Abstract
Immunization of macaques with attenuated simian immunodeficiency virus (SIV) with deletions in nef (SIVΔnef) is shown to elicit protective immunity to infection by pathogenic SIV, yet the mechanisms that orchestrate protection and prevent pathogenesis remains unknown. We utilized whole-genome transcriptional profiling to reveal molecular signatures of protective immunity in circulating CD8+ T cells of rhesus macaques vaccinated with SIVmac239Δnef and challenged with pathogenic SIVmac251. Our findings suggest that protective immunity to pathogenic SIV infection induced by SIVmac239Δnef is associated with balanced induction of T cell activation and immunoregulatory mechanisms and dampened activation of interferon-induced signaling pathways and cytolytic enzyme production as compared with pathogenic SIVmac251 infection of unvaccinated controls. We provide evidence that protective immunity to SIVmac251 correlates with induction of biomarkers of T cell activation, differentiation, signaling, and adhesion that were down regulated in unvaccinated controls. The study highlights potential immunomodulatory networks associated with protective immunity against the virus.
Keywords: HIV, SIV, CD8+ T cell response, gene expression, microarray, immunization, vaccine, immune activation, immune regulation
Introduction
Although development of a vaccine that provides immunity to human immunodeficiency virus (HIV) has not yet been achieved, a growing body of evidence suggests that complete protection requires an appropriate balance of cellular and humoral responses that effectively target a broad range of viral antigens (Friedrich and Watkins, 2008; Miller and Lu, 2003; Watkins, 2008). While recent clinical HIV vaccine trials have provided mixed results (Buchbinder et al., 2008; McElrath et al., 2008; Rerks-Ngarm et al., 2009), vital details of the molecular mechanisms that collectively contribute to protective immune responses continue to be unraveled through studies of non-human primate models. Vaccination of rhesus macaques with live attenuated strains of simian immunodeficiency virus (SIV) has been shown to elicit immunity to subsequent infection by pathogenic SIV strains (Connor et al., 1998; Daniel et al., 1992; Wyand et al., 1999). Studies of cytotoxic T-cell (CTL) responses in the rhesus macaque model show that the protection conferred by attenuated SIV is associated with establishment of a pool of antigen-specific memory cells that display rapid and vigorous ex vivo cytopathic responses (Johnson et al., 1997). Presumably however, efficient CTL killing of virally infected cells must be counterbalanced by mechanisms that regulate effector functions appropriately in order to limit bystander destruction and T cell exhaustion while maintaining optimal pools of functional memory cells (Roth and Pircher, 2004; Xiao et al., 2007; Youngblood et al., 2012). To that end, regulation of the systemic CD8+ T cell response is likely to play a critical role in the ability of the host to control pathogenesis. However, the characteristics of peripheral blood CD8+ T cell responses to attenuated SIV vaccines and pathogenic SIV and their potential impact on disease progression remain poorly understood.
In the current study, we utilize longitudinal transcriptional profiling to characterize the development of circulating CD8+ T cell responses to a live attenuated SIVΔnef vaccine in 6 rhesus macaques over a 40 week period, and 3 weeks following challenge with pathogenic SIVmac251. Two of the 6 vaccinated animals in the study displayed apparently complete protection to the pathogenic challenge while the remaining 4 showed robust control of viremia compared to SIVmac251 infection of unvaccinated controls. The CD8+ T cell molecular signatures associated with protection from SIV disease progression were elucidated through comparison of transcriptional profiles from vaccinated animals and unvaccinated SIVmac251-infected controls. In contrast to SIVmac251 infection of naïve animals, SIVΔnef vaccination resulted in a modest systemic induction of cytolytic enzymes and interferon-induced pathways that coincided with the induction of immunoregulatory factors. In addition, animals that displayed apparent protective immunity to pathogenic SIVmac251 challenge showed increased expression of a distinct set of genes associated with T cell activation, differentiation, signaling, and adhesion that were, by contrast, markedly down regulated in unvaccinated controls. Our findings provide novel insights into the molecular mechanisms of systemic CD8+ T cell responses that are associated with protective immunity to pathogenic SIV infection elicited by SIVΔnef vaccination.
Materials and Methods
Animal groups and study design
Colony-bred, Indian-origin adult rhesus macaques (Macaca mulatta) utilized for the study were housed at the New England Primate Research Center in accordance with American Association for Accreditation of Laboratory Animal Care (AAALAC) guidelines. Samples were collected under experimental protocols approved by the Harvard Medical Area Standing Committee on Animals, and conducted in accordance to the Guide for the Care and Use of Laboratory Animals. Six animals were vaccinated intravenously with live attenuated SIVmac239Δnef as described previously (Alpert et al., 2012) and immune responses were allowed to develop over a period of 40 weeks before intra-vaginal infection with SIVmac251. Peripheral blood samples were collected at 3, 20, and 40 weeks following vaccination, and at 3 weeks following SIVmac251 infection. Six unvaccinated healthy age-matched macaques were intra-vaginally infected with SIVmac251 and served as positive controls. A summary of the animals utilized in the study and their clinical characteristics is presented in Table 1. A comprehensive description of immunologic and virologic data for both SIVΔnef-vaccinated animals and naïve controls will be reported separately (R.K. Reeves, manuscript in preparation).
Table 1.
Clinical charactristics of the animals used in the study. Viral loads are reported as #Gag RNA copy equivalents per ml of blood.
| SIVΔnef Viral Load | WT SIV Viral Load | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Animal | Vaccine | Infection | 3 wks | 20 wks | 40 wks | 3 wks pc | 8 wks pc |
| 211-02 | Δnef | Yes | 140,000 | 10 | 10 | 11,000 | 350 |
| 225-97 | Δnef | No | 15,000 | 3,700 | 4,400 | 10 | 10 |
| 256-00 | Δnef | Yes | 3,300 | 190 | 70 | 12,000 | 460 |
| 286-07 | Δnef | Yes | 24,000 | 10 | 10 | 7,700 | 7,500 |
| 290-07 | Δnef | Yes | 18,000 | 10 | 10 | 1,700 | 340 |
| 292-07 | Δnef | No | 1,300 | 10 | 10 | 10 | 10 |
| 100-98 | Control | Yes | NA | NA | NA | 23,000,000 | 6,100,000 |
| 120-05 | Control | Yes | NA | NA | NA | 3,300,000 | 31,000 |
| 289-07 | Control | Yes | NA | NA | NA | 20,000,000 | 4,500,000 |
| 291-04 | Control | Yes | NA | NA | NA | 5,900,000 | 2,300,000 |
| 321-04 | Control | Yes | NA | NA | NA | 18,000,000 | 950,000 |
| 401-00 | Control | Yes | NA | NA | NA | 8,800,000 | 48,000 |
Viral Loads
Quantitation of viral burden in peripheral blood samples from SIV infected macaques was accomplished by real-time quantitative PCR as previously described (Cline et al., 2005; Salisch et al., 2010). Plasma samples (1.5 ml) were processed to yield a threshold limit of detection of 10 Gag RNA copy equivalents per ml of blood. Challenge virus was detected using oligonucleotide primers specific for the nef sequences of SIVmac251 within the deletion in SIVmac239Δnef. The SIVmac251-specific primers were GAATACPCCATGGAKAAACCCAGC and TGCCAATTTGTAA(C,T,G)TCATTGPTCTTAGG. To reflect the polymorphic nature of the uncloned SIVmac251 virus stocks, the primer set designed to amplify SIVmac251 contained degenerate bases P and K, which mimic mixtures of C and T or A and G, respectively (GlenResearch). The primers specific for SIVmac239Δnef were GAATACTCCATGGAGAAACCCAGC and ATTGCCAATTTGTAACTCATTGTTCTTAG. Complete protection was defined as the absence of detectable wild-type viral RNA from plasma at the post-challenge time point using the above primer/probe sets in a real-time RT-PCR assay with a nominal threshold of detection of 10–30 copies of RNA per ml of blood.
