Abstract
We describe here the metagenomics-derived viral sequences detected in beef, pork, and chicken purchased from stores in San Francisco. In beef we detected four previously reported viruses (two parvoviruses belonging to different genera, an anellovirus, and one circovirus-like virus) and one novel bovine polyomavirus species (BPyV2-SF) whose closest relatives infect primates. Detection of porcine hokovirus in beef indicated that this parvovirus can infect both ungulate species. In pork we detected four known parvoviruses from three genera, an anellovirus, and pig circovirus 2. Chicken meat contained numerous gyrovirus sequences including those of chicken anemia virus and of a novel gyrovirus species (GyV7-SF). Our results provide an initial characterization of some of the viruses commonly found in US store-bought meats which included a diverse group of parvoviruses and viral families with small circular DNA genomes. Whether any of these viruses can infect humans will require testing human sera for specific antibodies.
Keywords: Viral metagenomics, Meat, Polyomaviridae, Gyrovirus, Parvoviridae
Highlights
-
•
Eukaryotic viral genomes in store-bought beef, pork, and chicken are identified.
-
•
A novel bovine polyomavirus genome, closest to a group of viruses from primates, is sequenced.
-
•
Porcine hokovirus is detected in beef samples.
-
•
A small circovirus-like circular DNA genome in beef is genetically characterized.
-
•
Several species of gyrovirus including a novel species are detected in chicken meat.
Introduction
Viral metagenomics characterizes known and divergent viruses in sample preparations enriched for viral nucleic acids. Host DNA or RNA concentration is reduced by nuclease treatment while viral nucleic acids are protected within filterable viral particles (Allander et al., 2001). Following non-specific amplification and deep sequencing the viral sequences are recognized through their translated protein sequence similarity to known viral proteins. Here we characterize viral sequences in beef, pork, and chicken bought from stores in San Francisco. Such viruses are of interest given the enormous scale of their exposure to humans consuming meat products.
Meat and meat products are the source of numerous enteric infections in humans. Enteric viruses harbored by farmed animals may contaminate meat samples during slaughter and meat preparation (Machnowska et al., 2014). Hepatitis E virus (HEV) replicating in domesticated or wild-pigs can also be transmitted to human through undercooked pork consumption (Christou and Kosmidou, 2013, Meng, 2013). At slaughter time (22–29 weeks), HEV was present in 12% and 41% of the plasmas and stools from Canadian pigs, respectively (Leblanc et al., 2007). Viral transmission between farm animal species or between domesticated and wild animals may also occur increasing risk of adaptation to new host and changes in viral tropism (Ma et al., 2009, Sonnberg et al., 2013). Transmission of viruses in meat or blood may also occur through non-enteric routes such as skin cuts occurring during hunting and butchering as seen for simian T-lymphotropic and simian foamy virus (Betsem et al., 2011, Gessain et al., 2013, Switzer et al., 2004). Cross-species transmissions, likely due to hunting and/or butchering non-human primates, initiated various HIV epidemics (HIV-1 groups M, N, O, P and HIV-2 groups A–H) (Sharp and Hahn, 2011) and Ebola virus outbreaks (Leroy et al., 2004). The possibility of bovine viruses playing a role in carcinogenesis has been raised based on the association of colorectal cancer with consumption of under-cooked beef (Zur Hausen, 2012).
Results
Overview of virome
The Illumina MiSeq 2×250 bases run generated a total of 4,944,932 unique sequence reads. Sequence reads were assembled de novo and compared to the GenBank non-redundant protein database using BLASTx. 475 sequence reads showed similarity to known eukaryotic viral sequences with an E-value cutoff 10−5. Table 1 presents the characterization of the eukaryotic viral sequences detected in the meat samples.
Table 1.
Characterization of the eukaryotic viral sequence reads in meat samples.
| Name of the match (s) | GenBank no. of the match (s) | Aa identities with |the match (s; %) | Total reads | |
|---|---|---|---|---|
| Beef (total 2,735,653 unique reads) | Ungulate tetraparvovirus 2 (Porcine hokovirus) | JQ700068, FJ982248 | 95–100 | 92 |
| Polyomavirus | JQ178241, NC_020068, HQ385746, JQ479320 | 47–71 | 31 | |
| Ungulate erythroparvovirus 1 | AF406967 | 90–100 | 32 | |
| Torque teno virus | JX173481, NC_014070, HM633249 | 78–92 | 8 | |
| Circovirus-like PorkNW2/USA/2009 | HQ738638 | 98 | 2 | |
| Pork (total of 618,596 unique reads) | Torque teno virus | AY823990, HM633244 | 99–100 | 20 |
| Ungulate copiparvovirus 2 | GQ387500 | 99–100 | 18 | |
| Ungulate tetraparvovirus 2 (Porcine hokovirus) | KC211901, KF225549, KF225541 | 99–100 | 35 | |
| Ungulate protoparvovirus 1 | FJ853420, JX568158 | 98–100 | 19 | |
| Copiparvovirus (Porcine parvovirus 6) | NC_023860 | 88–92 | 12 | |
| Porcine cirvovirus 2 | EU483630 | 99–100 | 5 | |
| Chicken (total of 1,590,683 unique reads) | Chicken anemia virus | DQ360826, FR850022, AF242190 | 100 | 177 |
| Chicken Gyrovirus 4 | NC_018401 | 99 | 8 | |
| Chicken Gyrovirus 2 | KF436510.1 | 100 | 7 | |
| Chicken Gyrovirus 3 | NC_017091 | 92–100 | 6 | |
| Human Gyrovirus 1 | NC_015630 | 100 | 1 | |
| Novel Gyrovirus | NC_022789 | 57 | 2 | |
In the beef samples we detected eukaryotic viral sequences related to parvoviruses, polyomaviruses, anelloviruses and circoviruses (Table 1). In the pork samples we identified viral sequences related to parvoviruses, anelloviruses and circoviruses (Table 1). In chicken diverse gyroviruses were detected (Table 1).
