Abstract
Nef plays a major role in HIV-1 pathogenicity. We studied HIV-1 subtype C infected individuals in acute/early (n=120) or chronic (n=207) infection to investigate the relationship between Nef-mediated CD4/HLA-I down-regulation activities and disease progression, and the influence of immune-driven sequence variation on these Nef functions. A single Nef sequence per individual was cloned into an expression plasmid, followed by transfection of a T cell line and measurement of CD4 and HLA-I expression. In early infection, a trend of higher CD4 down-regulation ability correlating with higher viral load set point was observed (r=0.19, p=0.05), and higher HLA-I down-regulation activity was significantly associated with faster rate of CD4 decline (p=0.02). HLA-I down-regulation function correlated inversely with the number HLA-associated polymorphisms previously associated with reversion in the absence of the selecting HLA allele (r=−0.21, p=0.0002). These data support consideration of certain Nef regions in HIV-1 vaccine strategies designed to attenuate the infection course.
Keywords: HIV-1 Nef, HIV-1 subtype C, CD4 down-regulation, HLA-I down-regulation, HIV-1 disease progression, HLA-associated polymorphisms, immune evasion
Introduction
HIV-1 Nef is a small accessory protein that plays a major role in viral replication and pathogenesis (Foster et al., 2011). Nef displays multiple functions in vitro, including down-regulation of cell-surface CD4 (to enhance viral spread and possibly evade antibody-dependent cell-mediated cytotoxicity of infected cells), down-regulation of HLA class I (HLA-I) A and B molecules (to evade CD8+ T cell responses), activation of T cells and CD4-independent enhancement of virion infectivity (Foster and Garcia, 2008; Veillette et al., 2014). The relative importance of these Nef activities in vivo remain incompletely understood, although some studies suggest that Nef-mediated CD4 down-regulation, enhancement of infectivity, and one or more unknown Nef functions represent major pathogenic contributors (Iafrate et al., 2000; Watkins et al., 2013).
Nef’s influence on HIV-1 pathogenesis is clearly demonstrated by an attenuated disease course in individuals infected with HIV-1 harbouring gross genetic Nef defects (Deacon et al., 1995; Kestler et al., 1991; Kirchhoff et al., 1995). However, more subtle, naturally-occurring variation in Nef may also influence disease outcomes. For example, Nef proteins derived from HIV-1 subtype B infected elite controllers displayed relative functional impairments compared to those from progressors in the absence of major genetic defects (Mwimanzi et al., 2011c; Mwimanzi et al., 2013). Studies linking Nef function and disease outcomes in HIV-1 subtype C, the predominant subtype of the HIV pandemic, are however lacking.
Nef is also a highly immunogenic protein (Lichterfeld et al., 2004). Cross-sectional studies have shown that the overall breadth of Nef-specific CD8+ T cell responses does not correlate with viral control (Kiepiela et al., 2007) and in some studies a higher magnitude of these responses correlated with higher viral loads (Novitsky et al., 2003; Radebe et al., 2011). Nevertheless, the observed associations between lower viral loads and CD8+ T cell targeting of certain Nef epitopes in humans (Adland et al., 2013) and non-human primate models (Budde et al., 2012; Mudd et al., 2012), suggests that CD8+ T cell responses to specific Nef regions may be beneficial.
There is also somewhat limited evidence that immune-driven escape mutations can impair Nef function. Two HLA-B*35-associated CD8+ T cell escape mutations in a conserved proline-rich region of Nef impaired Nef-mediated HLA class I down-regulation and virus replication when occurring in combination (Mwimanzi et al., 2011a; Ueno et al., 2008). Accumulation of novel HLA-B*57-associated polymorphisms largely unique to elite controllers was associated with reduced Nef function in these individuals (Mwimanzi et al., 2013). Furthermore, a recent study predicted that over 50% of HLA-associated Nef polymorphisms would revert on transmission to an HLA-mismatched recipient, supporting biologically-relevant fitness costs of certain immune-driven mutations in Nef (Adland et al., 2013). Identification of immune-driven Nef mutations influencing its pathogenic functions could therefore reveal regions of potential relevance for inclusion in an HIV-1 vaccine designed to attenuate the infection course (Allen and Altfeld, 2008). Thus, studies to investigate this are warranted.
Also important to vaccine design is the identification of specific features of viruses that are transmitted successfully (i.e. understanding the transmission bottleneck). Transmitted/early viruses tend to exhibit particular Env characteristics, including shorter variable loops with fewer glycosylation sites (Derdeyn et al., 2004; Sagar, 2010) and near-exclusive CCR5 co-receptor usage (Sagar, 2010); however, there is little knowledge regarding whether Nef characteristics of early viruses may differ from those from later infection stages (Noviello et al., 2007). Similarly, envelope length and number of glycosylation sites may increase during the infection course (Sagar et al., 2006) (though this remains somewhat controversial (Novitsky et al., 2009a)); however, few studies have examined early sequence and/or functional changes in Nef in large numbers of patients, particularly in a subtype C context (Brumme et al., 2008; Kuang et al., 2014; Noviello et al., 2007).
To address these knowledge gaps, we assessed two Nef functions implicated in pathogenesis - down-regulation of CD4 and HLA-I - in clonal plasma HIV RNA-derived Nef sequences isolated from untreated HIV-1 subtype C infected individuals from different disease stages. We studied a single representative Nef clone from each patient, including 120 individuals in early infection (of whom a subset of 68 individuals were additionally characterized for Nef function one year later), and an independent group of 207 individuals in the chronic phase of infection. We investigated (i) the relationship between Nef function in early infection and subsequent rate of HIV-1 disease progression, and (ii) the relationship between Nef function and sequence, in particular immune-driven HLA-associated Nef mutations. In addition, as an exploratory analysis we assessed differences in the genetic characteristics of Nef in early and chronic infection as well as changes in Nef function and sequence over the first year of infection.
Methods
Study subjects
Antiretroviral naïve patients with acute/early and chronic HIV-1 subtype C infection were recruited from established observational cohorts and clinical trial sites in South Africa and Botswana.
Study participants with acute/early infection (n=120) comprised 39 individuals from the HIV Pathogenesis Programme (HPP) Acute Infection Cohort in Durban, South Africa (Radebe et al., 2011; Wright et al., 2011), 31 from the Tshedimoso Study in Botswana (Novitsky et al., 2009b; Novitsky et al., 2009c; Wright et al., 2011), and 50 participants of the Tenofovir Gel Research for AIDS Prevention Science (TRAPS) Cohort (in KwaZulu-Natal, South Africa) who seroconverted in the CAPRISA 004 trial and were from the placebo arm (Abdool Karim et al., 2010; Chopera et al., 2013). For acute/early patients, plasma samples used were collected at a median of 49 days post-infection (interquartile range [IQR], 33 to 66 days post-infection). For 68 of these patients, for whom plasma was available, an additional plasma sample collected at a median of 374 days post-infection [IQR, 363 to 381 days post-infection] was analysed. Viral load, CD4 count and high resolution HLA class I data were available for all study participants with acute/early infection. Study participants with chronic infection (n=207) were from the Sinikithemba Cohort in Durban, South Africa (Kiepiela et al., 2004; Mann et al., 2013; Wright et al., 2010). Viral loads, CD4 counts, and high resolution HLA class I data were available for these chronically infected patients as previously described (Kiepiela et al., 2004).
Written informed consent was obtained from all study participants and ethical approval was obtained from the relevant institutional committees.
Generation and sequencing of Nef clones
A single Nef clone per study participant was analysed. A subset of the Nef clones studied here were previously published, including 85 from HIV-1 subtype C chronically infected individuals (Mann et al., 2013) and 48 from individuals in acute/early infection who were in the placebo arm of the CAPRISA 004 trial (Chopera et al., 2013). Nef clones were generated from plasma and sequenced as previously described (Mann et al., 2013) Briefly, nef was amplified from plasma-derived HIV-1 RNA using primers containing AscI and SacII restriction enzyme sites. The nef gene was cloned into a pSELECT-GFPzeo expression plasmid (Invivogen, San Diego, CA, USA), which was engineered with the corresponding restriction enzyme sites and had separate promoters for the nef gene and green fluorescent protein (GFP) expression. Bulk nef polymerase chain reaction (PCR) products as well as nef clonal sequences were sequenced using the ABI Prism Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). For each study participant, a single representative Nef clone with an open reading frame and that clustered with the corresponding bulk sequence in a phylogenetic tree (Guindon and Gascuel, 2003) was selected for CD4 and HLA-I down-regulation analyses. Nef sequences were aligned to HXB2 and stripped of insertions with respect to HXB2 using HyPhy (Kosakovsky Pond et al., 2005). All nef clones were confirmed to be HIV-1 subtype C using the recombinant identification program (RIP; http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html). Maximum-likelihood phylogenetic trees were constructed using Phyml (Guindon and Gascuel, 2003) and pairwise genetic distances from the 2004 consensus C Nef sequence (available at http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html) were calculated using PATRISTIC (Fourment and Gibbs, 2006). VESPA (http://www.hiv.lanl.gov/content/sequence/VESPA/vespa.html) was used to identify signature differences in consensus amino acids between groups of sequences. Sequences analysed in this study are available under Genbank accession numbers KF208819, KF208821-3, KF208825-8, KF208831-4, KF208836, KF208838-9, KF208842-3, KF208845, KF208847-208853, KF208855, KF208857-208861, KF208863-5, KF208867, KF208870, KF208872-3, KF208878-9, KF208886, KF208889, KF208893-5, KC906734-906805, KC906991-2, and KM262907-263141.