Isolation of CD8+ T cells from blood
CD8+ T cells were purified from peripheral blood samples for downstream transcriptional profiling utilizing the protocols and reagents described in the CD8 negative selection kit for non-human primates (StemCell Technologies). Briefly, PBMCs were incubated with an enrichment cocktail designed for negative selection of CD8+T cells and then loaded onto a StemSep column to allow magnetic separation of CD8+ T cells, which were eluted by gravity feed.
DNA Microarray analysis
Total RNA was isolated from purified CD8+ T cells utilizing protocols and reagents in the RNeasy RNA isolation kit (Qiagen). Messenger RNA amplification, labeling, hybridization to Rhesus macaque GeneChips© (Affymetrix) staining and scanning were performed as previously described (George et al., 2005; Verhoeven et al., 2014) utilizing kits and protocols described in the Affymetrix Gene Expression Analysis Technical Manual.
Microarray data were analyzed longitudinally in individual vaccinated animals and cross-sectionally amongst all groups using BRB Array Tools (http://linus.nci.nih.gov/BRB-ArrayTools.html) and dChip (http://biosun1.harvard.edu/complab/dchip) software programs. For cross-sectional analyses, a minimum fold-change of 1.5 and a p-value ≤ 0.05 between groups were established as the cut off criteria for statistical validation as measured by Student t-test. Peripheral blood samples collected prior to vaccination (0 wk) provided healthy baseline values for subsequently identifying differentially expressed genes (DEG) in vaccinated animals during the 40-week vaccination period and in unvaccinated control animals 3 weeks following SIVmac251 infection. Baseline values for identifying DEG 3 weeks following SIVmac251 challenge of vaccinated animals were established from the 40-week post-vaccination time point for cross sectional analysis, and from the latest vaccination period sample available (either 40wk post-vaccination, or when not available, 20-week post-vaccination) for longitudinal analysis. Assignment of genes to functional categories and pathway analysis was performed using DAVID (Huang da et al., 2009a, b) Ingenuity Pathway Analysis (IPA) web-based software (http://www.ingenuity.com). Raw microarray data files are deposited at the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo) under the following accession number: GDSXXXX (# supplied upon acceptance for publication).
Quantitative RT- PCR
Complementary DNA (cDNA) was synthesized from CD8+ T cell derived mRNA and subjected to real-time-PCR (RT-PCR) analysis as previously described (Guadalupe et al., 2006; Verhoeven et al., 2014) to compare transcript levels of selected genes associated with CD8+ T cell cytolytic functions (granzyme B, and perforin 1). Fluorescence was detected with an ABI Prism 7700 sequence detector (PE Applied Biosystems) and data were analyzed with Sequence Detector Software (SDS). Internal normalization of CT values was performed based on housekeeping genes GAPDH and 18S ribosomal RNA. Differential gene expression was calculated by comparison of normalized CT values between experimental and control samples.
Statistical Analysis
Statistical analysis of RT-PCR assays was performed using GraphPad Prism version 6.00 (GraphPad Software, San Diego, CA). The significance of the differences in expression of granzyme B and perforin-1 between SIV infected and baseline control animals was determined using Student t-tests. Correlations between the fold-changes in transcription identified by RT-PCR and microarray analyses were determined using Pearson's coefficient and 2-tailed p-values.
Results
Replication characteristics of SIVΔnef and SIVmac251 in peripheral blood compartment
The objective of the study was to characterize the molecular signature of peripheral CD8+ T cell responses in rhesus macaques to a live attenuated SIVmac239Δnef vaccine (referred to hereafter as SIVΔnef) and subsequent challenge with the pathogenic strain SIVmac251. In addition, we sought to explore potential correlates of protection in the transcriptional profiles of two vaccinated macaques that displayed apparent sterile protection to SIVmac251 challenge. A total of 6 healthy adult female macaques were inoculated intravenously with SIVΔnef and peripheral blood samples collected prior to vaccination, at 3, 20, and 40 weeks post-vaccination (PV), and at 3 weeks post-vaginal challenge (PC). Six unvaccinated age matched animals were vaginally challenged with SIVmac251 and served as positive controls. A summary of the clinical parameters of the animals utilized in the study is shown in Table 1.
At 3 weeks PV, 4 of the vaccinated animals displayed viral loads >104 SIVΔnef copies/mL of blood, and the remaining 2 animals showed >103 copies/mL. At 20 and 40 weeks PV, viral replication was below 10 copies/mL (limit of detection) in 4 animals and had decreased to <200 copies/mL in macaque 256-00. Only one animal in the study (225-97) maintained peripheral viral loads >103 copies/mL throughout the vaccination period.
Forty weeks after SIVΔnef vaccination, all 6 vaccinated animals and the 6 unvaccinated controls were infected intravaginally with SIVmac251, and viral replication was measured in peripheral blood samples collected after 3 and 8 weeks. At 3 weeks PC, viral replication was below the limit of detection (10 copies/mL) in 2 of the 6 vaccinated animals (225-97 and 292-07), and the remaining 4 displayed substantially reduced viremia in comparison to unvaccinated controls (Table 1). By 8 weeks PC, SIVmac251 burden had dropped to <500 SIV copies/mL blood in all vaccinated macaques except 286-07. In comparison, SIV burden ranged from >104 to >106 copies/mL blood at 8 weeks PC in unvaccinated controls.
SIVΔnef and SIVmac251 infections alter the transcriptional programming of circulating CD8+ T cells
To characterize development of anti-SIVΔnef and -SIVmac251 responses in circulating CD8+ T cells, we purified CD8+ T cells from the blood by magnetic bead isolation and analyzed the transcriptomes using DNA microarrays. Interestingly, at 3 and 20 weeks PV, relatively few changes in gene expression were detected in circulating CD8+ T cells in comparison to healthy pre-vaccination time points (Figure 1), suggesting that SIVΔnef vaccination did not result in a broad reprogramming of systemic CD8+ T cell activity. However, at 40 weeks PV, a substantial increase was detected in the number of genes up and down regulated. The increase in transcriptional modulations at this later vaccination time point was unanticipated and surprising in light of the paucity of changes detected at 3 and 20 weeks PV. An extended analysis of these genes indicated that most were not associated with immune functions, but rather with general metabolic and transport functions, proliferation, and cell death (data not shown).
Figure 1. Hierarchical clustering of genes regulated in CD8+ T cells during the vaccination period and challenge.
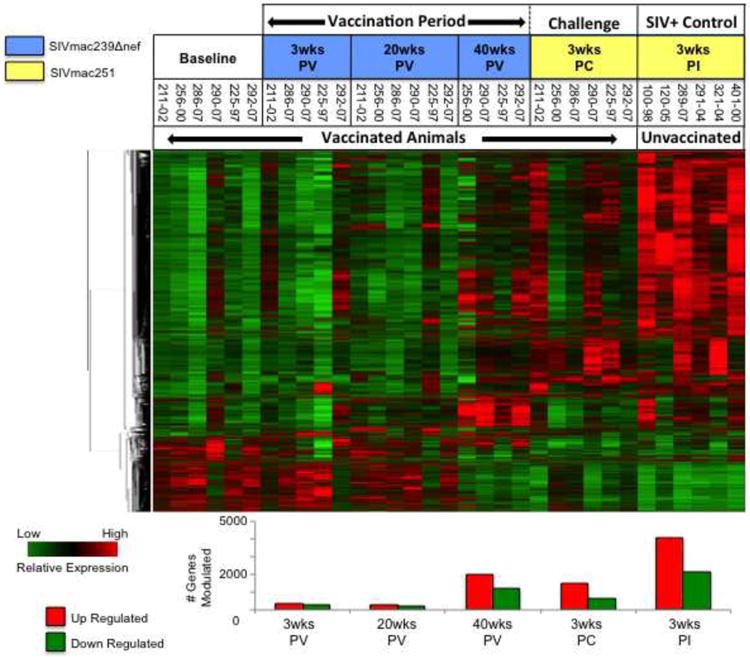
Genes up or down modulated by at least 1.5-fold (p value ≤ 0.05) in vaccinated animals and unvaccinated controls were clustered hierarchically to identify common patterns of regulation following vaccination with SIVΔnef and infection of vaccinated animals and unvaccinated controls with SIVmac251. The number of genes up (red bars) and down (green bars) regulated at each time point is depicted graphically below the heat map.