Parvoviruses in beef samples
Sequences identical to the previously described bovine parvovirus 3 (BPV3: prototype of the ungulate erythroparvovirus 1 species) were identified. Here we used the recently described taxonomy for the family Parvoviridae (Cotmore et al., 2014). BPV3 was one of two parvoviruses previously identified from bovine serum (Allander et al., 2001). Sequences identical to porcine hokovirus (P-PARV4 in the ungulate tetraparvovirus 2 species), first sequenced from porcine tissues (Lau et al., 2008), were also identified in beef. Porcine hokovirus is distinct from bovine hokovirus (B-PARV4-1 in the distinct ungulate tetraparvovirus 1 species) with only 63–65% nucleotide identity between them in ORF1 and ORF2 (Lau et al., 2008).
In order to confirm the existence of porcine hokovirus in beef samples, PCR screening was performed using two sets of nested primers. Twenty-two beef samples, including the 10 samples analyzed here and 12 beef samples collected from stores in San Francisco in 2010 (Li et al., 2011), were tested. PCR results indicated two beef samples were positive, one from a pool of five samples and the other from a beef sample collected in 2010. The PCR products were sequenced confirmed, sharing 99% sequence identity with a porcine hokovirus strain in GenBank (JQ700068).
A novel bovine polyomavirus
Polyomaviruses are small, circular DNA viruses that can cause persistent infections in numerous mammals and can be oncogenic in some hosts. The one previously reported bovine polyomavirus (BPyV) was originally isolated from bovine serum (Schuurman et al., 1990). Due to its high prevalence in cows BPyV is also a frequent environmental contaminant (Hundesa et al., 2006). A high sero-prevalence to BPyV has been reported in people with high exposure to cattle (Parry and Gardner, 1986) but no specific illness in either cows or humans has been attributed to BPyV.
In one beef samples pool 31 sequence reads generating eight different contigs were found to have strong amino acid similarity to polyomaviruses. PCR screening of the individual samples with a set of nested primers designed on the LT-Ag sequence indicated that a single beef sample was positive. Using inverse PCR the complete genome was acquired and Sanger sequenced (GenBank KM111535). The sequences of the primers used for PCR screening and inverse PCR are shown in “Materials and methods”.
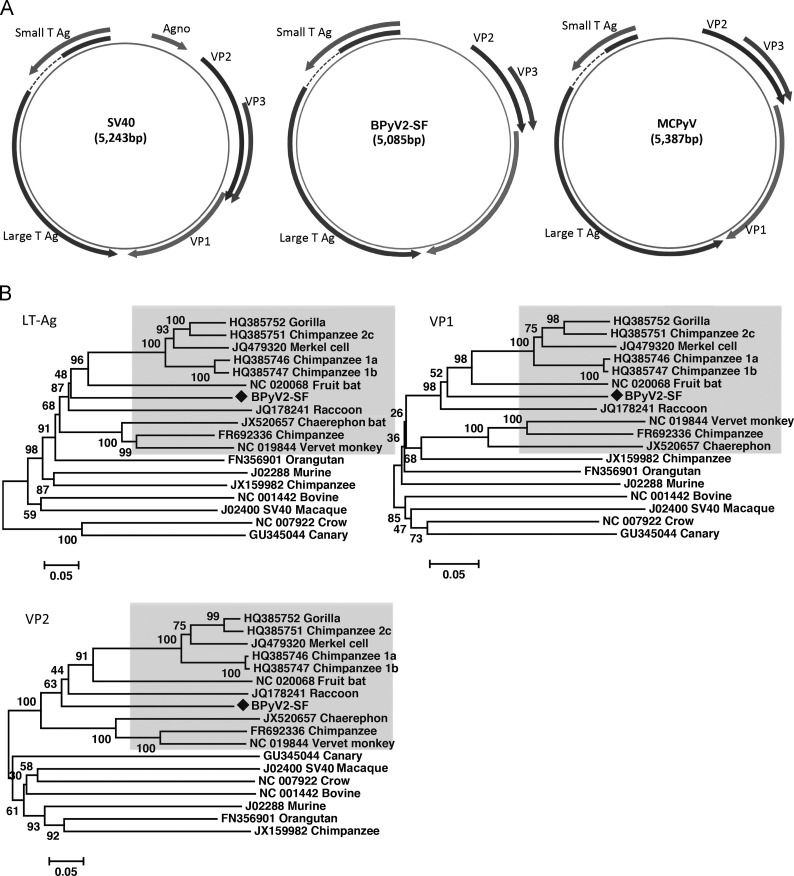
The circular genome of the bovine polyomavirus (named BPyV2-SF) was 5085 bases encoding five major proteins including an early region with regulatory ST-Ag and LT-Ag and a late region coding for the VP1, VP2, and VP3 structural proteins ( Fig. 1A), and a 214 aa alternative Large T ORF (ALTO) protein lying in the second exon of LT-Ag gene. The early region and late region were separated by a regulatory region that contained typical polyomavirus features (Pipas, 1992) including an AT-rich region on the left side of the putative replication origin. Four repeats of the consensus pentanucleotide LT-Ag binding site GAGGC were present. In polyomaviruses VP3 is translated by a leaky ribosomal scanning starting at an internal methionine codon of the larger VP2 ORF. The known polyomaviruses can be divided into “VP3+” species with a clear homolog of the VP3 N-terminal MALXXΦ motif (Φ is an aromatic residue) and “VP3-less” species lacking the motif (Schowalter and Buck, 2013). Phylogenetic analysis based on complete genome of known polyomaviruses revealed that VP3-less species cluster together into a monophyletic clade to which BpyV2-SF also belongs (Fig. 1B). Sequence analysis reveals that BpyV2-SF fits the characteristics of VP3-less polyomavirus species, such as having an ALTO protein within the LT-Ag frame, lacking the VP3 N-terminal MALXXΦ motif, and missing ~80 aa near the carboxyl terminus of the VP2 compared with the longer VP3+ species (Schowalter and Buck, 2013).
Fig. 1.