CD4 and HLA-I down-regulation
The CD4 and HLA-I down-regulation activities of Nef clones were measured as previously described using a CEM-derived T cell line expressing CD4 and engineered to stably express HLA-A*02:01 (Mann et al., 2013). Briefly, 300,000 cells were electroporated at 250V and 950μF with 4μg Nef clone and incubated for 20–24 hours, followed by staining using APC-labelled anti-CD4 and PE-labelled anti-HLA-A*02 antibodies (BD Biosciences, San Jose, CA, USA). CD4 and HLA-I cell surface expression was measured by flow cytometry in successfully transfected (i.e GFP-expressing) cells. The CD4 and HLA-I down-regulation activities (indicated by median fluorescence intensity [MFI] of CD4/HLA-I expression) of Nef clones were expressed relative to that of the positive control (SF2 Nef, set at 100% function) and the negative control (empty pSELECT plasmid, set at 0% function), as follows: (MFInegative control − MFIpatient Nef)/(MFInegative control − MFIpositive control). For the majority of Nef clones (n=297), a single transfection per Nef clone was performed and stained in duplicate, with duplicate measurements in excellent agreement (Spearman’s, r=0.98 and p<0.0001 for both Nef functions). For a subset of Nef clones (n=98), duplicate independent transfections were performed and also correlated well (Spearman’s, r=0.8 and p<0.0001 for CD4 down-regulation and r=0.9 and p<0.0001 for HLA-I down-regulation). Averaged results were analysed.
Nef expression
A minority (47 of 395, 12%) of Nef clones exhibited functions <45% of that of the reference strain SF2 for both CD4 and HLA-I down-regulation. For these, as well as a random selection of 78 Nef clones with functions above this threshold, Nef protein levels were measured by Western blot as previously described (Mann et al., 2013; Mwimanzi et al., 2011b). Briefly, following transfection of 1 million CEM cells with 10μg of Nef clones and a 24 hour incubation, cells were lysed. Following SDS-PAGE of cell lysates, proteins were electroblotted onto a polyvinylidene difluoride membrane and the presence of Nef protein was detected using rabbit polyclonal anti-HIV-1 Nef primary antibody (NIH AIDS Research and Reference Reagent Program, USA) and horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG secondary antibody (Amersham Biosciences, Little Chalfont, UK). Samples that failed Nef detection with the rabbit antibody were next probed using sheep polyclonal anti-HIV-1 Nef primary antibody (NIBSC Center for AIDS Reagents, UK) and HRP donkey anti-sheep IgG secondary antibody (Jackson ImmunoResearch Europe Ltd, Suffolk, UK). Actin expression was quantified simultaneously using monoclonal mouse anti-actin primary antibody (Sigma, St. Louis, Missouri, USA) and HRP goat anti-mouse secondary antibody (Jackson ImmunoResearch Europe Ltd, Suffolk, UK). Chemiluminescence of bands was measured using ImageQuant LAS 4000 (GE Healthcare Life Sciences, Little Chalfont, UK). All Nef bands, including that of the SF2 control, were quantified relative to actin expression. These values were then normalised to that of SF2 to obtain percentage expression.
Definition of HLA-associated polymorphisms in HIV-1 subtype C Nef
HLA-associated polymorphisms are HIV amino acid variants that are significantly over- (or under) represented in the presence of a particular HLA allele, as identified in statistical association studies of linked host (HLA) and HIV sequence datasets. A comprehensive list of HLA-associated polymorphisms was previously generated based on 1,336 subtype C Nef sequences from individuals from southern Africa using methods correcting for HIV phylogeny, amino acid co-variation, and HLA linkage disequilibrium (Carlson et al., 2014). HIV Nef polymorphisms in this list that were significantly positively associated with the expression of a particular HLA allele at q<0.05 were defined as “HLA-associated polymorphisms” in the present analysis (Supplementary Table 1).
The total number of HLA-associated polymorphisms was counted in each Nef clonal sequence. This was done without taking into account the HLA alleles expressed by the patient, since the aim of this analysis was to investigate the relationship between the number of HLA-associated polymorphisms in Nef (irrespective of whether they were transmitted or selected in the present host) and Nef function.
This analysis was repeated by summing only the subset of HLA-associated polymorphisms inferred to be “reverting” (defined as those where the absence of the restricting HLA allele is significantly associated with over-representation of the consensus amino acid or under-representation of the polymorphism in statistical association studies), suggesting that they confer a fitness cost. In this analysis, non-consensus polymorphisms at sites previously defined as “reverting” ((Adland et al., 2013), listed in Supplementary Table 1), were counted in each Nef clonal sequence.
Statistical analysis
Nef function versus disease progression analyses
Nef-mediated CD4 and HLA-I down-regulation functions in acute/early infection were correlated with markers of subsequent disease progression, namely viral load set point and rate of CD4 decline, using Spearman’s correlation. Viral load set point was calculated as the average viral load from 3 to 12 months post-infection. Simple linear regression was used to compute the rate of CD4 count decline for each individual over the treatment-free follow up period, where rate of decline was defined as the estimated slope of the fitted regression line (Wright et al., 2010). Multiple linear regression was also performed to assess the relationship between Nef functions and rate of CD4 decline while controlling for baseline CD4 count and length of follow-up. The nature of the CD4 decline data (a mixture of positive and negative values) prevented the use of traditional data transformations (e.g. square root, log, and inverse transformations). We thus limited the analysis to CD4 decline values within the range of −50 to 50 cells/mm3 per month since this ensured that the assumptions of normality were sufficiently met and included >95% of the data. Upon fitting the final model we tested the assumptions of normally distributed residuals using the Shapiro Wilk test, and validated that there was no relationship between the residuals and fitted values using Spearman’s correlation.
Nef sequence-function analyses
Comparisons of Nef sequence and/or Nef function were assessed using the Mann-Whitney U-test (with the exception of patients for whom paired early versus 1-year Nef clones were compared, in which case the Wilcoxon matched-pairs test was used), and Spearman’s correlation as appropriate. Significant differences in cohort consensus amino acids between acute/early and chronic Nef sequences were identified using Fisher’s exact test. Unless indicated otherwise, the significance cut-off for all statistical analyses was p<0.05. Specific Nef amino acids (present at a frequency of n≥5 in our dataset) associated with differences CD4 or HLA-I down-regulation function were assessed using the Mann-Whitney U test. Multiple comparisons were addressed using q-values, the p-value analogue of the false-discovery rate (Storey and Tibshirani, 2003). Here, associations with p<0.05 and q≤0.4 are reported.
Results
Nef clones: Function and expression
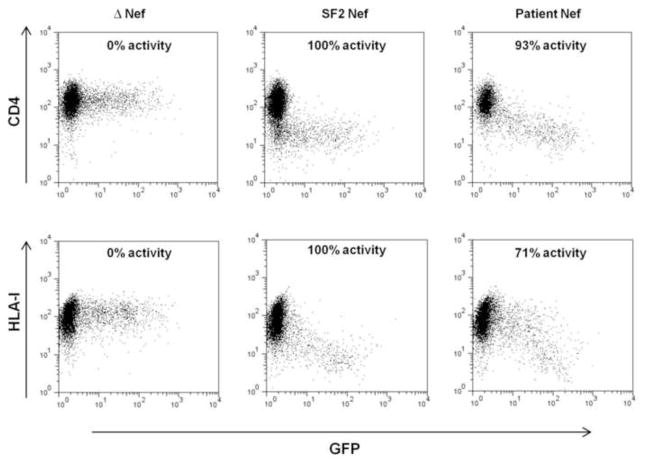
Nef-mediated CD4 and HLA-I down-regulation were measured for 395 Nef clones from 327 individuals (120 with acute/early infection, for whom 68 also had a follow-up clone at 1 year post-infection, as well as 207 with chronic infection) in a CEM T cell line expressing HLA-A*02:01. It should be noted that we previously observed an excellent correlation between the relative ability of Nef clones to down-regulate HLA-A*02 and HLA-B*07, justifying that HLA-A*02 down-regulation is in general representative of HLA-I down-regulation (Mann et al., 2013). The functions of patient-derived Nef sequences were expressed relative to that of the subtype B reference Nef sequence SF2, which represented 100% activity (Supplementary Table 2, representative flow plots in Figure 1). Overall, patient-derived Nef clones showed a median CD4 down-regulation ability of 96% (IQR, 77.6 to 99.7%) and a median HLA-I down-regulation ability of 76% (IQR, 53.4 to 87%) compared to the control subtype B SF2 strain. A minority of Nef clones (n=47, 12%) were poorly functional (<45% for both Nef functions). To confirm their Nef protein expression, Western blots were performed on this subset as well a randomly-selected set of 78 Nef clones that displayed >50% CD4 or HLA-I down-regulation ability relative to SF2 (indicating that Nef was expressed). Of the 47 poorly functional clones, 41 were not readily detectable by Western blot using polyclonal anti-Nef sera from rabbit or sheep (defined as <30% of SF2 control, Supplementary Table 2), while 8 of the 78 functional clones were below this threshold of detection. Since we cannot exclude the possibility that Nef clones with poor expression and function are attributable to cloning or other in vitro artifact, we excluded these 41 isolates from all further analyses. This left a total of 354 Nef clones for analysis: 107 from acute/early infection, 61 from 1 year post-infection (including 56 paired clones at baseline and 1 year post-infection), and 186 from chronic infection. The Western blot band intensities of the remaining Nef clones did not correlate significantly with CD4 down-regulation (Spearman’s, r=0.16 and p=0.15) or HLA-I down-regulation (Spearman’s, r=0.18 and p=0.1) (Supplementary Figure 1).
