Abstract
Dysregulation of cholesterol metabolism in the brain has been associated with many neurodegenerative disorders such as Alzheimer’s disease, Niemann-Pick type C disease, Smith-Lemli-Opitz syndrome, Hungtington’s disease and Parkinson’s disease. Specifically, genes involved in cholesterol biosynthesis (24-dehydrocholesterol reductase, DHCR24) and cholesterol efflux (ATP-binding cassete transporter, ABCA1, and apolipoprotein E, APOE) have been associated with developing Alzheimer’s disease. Indeed, APOE was the first gene variation found to increase the risk of Alzheimer’s disease and remains the risk gene with the greatest known impact. Mutations in another cholesterol biosynthetic gene, 7-dehydrocholesterol reductase (DHCR7), cause Smith-Lemli-Opitz syndrome and impairment in cellular cholesterol trafficking caused by mutations in the NPC1 protein results in Niemann-Pick type C disease. Taken together, these findings provide strong evidence that cholesterol metabolism needs to be controlled at very tight levels in the brain. Recent studies have implicated microRNAs (miRNAs) as novel regulators of cholesterol metabolism in several tissues. These small non-coding RNAs regulate gene expression at the post-transcriptional level by either suppressing translation or inducing mRNA degradation. This review article focuses on how cholesterol homeostasis is regulated by miRNAs and their potential implication in several neurodegenerative disorders, such as Alzheimer’s disease. Finally, we also discuss how antagonizing miRNA expression could be a potential therapy for treating cholesterol related diseases.
Keywords: miRNAs, cholesterol metabolism, neurodegenerative disorders
INTRODUCTION
In this article, we discuss the importance of cholesterol homeostasis in the central nervous system and how alterations in cholesterol metabolism result in major neurodegenerative disorders, including Alzheimer’s disease, Niemann-Pick type C disease, Smith-Lemli-Opitz syndrome, Hungtington’s disease and Parkinson’s disease. We also review the most recent discoveries of miRNAs as novel regulators of cholesterol metabolism, focusing specifically on how miRNAs could contribute to the pathogenesis of Alzheimer’s disease. Finally, we discuss the potential therapeutic use of antisense oligonucleotides to inhibit miRNAs in metabolic diseases and how this technology might be applicable for treating neurodegenerative disorders.
CHOLESTEROL METABOLISM AND NEURODEGERATIVE DISORDERS
Cholesterol is an essential constituent of eukaryotic membranes that modulate membrane fluidity and permeability (Yeagle, 1991). Cholesterol is also the precursor of all steroid hormones and bile acids and plays a key role in membrane trafficking and trans-membrane signaling trafficking (Schroeder et al, 1991). Tight regulation of cholesterol metabolism is necessary to maintain neurological functions and dysregulation of cholesterol homeostasis in the brain has been linked to chronic neurodegenerative disorders, including Alzheimer’s disease, Huntington’s disease, Parkinson’s disease, Niemann Pick type C disease and Smith-Lemli-Opitz syndrome (Karasinska & Hayden, 2011; Liu et al, 2010; Maulik et al, 2013; Nowaczyk & Irons, 2012; Vance, 2006; Vance, 2012). In the mature brain, the highest cholesterol content is found in myelin. As such, cholesterol depletion leads to synaptic and dendritic spine degeneration, failed neurotransmission, and decreased synaptic plasticity (Dupree & Pomicter, 2010; Saher & Simons, 2010).