Genome organization (A) and amino acid-based neighbor-joining analysis of BPyV2-SF (GenBank KM111535) (B). Genome organization of MCPyV and SV40 is included for comparison. The vertebrate hosts are shown for each taxon. BPyV2-SF taxa are labeled by a black diamond. Shaded area indicated the VP3-less polyomaviruses.
To determine the divergence in sequence between BPyV2-SF and other VP3-less polyomavirus species, amino acid sequence alignments of LT-Ag, VP1, and VP2 were performed, and neighbor-joining trees generated. The sequence alignments include BPyV2-SF, 10 representative VP3-less polyomavirus strains, and seven representative VP3+ strains. Phylogenetic analysis indicated that BpyV2-SF is highly divergent from these other polyomaviruses and that it fell within the VP3-less polyomavirus monophyletic clade together with polyomaviruses from gorilla, chimpanzee, humans, eidolon (megabat), and raccoon (Fig. 1B). Table 2 presents the amino acid distance between BPyV2-SF and the closest relative in LT-Ag, ST-Ag, VP1 and VP2. The high divergence of protein sequences confirms that BPyV2-SF is a novel polyomavirus species.
Table 2.
Comparison of viral protein sequence identity (%) between BPyV2-SF and most closely related strains.
| GenBank no. (host species) | Amino acid sequence identity (%) |
|||
|---|---|---|---|---|
| LT-Ag | ST-Ag | VP1 | VP2 | |
| HQ385752 (Gorilla) | 56.2 | 52.5 | 65.4 | 50.4 |
| HQ385751 (Chimpanzee) | 57.0 | 52.5 | 65.7 | 49.6 |
| JQ479320 (Merkel cell) | 54.5 | 51.9 | 66.3 | 49.4 |
| HQ385746 (Chimpanzee) | 55.8 | 51.9 | 66.8 | 48.3 |
| HQ385747 (Chimpanzee) | 54.9 | 47.0 | 66.6 | 48.3 |
| NC_020068 (Fruit bat) | 48.8 | 44.9 | 67.8 | 43.6 |
| JQ178241 (Raccoon) | 49.0 | 40.3 | 65.9 | 48.9 |
Circovirus-like sequence in the beef
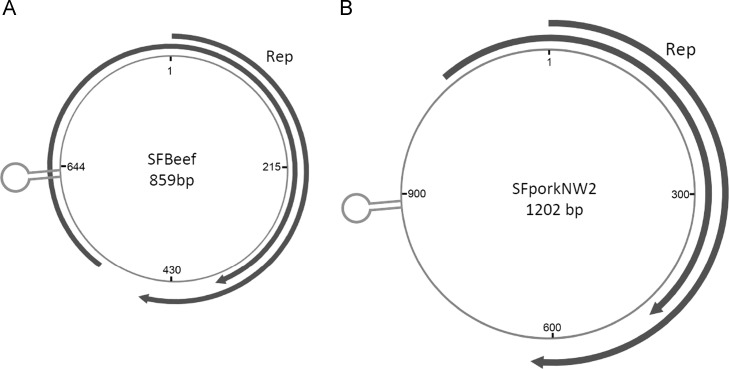
In a beef pools two sequence reads showed high sequence identity with a previously reported circovirus-like virus PorkNW2 sequence (GenBank accession HQ738638), previously found in both pork and beef samples from San Francisco stores using Circoviridae consensus primers (Li et al., 2011). PCR screening with a set of nested primers designed on the sequence detected in this study confirmed that two out of the 10 beef samples were positive. In order to clarify whether this sequence was from a circular DNA molecular, inverse PCR was performed and the amplicon sequenced showing that this circular DNA was 859 bases (named mini PCV-like virus strain MPCLV-SFBeef), shorter than the 1202 bases sequence of the original PorkNW2 genome ( Fig. 2). Although BLASTx search revealed a partial putative Rep protein (153 aa) with 97% amino acid sequence similarity to SFPorkNW2 [30], no suitably located ATG initiator codon was identified. Protein analysis indicated that the Rep protein of SFBeef is 68Aa shorter than that of SFPorkNW2, missing the RNA helicase domain. Nucleotide sequence analysis showed that both genomes contain a identical stem-loop lying upstream of Rep open reading frame with the conserved origin of replication NANTATTAC nonamer atop the stem-loop found in members of the Circoviridae, Nanoviridae and Geminiviridae families. Another ORF potentially encoding 243 aa protein arranged in the same coding direction as Rep protein was also found yielding an ORF map similar to that of NW2 (Fig. 2A and B). BLASTp search indicated part of the putative protein (60 aa) shared 47% amino acid sequence identity with the M169 protein of unknown function of Murid herpesvirus (GenBank no. HE610454).
Fig. 2.
The ORF map of MPCLV-SFBeef (GenBank KM111537) (A) and SFPorkNW2 (B).
Viruses in pork
We identified sequences from pork-derived viral particles that were identical to porcine parvovirus 4 (PPV4 in the ungulate copiparvovirus 2 species). PPV4 was initially identified in a lung lavage from a pig co-infected with pig circovirus 2 (Cheung et al., 2010). We also detected ungulate tetraparvovirus 2 (aka porcine hokovirus, the same virus detected in beef samples) initially reported to infect pigs in Honk Kong at high frequency (Lau et al., 2008). Sequences from a third parvovirus species in a third genus (ungulate protoparvovirus 1) were also detected. Last, a fourth set of parvovirus-like sequences closest to a still unclassified copiparvoviruses labeled porcine parvovirus 6 (PPV6 deposited March 31, 2014 GenBank NC_023860) from pigs in China were also detected. Because PPV6 NS1 shows closest identity to PPV4 but with only 58% identity it likely represents a new copriparvovirus species (proposed name ungulate copiparvovirus 3) (Cotmore et al., 2014). Four parvovirus species in three distinct Parvoviridae genera were therefore detected in these few pork samples.
Also detected in both pork and beef samples were anelloviruses which have been reported in the blood of numerous mammals including cows and pigs (Brassard et al., 2008). Pig circovirus 2 (PCV2) a frequent infection of pigs occurring world-wide was present.