Figure 1. Nef-mediated down-regulation of cell-surface CD4 and HLA-I.
Representative flow cytometry plots show cell-surface expression of CD4 or HLA-I (y axis) in cells transfected with empty plasmid (Δ Nef, negative control), wild-type Nef plasmid (SF2 Nef, positive control), and plasmid expressing a patient-derived Nef. Green fluorescent protein (GFP) expression (x axis) was used as a marker of cells transfected with plasmids.
Similar Nef function and sequence in different cohorts and laboratories
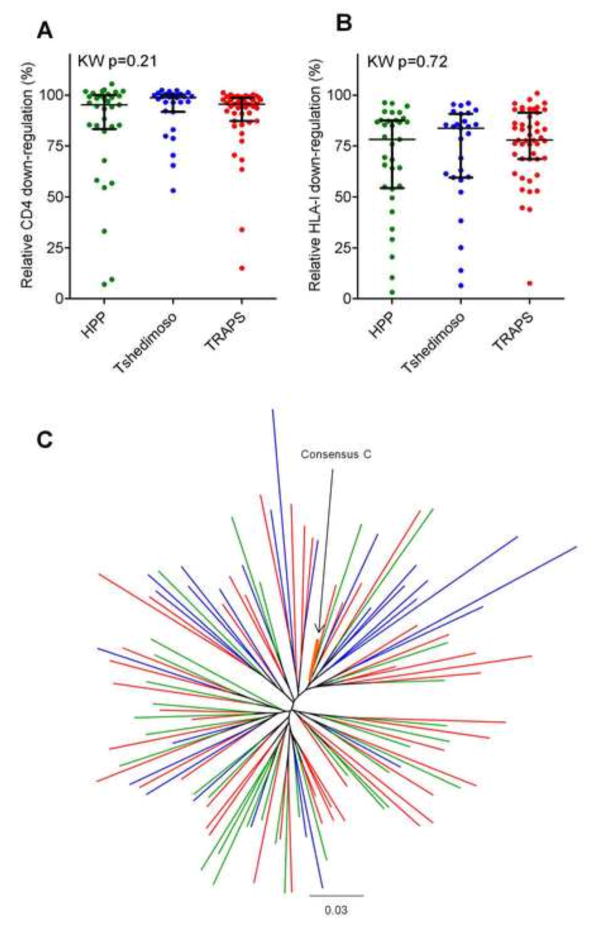
Our Nef sequences from acute/early infection were derived from various cohort and clinical studies in Botswana (Tshedimoso [n=27]) and South Africa (HPP acute infection [n=33] and TRAPS [n=47] cohorts). In addition, samples from the HPP acute infection cohort and Tshedimoso cohort were assayed in the HIV Pathogenesis Programme Laboratory at the University of KwaZulu-Natal while those from the TRAPS cohort were assayed in the HIV/AIDS Molecular Epidemiology Laboratory at Simon Fraser University. As such, we confirmed that there were no significant cohort- or laboratory-specific biases in Nef function before pooling data for analysis. Both Nef-mediated CD4 down-regulation and HLA-I down-regulation activities of Nef clones from recent infection were comparable across cohorts (Kruskal-Wallis, p>0.2) and laboratories (Mann-Whitney p>0.2) (Figure 2A and B). In addition, no gross clustering of Nef clones by site was observed in a combined phylogeny (Figure 2C), and no significant differences in the average pairwise distance between patient Nef clone and published Nef consensus C sequence was observed by cohort (Kruskal Wallis, p=0.84). Thus, all Nef sequences from acute/early infection were combined in subsequent analyses.
Figure 2. Functional and sequence comparison of acute/early infection Nef clones derived from different cohorts and assayed in different laboratories.
Panel A and B: Graphs show no significant differences in CD4 (A) and HLA-I (B) down-regulation function between Nef clones derived from the HPP (green), Tshedimoso (blue) and TRAPS (red) cohorts, as tested by Kruskal-Wallis (KW). No significant differences in either Nef function were found between Nef clones constructed and assayed in different laboratories (HPP and Tsedimoso versus TRAPS) using the Mann-Whitney U test (p>0.2). Bars indicate the median and whiskers represent the inter-quartile range. Panel C: The maximum-likelihood phylogenetic tree shows good intermixing of Nef clonal sequences from HPP (green), Tshedimoso (blue) and TRAPS (red) cohorts. The reference 2004 consensus C Nef sequence from the Los Alamos HIV sequence database is highlighted in orange.
Association of Nef-mediated CD4 and HLA-I down-regulation function in acute/early infection with subsequent disease progression
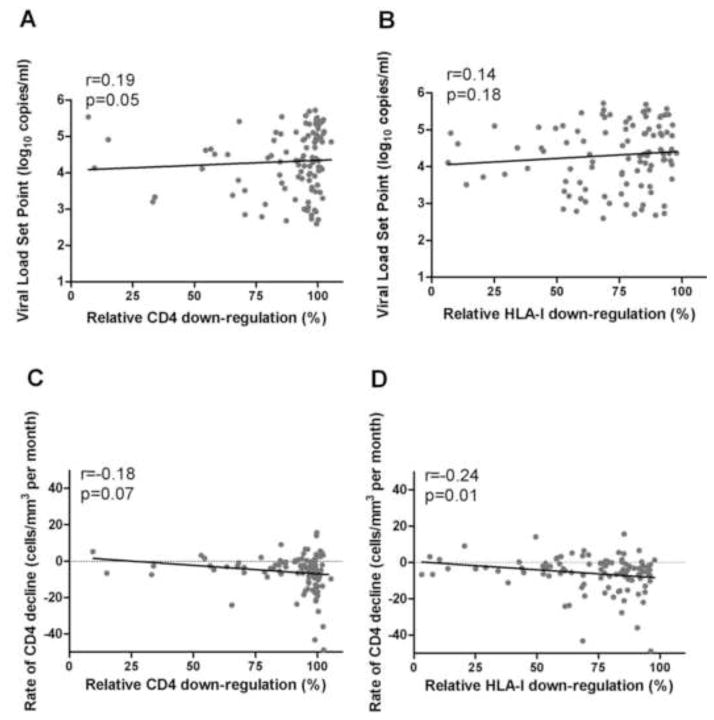
To assess the relationship between Nef function in acute/early infection and subsequent disease progression, the CD4 and HLA-I down-regulation abilities of Nef clones from the earliest time-point post-infection were correlated with subsequent viral load set point and rate of CD4 decline. Viral load set point (median of 4.4 log10 copies/ml; IQR, 3.7 to 5 log10 copies/ml) and rate of CD4 decline (median of −4 cells/mm3 per month; IQR, −10 to −1 cells/mm3 per month; were calculated for 102 and 107 individuals, respectively, over a median treatment-free follow-up of 672 days; IQR, 346 to 1,304 days).
A trend of increased Nef-mediated CD4 down-regulation function at baseline correlating with higher viral load set point was observed (Spearman’s, r=0.19 and p=0.05), but no relationship was observed between HLA-I down-regulation and viral load set point (Spearman’s, r=0.14 and p=0.18) (Figure 3A and B). Furthermore, both CD4 and HLA-I down-regulation functions were associated inversely with rate of CD4 decline, the latter significantly so (Spearman’s, r=−0.18, p=0.07 and r=−0.24, p=0.01 respectively). However, baseline CD4 count and length of follow-up time also significantly correlated with CD4 decline in our cohorts (Spearman’s, r=−0.25, p=0.01 and r=0.31, p=0.001 respectively); therefore, a multiple linear regression was performed to assess the relationship between Nef function and rate of CD4 decline while controlling for these potential confounders (see methods and Table 1). After accounting for these and other baseline parameters such as viral load, the association between higher Nef-mediated HLA-I down-regulation and faster rate of CD4 decline remained statistically significant (p=0.02) (Table 1). In a multivariate analysis additionally adjusting for cohort site, HLA-I down-regulation still remained significantly associated with rate of CD4 decline (p=0.048; data not shown).
Figure 3. Relationship between Nef functions in acute/early infection and markers of disease progression.
Panel A and B: A trend for a positive correlation between higher CD4 down-regulation in acute/early infection and subsequently higher viral load set point is shown (A), while no significant relationship was observed between HLA-I down-regulation and viral load set point (B) (n=102). Panel C and D: Associations between faster rate of CD4 decline and higher Nef-mediated CD4 (C) or HLA-I (D) down-regulation in acute/early infection are shown, but this was statistically significant for HLA-I down-regulation only (n=107). Data in graphs C and D are restricted to −50 to +50 cells/mm3 per month, and thus do not include 5 outlier data points. Spearman’s correlation was used to assess significance of relationships.
Table 1.
