imidazolyl-decanoic acid. This motion allows the acyl chain of the substrate to access a binding channel running parallel to the I-helix just above Phe 298, which presumably accounts for the larger active site volume in the presence of this substrate. It is conceivable that π-interactions between unsaturated aldehydes and the 2E1 active site may have an analogous effect on Phe 298, essentially holding this door open so that longer chain aldehydes can gain access to the more remote areas of the active site. Alternatively, computational studies by Li et al. [17] suggest that the phenyl ring of Phe 478 can rotate to become almost perpendicular to the heme as a substrate binds to an “effector” site, which lies along the substrate access channel, directly above the γ-meso position on the heme. This motion repositions both the active substrate and a key catalytic residue T303, resulting in negative cooperativity. Li et al. [17] indicate the orientation of Phe 478 is significantly altered via π-π stacking interactions when an aromatic ligand occupies the effector site. As the saturated aldehydes would not be expected to bind to this effector site, it is also reasonable to suggest that the π-system of the 2-unsaturated aldehyde allows it to gain access to the effector site, thus increasing the available active site volume for unsaturated aldehydes. The lack of a hydrogen bonding group analogous to Asn-297 in 2A6 appears to allow the carbonyl group of the substrate more freedom to explore the active site, resulting in its oxidation by 2E1, but not 2A6.
Figure 4.
Active site structure of P4502E1 showing the position of key Phe residues in the presence of the substrates (A) pilocarpene (PDB: 3T3Z), (B) indazole (PDB: 3E6I) and (C) imidazolyl-dodecanoic acid (PDB: 3LC4). Images were prepared using RasWin 2.7.5.2 Molecular Graphics software.
5. Conclusions
In summary, the current work demonstrates that a series of saturated and unsaturated aliphatic aldehydes are effective competitive inhibitors of both human P450 2E1 and 2A6. The relatively small size of the 2E1 and 2A6 active sites result in a preference for binding the 7–10 carbon saturated aldehydes. In addition, 2-unsaturated aldehydes display 5 to 30-fold higher affinity for both human enzymes than their corresponding unsaturated counterparts, which is likely due to the potential for π-stacking interactions in Phenylalanine-rich active site of these enzymes. This study also provides direct experimental support for the flexibility model of substrate binding to 2E1, and suggests that an extended π-system may be a key determinant in active site expansion.
Supplementary Material
Highlights.
Human P450 2E1 and 2A6 active sites were probed using a series of small aldehydes
Both enzymes preferred α,β-unsaturated aldehydes to corresponding saturated forms
Branching in the aldehyde reduced binding to both isoforms
The 2E1 active site appeared to expand in response to α,β-unsaturation
The aldehydes were oxidized by 2E1, but not 2A6
Acknowledgments
Funding for this research was provided by The National Science Foundation (#0414301), Research Corporation (CC4924) and National Center for Complementary and Alternative Medicine (R15 AT007860-01A1) to G.M.R.