Gyroviruses in chicken
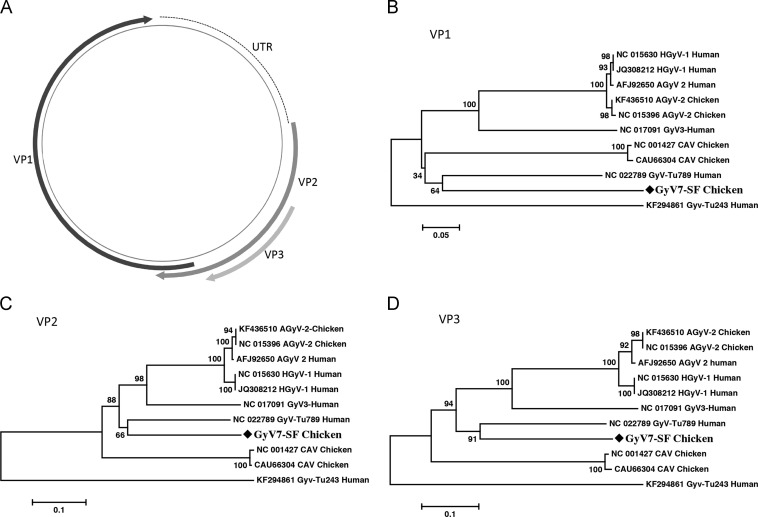
Gyroviruses are small nonenveloped DNA viruses with a negative sense single-stranded circular DNA of about 2.3 kb. The genus Gyrovirus is currently in the Circoviridae family but closer (although still distinct) in genetic organization to members of the Anelloviridae (Hino and Prasetyo, 2009). Here sequences from six different gyroviruses were detected in the chicken meat samples: chicken anemia virus (CAV), human gyrovirus 1 (HGyV1) and the very closely related gyrovirus 2 (GyV2), GyV3, GyV4 and a novel gyrovirus here named GyV7. BLASTx search indicated that the known gyroviruses found here shared high protein sequence identities (>92%) with their best matches in the GenBank, while the novel gyrovirus (GyV7-SF) showed only 57% protein sequence similarity to its closest matches (Table 1). PCR screening with a set of nested primers designed on the novel sequences indicated that only one chicken sample was positive for GyV7-SF. The complete circular genome of GyV7-SF1 was acquired using inverse PCR and Sanger sequencing (GenBank KM111536). The GyV7-SF genome is 2439 bases, with a 514 base non-translated region (NTR) and a polyadenylation signal [AATAAA]. ORF prediction revealed three overlapping ORFs for VP1 (465-aa), VP2 (238-aa) and VP3/Apoptin (130-aa). The genome organization of GyV7-SF was similar to that of the other gyroviruses ( Fig. 3A). BLASTp search based on VP1, VP2, and VP3 all revealed the best match with GyV6 (NC_022789), with protein sequence identities of 49%, 53%, and 63%, respectively. Phylogenetic based the three major protein sequence all indicated that GyV7-SF clustered loosely with GyV6 (Fig. 3B–D), which was detected in diarrhea samples from Tunisian children (Gia Phan et al., 2013).
Fig. 3.
Genome organization (A) and amino acid-based neighbor-joining analysis of GyV-SF1 (GenBank KM111536) (B–D). Included with each taxon is the species in which each sequence was found. GyV-SF1 identified in this study is marked with black diamond.
Discussion
Our results indicate that diverse parvoviruses are readily found in meat samples from hoofed farm animals. Parvoviruses are frequently detected in domesticated and wild animal as well as human feces (Bodewes et al., 2013, Canuti et al., 2011, Handley et al., 2012, Phan et al., 2012, Shan et al., 2011, Sharp et al., 2010, Väisänen et al., 2014). Some parvovirus can be highly pathogenic, notoriously canine parvovirus which recently adapted from cats to dogs possibly through intermediate species (Stucker et al., 2012) and is thought to be readily transmitted across carnivore species (Allison et al., 2013). Parvoviruses can also be detected in blood as exemplified by PARV4 (primate tetraparvovirus 1) and parvovirus B19 (primate erythroparvovirus 1) which is typically a benign infection but which can cause serious anemia or fetal hydrops in susceptible individuals or when infecting fetus (Feldman et al., 2010, Morinet et al., 2011). Parvovirus B19 can also be deposited in muscles and other tissues where it can be PCR amplified even decades after primary infection (Norja et al., 2006). The detection of ungulate erythroparvovirus 1 virus in beef may therefore reflect either active replication or particle deposition into tissues. Porcine hokovirus (ungulate tetraparvovirus 2) was also found in beef indicating that the tropism of this viral species may include both pigs and cattle.
As expected from its previously reported high frequency in pigs (Lau et al., 2008) porcine hokovirus was also found in the pork samples analyzed here. The lower number of sequence reads in pork than in beef (Table 2) indicates that laboratory contamination from the pork to the beef samples was unlikely. The possibility that the deep sequencing and PCR detection of parvoviruses in beef or pork were due to cross-contamination during slaughter, shipping, or on store shelves cannot be excluded and will require confirmation in independent studies.
Infection of polyomaviruses in mammals generally causes persistent asymptomatic infection. Polyomaviruses have also been associated with a broad spectrum of diseases in humans and animals, such as progressive multifocal leukoencephalopathy in immunosuppressed patients (JCV) (Hou and Major, 2000), nephropathy (BKV)(White et al., 2013), Merkel cell cancer (MCV) (Feng et al., 2008, Houben et al., 2010), trichodysplasia spinulosa (TSPyV) (Van der Meijden et al., 2010) and retinal blindness/vasculitic myopathy (Mishra et al., 2014) in humans, fibroma in African elephant (AelPyV-1) (Stevens et al., 2013), brain tumors in raccoons (RacPyV) (Dela Cruz et al., 2013), and sarcoma in laboratory rodents (SV40) (Kirschstein and Gerber, 1962). With the increasing utilization of deep sequencing numerous polyomaviruses have been identified in human and in non-human primates (Ehlers and Moens 2014, Scuda et al., 2013, White et al., 2013) and it is anticipated that a similar expansion will occur for other host species. BPyV2-SF proteins, while still highly distinct (57–47% protein identity), are the most closely related protein sequences (except VP1) to those of primate polyomaviruses infecting human, chimpanzees, and gorillas (Table 2).