Multiple linear regression models investigating the relationship between rate of CD4 decline and Nef functions.
| Variables | CD4 decline, model 1a | Variables | CD4 decline, model 2b | ||
|---|---|---|---|---|---|
| Co-efficient | p-value | Co-efficient | p-value | ||
| CD4 down-regulation | −7.7 | 0.12 | HLA-I down-regulation | −9.07 | 0.02 |
| Square-root baseline CD4 | −1.02 | <0.0001 | Square-root baseline CD4 | −0.99 | <0.0001 |
| Follow-up time | 0.01 | <0.0001 | Follow-up time | 0.01 | <0.0001 |
| Log baseline viral load | −0.87 | 0.32 | Log baseline viral load | −1.04 | 0.22 |
Model for assessing relationship between CD4 decline and CD4 down-regulation: adjusted r2 = 0.25 p = <0.0001 n = 102
Model for assessing relationship between CD4 decline and HLA-I down-regulation: adjusted r2 = 0.28 p = <0.0001 n = 102
Sequence-function analysis part 1: Lack of genetic differences between acute/early and chronic Nef sequences
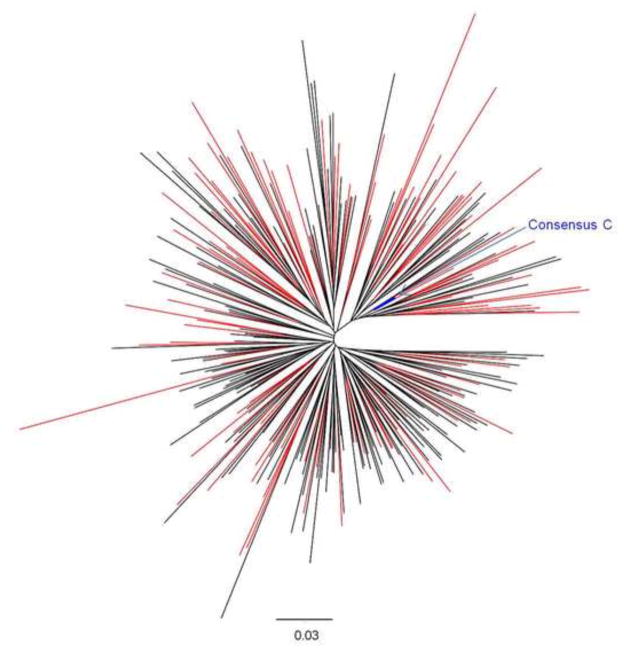
We next wished to identify sequence determinants of Nef function using a maximally-powered dataset. To do this, we first compared Nef sequences from acute/early (n=107) and chronic infection (n=186) to investigate whether the former possess features that distinguish them from the latter and whether combining Nef sequence data across different disease stages for our sequence-function analysis would introduce major biases. No significant differences in Nef sequence length (Mann-Whitney, p=0.35) and no significant differences in the frequency of the predominant (consensus) amino acids were observed at any Nef codon between clones collected during acute/early and chronic infection (Fisher’s exact test, p>0.09; data not shown). Moreover, acute/early and chronic sequences were interspersed in a phylogenetic tree (Figure 4). There were, however, marginally greater pairwise distances from Nef consensus C sequence in early infection clones (median 11.9% [IQR, 10.6 to 13.9%]) compared to 11.1% for chronic infection clones [IQR, 10 to 12.6%], Mann-Whitney, p=0.007). In summary, although we cannot exclude the possibility that there is some transmission bottleneck in the Nef gene resulting in increased transmission of non-consensus sequences, we found overall that there were no major differences between Nef sequences from acute/early and chronic infection. Given the lack of major differences between Nef sequences from acute/early and later infection and phylogenetic intermixing of these groups of sequences, we combined these into a single dataset to increase power. Therefore, subsequent sequence/function analyses were performed on a set of n=298 Nef clones from unique patients as follows: 107 from acute/early infection, 5 from 1 year post-infection and 186 from chronic infection (the acute/early sequence was used in the event where both acute/early and 1 year post-infection sequences were available for the same individual).
Figure 4. Phylogenetic tree of HIV-1 subtype C Nef sequences from acute/early and chronic infection.
The maximum-likelihood phylogenetic tree shows good intermixing of Nef clonal sequences derived from acute/early (red) and chronic (black) infection. The reference 2004 consensus C Nef sequence from the Los Alamos HIV sequence database is shown in blue.
Sequence-function analysis part 2: Nef amino acids associated with increased or decreased HLA-I down-regulation function
A Nef sequence-function analysis for CD4 down-regulation identified no Nef amino acids significantly associated with variations in this Nef function. However, a similar analysis for HLA-I down-regulation identified 31 amino acid variants at 22 different codons significantly associated with this function (Table 2). Several of these associations corroborated those identified in previous studies: amino acid associations with differential HLA-I down-regulation were identified at codons 3, 8, 9, 40, 102, 105, and 108 in a study of Nef subtypes A, B, C and D that included 74 subtype C Nef clonal sequences analysed in the present study (Mann et al., 2013) and similarly at codon 108 in HIV subtype B Nef sequences (Mwimanzi et al., 2013). Of note, 3 codons identified in this analysis, viz. 20, 62 and 64, are codons previously identified by mutational analysis to play a role in HLA-I down-regulation (Akari et al., 2000; Foster et al., 2011), while non-consensus variants 102H, 105R, 108D and 199Y, associated with reduced HLA-I down-regulation (Table 2), are HLA*B44, C*07:01, B*44 and B*18, and C*16-associated polymorphisms respectively (Supplementary Table 1). In contrast, the consensus 108E, which is enriched in individuals with HLA-B*57:02 (Supplementary Table 1), is linked to higher HLA-I down-regulation (Table 2). Together, these results suggest that HLA-I-restricted CD8+ T cell responses could select polymorphisms which impact Nef-mediated HLA-I down-regulation.
Table 2.
Amino acids (n≥5) associated with increased or decreased Nef HLA-I down-regulation function
| Codona | AAb | Cons.c | Relative Nef function (%)d | No. of samplese | p-value | q-value | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| With AA | Without AA | With AA | Without AA | |||||
| 3 | N | G | 85.0 | 77.8 | 32 | 266 | 0.03 | 0.4 |
| 8 | S | S | 77.8 | 84.7 | 256 | 35 | 0.02 | 0.3 |
| 9 | S | S | 76.2 | 90.1 | 250 | 20 | <0.0001 | 0.01 |
| 9 | R | S | 85.5 | 77.1 | 11 | 259 | 0.007 | 0.2 |
| 12 | G | G | 77.0 | 84.0 | 262 | 14 | 0.01 | 0.2 |
| 15 | E | A | 84.9 | 77.1 | 47 | 251 | 0.03 | 0.3 |
| 20 | M | I | 83.0 | 77.6 | 88 | 210 | 0.03 | 0.4 |
| 20 | I | I | 76.9 | 81.1 | 179 | 119 | 0.04 | 0.4 |
| 23 | T | T | 76.3 | 83.9 | 186 | 112 | 0.009 | 0.2 |
| 23 | A | T | 83.9 | 76.8 | 106 | 192 | 0.01 | 0.2 |
| 40 | Y | H | 74.8 | 82.0 | 126 | 172 | 0.002 | 0.1 |
| 40 | H | H | 82.1 | 74.9 | 165 | 133 | 0.003 | 0.1 |
| 51 | S | N | 64.7 | 79.3 | 9 | 289 | 0.02 | 0.3 |
| 62 | E | E | 79.1 | 37.7 | 293 | 5 | 0.005 | 0.2 |
| 64 | E | E | 80.2 | 71.1 | 253 | 45 | 0.01 | 0.3 |
| 64 | D | E | 71.1 | 79.9 | 27 | 271 | 0.02 | 0.3 |
| 79 | M | M | 79.1 | 44.6 | 293 | 5 | 0.02 | 0.3 |
| 102 | H | Y | 71.8 | 79.9 | 45 | 253 | 0.01 | 0.2 |
| 105 | R | K | 74.1 | 80.2 | 46 | 252 | 0.03 | 0.4 |
| 108 | E | E | 82.7 | 74.2 | 159 | 139 | 0.001 | 0.1 |
| 108 | D | E | 74.3 | 82.7 | 138 | 160 | 0.002 | 0.1 |
| 133 | V | V | 80.2 | 68.6 | 265 | 33 | 0.005 | 0.2 |
| 146 | V | V | 79.1 | 53.1 | 293 | 5 | 0.02 | 0.3 |
| 156 | N | A | 89.1 | 78.1 | 6 | 292 | 0.02 | 0.3 |
| 161 | D | N | 84.7 | 77.0 | 53 | 244 | 0.007 | 0.2 |
| 161 | N | N | 76.9 | 83.2 | 235 | 62 | 0.02 | 0.3 |
| 178 | K | R | 61.1 | 79.2 | 14 | 284 | 0.02 | 0.3 |
| 178 | G | R | 84.9 | 77.9 | 40 | 258 | 0.04 | 0.4 |
| 188 | N | S | 21.1 | 79.1 | 5 | 293 | 0.03 | 0.3 |
| 199 | H | H | 79.4 | 47.8 | 290 | 8 | 0.002 | 0.1 |
| 199 | Y | H | 51.8 | 79.3 | 7 | 291 | 0.005 | 0.2 |
Numbered according to HXB2.
Amino acid associated with increased or decreased HLA-I down-regulation function.
Cohort consensus amino acid at that particular codon.
Median percentage HLA-I down-regulation function (expressed relative to SF2 control) of Nef clones with and without the amino acid.
Number of sequences with and without the amino acid. Amino acid totals vary, as gaps in the alignment are considered missing data.