Abbreviations
- NADPH
nicotinamide adenine dinucleotide
- HLM
human liver S9 fractions
- 2A6
cytochrome P4502A6
- 2E1
cytochrome P4502E1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guengerich FP, Kim DH, Iwasaki M. Chem Res Toxicol. 1991;4:168–179. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhury G, Calcutt MW, Guengerich FP. J Biol Chem. 2010;285:8031–8044. doi: 10.1074/jbc.M109.088039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Glatt H. Mutat Res. 2008;643:64–69. doi: 10.1016/j.mrfmmm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Castillo T, Koop DR, Kamimura S, Triadafilopoulos G, Tsukamoto H. Hepatology. 1992;16:992–996. doi: 10.1002/hep.1840160423. [DOI] [PubMed] [Google Scholar]
- 5.Tung YT, Wu JH, Huang CC, Peng HC, Chen YL, Yang SC, Chang ST. Food Chem Toxicol. 2009;47:1385–1392. doi: 10.1016/j.fct.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Karamanakos PN, Trafalis DT, Geromichalos GD, Pappas P, Harkitis P, Konstandi M, Marselos M. Arch Toxicol. 2009;83:571–580. doi: 10.1007/s00204-008-0350-6. [DOI] [PubMed] [Google Scholar]
- 7.von Weymarn LB, Chun JA, Hollenberg PF. Carcinogenesis. 2006;27:782–90. doi: 10.1093/carcin/bgi301. [DOI] [PubMed] [Google Scholar]
- 8.Yano JK, Denton TT, Cerny MA, Zhang X, Johnson EF, Cashman JR. J Med Chem. 2006;49:6987–7001. doi: 10.1021/jm060519r. [DOI] [PubMed] [Google Scholar]
- 9.Porubsky PR, Meneely KM, Scott EE. J Biol Chem. 2008;283:33698–707. doi: 10.1074/jbc.M805999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang MH, Wade D, Chen L, White S, Yang CS. Arch Biochem Biophys. 1995;317:299–304. doi: 10.1006/abbi.1995.1166. [DOI] [PubMed] [Google Scholar]
- 11.Anzenbacherova E, Hudecek J, Murgida D, Hildebrandt P, Marchal S, Lange R, Anzenbacher P. Biochem Biophys Res Commun. 2005;338:477–482. doi: 10.1016/j.bbrc.2005.08.063. [DOI] [PubMed] [Google Scholar]
- 12.Porubsky PR, Battaile KP, Scott EE. J Biol Chem. 2010;285:22282–22290. doi: 10.1074/jbc.M110.109017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeVore NM, Meneely KM, Bart AG, Stephens ES, Battaile KP, Scott EE. FEBS J. 2012;279:1621–1631. doi: 10.1111/j.1742-4658.2011.08412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skopalík J, Anzenbacher P, Otyepka M. J Phys Chem B. 2008;112:8165–8173. doi: 10.1021/jp800311c. [DOI] [PubMed] [Google Scholar]
- 15.Hendrychova T, Berka K, Navratilova V, Anzenbacher P, Otyepka M. Curr Drug Metab. 2012;13:177–189. doi: 10.2174/138920012798918408. [DOI] [PubMed] [Google Scholar]
- 16.Ping J, Wang YJ, Wang JF, Li X, Li YX, Hao P. Curr Drug Metab. 2012;13:1024–1031. doi: 10.2174/138920012802138606. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Wei DQ, Wang JF, Li YX. J Chem Inf Model. 2011;51:3217–3225. doi: 10.1021/ci2004016. [DOI] [PubMed] [Google Scholar]
- 18.Raner GM, Chiang EW, Vaz AD, Coon MJ. Biochemistry. 1997;36:4895–4902. doi: 10.1021/bi9630568. [DOI] [PubMed] [Google Scholar]
- 19.Yang SP, Medling T, Raner GM. Comp Biochem Physiol C. 2003;136:297–308. doi: 10.1016/j.cca.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Waxman DJ, Chang TK. Methods Mol Biol. 2006;320:91–96. doi: 10.1385/1-59259-998-2:91. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz de Montellano PR, Beilan HS, Kunze KL, Mico BA. J Biol Chem. 1981;256:4395–4399. [PubMed] [Google Scholar]
- 22.Ortiz de Montellano PR, Beilan HS, Kunze KL. J Biol Chem. 1981;256:6708–6713. [PubMed] [Google Scholar]
- 23.Ortiz de Montellano PR, Beilan HS, Kunze KL. Proc Natl Acad Sci U S A. 1981;78:1490–1494. doi: 10.1073/pnas.78.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo CL, Raner GM, Vaz AD, Coon MJ. Biochemistry. 1999;38:10511–10518. doi: 10.1021/bi9904712. [DOI] [PubMed] [Google Scholar]
- 25.Vaz AD, Pernecky SJ, Raner GM, Coon MJ. Proc Natl Acad Sci U S A. 1996;93:4644–4648. doi: 10.1073/pnas.93.10.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Shen Z, Shen J, Liu G, Li W, Tang Y. J Mol Graph Model. 2011;30:1–9. doi: 10.1016/j.jmgm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.