Pork samples also contained parvovirus sequences from three genera which had been previously described in other pig samples. Surprisingly we did not detect any sequences belonging to the Bocaparvovirus genus frequently detected in pig fecal samples (Jiang et al., 2014, Shan et al., 2011) possibly due to a tropism generally limited to respiratory or gut tissues.
The presence of porcine circovirus 2 in pork may be due to either inoculation with a live attenuated vaccine frequently administered to piglets or from a natural infection (Zhai et al., 2014). A recent study did not detect human antibodies to PCV2 despite frequent consumption of infected pork therefore reducing concern about zoonosis to humans (Burbelo et al., 2013, Victoria et al., 2010).
The 0.8 kb circular genome found in beef reassembled a 1.2 kb genome previously sequenced in both pork and beef from the US (Li et al., 2011). Short, PCV2-like, rearranged genomes (483–896 bases) have also been reported in Chinese pigs (Wen et al., 2012, Wen et al., 2012, Zhai et al., 2014). The generation of such truncated and rearranged genomes may be related to that seen when viruses are passaged at high multiplicity of infection leading to defective interfering viral particles retaining only essential Cis elements required for genome replication and requiring trans-complementation from a helper virus (Mankertz et al., 1997). A related phenomenon may be the generation of truncated ssDNA circular geminivirus and anellovirus genome when passaged in vitro (De Villiers et al., 2011).
Chicken meat showed the presence of numerous gyroviruses including the highly prevalent CAV reported world-wide (Schat, 2009, Toro et al., 2006). The gyrovirus genus includes several species, namely avian gyrovirus 2 (AGV2) from chicken (Rijsewijk et al., 2011), the closely related HGyV1 from human skin (Sauvage et al., 2011), GyV3 and GyV4 from children feces and chicken meat (Chu et al., 2012, Phan et al., 2012), and GyV5 and GyV6 from children feces (Gia Phan et al., 2013), many of which were also detected in chicken and children feces from South Africa (Smuts, 2014). The characterization of a seventh species of gyrovirus (GyV7) in this limited sampling of chicken indicates that still more gyroviruses from chicken and other birds are likely to be identified. The current lack of report describing gyrovirus sero-conversion in human or replication in mammalian cells indicates that a possible explanation for the frequent detection of gyrovirus DNA in human feces (gyrovirus species GyV3–GyV6) (Chu et al., 2012, Gia Phan et al., 2013, Phan et al., 2012, Phan et al., 2012) and recently cat feces (CAT-CAV) (X. Zhang et al., 2014) may reflect their dietary origin from eating chicken or other birds.
Only DNA viral sequences were identified in this limited sampling of muscle tissues from presumably healthy animals while RNA viruses such as picornaviruses, caliciviruses, coronaviruses, and astroviruses tend to predominate in the feces of farm animals (Sachsenröder et al., 2014, Shan et al., 2011, Zhang et al., 2014). The absence of detected RNA viruses in the muscle tissues tested here may reflect a low level of contamination with fecal material, a general preference of RNA viruses for enteric rather than systemic infections, and/or as recently reported (Sachsenröder et al., 2014) the older age of the animals used for meat production.
Materials and methods
Sample preparation and metagenomic analysis
Raw pork (7 pieces and 3 ground), beef (7 pieces and 3 ground), chicken (6 pieces and 4 ground) were purchased from six randomly selected meat markets in the Sunset district of San Francisco, USA from Jan 12–28, 2014. Two samples of beef, pork, and chicken were purchased from each of five stores except for two stores in which only a single sample of chicken was purchased. The remaining two chicken samples were purchased from a sixth store. All ground meats were purchased already wrapped in plastic labeled with name of national conglomerates. Neat piece of meats was collected and immediately placed in zippered plastic bags. All samples were frozen the same day at −80 °C until further processing. None of the meat was butchered or ground in the shops. In order to avoid potential contamination, small inner section of the meat pieces was cut out and used. Meat pieces (~25 mg) were frozen and thawed on dry ice three times, homogenized, filtered, and nuclease treated as previously described to enrich for nucleic acids within viral particles (Li et al., 2013). Remaining total nucleic acid was then isolated using MagMAX™ Viral RNA Isolation Kits (Qiagen) according to manufacturer׳s protocol. A library of randomly amplified PCR products from viral RNA and DNA was prepared by using a Nextera DNA Sample Prep Kit and sequenced on the MiSeq Illumina platform (Illumina, San Diego, CA, USA). Six separate pools of nucleic acids from the 30 meat specimens, each pool contained five specimens from the same farm animal species, were generated. The viral nucleic acid pools, containing both DNA and RNA viral sequences, were subjected to reverse transcription reactions with SuperScript III reverse transcriptase (Invitrogen) and 100 picomoles of random hexamer primer, followed by a single round of DNA synthesis using Klenow fragment polymerase (New England BioLabs). A library was then constructed using Nextera XT DNA Sample Preparation Kit (Illumina) and then sequenced using the Miseq Illumina platform with 250 bases paired ends with a distinct molecular tag for each pool. Resulting raw reads were trimmed for quality and primer, and de novo assembled into contigs. Sequences and contigs were compared to the GenBank non-redundant protein database using BLASTx with an E-value cutoff of <10−5.
PCR screening and genome sequencing
PCR screening was performed for the bovine polyomavirus, circovirus-like genome, the porcine hokovirus in beef, and the novel gyrovirus in chicken. Inverse PCR was used to generate the complete genome of the novel polyomavirus and the circovirus-like sequence in beef. PCR to bridge sequence gaps was used to acquire the whole genome of the novel gyrovirus in chicken. Sequences and characteristics of the primers used in PCR screening and inverse PCR are shown in Supplemental Table 1. Sanger method was used for sequencing complete viral genomes and to confirm the sequence of PCR products.