Sequence-function analysis part 3: Increased numbers of HLA-associated polymorphisms correlate with reduced HLA-I down-regulation
To further explore the effect of HLA-associated polymorphisms on Nef function, the number of such polymorphisms present in each Nef sequence (defined according to an independently-derived reference list of HLA-associated polymorphisms in HIV subtype C; Supplementary Table 1) was correlated with Nef function (see methods). No significant relationship between CD4 down-regulation and number of polymorphisms was observed (Spearman’s, r=0.008 p=0.89). However, a weak trend of inverse correlation between the number of HLA-associated Nef polymorphisms and HLA-I down-regulation function was observed (Spearman’s, r=−0.11 and p=0.06) (Figure 5A).
Figure 5. Relationship between HLA-associated Nef polymorphisms and Nef-mediated HLA-I down-regulation activity.
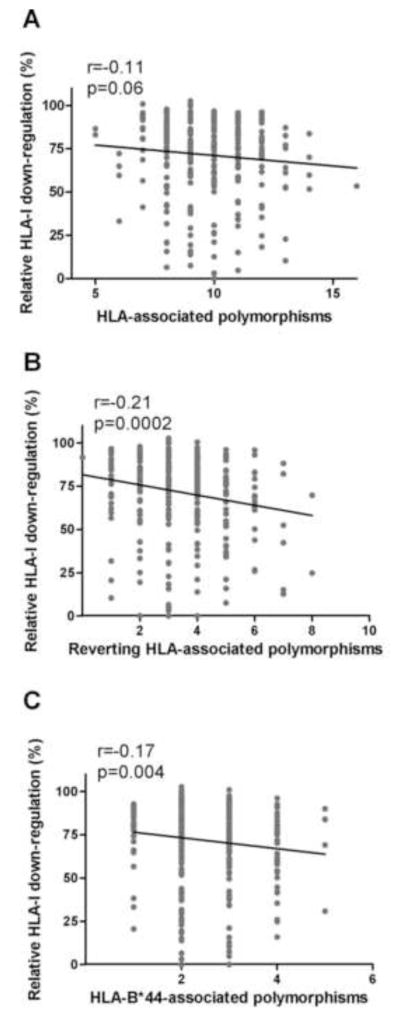
Panel A: A trend for lower HLA-I down-regulation activity with increasing numbers of HLA-associated polymorphisms is shown. Panel B: A significant negative correlation between the number of non-consensus polymorphisms at HLA-associated codons previously associated with reversion (Adland et. al, 2013) and HLA-I down-regulation ability is shown. Panel C: The number of HLA-B*44-associated polymorphisms present correlates negatively with HLA-I down-regulation function. Spearman’s correlation was used to assess significance of relationships (n=298 for all analyses).
Several HLA-associated mutations in Nef have been predicted to revert upon transmission to a host lacking the original restricting HLA, suggesting they may have fitness costs (Adland et al., 2013). To investigate the relationship between such “reverting” polymorphisms and Nef HLA-I down-regulation function (n=21 at 15 sites previously associated with reversion, (Adland et al., 2013) and Supplementary Table 1), we repeated our analysis considering only these polymorphisms. Of note, we observed a stronger inverse correlation with HLA-I down-regulation function (Spearman’s, r=−0.21 and p=0.0002) (Figure 5B) compared to the original analysis undertaken on all HLA-associated polymorphisms (Figure 5A). Collectively, these results suggest that HLA alleles can drive the selection of certain polymorphisms in Nef that impair its function.
Sequence-function analysis part 4: Polymorphisms associated with HLA-B*44, but not protective alleles B*57, B*58:01 and B*81, correlate with reduced HLA-I down-regulation
Previous studies have shown correlations between the presence of polymorphisms associated with protective HLA alleles and reduced Gag-protease function (Brockman et al., 2007; Crawford et al., 2009; Schneidewind et al., 2007; Wright et al., 2011), and between increasing numbers of B*57-associated Nef polymorphisms largely unique to elite controllers and reduced Nef CD4 down-regulation function (Mwimanzi et al., 2013). Therefore, we investigated whether Nef sequences harbouring polymorphisms associated with HIV subtype C-specific protective HLA alleles B*57, B*58:01 and B*81 (Kiepiela et al., 2004) exhibited decreased Nef function. No relationship between the numbers of these polymorphisms and either Nef function was observed (Spearman’s, p>0.17; data not shown). In fact, the number of polymorphisms specifically associated with HLA-B*57 (83G, 83E, 108E, and 116N) was positively associated with HLA-I down-regulation function (Spearman’s, r=0.12 and p=0.04) (data not shown). This result appeared to be driven by 108E, the consensus amino acid at codon 108, which alone was associated with higher HLA-I down-regulation (Mann-Whitney, p=0.001).
HLA-B*44:03, the most common subtype of B*44 in South Africa, is associated with a modest (≈0.25 log) reduction in viral load (Kiepiela et al., 2004; Leslie et al., 2010) and restricts a CD8+ T cell epitope (KY11, codons 92–102) within a “vulnerable” Nef region (codons 88-105) to which CD8+ T cell responses have been associated with a protective effect (Adland et al., 2013). There are also more Nef polymorphisms (n=7; 65E, 71R, 93D, 102H, 108D, 176D, 176V) associated with HLA-B*44 than any other allele (Supplementary Table 1), five of which are predicted to revert in the absence of HLA-B*44 (Adland et al., 2013). Consistent with a cumulative effect of HLA-B*44-driven polymorphisms on Nef function, we observed a significant inverse correlation between the number of such polymorphisms present and HLA-I down-regulation function (Spearman’s, r=−0.17 and p=0.0042) (Figure 5C).
Nef sequence and function in the first year of infection: adaptation to host environment and lack of consistent functional change
Exploratory analyses of 56 paired acute/early and 1 year post-infection Nef clones were next performed to investigate immune-driven evolution in Nef and functional changes over the first year of infection.
Firstly, sequences of matched clones were analysed to identify common sequence changes away from or towards a particular amino acid, that were observed in ≥5 individuals, between baseline and 1 year post-infection. Five such changes were identified: X83A, X102Y, X105K, E108D, and H116N, where X indicates any amino acid (Table 3); all represent codons at which HLA-associated polymorphisms have been identified (Supplementary Table 1). Changes towards consensus (X83A, X102Y, and X105K) may thus represent reversions of escape mutations, suggesting fitness costs for these (Matthews et al., 2008). Indeed, reversions at codons 83 and 102 were predicted in a recent study (Adland et al., 2013) and non-consensus amino acids at codons 83, 102 and 105 have been statistically associated with reduced HLA-I down-regulation previously (Mann et al., 2013; Mwimanzi et al., 2013) and/or in the present study (Table 2). A minority of these reversions to consensus, particularly X102Y, occurred in the presence of the associated HLA allele, however such an occurrence has previously been documented (Allen et al., 2004). In contrast, HLA-associated mutations from consensus (E108D and H116N) may represent HLA-driven escape. Indeed, H116N is an HLA-B*57 escape mutation (Frater et al., 2007; Pillay et al., 2005) that has not been associated with functional costs, and 108D, which associated with decreased HLA-I down-regulation in the present and previous studies (Mann et al., 2013), was selected four out of five times in patients harbouring the associated HLA alleles B*44 and B*18. Thus, common Nef sequence changes in the first year of infection appear to be largely driven by adaptation to host HLA.
Table 3.
Common sequence differences (n≥5) occurring from acute/early to 1 year post-infection in 56 pairs of Nef clones
| Change | Con.a | HLA-associationb | HLA presentc | Function associationd |
|---|---|---|---|---|
| X83A | A | 83G – B*57, B*58:01, A*03:01, A*34 83E – B*57 |
0/5 | 83G decreased HLA-I down-regulation (Mann et al., 2013) |
| X102Y | Y | 102H – B*44 | 3/5 | 102H decreased HLA-I down-regulation (Mann et al., 2013) |
| X105K | K | 105R – C*07:01 | 1/5 | 105K increased HLA-I down-regulation (Mann et al., 2013) 105R and Q decreased HLA-I down-regulation (Mann et al., 2013) 105R decreased CD4 down-regulation (Mwimanzi et al., 2013) |
| E108D | E | 108D – B*44, B*18 108E –B*57:02 |
4/5 | 108D decreased HLA-I down-regulation (Mann et al., 2013; Mwimanzi et al., 2013) |
| H116N | H | 116N – B*57:03 | 0/5 | No association |
Consensus amino acid.
HLA alleles positively associated with an amino acid at the relevant codon at p<0.05, q<0.05 (Supplementary Table 1).
Indicates the number of individuals who expressed the relevant HLA allele (associated with variants at the codon at which the sequence change occurred).
Statistical associations between Nef amino acid variants and Nef function as identified in previous studies.
Some of these common early sequence changes are expected to increase Nef function and some are expected to decrease Nef function. As such, it is perhaps unsurprising that in a longitudinal analysis, no overall consistent changes in Nef-mediated CD4 or HLA-I down-regulation functions were observed during the first year of infection (Wilcoxon matched-pairs signed-ranks test, p=0.54 and p=0.56, respectively) (Figure 6A and B).
Figure 6. Nef functions over the first year of infection.