In contrast to the small number of genes impacted at 3 weeks PV by SIVΔnef, the expression of >2000 genes was up regulated, and >600 down regulated in the same animals 3 weeks following SIVmac251 challenge. It was notable however, that the number of transcriptional perturbations in circulating CD8+ T cells caused by SIVΔnef vaccination and subsequent SIVmac251 challenge was substantially smaller than the number resulting from SIVmac251 infection of unvaccinated controls. More than twice as many genes were up (4098) and down (2121) regulated in peripheral CD8+ T cells at 3 weeks post-infection than at any time point sampled from the vaccinated animals, and this discrepancy is reflected in the clustering pattern of the hierarchically generated heat map shown in Fig. 1.
Regulation of systemic CD8+ T cell biofunctions in vaccinated and unvaccinated animals
Pathway analysis tools were utilized to identify statistically enriched biological themes in the genes that transcriptionally modulated in circulating CD8+ T cells during the 40 week vaccination period (Figure 2A), at 3 weeks PC of vaccinated animals (Figure 2B), and at 3 weeks post-SIVmac251 infection (PI) in unvaccinated controls (Figure 2C). The results provide a broad overview of processes and pathways that were commonly impacted by SIVΔnef vaccination and SIVmac251 infection (e.g. redox homeostasis, apoptosis, cell cycle, etc.), as well as processes that were differentially regulated between experimental conditions (e.g. lymphocyte activation, cell adhesion, antiviral defense, Wnt signaling, etc.). A complete list of fold-changes and corresponding p-values for the genes associated with each biofunction is presented in Supplementary Table I (S1).
Figure 2. Pathway analysis of genes regulated in circulating CD8+ T cells.
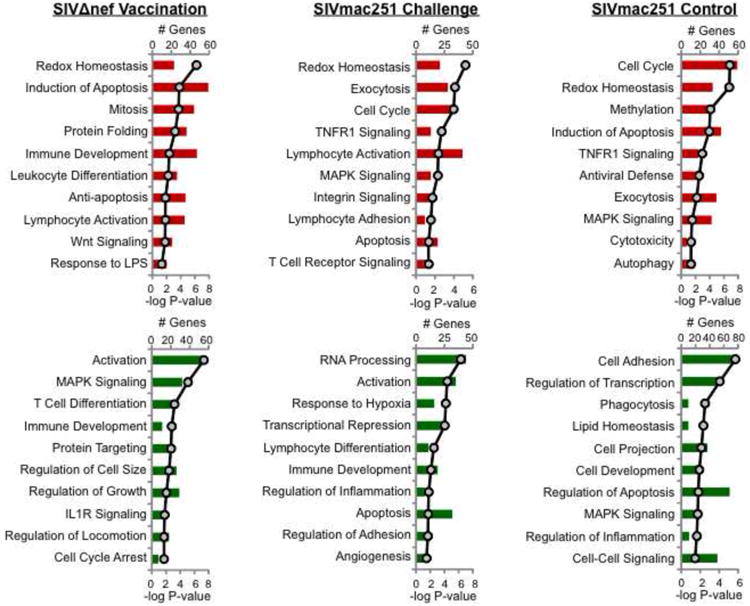
The top 10 pathways and processes statistically enriched in the lists of up and down regulated genes generated through hierarchical clustering were identified (A) during the vaccination period, (B) 3 weeks following SIVmac251 challenge of vaccinated animals, and (C) 3 weeks following SIVmac251 infection of unvaccinated controls. The number of genes modulated is depicted in bar graphs (red=up regulated genes; green=down regulated genes) on the primary x-axis, and statistical confidence (P-value) is depicted linearly on the secondary x-axis.
As expected, functional categories were highlighted during the 40-week vaccination period (Fig. 2A) that reflect the development of CD8+ T cell responses to SIVΔnef during that time, including immune development, leukocyte differentiation, and lymphocyte activation. Interestingly, however, antiviral response pathways were highlighted amongst the top 10 up regulated functional categories only in the unvaccinated controls, and not in SIVΔnef vaccinated animals. This comparatively dampened antiviral response was again observed when vaccinated animals were challenged with pathogenic SIVmac251.
Impact of SIVΔnef vaccination on CD8+ T cell differentiation in peripheral blood
A closer evaluation of the genes associated with the biofunctions depicted in Fig. 2 revealed evidence of discordance in the expression of T cell activation and memory biomarkers between SIVΔnef-vaccinated animals and unvaccinated controls. Both groups displayed down modulation of CD28 (Figure 3A) and Tob-1 (Figure 3D) expression 3 weeks following SIVmac251 infection, indicating comparable levels of CD8+ T cell activation occurred in the peripheral blood compartment. However, we noted that CCR7, a biomarker of memory T cell differentiation, was significantly down regulated in unvaccinated controls (Figure 3B), but not in the vaccinated animals at any of the time points sampled. Moreover, we found that CCR7 was, in contrast, up regulated at 20 weeks following SIVΔnef inoculation. Expression of another marker of memory cell differentiation, CD44, was unaffected by SIVmac251 infection of naïve controls, but displayed a pattern trending toward increasing expression through the vaccination period that was continued in response to SIVmac251 challenge (Figure 3C). Taken together with CCR7 data, the increase in CD44 expression indicates that SIVΔnef vaccination may lead to development of memory CD8+ T cell populations that are otherwise inhibited during pathogenic SIV infection of naïve animals. In contrast to the down regulation of memory T cell biomarkers, expression of T-bet, a transcription factor associated with effector CD8+ T cell differentiation (Kallies, 2008; Naito et al., 2011), was increased in naïve controls at 3 weeks PI, but not in SIVΔnef-vaccinated animals (Figure 3E), consistent with the higher levels of viremia in that group. A progressive increase in T-bet expression was observed in the vaccinated group, reflecting a potential maturation of CD8+ T cell responses that appeared to be associated with better protection against challenge at later time points.
Figure 3. Analysis of CD8+ T cell activation and memory subset biomarkers.