Phylogenetic analysis
Phylogenetic analyses were performed based on the predicted amino acid or nucleotide sequences in the present study, their closest viral relatives based on best BLASTx hits, and representative members of related viral species or genera. Sequence alignment was performed using CLUSTAL W with the default settings. Aligned sequences were trimmed to match the genomic regions of the viral sequences obtained in the study. A phylogenetic tree with 100 bootstrap resamples of the alignment data sets was generated using the neighbor-joining method based on the Jones–Taylor–Thornton matrix-based model in MEGA5.0 (Tamura et al., 2011). Bootstrap values (based on 100 replicates) for each node are given. Putative ORFs in the genome were predicted by NCBI ORF finder. Putative exon and intron were predicted by Netgenes2 at http://www.cbs.dtu.dk/services/NetGene2/.
Nucleotide sequence accession numbers
The genomes of known and new viruses described in detail here were deposited in GenBank under the following accession numbers: KM111535–KM111537. The raw sequence reads from the metagenomic library were deposited in the Short Read Archive of GenBank database under accession number SRX652341.
Acknowledgments
We thank Christopher Buck for helpful discussions. The work was supported by the Blood Systems Research Institute and National Institutes of Health R01 HL105770 to E.D., and China Scholarship Council No. 201208320503 to W.Z.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2014.08.025.
Appendix A. Supporting information
Supplementary data
References
- Allander T., Emerson S.U., Engle R.E., Purcell R.H., Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. USA. 2001;98(20):11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison A.B., Kohler D.J., Fox K.A., Brown J.D., Gerhold R.W., Shearn-Bochsler V.I., Holmes E.C. Frequent cross-species transmission of parvoviruses among diverse carnivore hosts. J. Virol. 2013;87(4):2342–2347. doi: 10.1128/JVI.02428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsem E., Rua R., Tortevoye P., Froment A., Gessain A. Frequent and recent human acquisition of simian foamy viruses through apes׳ bites in central Africa. PLoS Pathog. 2011;7(10):e1002306. doi: 10.1371/journal.ppat.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., van der Giessen J., Haagmans B.L., Osterhaus A.D.M.E., Smits S.L. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J. Virol. 2013;87(13):7758–7764. doi: 10.1128/JVI.00568-13. 〈http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3700315&tool=pmcentrez&rendertype=abstract〉 (Retrieved from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassard J., Gagné M.-J., Lamoureux L., Inglis G.D., Leblanc D., Houde A. Molecular detection of bovine and porcine Torque teno virus in plasma and feces. Vet. Microbiol. 2008;126(1–3):271–276. doi: 10.1016/j.vetmic.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Burbelo P.D., Ragheb J.A., Kapoor A., Zhang Y. The serological evidence in humans supports a negligible risk of zoonotic infection from porcine circovirus type 2. Biologicals. 2013;41(6):430–434. doi: 10.1016/j.biologicals.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canuti M., Eis-Huebinger A.M., Deijs M., de Vries M., Drexler J.F., Oppong S.K., van der Hoek L. Two novel parvoviruses in frugivorous New and Old World bats. PloS One. 2011;6(12):e29140. doi: 10.1371/journal.pone.0029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.K., Wu G., Wang D., Bayles D.O., Lager K.M., Vincent A.L. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 2010;155(5):801–806. doi: 10.1007/s00705-010-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christou L., Kosmidou M. Hepatitis E virus in the Western world—a pork-related zoonosis. Clin. Microbiol. Infect. 2013;19(7):600–604. doi: 10.1111/1469-0691.12214. [DOI] [PubMed] [Google Scholar]
- Chu D.K.W., Poon L.L.M., Chiu S.S.S., Chan K.H., Ng E.M., Bauer I., Peiris J.S.M. Characterization of a novel gyrovirus in human stool and chicken meat. J. Clin. Virol. 2012;55(3):209–213. doi: 10.1016/j.jcv.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S.F., Agbandje-McKenna M., Chiorini J.A., Mukha D.V., Pintel D.J., Qiu J., Davison A.J. The family Parvoviridae. Arch. Virol. 2014;159(5):1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Villiers E.-M., Borkosky S.S., Kimmel R., Gunst K., Fei J.-W. The diversity of torque teno viruses: in vitro replication leads to the formation of additional replication-competent subviral molecules. J. Virol. 2011;85(14):7284–7295. doi: 10.1128/JVI.02472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela Cruz F.N., Giannitti F., Li L., Woods L.W., Del Valle L., Delwart E., Pesavento P.A. Novel polyomavirus associated with brain tumors in free-ranging raccoons, western United States. Emerg. Infect. Dis. 2013;19(1):77–84. doi: 10.3201/eid1901.121078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers, B., Moens, U., Genome analysis of non-human primate polyomaviruses, Infect. Genet. Evol. 2014 Aug;26C:283–294, 10.1016/j.meegid.2014.05.030. [DOI] [PubMed]
- Feldman D.M., Timms D., Borgida A.F. Toxoplasmosis, parvovirus, and cytomegalovirus in pregnancy. Clin. Lab. Med. 2010;30(3):709–720. doi: 10.1016/j.cll.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessain A., Rua R., Betsem E., Turpin J., Mahieux R. HTLV-3/4 and simian foamy retroviruses in humans: discovery, epidemiology, cross-species transmission and molecular virology. Virology. 2013;435(1):187–199. doi: 10.1016/j.virol.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gia Phan T., Phung Vo N., Sdiri-Loulizi K., Aouni M., Pothier P., Ambert-Balay K., Delwart E. Divergent gyroviruses in the feces of Tunisian children. Virology. 2013;446(1–2):346–348. doi: 10.1016/j.virol.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley S.A., Thackray L.B., Zhao G., Presti R., Miller A.D., Droit L., Virgin H.W. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151(2):253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino S., Prasetyo A.A. Relationship of Torque teno virus to chicken anemia virus. Curr. Top. Microbiol. Immunol. 2009;331:117–130. doi: 10.1007/978-3-540-70972-5_8. 〈http://www.ncbi.nlm.nih.gov/pubmed/19230561〉 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Hou J., Major E.O. Progressive multifocal leukoencephalopathy: JC virus induced demyelination in the immune compromised host. J. Neurovirol. 2000;6(Suppl 2):S98–S100. 〈http://www.ncbi.nlm.nih.gov/pubmed/10871795〉 (Retrieved from) [PubMed] [Google Scholar]