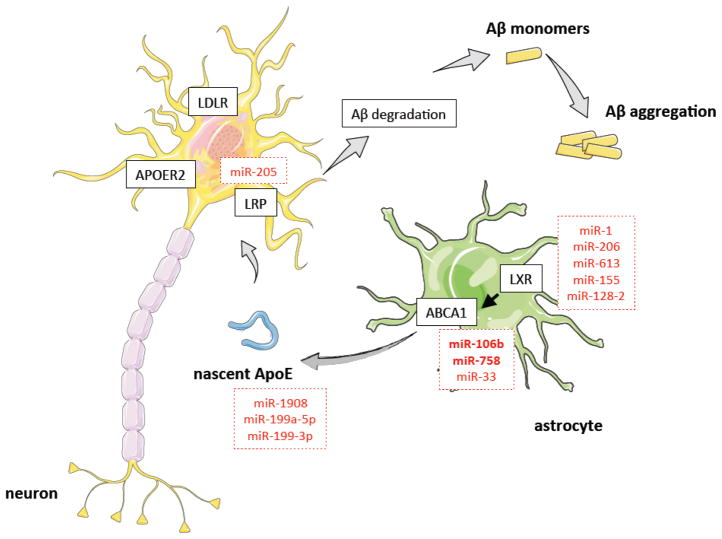
In contrast to the peripheral tissues, most cholesterol in the brain is synthesized in situ because the plasma lipoproteins do not cross the intact blood-brain barrier (BBB) (Danik et al, 1999; Dietschy & Turley, 2001; Dietschy & Turley, 2004). For this reason, mutations in the cholesterol biosynthetic enzymes cause dramatic neurological disorders including desmosterolosis and Smith-Lemli-Opitz syndrome. In addition to genes involved in cholesterol biosynthesis, mutations in genes associated with intracellular cholesterol transport, such as NPC1 and cellular cholesterol uptake and efflux, including apolipoprotein E (APOE) and ATP-binding cassette A1 (ABCA1), have been associated with neurological disorders. The pool of cholesterol in the brain has a remarkably long half-life (6 months in rodents and up to 5 years in humans) (Dietschy & Turley, 2001). In addition to de novo cholesterol synthesis, neurological cells can uptake cholesterol from lipoproteins synthesized within the brain. In this regard, astrocytes play an important role by generating ApoE-lipidated particles, which have a similar size to the circulating high-density lipoproteins (HDL). ABCA1, ABCG1 and ABCG4 regulate the lipidation of nascent ApoE particles. The uptake of these particles by neurons is mediated by the low-density lipoprotein receptor (LDLR), LDL-receptor-related protein (LRP) and APOE receptor 2 (ApoER2) (Boyles et al, 1989; Herz, 2001). In addition to the uptake of lipoproteins, the interaction of these particles with the neuronal lipoprotein receptors is important for maintaining neuronal function. Indeed, it has previously been reported that APOE-containing lipoproteins stimulate neuronal survival and synaptogenesis, as well enhance axonal growth (Hayashi et al, 2004; Hayashi et al, 2007; Mauch et al, 2001). The importance of APOE function in the central nervous system was highlighted when linkage studies followed by association analysis found the APOE4 allele as a strong genetic risk factor for Alzheimer’s disease (Corder et al, 1993; Reiman et al, 1996; Strittmatter et al, 1993). APOE is polymorphic with three major isoforms APOE2 (cys112, cys158), APOE3 (cys112, arg158) and APOE4 (arg112, arg158). The E4 variant is the largest known risk factor for late-onset sporadic Alzheimer disease in a variety of ethnic groups (Corder et al, 1993; Reiman et al, 1996; Strittmatter et al, 1993). Caucasian and Japanese carriers of 2 E4 alleles have between 10 and 30 times the risk of developing Alzheimer disease by 75 years of age, as compared to those not carrying any E4 allele. The current understanding for the role of APOE during the progression of Alzheimer’s disease associates this allele with altered β-amyloid metabolism, synaptic plasticity, neuroinflammation and tau pathology (Brecht et al, 2004; Huang et al, 2001; Reiman et al, 2004).