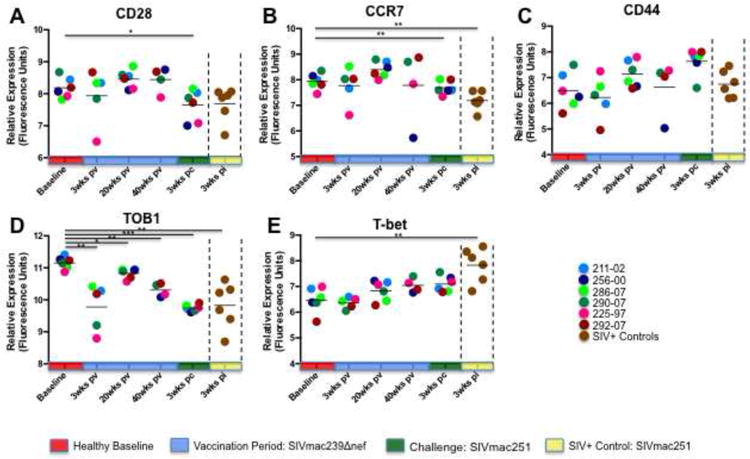
Although genes mediating CD8+ T cell activation (CD28, TOB1) were expressed at similar levels in the blood of vaccinated animals and unvaccinated controls, expression of biomarkers of differentiation of memory (CCR7, CD44) and terminal effector CD8+ T cell subsets was discordant between the two groups. SIVΔnef vaccinated animals up regulated expression of memory-specific biomarkers and down modulated factors associated with terminal effector functions, while unvaccinated animals displayed the opposite profile. Note: * denotes p-value < 0.05; ** denotes p-value ≤ 0.01; *** denotes p-value ≤ 0.001.
To assess and compare the extent of CD8+ T cell effector activity in response to SIVΔnef and SIVmac251, we evaluated expression of cytolytic enzymes in vaccinated and unvaccinated macaques (Figure 4). Transcriptional profiling revealed that granzymes A (Fig. 4A; 42.6-fold) and B (Fig. 4B; 5.3-fold) and perforin (Fig 4C; 6.7-fold) were significantly up regulated at 3 weeks PI in circulating CD8+ T cells of unvaccinated animals in response to SIVmac251. In contrast however, the only statistically significant increase in cytolytic enzymes detected in vaccinated animals was a modest induction (2.4-fold) of perforin 3 weeks following SIVmac251 challenge. Notably, these data were in agreement with the pathway analysis and enumeration of tetramer+ SIV-specific cells, and indicated that higher expression of cytolytic enzymes was not associated with better control of viral replication.
Figure 4. Systemic expression of cytolytic enzymes in response to SIVΔnef and SIVmac251.
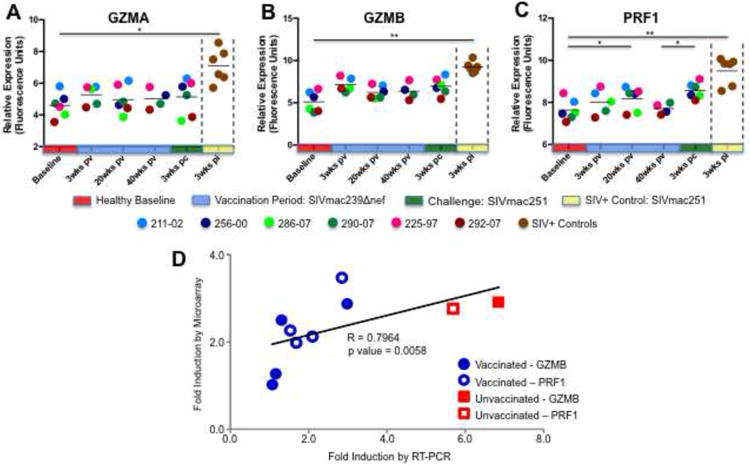
The transcriptional profiles of granzymes A (A) and B (B) and perforin (C) in peripheral CD8+ T cells of SIVΔnef vaccinated animals and unvaccinated controls as determined by microarray are shown (* denotes p-value ≤ 0.05; ** denotes p-value ≤ 0.01). (D) The magnitude of increase (fold-induction, in comparison to unvaccinated uninfected animals) in perforin and granzyme A expression was compared in vaccinated animals and unvaccinated controls by quantitative real-time PCR (qRT-PCR) and compared to induction levels generated by microarray analysis. The strength and statistical significance of the relationship was determined through Spearman's rank correlation coefficient analysis.
To validate the DNA microarray findings, we measured the expression of granzyme B and perforin at 3 weeks following SIVmac251 infection by quantitative real-time PCR (RT-PCR) in 5 of the animals. As expected, the microarray and RT-PCR methodologies showed similar fold-increases over baseline expression values and highly compatible overall induction profiles (Fig. 4D). These confirmatory findings support our microarray-based observations of relatively modest systemic granzyme and perforin induction in SIVΔnef vaccinated animals.
Comparison of interferon signaling pathways in vaccinated and unvaccinated macaques
Previous studies have demonstrated that the induction of interferon (IFN) signaling pathways in CD8+ T cells is associated with their clonal expansion in response to viral infection (Manfras et al., 2004; Tough et al., 1996), and that signal transducer and activator of transcription-1 (STAT1) function appears to play an important role in mediating this effect (Hardy et al., 2009; Kohlmeier et al., 2010; Kotelkin et al., 2006). To evaluate the impact of IFN induction in the CD8+ T cell transcriptional responses of vaccinated and unvaccinated animals, we examined expression of IFNα, IFNγ, downstream IFN-induced molecules, and upstream IFN signaling transducers. Although significant changes in transcription of IFNα were not detected during the vaccination period or following challenge of the vaccinated animals, numerous IFN-induced (IFI) factors were up regulated in the circulating CD8+ T cells of unvaccinated animals at 3 weeks PI, including IFITM3, IFI6, IFI27, IFI35, and IFI44 (Figure 5). Similar induction of IFN associated genes was comparatively absent at all time points during the vaccination period and again at 3 weeks PC in SIVΔnef inoculated animals. Upstream IFN signaling machinery exhibited a similar transcriptional signature. Janus kinase-1 and -2 (JAK1, JAK2), STAT1, and STAT2 were all significantly up regulated in unvaccinated controls, but not in vaccinated animals (Figure 6).
Figure 5. Expression of interferon-induced genes in CD8+ T cells of vaccinated and unvaccinated animals.
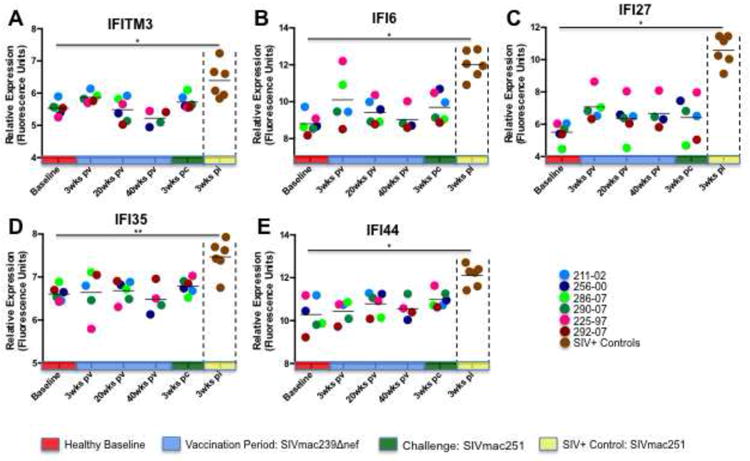
With the exception of IFI6 at 20 weeks PV, no statistically significant increases in expression of interferon-induced genes were detected in the peripheral CD8+ T cells of SIVΔnef inoculated animals during the vaccination period or following SIVmac251 challenge. In contrast, interferon-induced genes were strongly up regulated in the CD8+ T cells of unvaccinated controls 3 weeks following SIVmac251 infection. Note: * denotes p-value ≤ 0.05; ** denotes p-value ≤ 0.01.
Figure 6. Analysis of type I and II interferon signaling factors.