- Houben R., Shuda M., Weinkam R., Schrama D., Feng H., Chang Y., Becker J.C. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J. Virol. 2010;84(14):7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundesa A., Maluquer de Motes C., Bofill-Mas S., Albinana-Gimenez N., Girones R. Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Appl. Environ. Microbiol. 2006;72(12):7886–7893. doi: 10.1128/AEM.01090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.-H., Xiao C.-T., Yin S.-H., Gerber P.F., Halbur P.G., Opriessnig T. High prevalence and genetic diversity of porcine bocaviruses in pigs in the USA, and identification of multiple novel porcine bocaviruses. J. Gen. Virol. 2014;95(Pt 2):453–465. doi: 10.1099/vir.0.057042-0. [DOI] [PubMed] [Google Scholar]
- Kirschstein R.L., Gerber P. Ependymomas produced after intracerebral inoculation of SV40 into new-born hamsters. Nature. 1962;195:299–300. doi: 10.1038/195299b0. 〈http://www.ncbi.nlm.nih.gov/pubmed/14456349〉 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Tse H., Fu C.T.Y., Au W.-K., Chen X.-C., Yuen K.-Y. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 2008;89(Pt 8):1840–1848. doi: 10.1099/vir.0.2008/000380-0. [DOI] [PubMed] [Google Scholar]
- Leblanc D., Ward P., Gagné M.-J., Poitras E., Müller P., Trottier Y.-L., Houde A. Presence of hepatitis E virus in a naturally infected swine herd from nursery to slaughter. Int. J. Food Microbiol. 2007;117(2):160–166. doi: 10.1016/j.ijfoodmicro.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Leroy E.M., Rouquet P., Formenty P., Souquière S., Kilbourne A., Froment J.-M., Rollin P.E. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303(5656):387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- Li L., McGraw S., Zhu K., Leutenegger C.M., Marks S.L., Kubiski S., Pesavento P.A. Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg. Infect. Dis. 2013;19(4):534–541. doi: 10.3201/eid1904.121390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Shan T., Soji O.B., Alam M.M., Kunz T.H., Zaidi S.Z., Delwart E. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J. Gen. Virol. 2011;92(Pt 4):768–772. doi: 10.1099/vir.0.028704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Lager K.M., Vincent A.L., Janke B.H., Gramer M.R., Richt J.A. The role of swine in the generation of novel influenza viruses. Zoonoses Public Health. 2009;56(6–7):326–337. doi: 10.1111/j.1863-2378.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- Machnowska P., Ellerbroek L., Johne R. Detection and characterization of potentially zoonotic viruses in faeces of pigs at slaughter in Germany. Vet. Microbiol. 2014;168(1):60–68. doi: 10.1016/j.vetmic.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Mankertz A., Persson F., Mankertz J., Blaess G., Buhk H.J. Mapping and characterization of the origin of DNA replication of porcine circovirus. J. Virol. 1997;71(3):2562–2566. doi: 10.1128/jvi.71.3.2562-2566.1997. 〈http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=191374&tool=pmcentrez&rendertype=abstract〉 (Retrieved from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.-J. Zoonotic and foodborne transmission of hepatitis E virus. Semin. Liver Dis. 2013;33(1):41–49. doi: 10.1055/s-0033–1338113. [DOI] [PubMed] [Google Scholar]
- Mishra, N., Pereira, M., Rhodes, R.H., An, P., Pipas, J.M., Jain, K., Lipkin, W.I., et al., Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy, J. Infect. Dis. 2014 May 1. pii: jiu250. [Epub ahead of print], 10.1093/infdis/jiu250 [DOI] [PMC free article] [PubMed]
- Morinet F., Leruez-Ville M., Pillet S., Fichelson S. Concise review: anemia caused by viruses. Stem Cells. 2011;29(11):1656–1660. doi: 10.1002/stem.725. [DOI] [PubMed] [Google Scholar]
- Norja P., Hokynar K., Aaltonen L.-M., Chen R., Ranki A., Partio E.K., Hedman K. Bioportfolio: lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc. Natl. Acad. Sci. USA. 2006;103(19):7450–7453. doi: 10.1073/pnas.0602259103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry J.V., Gardner S.D. Human exposure to bovine polyomavirus: a zoonosis? Arch. Virol. 1986;87(3–4):287–296. doi: 10.1007/BF01315306. 〈http://www.ncbi.nlm.nih.gov/pubmed/3004390〉 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Phan T.G., Li L., O’Ryan M.G., Cortes H., Mamani N., Bonkoungou I.J.O., Delwart E. A third gyrovirus species in human faeces. J. Gen. Virol. 2012;93(Pt 6):1356–1361. doi: 10.1099/vir.0.041731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Vo N.P., Bonkoungou I.J.O., Kapoor A., Barro N., O’Ryan M., Delwart E. Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J. Virol. 2012;86(20):11024–11030. doi: 10.1128/JVI.01427-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas J.M. Common and unique features of T antigens encoded by the polyomavirus group. J. Virol. 1992;66(7):3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. 〈http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=241200&tool=pmcentrez&rendertype=abstract〉 (Retrieved from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijsewijk F.A.M., Dos Santos H.F., Teixeira T.F., Cibulski S.P., Varela A.P.M., Dezen D., Roehe P.M. Discovery of a genome of a distant relative of chicken anemia virus reveals a new member of the genus Gyrovirus. Arch. Virol. 2011;156(6):1097–1100. doi: 10.1007/s00705-011-0971-6. [DOI] [PubMed] [Google Scholar]