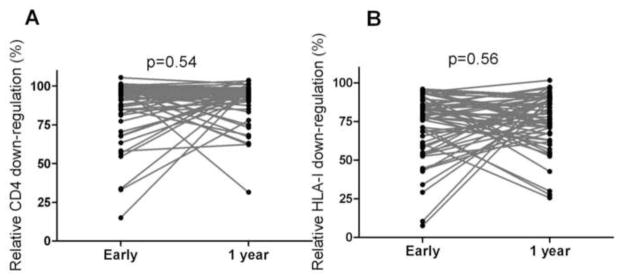
Panel A and B: Graphs show no consistent increase or decrease in Nef-mediated CD4 (A) and HLA-I (B) down-regulation over the first year of infection. Functions of 56 pairs of matched Nef clones derived from acute/early infection and 1 year post-infection from the same individuals are shown. P values from the Wilcoxon matched-pairs test are shown.
Discussion
There is limited direct evidence that CD8+ T cell responses to HIV-1 Nef impose a functional cost on the virus (Adland et al., 2013; Mwimanzi et al., 2013; Ueno et al., 2008) and the contribution of Nef-specific CD8+ T cell responses to viral load control remains debated (Adland et al., 2013; Kiepiela et al., 2007). Furthermore, while gross genetic defects in Nef can contribute to long-term non-progression (Deacon et al., 1995; Kirchhoff et al., 1995) and attenuation of Nef functions in elite controllers have been demonstrated (Mwimanzi et al., 2013), large population-based studies correlating Nef functions with disease progression in HIV-1 subtype C infection are lacking. Therefore, we investigated the relationships between Nef-mediated CD4 and HLA-I down-regulation functions and disease progression, as well as the potential influence of immune-driven Nef sequence variation on these functions, in HIV-1 subtype C infection.
As previously observed by our group (Mann et al., 2013), Nef-mediated CD4 down-regulation function was considerably more well-conserved than HLA-I down-regulation function in our study, suggesting the former function as particularly important for the virus to conserve. This is consistent with the critical role ascribed to Nef-mediated CD4 down-regulation in mediating viral replication and pathogenesis in humanised mouse and macaque models in early infection, while Nef-mediated HLA-I down-regulation is neither sufficient nor required for these pathogenic effects (Iafrate et al., 2000; Watkins et al., 2013). Despite the fairly narrow spread of observed CD4 down-regulation values in patient sequences (that could reduce our ability to identify significant correlations between this Nef function and clinical/sequence parameters), we observed a modest positive association (p=0.05) between Nef-mediated CD4 down-regulation function in early infection and viral load set point. This result is consistent with reports that CD4 down-regulation directly enhances viral replication through promoting budding of infectious virions (Argañaraz et al., 2003; Lundquist et al., 2002).
On the other hand, we observed no evidence linking Nef-mediated HLA-I down-regulation function in early infection and viral load set point. This is consistent with studies showing a lack of effect on SIV viral loads following knockout of HLA-I down-regulation function in the macaque model (Swigut et al., 2004) and a lack of correlation between this Nef function and viral loads in a small study of HIV-1 infected individuals (Lewis et al., 2008), indicating that this function does not directly affect viral replication. However, we observed that increased Nef-mediated HLA-I down-regulation in early infection was linked with a subsequent faster rate of CD4 decline. This suggests that variability in this Nef function may affect disease progression following the initial CD4 depletion, supporting its relevance in vivo. The biological relevance of Nef-mediated HLA-I down-regulation is also supported by strong in vivo selection pressure to restore this function following mutational knock-out in an SIV model (Swigut et al., 2004). As such, the relationship between HLA-I down-regulation ability and CD4 decline may be mediated via enhanced evasion of CD8+ T cell responses by HLA-I down-regulation (Lewis et al., 2008; Swigut et al., 2004), though why this is not additionally manifested in higher plasma viral load remains an open question.
In this study, we found evidence that mutations selected by HLA alleles, likely CD8+ T cell escape mutations, may reduce Nef-mediated HLA-I down-regulation ability, but not CD4 down-regulation function (which may be related to the highly conserved nature of that function). The presence of certain HLA-associated polymorphisms (102H, 105R, 108D and 199Y) were individually significantly associated with reduced HLA-I down-regulation. These results extend those of a previous study demonstrating a reduction in HLA-I down-regulation activity of Nef sequences harbouring a combination of escape mutations in the proline-rich region (Ueno et al., 2008). Furthermore, consistent with an inferred fitness cost of Nef HLA-associated polymorphisms associated with reversion (Adland et al., 2013); we observed that increasing numbers of these polymorphisms in our patient Nef sequences correlated strongly with reduced HLA-I down-regulation.
We additionally observed that increasing numbers of mutations associated with HLA-B*44, which has been linked to a modest reduction in viral loads (Kiepiela et al., 2004; Leslie et al., 2010), corresponded with decreased HLA-I down-regulation. Furthermore, we found that the number of polymorphisms within two overlapping peptides (codons 88-105 and 134-148), to which CD8+ T cell responses have been associated with lower viral loads in HIV-1 infection (Adland et al., 2013), correlated significantly and negatively with HLA-I down-regulation function (data not shown). These observations suggest that functional constraints may contribute to the modest benefit associated with HLA-B*44 and the beneficial effects of CD8+ targeting of Nef regions 88-105 and 134-148, either directly (via fitness-costly escape) or indirectly (via sustained CD8+ T cell responses to a mutationally-constrained region i.e. delayed escape). In fact, there is much evidence that a balance between these two factors influences viral load (Crawford et al., 2009; Kawada et al., 2006; Wright et al., 2011). Interestingly, we found no evidence that Nef polymorphisms commonly associated with the protective HLA-B*57 allele reduce Nef function, while others have found that the number of non-canonical HLA-B*57-associated Nef polymorphisms particular to elite controllers, but not those commonly associated with B*57, correlated negatively with several Nef functions in elite-controller-derived Nef sequences (Mwimanzi et al., 2013). Nevertheless, it is possible that HLA-B*57 responses to Nef epitopes may contribute to slower progression in B*57 individuals who are not elite controllers, considering that the B*57-restricted Nef epitopes overlap considerably with Nef regions associated with viral control (Adland et al., 2013) and that maintenance of Nef HW9 responses were associated with long-term non-progression in HLA-B*57-expressing individuals (Navis et al., 2008).
Of relevance to HIV vaccine design is whether viruses in early infection differ from those in chronic infection (which might indicate preferential transmission of certain variants). Although Nef sequences from early infection were marginally less similar to the consensus C sequence, we found no features that clearly distinguished early from chronic Nef sequences. This is consistent with a previous report describing similar diversity in donor and acutely-infected recipient Nef sequences as well as a lack of optimization of specific Nef functions during transmission (Noviello et al., 2007). Our longitudinal analysis of individuals from early to 1 year post-infection revealed no consistent functional changes and that common sequence changes occurring during this period were reversions and selections of HLA-associated mutations. Similarly, it was previously found that the differences between HIV sequences from early and later infection stages largely represented reversion of mutations and escape mutations occurring post-transmission in adaptation to the specific host environment (Treurnicht et al., 2010). Also in line with these findings, Noviello et al. (2007) observed that CD8+ T cell responses appeared to be a major driver of Nef evolution during early HIV-1 infection (Noviello et al., 2007).
In summary, this study provides evidence that variability in Nef-mediated CD4 and HLA-I down-regulation ability among subtype C viruses in the absence of obvious Nef genetic defects may influence disease progression. Furthermore, immune pressure on certain regions of Nef may result in selection of mutations that reduce the HLA-I down-regulation function of Nef. Together with previous studies showing clinical benefit associated with CD8+ T cell responses to particular epitopes in Nef, results support certain Nef epitopes for consideration as components of an HIV vaccine designed to attenuate the infection course.
Supplementary Material
Panel A and B: Nef expression was measured by Western blot and expressed relative to the SF2 Nef positive control for Nef clones that were poorly functional (<45% relative to SF2 Nef for both Nef functions, n=47) as well a randomly-selected set of 78 Nef clones that displayed >50% CD4 or HLA down-regulation ability (indicating that Nef was expressed). 41 Nef clones for which expression could not be confirmed by either Western blot (<30% of SF2 control) or functional measurements were removed from further analyses as in vitro artifact could not be excluded. The Western blot band intensities of the remaining Nef clones (n=84) did not correlate significantly with CD4 down-regulation (A) or HLA-I down-regulation (B) by Spearman’s correlation.
We measured CD4/HLA-I down-regulation by 395 patient-derived HIV-1C Nef clones.
HLA-I down-regulation in early infection correlated positively with CD4 decline.
CD4 down-regulation in early infection associated positively with viral set point.
Certain immune-driven mutations correlated with reduced HLA-I down-regulation.
Certain Nef epitopes could be a component of an HIV-1 attenuation-based vaccine.
Acknowledgments
JKM received a pilot grant from the Canada-Sub Saharan Africa (CANSSA) HIV/AIDS Network. DRC is a recipient of the Canada-HOPE fellowship from the Canadian Institutes for Health Research (CIHR) and Sanofi-Aventis, the Clinical Infectious Diseases Research Initiative fellowship and the Claude Leon Foundation fellowship. XTK is funded by a CAHR/BMS Master’s Scholarship in Basic Science. AQL is funded by a CIHR Frederick Banting and Charles Best Masters Award. RD is supported by the Merck-Canada Training of Aboriginal Youth in Biomedical Labs program at Simon Fraser University. PM is the recipient of Postdoctoral fellowships from the Michael Smith Foundation for Health Research (MSFHR) and the CIHR. MAB holds a Canada Research Chair, Tier 2, in Viral Pathogenesis and Immunity. ZLB is the recipient of a CIHR New Investigator Award and a Scholar Award from the MSFHR. TN holds the South African DST/NRF Research Chair in Systems Biology of HIV/AIDS, the Victor Daitz Chair in HIV/TB Research and an International Early Career Scientist Award from the Howard Hughes Medical Institute.