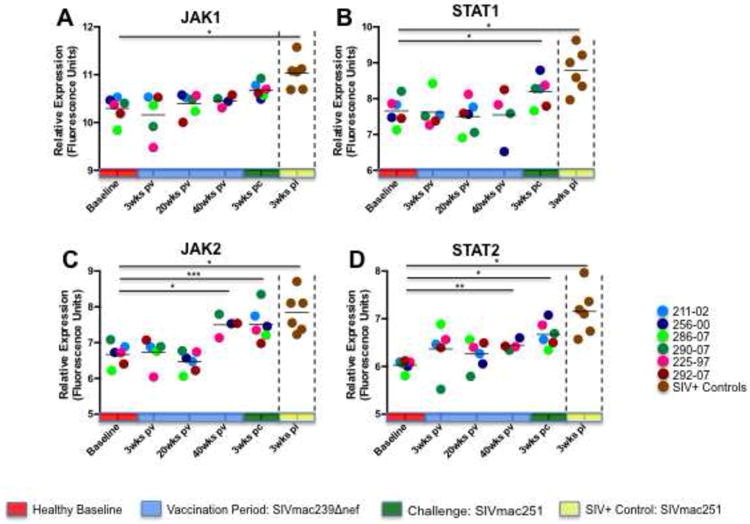
The expression JAK and STAT signaling factors linked to type I and type II IFN signaling in the peripheral CD8+ T cells of SIVΔnef vaccinated animals and unvaccinated controls is shown. JAK2 and STAT2 were modestly up regulated at 40 weeks post-vaccination, and again following SIVmac251 challenge. STAT1 displayed modest induction following SIVmac251 challenge but showed no statistically significant increases during the vaccination period. In contrast, and JAK-STAT signaling molecules were all strongly up regulated in unvaccinated controls 3 weeks following SIVmac251 infection. Note: * denotes p-value ≤ 0.05; ** denotes p-value ≤ 0.01; *** denotes p-value ≤ 0.001.
Interestingly, we found that transcription of IFNγ was significantly down regulated at 3 weeks (3.7-fold; P= 0.009), 20 weeks (3.8-fold: P=0.002), and 40 weeks (8.9-fold; P=0.036) following SIVΔnef vaccination compared to pre-vaccination time points (data not shown). However, at 3 weeks PC, a 2-fold increase (P=0.021) in IFNγ was detected in the vaccinated group. A similar increase (1.7-fold) in IFNγ was detected in the unvaccinated controls at 3 weeks PI, but this change did not reach statistical significance (P=0.31). Taken together, these data indicate that SIVmac251 infection led to a broad activation of IFN signaling pathways in unvaccinated controls, but not in vaccinated animals, and suggest that activation of IFN responses may require high and sustained levels of viral replication such as observed in the SIVmac251-infected naïve controls.
SIVΔnef vaccination induces expression of immunoregulatory factors in peripheral CD8+ T cells
Our findings of subdued effector CD8+ T cell responses in the peripheral blood of vaccinated animals prompted us to further evaluate the potential role of immunoregulation in the systemic CD8+ T cell responses to SIVΔnef vaccine and subsequent challenge with SIVmac251. We found that by 20 weeks PV, all of the vaccinated animals displayed significant induction of cytotoxic T lymphocyte associated factor 4 (CTLA4), interleukin 10 (IL10) and transforming growth factor beta (TGFB) (Figure 7). Collectively, these data provide evidence that the development of circulating CD8+ T cell responses following vaccination with SIVΔnef was marked by coordinated stimulation of immunoregulatory molecules during chronic stage, potentially impacting the expression of interferon associated genes and cytolytic enzymes.
Figure 7. SIVΔnef up regulates expression of immunoregulatory molecules.
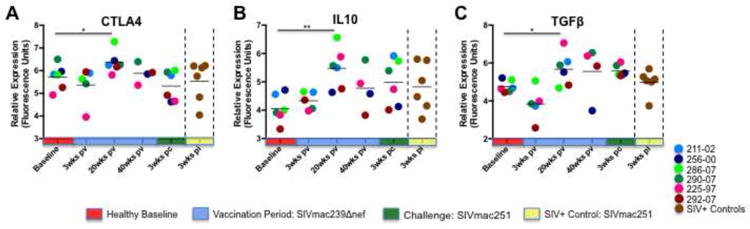
Increased expression of (A) CD25, (B) CD264, and (C) IL-37 was detected in circulating CD8+ T cells at 3 weeks post-vaccination. Macaques that ultimately displayed protective immunity to SIVmac251 challenge (225-97 and 292-07) displayed the most consistent and often the largest magnitudes of induction at 3 weeks PV, and these changes are highlighted in the graphs by connecting lines. Statistically significant (p value ≤ 0.05) increases in the expression of CTLA4, IL-10, and TGFβ were observed in all vaccinated animals at 20 weeks PV. In contrast, no similar increases were detected in the expression of immunoregulatory molecules in unvaccinated SIVmac251 infected animals. Note: * denotes p-value ≤ 0.05; ** denotes p-value ≤ 0.01.
Analysis of CD8+ T cell transcriptional signatures associated with protective immunity
As an initial step to gain further insights into putative mechanisms of protective immunity, we focused on the analysis on CD8+ T cell transcriptional profiles of macaques 225-97 and 292-07. We discovered that the number of genes that were commonly up and down regulated in these two animals increased steadily throughout the 40-week vaccination period, and that transcriptional modulations decreased substantially after SIVmac251 challenge (Figure 8). Functional analysis of the genes with altered expression at 3 weeks PC revealed that the ability to control SIVmac251 replication coincided with the induction of a distinct set of genes associated with cell adhesion, activation, and regulation of cytolytic activity. In contrast, these genes were largely down regulated in unvaccinated controls. Cell adhesion biomarkers included activated leukocyte adhesion molecule (ALCAM), CD9, fibronectin (FN), beta integrins 4 and 6, and junctional adhesion molecule 3 (JAM3) (Figure 9A). While the consequences of the collective induction in expression of these genes are not immediately clear, an increased production of JAM3 may have provided an important trafficking advantage to circulating CD8+ T cells. JAM3 has been identified as a highly specific T and NK cell receptor for vascular endothelial-junctional adhesion molecule (VE-JAM) expressed in the endothelium of lymph nodes and inflammatory foci (Liang et al., 2002). Thus, the induction of JAM3 in macaques 225-97 and 292-07 could reflect a greater capacity of their circulating CD8+ T cells to infiltrate mucosal and lymphoid compartments as compared to unvaccinated controls.
Figure 8. Analysis of genes up and down regulated in macaques with protective immunity to SIVmac251.
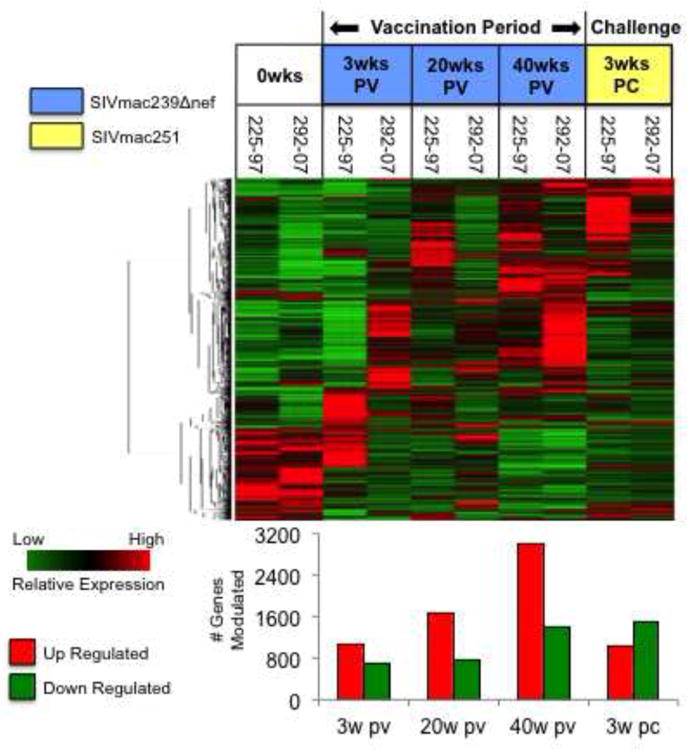
Genes up or down modulated by at least 1.5-fold in the two vaccinated macaques that ultimately displayed protective immunity to SIVmac251 challenge were identified and clustered hierarchically. The number of genes commonly up (red bars) and down (green bars) regulated at each time point is depicted graphically below the heat map.