- Sachsenröder J., Twardziok S.O., Scheuch M., Johne R. The general composition of the faecal virome of pigs depends on age, but not on feeding with a probiotic bacterium. PloS One. 2014;9(2):e88888. doi: 10.1371/journal.pone.0088888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage V., Cheval J., Foulongne V., Gouilh M.A., Pariente K., Manuguerra J.C., Eloit M. Identification of the first human gyrovirus, a virus related to chicken anemia virus. J. Virol. 2011;85(15):7948–7950. doi: 10.1128/JVI.00639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat K.A. Chicken anemia virus. Curr. Top. Microbiol. Immunol. 2009;331:151–183. doi: 10.1007/978-3-540-70972-5_10. 〈http://www.ncbi.nlm.nih.gov/pubmed/19230563〉 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Schowalter R.M., Buck C.B. The Merkel cell polyomavirus minor capsid protein. PLoS Pathog. 2013;9(8):e1003558. doi: 10.1371/journal.ppat.1003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurman R., Sol C., van der Noordaa J. The complete nucleotide sequence of bovine polyomavirus. J. Gen. Virol. 1990;71(Pt 8):1723–1735. doi: 10.1099/0022-1317-71-8-1723. 〈http://www.ncbi.nlm.nih.gov/pubmed/2167926〉 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Scuda N., Madinda N.F., Akoua-Koffi C., Adjogoua E.V., Wevers D., Hofmann J., Ehlers B. Novel polyomaviruses of nonhuman primates: genetic and serological predictors for the existence of multiple unknown polyomaviruses within the human population. PLoS Pathog. 2013;9(6):e1003429. doi: 10.1371/journal.ppat.1003429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T., Lan D., Li L., Wang C., Cui L., Zhang W., Delwart E. Genomic characterization and high prevalence of bocaviruses in swine. PloS One. 2011;6(4):e17292. doi: 10.1371/journal.pone.0017292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T., Li L., Simmonds P., Wang C., Moeser A., Delwart E. The fecal virome of pigs on a high-density farm. J. Virol. 2011;85(22):11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp C.P., LeBreton M., Kantola K., Nana A., Diffo J.L.D., Djoko C.F., Simmonds P. Widespread infection with homologues of human parvoviruses B19, PARV4, and human bocavirus of chimpanzees and gorillas in the wild. J. Virol. 2010;84(19):10289–10296. doi: 10.1128/JVI.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 2011;1(1):a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smuts H.E.M. Novel Gyroviruses, including Chicken Anaemia Virus, in Clinical and Chicken Samples from South Africa. Adv. Virol. 2014;2014:321284. doi: 10.1155/2014/321284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnberg S., Webby R.J., Webster R.G. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013;178(1):63–77. doi: 10.1016/j.virusres.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens H., Bertelsen M.F., Sijmons S., Van Ranst M., Maes P. Characterization of a novel polyomavirus isolated from a fibroma on the trunk of an African elephant (Loxodonta africana) PloS One. 2013;8(10):e77884. doi: 10.1371/journal.pone.0077884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucker K.M., Pagan I., Cifuente J.O., Kaelber J.T., Lillie T.D., Hafenstein S., Parrish C.R. The role of evolutionary intermediates in the host adaptation of canine parvovirus. J. Virol. 2012;86(3):1514–1521. doi: 10.1128/JVI.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer W.M., Bhullar V., Shanmugam V., Cong M.-E., Parekh B., Lerche N.W., Heneine W. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 2004;78(6):2780–2789. doi: 10.1128/JVI.78.6.2780-2789.2004. 〈http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=353775&tool=pmcentrez&rendertype=abstract〉 (Retrieved from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro H., Ewald S., Hoerr F.J. Serological evidence of chicken infectious anemia virus in the United States at least since 1959. Avian Dis. 2006;50(1):124–126. doi: 10.1637/7442-092205R.1. 〈http://www.ncbi.nlm.nih.gov/pubmed/16617995〉 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Väisänen E., Kuisma I., Phan T.G., Delwart E., Lappalainen M., Tarkka E., Söderlund-Venermo M. Bufavirus in feces of patients with gastroenteritis, Finland. Emerg. Infect. Dis. 2014;20(6):1078–1080. doi: 10.3201/eid2006.131674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meijden E., Janssens R.W.A., Lauber C., Bouwes Bavinck J.N., Gorbalenya A.E., Feltkamp M.C.W. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6(7):e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria J.G., Wang C., Jones M.S., Jaing C., McLoughlin K., Gardner S., Delwart E.L. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J. Virol. 2010;84(12):6033–6040. doi: 10.1128/JVI.02690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., He K., Li B., Wang X., Guo R.L., Yu Z., Jiang J. In vitro and in vivo isolation of a novel rearranged porcine circovirus type 2. J. Virol. 2012;86(23):13120. doi: 10.1128/JVI.02392-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., He K., Ni Y., Zhang X., Li B., Wang X., Jiang J. Complete genome sequence of the rearranged porcine circovirus type 2. J. Virol. 2012;86(10):5963. doi: 10.1128/JVI.00494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.K., Gordon J., Khalili K. The rapidly expanding family of human polyomaviruses: recent developments in understanding their life cycle and role in human pathology. PLoS Pathog. 2013;9(3):e1003206. doi: 10.1371/journal.ppat.1003206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai S.-L., Chen S.-N., Xu Z.-H., Tang M.-H., Wang F.-G., Li X.-J., Wei W.-K. Porcine circovirus type 2 in China: an update on and insights to its prevalence and control. Virol. J. 2014;11(1):88. doi: 10.1186/1743-422X-11–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Tang C., Yue H., Ren Y., Song Z. Viral metagenomics analysis demonstrates the diversity of viral flora in piglet diarrheic feces in China. J. Gen. Virol. 2014 doi: 10.1099/vir.0.063743-0. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu Y., Ji J., Chen F., Sun B., Xue C., Xie Q. Identification of a chicken anemia virus variant-related gyrovirus in stray cats in china, 2012. BioMed Res. Int. 2014:313252. doi: 10.1155/2014/313252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Hausen H. Red meat consumption and cancer: reasons to suspect involvement of bovine infectious factors in colorectal cancer. Int. J. Cancer. 2012;130(11):2475–2483. doi: 10.1002/ijc.27413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data