We thank Dr. Bemuluyigza Baraki for technical assistance; Dr. Johannes Viljoen and the Africa Centre laboratory for providing access to tissue culture and sequencing facilities; and all study participants and support staff.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, Kharsany AB, Sibeko S, Mlisana KP, Omar Z, Gengiah TN, Maarschalk S, Arulappan N, Mlotshwa M, Morris L, Taylor D CAPRISA 004 Trial Group. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329(5996):1168–1174. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adland E, Carlson JM, Paioni P, Kløverpris H, Shapiro R, Ogwu A, Riddell L, Luzzi G, Chen F, Balachandran T, Heckerman D, Stryhn A, Edwards A, Ndung’u T, Walker BD, Buus S, Goulder P, Matthews PC. Nef-specific CD8+ T cell responses contribute to HIV-1 immune control. PLoS One. 2013;8(9):e73117. doi: 10.1371/journal.pone.0073117. doi:73110.71371/journal.pone.0073117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akari H, Arold S, Fukumori T, Okazaki T, Strebel K, Adachi A. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J Virol. 2000;74(6):2907–2912. doi: 10.1128/jvi.74.6.2907-2912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T, Altfeld M, Yu XG, O’Sullivan KM, Lichterfeld M, Le Gall S, John M, Mothe BR, Lee PK, Kalife ET, Cohen DE, Freedberg KA, Strick DA, Johnston MN, Sette A, Rosenberg E, Mallal SA, Goulder PJR, Brander C, Walker BD. Selection, Transmission, and Reversion of an Antigen-Processing Cytotoxic T-Lymphocyte Escape Mutation in Human Immunodeficiency Virus Type 1 Infection. J Virol. 2004;78(13):7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, Altfeld M. Crippling HIV one mutation at a time. J Exp Med. 2008;205(5):1003–1007. doi: 10.1084/jem.20080569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argañaraz ER, Schindler M, Kirchhoff F, Cortes MJ, Lama J. Enhanced CD4 down-modulation by late stage HIV-1 nef alleles is associated with increased Env incorporation and viral replication. J Biol Chem. 2003;278(36):33912–33919. doi: 10.1074/jbc.M303679200. [DOI] [PubMed] [Google Scholar]
- Brockman MA, Schneidewind A, Lahaie M, Schmidt A, Miura T, DeSouza I, Ryvkin F, Derdeyn CA, Allen S, Hunter E, Mulenga J, Goepfert PA, Walker BD, Allen TM. Escape and Compensation from Early HLA-B57-Mediated Cytotoxic T-Lymphocyte Pressure on Human Immunodeficiency Virus Type 1 Gag Alter Capsid Interactions with Cyclophilin A. J Virol. 2007;81(22):12608–12618. doi: 10.1128/JVI.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme ZL, Brumme CJ, Carlson J, Streeck H, John M, Eichbaum Q, Block BL, Baker B, Kadie C, Markowitz M, Jessen H, Kelleher AD, Rosenberg E, Kaldor J, Yuki Y, Carrington M, Allen TM, Mallal S, Altfeld M, Heckerman D, Walker BD. Marked Epitope- and Allele-Specific Differences in Rates of Mutation in Human Immunodeficiency Type 1 (HIV-1) Gag, Pol, and Nef Cytotoxic T-Lymphocyte Epitopes in Acute/Early HIV-1 Infection. J Virol. 2008;82 (18):9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde ML, Greene JM, Chin EN, Ericsen AJ, Scarlotta M, Cain BT, Pham NH, Becker EA, Harris M, Weinfurter JT, O’Connor SL, Piatak MJ, Lifson JD, Gostick E, Price DA, Friedrich TC, O’Connor DH. Specific CD8+ T cell responses correlate with control of simian immunodeficiency virus replication in Mauritian cynomolgus macaques. J Virol. 2012;86(14):7596–7604. doi: 10.1128/JVI.00716-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin TH, Peng J, Seese AM, Shapiro R, Frater J, Ndung’u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJ, Allen TM, Allen S, Hunter E. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345(6193):1254031. doi: 10.1126/science.1254031. doi:1254010.1251126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopera DR, Mann JK, Mwimanzi P, Omarjee S, Kuang XT, Ndabambi N, Goodier S, Martin E, Naranbhai V, Abdool-Karim S, Abdool-Karim Q, Brumme ZL, Ndung’u T, Williamson C, Brockman MA the CAPRISA 004 TRAPS Team. No evidence for selection of HIV-1 with enhanced Gag-Protease or Nef function among breakthrough infections in the CAPRISA 004 tenofovir microbicide trial. PLoS One. 2013;8(8):e71758. doi: 10.1371/journal.pone.0071758. doi:71710.71371/journal.pone.0071758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, Tang J, Farmer P, Ndung’u T, Lakhi S, Gilmour J, Goepfert P, Walker BD, Kaslow R, Mulenga J, Allen S, Goulder PJR, Hunter E. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206:909–919. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, Lawson VA, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan JS, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270(5238):988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Foster JL, Denial SJ, Temple BRS, Garcia JV. Mechanisms of HIV-1 Nef Function and Intracellular Signaling. J Neuroimmune Pharmacol. 2011;6:230–246. doi: 10.1007/s11481-011-9262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JL, Garcia JV. HIV-1 Nef: at the crossroads. Retrovirology. 2008;5:84. doi: 10.1186/1742-4690-1185-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourment M, Gibbs MJ. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol Biol. 2006;6:1. doi: 10.1186/1471-2148-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frater AJ, Brown H, Oxenius A, Günthard HF, Hirschel B, Robinson N, Leslie AJ, Payne R, Crawford H, Prendergast A, Brander C, Kiepiela P, Walker BD, Goulder PJR, McLean A, Phillips RE the Swiss HIV-Cohort Study. Effective T-cell Responses Select Human Immunodeficiency Virus Mutants and Slow Disease Progression. J Virol. 2007;81(12):6742–6751. doi: 10.1128/JVI.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Carl S, Bronson S, Stahl-Hennig C, Swigut T, Skowronski J, Kirchhoff F. Disrupting surfaces of nef required for downregulation of CD4 and for enhancement of virion infectivity attenuates simian immunodeficiency virus replication in vivo. J Virol. 2000;74(21):9836–9844. doi: 10.1128/jvi.74.21.9836-9844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada M, Igarashi H, Takeda A, Tsukamoto T, Yamamoto H, Dohki S, Takiguchi M, Matano T. Involvement of Multiple Epitope-Specific Cytotoxic T-Lymphocyte Responses in Vaccine-Based Control of Simian Immunodeficiency Virus Replication in Rhesus Macaques. J Virol. 2006;80(4):1949–1958. doi: 10.1128/JVI.80.4.1949-1958.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestler HW, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65(4):651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJR. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–774. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332(4):228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Kuang XT, Li X, Anmole G, Mwimanzi P, Shahid A, Le AQ, Chong L, Qian H, Miura T, Markle T, Baraki B, Connick E, Daar ES, Jessen H, Kelleher AD, Little S, Markowitz M, Pereyra F, Rosenberg ES, Walker BD, Ueno T, Brumme ZL, Brockman MA. Impaired Nef function is associated with early control of HIV-1 viremia. J Virol. 2014 doi: 10.1128/JVI.01334-14. pii: JVI.01334-01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A, Matthews PC, Listgarten J, Carslon J, Kadie C, Ndung’u T, Coovadia H, Walker BD, Heckerman D, Goulder PJR. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol. 2010;84:9879–9888. doi: 10.1128/JVI.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Balamurugan A, Ohno A, Kilpatrick S, Ng HL, Yang OO. Functional adaptation of Nef to the immune milieu of HIV-1 infection in vivo. J Immunol. 2008;180(6):4075–4081. doi: 10.4049/jimmunol.180.6.4075. [DOI] [PubMed] [Google Scholar]
- Lichterfeld M, Yu XG, Cohen D, Addo MM, Malenfant J, Perkins B, Pae E, Johnston MN, Strick D, Allen TM, Rosenberg ES, Korber B, Walker BD, Altfeld M. HIV-1 Nef is preferentially recognized by CD8 T cells in primary HIV-1 infection despite a relatively high degree of genetic diversity. AIDS. 2004;18(10):1383–1392. doi: 10.1097/01.aids.0000131329.51633.a3. [DOI] [PubMed] [Google Scholar]