Figure 9. Characteristics of systemic CD8+ T cell response in animals with protective immunity to SIVmac251.
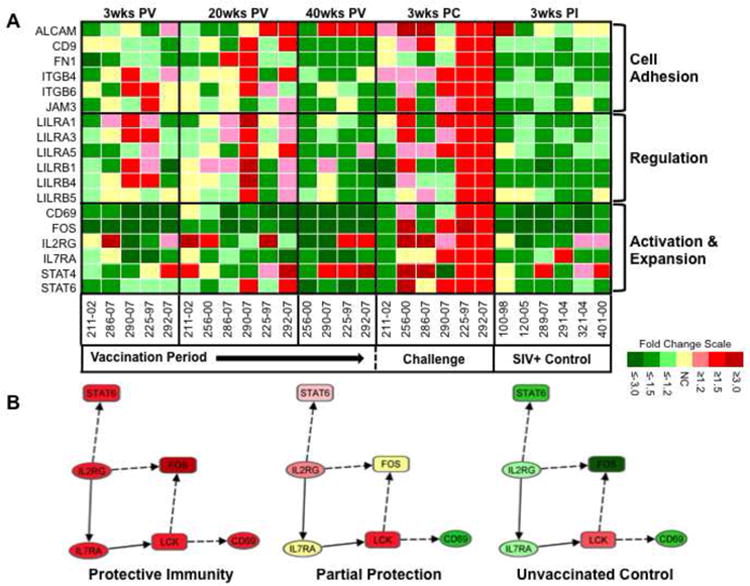
(A) A distinct set of cell adhesion molecules, leukocyte immunoglobulin-like receptors, and T cell activation biomarkers was up regulated predominantly in CD8+ T cells from SIVΔnef vaccinated macaques that displayed protective immunity to pathogenic SIVmac251 challenge (225-97 and 292-07). (B) Network diagram of known relationships between biomarkers of T cell activation that were up regulated in macaques 225-97 and 292-07 at 3 weeks PC. The network depicts co-stimulatory interactions and other functional relationships between IL2RG, IL7RA, FOS, LCK, STAT6, and CD69 that were identified utilizing the Ingenuity Pathway Analysis program.
CD8+ T cells from macaques with protective immunity to SIVmac251 also displayed increased expression of multiple leukocyte immunoglobulin-like receptors (LIRs); LILRA-1, -3, and -5, and LILRB-1, -4, and -5 following challenge (Fig. 9A). This finding was notable because LIRs are closely related to killer cell inhibitory receptors (KIRs) but are expressed on the surface of a broad range of lymphoid and myeloid cells (Borges and Cosman, 2000) and have recently been implicated to have a potential role in HIV-1 infection (Lichterfeld and Yu, 2012). Like KIRs, LIR family members are known to have activating and inhibitory functions. LIR family members of the B subclass regulate inflammatory and cytotoxic responses (Antrobus et al., 2005; Schleinitz et al., 2008), and A subclass members, although less characterized, appear to provide cellular activation functions. Thus, the induction of multiple A and B LIR family members in macaques 225-97 and 292-07 suggests that maintenance of an appropriate balance between activation and regulation may have played an important role in the manifestation of protective immunity.
Changes in the transcription of multiple biomarkers of CD8+ T cell activation provided further evidence of a balanced immune response to SIVmac251 challenge in the protected animals. Three weeks following challenge, macaques 225-97 and 292-07 up regulated expression of CD69, FOS, IL2RG, IL7RA, and STAT-4 and -6 (Fig. 9A). This was in striking contrast to the down modulation of these genes in unvaccinated controls at 3 weeks PI. To investigate the potential causes and consequences of the collective induction of these activation markers, we examined their interactions and functional relationships utilizing the Ingenuity Pathway Analysis© (IPA) tool (Figure 9B). This analytical approach revealed that all of these genes except STAT-4 form a functional gene network together with the lymphocyte-specific protein tyrosine kinase (LCK). The network connections represent previous experimental findings and suggest that increased expression of IL2RG in CD8+ T cells from macaques 225-97 and 292-07 could have contributed to the induction of IL7RA, and ultimately, enhanced up regulation of the T cell activating C-type lectin, CD69. Collectively, these findings provide further indication that protective immunity to SIVmac251 conferred by SIVΔnef vaccination was associated with a circulating CD8+ T cell transcriptional profile distinct from that of unprotected animals and unvaccinated controls.
Discussion
Vaccine-based protective immunity to HIV/SIV infection is characterized by robust antigen specific cellular and humoral responses that are appropriately regulated at the systemic level to limit bystander damage that leads to chronic immune activation and increases the availability of susceptible viral targets (Gougeon and Piacentini, 2009; Ipp and Zemlin, 2013). In the current study, we performed a comprehensive characterization of the development of systemic CD8+ T cell responses in rhesus macaques to an attenuated SIVΔnef vaccine and to a subsequent challenge with SIVmac251 through longitudinal analyses of transcriptional profiles. Our findings indicate that attenuated viral vaccine-induced protection from pathogenic SIVmac251 infection was associated with a peripheral CD8+ T cell response profile during the vaccination period and again following challenge that was compellingly distinct from the CD8+ T cell response of unvaccinated controls.
SIVΔnef vaccination led to alterations in peripheral CD8+ T cells in the expression of genes associated with a range of cellular activation, differentiation, signaling, homeostasis, apoptosis, and growth functions (Fig. 2). However, it is notable that more detailed information on the magnitude of transcriptional changes in different CD8+ T cell subsets (e.g. naïve vs. central memory vs. effector memory) could easily have been diluted in analysis of the bulk CD8+ T cell population. Moreover, since SIV-specific CD8+ T cells constitute only a fraction of all CD8+ T cells in SIVΔnef-vaccinated macaques (Fukazawa et al., 2012), transcriptional changes in CD8+ T cells specifically responding to SIVΔnef immunization would therefore be considerable muted in the bulk populations of CD8+ lymphocytes used for transcriptional profiling in this study. Differences in the magnitude of transcriptional changes observed between SIVΔnef-vaccinated animals and unvaccinated controls post-challenge were presumably reflective of log-scale discrepancies in viral burden (Table 1), differences in the numbers of SIV-specific cells and the significant immune activation induced by pathogenic SIV infection. Interestingly, the modest increases in granzymes and perforin during the vaccination period were counterbalanced at 20 weeks PV by an up regulation of immunoregulatory factors that included CTLA-4, IL-10, and TGFβ (Fig. 7).
Similarly, protective immunity to SIVmac251 challenge in vaccinated macaques 225-97 and 292-07 was associated with the induction of multiple immunoregulatory leukocyte immunoglobulin-like receptors that were, by contrast, markedly down regulated in unvaccinated controls (Fig. 9A). Previous studies have shown that LILRB-1 is expressed on CD8+ T cells as well as NK cells where it negatively regulates T cell effector functions through interaction with MHC I molecules (Anfossi et al., 2004). In support of these reports, we found that the increased expression of LILRB-1 in macaques 225-97 and 292-07 coincided with dampened systemic induction of cytolytic enzymes and IFN-associated molecules.