Cholesterol biosynthesis is tightly controlled by the expression and proteolytic activation of the sterol regulatory element-binding proteins (SREBPs). These transcription factors bind to the sterol regulatory element (SRE) located within the promoter regions of the cholesterol response genes. In cells with low levels of cholesterol, SREBP is activated and translocates to the nucleus where it activates the transcription of genes involved in cholesterol biosynthesis, such as HMGCR, and cholesterol uptake, including the LDLR. Interestingly, in brain-derived cells isolated from patients with mutations in huntingtin, a protein accumulated in patients with Huntington’s disease, the processing and expression of SREBP is markedly reduced (Valenza et al, 2005). As expected by the reduced SREBP activation, the cholesterol level in the cortex and striatum of a mouse model of Hungtington’s disease is significantly lower than in wild-type mice.
miRNAs AND CHOLESTEROL METABOLISM
In addition to the classic regulatory mechanisms that regulate intracellular cholesterol levels (i.e. SREBPs), work over the last few years has uncovered a critical role for miRNAs in controlling cholesterol homeostasis (Fernandez-Hernando et al, 2013; Fernandez-Hernando et al, 2011; Moore et al, 2010). miRNAs are small (~22 nucleotide) single-stranded, non-coding RNAs that regulate gene expression at the post-transcriptional level (Ambros, 2004; Bartel, 2004; Filipowicz et al, 2008). miRNAs are transcribed in the nucleus by RNA polymerase II and processed by the endonuclease, DROSHA, in the nucleus (Ambros, 2004; Bartel, 2004; Filipowicz et al, 2008). The pre-miRNA is then exported to the cytoplasm and processed by DICER. The resulting mature miRNA sequence is incorporated into the RNA silencing complex (RISC). Within the RISC, the miRNA guides the complex to its RNA target, thereby mediating its repression. The target selectivity is dictated by the miRNA “seed” sequence located within nts 2–8 at the 5′-end of the mature miRNA sequence. The seed sequence binds through Watson-crick base pairing within the 3′ untranslated region (3′UTR) of the target genes, leading to translational repression of the target mRNA by either transcript destabilization, translational inhibition or both (Ambros, 2004; Bartel, 2004; Filipowicz et al, 2008).
A number of miRNAs have been shown to regulate cholesterol metabolism including miR-122-5p, miR-370-5p, miR-143-3p, miR-27-3p, miR-106b-5p, miR-758-5p, miR-144-3p and miR-33-5p (Davalos et al, 2011; de Aguiar Vallim et al, 2013; Elmen et al, 2008; Esau et al, 2006; Gerin et al, 2010; Goedeke et al, 2013; Horie et al, 2013; Kim et al, 2012; Najafi-Shoushtari et al, 2010; Ramirez et al, 2011; Ramirez et al, 2013; Rayner et al, 2010). miR-122-5p is the most abundantly expressed miRNA in the liver and regulates plasma lipid levels by controlling the expression of multiple genes involved in cholesterol biosynthesis, lipoprotein export and fatty acid oxidation and synthesis (Elmen et al, 2008; Esau et al, 2006). Mice lacking miR-122 have a marked reduction in plasma cholesterol levels (Castoldi et al, 2011; Hsu et al, 2012). Similar results were obtained in mice and non-human primates treated with miR-122-5p antisense oligonucleotides. miR-143-3p and miR-27-3p regulate adipocyte differentiation by targeting extracellular signal-related kinase 5 (ERK5) and peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT/enhancer binding protein (C/EBP) alpha, respectively (Esau et al, 2004; Lin et al, 2009). miR-106b-5p, miR-758-5p, miR-144-3p, miR-145-5p and miR-33-5p have been shown to regulate the expression of ABCA1, a cholesterol transporter that plays a key role in HDL biogenesis and cholesterol efflux (de Aguiar Vallim et al, 2013; Kang et al, 2013; Kim et al, 2012; Marquart et al, 2010; Ramirez et al, 2011; Ramirez et al, 2013; Rayner et al, 2010). Moreover ABCA1 also controls brain lipid metabolism and the progression of Alzheimer’s disease (Koldamova et al, 2005a).