- Lundquist CA, Tobiume M, Zhou J, Unutmaz D, Aiken C. Nef-mediated downregulation of CD4 enhances human immunodeficiency virus type 1 replication in primary T lymphocytes. J Virol. 2002;76(9):4625–4633. doi: 10.1128/JVI.76.9.4625-4633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JK, Byakwaga H, Kuang XT, Le AQ, Brumme CJ, Mwimanzi P, Omarjee S, Martin E, Lee GQ, Baraki B, Danroth R, McCloskey R, Muzoora C, Bangsberg DR, Hunt PW, Goulder PJ, Walker BD, Harrigan PR, Martin JN, Ndung’u T, Brockman MA, Brumme ZL. Ability of HIV-1 Nef to downregulate CD4 and HLA class I differs among viral subtypes. Retrovirology. 2013;10(1):100. doi: 10.1186/1742-4690-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PC, Prendergast A, Leslie A, Crawford H, Payne R, Rousseau C, Rolland M, Honeyborne I, Carlson J, Kadie C, Brander C, Bishop K, Mlotshwa N, Mullins JI, Coovadia H, Ndung’u T, Walker BD, Heckerman D, Goulder PJR. Central Role of Reverting Mutations in HLA Associations with Human Immunodeficiency Virus Set Point. J Virol. 2008;82 (17):8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, Weisgrau KL, Furlott JR, Kim YI, Veloso de Santana MG, Rakasz E, Capuano Sr, Wilson NA, Bonaldo MC, Galler R, Allison DB, Piatak MJ, Haase AT, Lifson JD, Allen TM, Watkins DI. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491(7422):129–133. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwimanzi P, Hasan Z, Hassan R, Suzu S, Takiguchi M, Ueno T. Effects of naturally-arising HIV Nef mutations on cytotoxic T lymphocyte recognition and Nef’s functionality in primary macrophages. Retrovirology. 2011a;8:50. doi: 10.1186/1742-4690-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwimanzi P, Hasan Z, Hassan R, Suzu S, Takiguchi M, Ueno T. Effects of naturally-arising HIV Nef mutations on cytotoxic T lymphocyte recognition and Nef’s functionality in primary macrophages. Retrovirology. 2011b;8:50. doi: 10.1186/1742-4690-1188-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwimanzi P, Markle T, Otsuka H, Ogata Y, Tokunaga M, Miura T, Martin E, Pereyra F, Walker B, Brumme Z, Brockman M, Ueno T. Impairment of viral replication capacity by nef alleles from HIV elite controllers. Retrovirology. 2011c;8(Suppl2):P53. doi: 10.1186/1742-4690-1188-S1182-P1153. [DOI] [Google Scholar]
- Mwimanzi P, Markle TJ, Martin E, Ogata Y, Kuang XT, Tokunaga M, Mahiti M, Pereyra F, Miura T, Walker BD, Brumme ZL, Brockman MA, Ueno T. Attenuation of multiple Nef functions in HIV-1 elite controllers. Retrovirology. 2013;10(1):1. doi: 10.1186/1742-4690-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navis M, Schellens IM, van Swieten P, Borghans JA, Miedema F, Kootstra NA, van Baarle D, Schuitemaker H. A nonprogressive clinical course in HIV-infected individuals expressing human leukocyte antigen B57/5801 is associated with preserved CD8+ T lymphocyte responsiveness to the HW9 epitope in Nef. J Infect Dis. 2008;197(6):871–879. doi: 10.1086/528695. [DOI] [PubMed] [Google Scholar]
- Noviello CM, Pond SL, Lewis MJ, Richman DD, Pillai SK, Yang OO, Little SJ, Smith DM, Guatelli JC. Maintenance of Nef-mediated modulation of major histocompatibility complex class I and CD4 after sexual transmission of human immunodeficiency virus type 1. J Virol. 2007;81(9):4776–4786. doi: 10.1128/JVI.01793-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Gilbert P, Peter T, McLane MF, Gaolekwe S, Rybak N, Thior I, Ndung’u T, Marlink R, Lee TH, Essex M. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J Virol. 2003;77(2):882–890. doi: 10.1128/JVI.77.2.882-890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Lagakos S, Herzig M, Bonney C, Kebaabetswe L, Rossenkhan R, Nkwe D, Margolin L, Musonda R, Moyo S, Woldegabriel E, van Widenfelt E, Makhema J, Essex M. Evolution of proviral gp120 over the first year of HIV-1 subtype C infection. Virology. 2009a;383(1):47–59. doi: 10.1016/j.virol.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Wang R, Margolin L, Baca J, Kebaabetswe L, Rossenkhan R, Bonney C, Herzig M, Nkwe D, Moyo S, Musonda R, Woldegabriel E, van Widenfelt E, Makhema J, Lagakos S, Essex M. Timing Constraints of In Vivo Gag Mutations during Primary HIV-1 Subtype C Infection. PLoS One. 2009b;4(11):e7727. doi: 10.1371/journal.pone.0007727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Woldegabriel E, Kebaabetswe L, Rossenkhan R, Mlotshwa B, Bonney C, Finucane M, Musonda R, Moyo S, Wester C, van Widenfelt E, Makhema J, Lagakos S, Essex M. Viral Load and CD4+ T Cell Dynamics in Primary HIV-1 Subtype C Infection. J Acquir Immune Defic Syndr. 2009c;50:65–76. doi: 10.1097/QAI.0b013e3181900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay T, Zhang H, Drijfhout JW, Robinson N, Brown H, Khan M, Moodley J, Adhikari M, Pfafferott K, Feeney ME, St John A, Holmes EC, Coovadia HM, Klenerman P, Goulder PJR, Phillips RE. Unique Acquisition of Cytotoxic T-Lymphocyte Escape Mutants in Infant Human Immunodeficiency Virus Type 1 Infection. J Virol. 2005;79(18):12100–12105. doi: 10.1128/JVI.79.18.12100-12105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radebe M, Nair K, Chonco F, Bishop K, Wright JK, van der Stok M, Bassett IV, Mncube Z, Altfeld M, Walker BD, Ndung’u T. Limited Immunogenicity of HIV CD8+ T-Cell Epitopes in Acute Clade C Virus Infection. J Infect Dis. 2011;204(5):768–776. doi: 10.1093/infdis/jir394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M. HIV-1 Transmission Biology: Selection and Characteristics of Infecting Viruses. J Infect Dis. 2010;202(S2):S289–S296. doi: 10.1086/655656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Wu X, Lee S, Overbaugh J. Human Immunodeficiency Virus Type 1 V1–V2 Envelope Loop Sequences Expand and Add Glycosylate Sites over the Course of Infection, and These Modifications Affect Antibody Neutralization Sensitivity. J Virol. 2006;80(19):9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A, Brockman MA, Yang R, Adam I, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA, Kuntzen T, Tung CS, LaBute MX, Mueller SM, Harrer T, McMichael AJ, Goulder PJ, Aiken C, Brander C, Kelleher AD, Allen TM. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Alexander L, Morgan J, Lifson J, Mansfield KG, Lang S, Johnson RP, Skowronski J, Desrosiers R. Impact of Nef-mediated downregulation of major histocompatibility complex class I on immune response to simian immunodeficiency virus. J Virol. 2004;78(23):13335–13344. doi: 10.1128/JVI.78.23.13335-13344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treurnicht FK, Seoighe C, Martin DP, Wood N, Abrahams M-RdARD, Bredell H, Woodman Z, Hide W, Mlisana K, Abdool Karim S, Gray CM, Williamson C. Adaptive changes in HIV-1 subtype C proteins during early infection are driven by changes in HLA-associated immune pressure. Virology. 2010;396(2):213–225. doi: 10.1016/j.virol.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Motozono C, Dohki S, Mwimanzi P, Rauch S, Fackler OT, Oka S, Takiguchi M. CTL-Mediated Selective Pressure Influences Dynamic Evolution and Pathogenic Functions of HIV-1 Nef. J Immunol. 2008;180:1107–1116. doi: 10.4049/jimmunol.180.2.1107. [DOI] [PubMed] [Google Scholar]
- Veillette M, Désormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol. 2014;88(5):2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RL, Zou W, Denton PW, Krisko JF, Foster JL, Garcia JV. In vivo analysis of highly conserved Nef activities in HIV-1 replication and pathogenesis. Retrovirology. 2013;10(125) doi: 10.1186/1742-4690-1110-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JK, Brumme ZL, Carlson JM, Heckerman D, Kadie CM, Brumme CJ, Wang B, Losina E, Miura T, Chonco F, van der Stok M, Mncube Z, Bishop K, Goulder PJR, Walker BD, Brockman MA, Ndung’u T. Gag-Protease-Mediated Replication Capacity in HIV-1 Subtype C Chronic Infection: Associations with HLA Type and Clinical Parameters. J Virol. 2010;84(20):10820–10831. doi: 10.1128/JVI.01084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JK, Novitsky V, Brockman MA, Brumme ZL, Brumme CJ, Carlson JM, Heckerman D, Wang B, Losina E, Leshwedi M, van der Stok M, Maphumulo L, Mkhwanazi N, Chonco F, Goulder PJ, Essex M, Walker BD, Ndung’u T. Influence of Gag-protease-mediated replication capacity on disease progression in individuals recently infected with HIV-1 subtype C. J Virol. 2011;85:3996–4006. doi: 10.1128/JVI.02520-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panel A and B: Nef expression was measured by Western blot and expressed relative to the SF2 Nef positive control for Nef clones that were poorly functional (<45% relative to SF2 Nef for both Nef functions, n=47) as well a randomly-selected set of 78 Nef clones that displayed >50% CD4 or HLA down-regulation ability (indicating that Nef was expressed). 41 Nef clones for which expression could not be confirmed by either Western blot (<30% of SF2 control) or functional measurements were removed from further analyses as in vitro artifact could not be excluded. The Western blot band intensities of the remaining Nef clones (n=84) did not correlate significantly with CD4 down-regulation (A) or HLA-I down-regulation (B) by Spearman’s correlation.