It is also notable that baseline expression levels of LILRB-1 and LILRA-3 in uninfected control samples were significantly higher than all other LIRs, indicating that these two molecules may be the principle components of an LIR-based regulatory mechanism in peripheral CD8+ T cells. Previous studies have shown that, unlike other A subclass LIR family members, LILRA3 lacks transmembrane and cytoplasmic domains, suggesting it functions mainly as a soluble receptor (Borges et al., 1997). LILRA3 expression is associated with chronic inflammatory conditions such as rheumatoid arthritis (An et al., 2010), and may control the inhibitory immune response induced by LILRB1 (Ryu et al., 2011). Taken together, the evidence of LIR-based regulation of CD8+ T cell activity in animals with protective immunity to SIVmac251 may warrant further investigation into a potential role for these receptors in mediating systemic CTL responses to SIV/HIV infection.
It was intriguing to find that the expression of major CD8+ T cell activation biomarkers such as CD69 and IL7RA was down modulated in unvaccinated animals following SIVmac251 infection, and that a similar response was also observed in naïve animals following SIVΔnef vaccination (Fig 9A). Indeed, the SIVΔnef vaccination led to decreased expression of CD8+ T cell activation biomarkers at each time point tested during the 40-week vaccination period. This finding may have implications with regard to the mechanisms by which SIV (and HIV) can circumvent CD8+ T cell response. CD69 is known to play a vital role in stimulating early events of T cell activation (Mari et al., 1994; Radulovic et al., 2013), while IL-7 signaling is critical for the expansion and survival of memory CD8+ T cell subsets (Chandele and Kaech, 2005; Sereti et al., 2009). In the current study, it appears that the protective response to SIVmac251 elicited by SIVΔnef vaccination was characterized by efficient CD8+ T cell activation that was counterbalanced by the induction of appropriate regulatory mechanisms that presumably prevent excessive non-specific cytolytic activity.
It is possible that limiting chronic immune activation and T cell killing may have helped in maintaining a more optimal pool of functional memory cells in protected animals than in unvaccinated controls. Recent studies have reported that, in spite of robust virus specific CTL responses in vaccinated animals that show protection to SIV, they appear to generate low responses to the more prevalent immunodominant epitopes (Keckler et al., 2007). Our findings further highlight the importance of regulating systemic cytolytic responses and provide evidence that leukocyte immunoglobulin-like receptors may play an important role in this process. Additional investigations are warranted to fully dissect the mechanisms of the transcriptional reprogramming that occurs in circulating CD8+ T cells upon initial exposure to SIV by vaccination or infection.
Supplementary Material
Research Highlights.
Transcriptome of circulating CD8 T cells in macaques vaccinated with attenuated SIV
Contrasting pathogenic SIV infection, vaccine elicits regulated cytolytic response
Protection from pathogenic SIV challenge linked with systemic CD8 immunoregulation
Acknowledgments
We thank Mathew Rolston for his technical expertise in performing the microarray experiments, Dr. Ron Desrosiers for providing SIVΔnef, Dr. Chris Miller for providing SIVmac251, Jacqueline Gillis, Fay Eng Wong, and Elizabeth Curran for assistance with tissue processing, Dr. Angela Carville and other members of the NEPRC Division of Veterinary Resources for their expert animal care. This study was funded by NIH grants AI71306, AI095985, and RR00168 (currently OD011103).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alpert MD, Harvey JD, Lauer WA, Reeves RK, Piatak M, Jr, Carville A, Mansfield KG, Lifson JD, Li W, Desrosiers RC, Johnson RP, Evans DT. ADCC develops over time during persistent infection with live-attenuated SIV and is associated with complete protection against SIV(mac)251 challenge. PLoS pathogens. 2012;8:e1002890. doi: 10.1371/journal.ppat.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Chandra V, Piraino B, Borges L, Geczy C, McNeil HP, Bryant K, Tedla N. Soluble LILRA3, a potential natural antiinflammatory protein, is increased in patients with rheumatoid arthritis and is tightly regulated by interleukin 10, tumor necrosis factor-alpha, and interferon-gamma. The Journal of rheumatology. 2010;37:1596–1606. doi: 10.3899/jrheum.091119. [DOI] [PubMed] [Google Scholar]
- Anfossi N, Doisne JM, Peyrat MA, Ugolini S, Bonnaud O, Bossy D, Pitard V, Merville P, Moreau JF, Delfraissy JF, Dechanet-Merville J, Bonneville M, Venet A, Vivier E. Coordinated expression of Ig-like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J Immunol. 2004;173:7223–7229. doi: 10.4049/jimmunol.173.12.7223. [DOI] [PubMed] [Google Scholar]
- Antrobus RD, Khan N, Hislop AD, Montamat-Sicotte D, Garner LI, Rickinson AB, Moss PA, Willcox BE. Virus-specific cytotoxic T lymphocytes differentially express cell-surface leukocyte immunoglobulin-like receptor-1, an inhibitory receptor for class I major histocompatibility complex molecules. The Journal of infectious diseases. 2005;191:1842–1853. doi: 10.1086/429927. [DOI] [PubMed] [Google Scholar]
- Borges L, Cosman D. LIRs/ILTs/MIRs, inhibitory and stimulatory Ig-superfamily receptors expressed in myeloid and lymphoid cells. Cytokine & growth factor reviews. 2000;11:209–217. doi: 10.1016/s1359-6101(00)00007-1. [DOI] [PubMed] [Google Scholar]
- Borges L, Hsu ML, Fanger N, Kubin M, Cosman D. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN Step Study Protocol, T. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandele A, Kaech SM. Cutting edge: memory CD8 T cell maturation occurs independently of CD8alphaalpha. J Immunol. 2005;175:5619–5623. doi: 10.4049/jimmunol.175.9.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. Journal of medical primatology. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Connor RI, Montefiori DC, Binley JM, Moore JP, Bonhoeffer S, Gettie A, Fenamore EA, Sheridan KE, Ho DD, Dailey PJ, Marx PA. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. Journal of virology. 1998;72:7501–7509. doi: 10.1128/jvi.72.9.7501-7509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- Friedrich TC, Watkins DI. Wanted: correlates of vaccine-induced protection against simian immunodeficiency virus. Current opinion in HIV and AIDS. 2008;3:393–398. doi: 10.1097/COH.0b013e3282faa461. [DOI] [PubMed] [Google Scholar]
- Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, Reyes MD, 3rd, Swanson T, Legasse AW, Sylwester A, Hansen SG, Smith AT, Stafova P, Shoemaker R, Li Y, Oswald K, Axthelm MK, McDermott A, Ferrari G, Montefiori DC, Edlefsen PT, Piatak M, Jr, Lifson JD, Sekaly RP, Picker LJ. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nature medicine. 2012;18:1673–1681. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MD, Reay E, Sankaran S, Dandekar S. Early antiretroviral therapy for simian immunodeficiency virus infection leads to mucosal CD4+ T-cell restoration and enhanced gene expression regulating mucosal repair and regeneration. J Virol. 2005;79:2709–2719. doi: 10.1128/JVI.79.5.2709-2719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougeon ML, Piacentini M. New insights on the role of apoptosis and autophagy in HIV pathogenesis. Apoptosis : an international journal on programmed cell death. 2009;14:501–508. doi: 10.1007/s10495-009-0314-1. [DOI] [PubMed] [Google Scholar]
- Guadalupe M, Sankaran S, George MD, Reay E, Verhoeven D, Shacklett BL, Flamm J, Wegelin J, Prindiville T, Dandekar S. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J Virol. 2006;80:8236–8247. doi: 10.1128/JVI.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy GA, Sieg SF, Rodriguez B, Jiang W, Asaad R, Lederman MM, Harding CV. Desensitization to type I interferon in HIV-1 infection correlates with markers of immune activation and disease progression. Blood. 2009;113:5497–5505. doi: 10.1182/blood-2008-11-190231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ipp H, Zemlin A. The paradox of the immune response in HIV infection: when inflammation becomes harmful. Clinica chimica acta; international journal of clinical chemistry. 2013;416:96–99. doi: 10.1016/j.cca.2012.11.025. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Glickman RL, Yang JQ, Kaur A, Dion JT, Mulligan MJ, Desrosiers RC. Induction of vigorous cytotoxic T-lymphocyte responses by live attenuated simian immunodeficiency virus. Journal of virology. 1997;71:7711–7718. doi: 10.1128/jvi.71.10.7711-7718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A. Distinct regulation of effector and memory T-cell differentiation. Immunology and cell biology. 2008;86:325–332. doi: 10.1038/icb.2008.16. [DOI] [PubMed] [Google Scholar]
- Keckler MS, Hodara VL, Parodi LM, Giavedoni LD. Novel application of nonhuman primate tethering system for evaluation of acute phase SIVmac251 infection in rhesus macaques (Macaca mulatta) Viral immunology. 2007;20:623–634. doi: 10.1089/vim.2007.0068. [DOI] [PubMed] [Google Scholar]
- Kohlmeier JE, Cookenham T, Roberts AD, Miller SC, Woodland DL. Type I interferons regulate cytolytic activity of memory CD8(+) T cells in the lung airways during respiratory virus challenge. Immunity. 2010;33:96–105. doi: 10.1016/j.immuni.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelkin A, Belyakov IM, Yang L, Berzofsky JA, Collins PL, Bukreyev A. The NS2 protein of human respiratory syncytial virus suppresses the cytotoxic T-cell response as a consequence of suppressing the type I interferon response. Journal of virology. 2006;80:5958–5967. doi: 10.1128/JVI.00181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang TW, Chiu HH, Gurney A, Sidle A, Tumas DB, Schow P, Foster J, Klassen T, Dennis K, DeMarco RA, Pham T, Frantz G, Fong S. Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM 2 interacts with T, NK, and dendritic cells through JAM 3. J Immunol. 2002;168:1618–1626. doi: 10.4049/jimmunol.168.4.1618. [DOI] [PubMed] [Google Scholar]
- Lichterfeld M, Yu XG. The emerging role of leukocyte immunoglobulin-like receptors (LILRs) in HIV-1 infection. Journal of leukocyte biology. 2012;91:27–33. doi: 10.1189/jlb.0811442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfras BJ, Weidenbach H, Beckh KH, Kern P, Moller P, Adler G, Mertens T, Boehm BO. Oligoclonal CD8+ T-cell expansion in patients with chronic hepatitis C is associated with liver pathology and poor response to interferon-alpha therapy. Journal of clinical immunology. 2004;24:258–271. doi: 10.1023/B:JOCI.0000025447.23473.ab. [DOI] [PubMed] [Google Scholar]
- Mari B, Imbert V, Belhacene N, Far DF, Peyron JF, Pouyssegur J, Van Obberghen-Schilling E, Rossi B, Auberger P. Thrombin and thrombin receptor agonist peptide induce early events of T cell activation and synergize with TCR cross-linking for CD69 expression and interleukin 2 production. J Biol Chem. 1994;269:8517–8523. [PubMed] [Google Scholar]
- McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR Step Study Protocol, T. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894–1905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CJ, Lu FX. Anti-HIV and -SIV immunity in the vagina. International reviews of immunology. 2003;22:65–76. doi: 10.1080/08830180305230. [DOI] [PubMed] [Google Scholar]
- Naito T, Tanaka H, Naoe Y, Taniuchi I. Transcriptional control of T-cell development. International immunology. 2011;23:661–668. doi: 10.1093/intimm/dxr078. [DOI] [PubMed] [Google Scholar]
- Radulovic K, Rossini V, Manta C, Holzmann K, Kestler HA, Niess JH. The early activation marker CD69 regulates the expression of chemokines and CD4 T cell accumulation in intestine. PloS one. 2013;8:e65413. doi: 10.1371/journal.pone.0065413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH Investigators, M.-T. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. The New England journal of medicine. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Roth E, Pircher H. IFN-gamma promotes Fas ligand- and perforin-mediated liver cell destruction by cytotoxic CD8 T cells. J Immunol. 2004;172:1588–1594. doi: 10.4049/jimmunol.172.3.1588. [DOI] [PubMed] [Google Scholar]
- Ryu M, Chen Y, Qi J, Liu J, Fan Z, Nam G, Shi Y, Cheng H, Gao GF. LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence. PloS one. 2011;6:e19245. doi: 10.1371/journal.pone.0019245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisch NC, Kaufmann DE, Awad AS, Reeves RK, Tighe DP, Li Y, Piatak M, Jr, Lifson JD, Evans DT, Pereyra F, Freeman GJ, Johnson RP. Inhibitory TCR coreceptor PD-1 is a sensitive indicator of low-level replication of SIV and HIV-1. J Immunol. 2010;184:476–487. doi: 10.4049/jimmunol.0902781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleinitz N, Cognet C, Guia S, Laugier-Anfossi F, Baratin M, Pouget J, Pelissier JF, Harle JR, Vivier E, Figarella-Branger D. Expression of the CD85j (leukocyte Ig-like receptor 1, Ig-like transcript 2) receptor for class I major histocompatibility complex molecules in idiopathic inflammatory myopathies. Arthritis and rheumatism. 2008;58:3216–3223. doi: 10.1002/art.23871. [DOI] [PubMed] [Google Scholar]
- Sereti I, Dunham RM, Spritzler J, Aga E, Proschan MA, Medvik K, Battaglia CA, Landay AL, Pahwa S, Fischl MA, Asmuth DM, Tenorio AR, Altman JD, Fox L, Moir S, Malaspina A, Morre M, Buffet R, Silvestri G, Lederman MM Team, A.S. IL-7 administration drives T cell-cycle entry and expansion in HIV-1 infection. Blood. 2009;113:6304–6314. doi: 10.1182/blood-2008-10-186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tough DF, Borrow P, Sprent J. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science. 1996;272:1947–1950. doi: 10.1126/science.272.5270.1947. [DOI] [PubMed] [Google Scholar]
- Verhoeven D, George MD, Hu W, Dang AT, Smit-McBride Z, Reay E, Macal M, Fenton A, Sankaran-Walters S, Dandekar S. Enhanced innate antiviral gene expression, IFN-alpha, and cytolytic responses are predictive of mucosal immune recovery during simian immunodeficiency virus infection. Journal of immunology. 2014;192:3308–3318. doi: 10.4049/jimmunol.1302415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins DI. The hope for an HIV vaccine based on induction of CD8+ T lymphocytes--a review. Memorias do Instituto Oswaldo Cruz. 2008;103:119–129. doi: 10.1590/s0074-02762008000200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyand MS, Manson K, Montefiori DC, Lifson JD, Johnson RP, Desrosiers RC. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. Journal of virology. 1999;73:8356–8363. doi: 10.1128/jvi.73.10.8356-8363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Mescher MF, Jameson SC. Detuning CD8 T cells: down-regulation of CD8 expression, tetramer binding, and response during CTL activation. The Journal of experimental medicine. 2007;204:2667–2677. doi: 10.1084/jem.20062376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Wherry EJ, Ahmed R. Acquired transcriptional programming in functional and exhausted virus-specific CD8 T cells. Current opinion in HIV and AIDS. 2012;7:50–57. doi: 10.1097/COH.0b013e32834ddcf2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