miRNAs AND NEURODEGENERATION
In recent years, several studies have identified numerous miRNAs associated with neurodegenerative disorders including Alzheimer’s disease, Huntington’s disease, Parkinson’s disease and Amiotrophic Lateral Sclerosis. Intriguingly, most of the genes involved in the development of Alzheimer’s disease, including amyloid precursor protein (APP), beta-amyloid precursor protein-converting enzyme (BACE1), insulin growth factor (IGF1) and TAU are regulated by miRNAs. This intricate network of miRNAs that regulate gene expression is particularly complicated for certain genes, such as APP, which appears to be regulated by multiple miRNAs (miR-106a-5p, miR-520-5p, miR-20a-5p, miR-106a/b-5p, miR-16-5p, miR-101-3p, miR-147-5p and miR-655) (Fan et al, 2010; Kim et al, 2012; Liu et al, 2012; Long & Lahiri, 2011; Sodhi & Singh, 2013; Vilardo et al, 2010). Similarly, BACE-1, the protease that regulates APP processing is also regulated by many miRNAs, including miR-298 and miR-328-3p (Boissonneault et al, 2009). Both miRNAs are expressed in neuronal cells and have regulatory effects on BACE-1 protein expression. A similar complex scenario is observed in Parkinson’s disease where the expression of leucine-rich repeat kinase 2 (LRRK2), a gene associated with an increased risk of Parkinson’s disease, is also controlled by several miRNAs, including let-7-5p, miR-205-5p and miR-184-3p (Maciotta et al, 2013). More detailed information about miRNAs associated with Alzheimer’s disease, as well as other neurological disorders, can be found in some excellent recent review articles (Goodall et al, 2013; Maciotta et al, 2013; Schonrock & Gotz, 2012). As such, for the remainder of the review, we focus on describing the role of miRNAs in controlling cholesterol homeostasis in the brain and the potential implication of these miRNAs in regulating the progression of neurological disorders including Alzheimer’s disease.
ROLE OF ABCA1 TRANSPORTER AND miRNAs IN THE PATHOGENESIS OF ALZHEIMER’S DISEASE
Alzheimer’s disease is a progressive neurodegenerative disorder and the most common cause of dementia in elderly people. A major pathological hallmark of Alzheimer’s disease is the accumulation of amyloid beta peptide (Aβ) in senile plaques in the brain of Alzheimer’s disease patients (Blass, 2003; Jakob-Roetne & Jacobsen, 2009). While the exact mechanism by which Alzheimer’s disease takes place remains unknown, the role of ABC transporters in the pathology of Alzheimer’s disease has recently been recognized. In particular, many studies have highlighted the critical role of ABCA1 in the development of Alzheimer’s disease (Koldamova et al, 2010; Koldamova et al, 2005a).
ABCA1 is a membrane-associated lipid pump that plays a key role in maintaining cholesterol homeostasis by removing excess cholesterol from cells (Goedeke & Fernandez-Hernando, 2012). ABCA1 mediates cholesterol and phospholipid efflux from cells to lipid-poor apolipoprotein A1 (APOA1) and APOE, enabling the formation of nascent, discoidal HDL particles. In the brain, ABCA1 acts to lipidate APOE, which is essential for its interaction with Aβ and subsequent clearance (Kim et al, 2007; Koldamova et al, 2010; Koldamova & Lefterov, 2007; Koldamova et al, 2005a; Koldamova et al, 2005b). Although the molecular mechanisms by which ABCA1 impacts Aβ production and amyloid deposition is not fully understood, a growing body of evidence suggests that ABCA1 plays a critical role in Aβ metabolism and accumulation (Koldamova et al, 2010; Koldamova et al, 2005a). Specifically, several reports have demonstrated that treatment of neuronal and non-neuronal cell lines with liver X receptor (LXR) agonists causes an induction of ABCA1 expression and subsequent decrease in Aβ production (Koldamova & Lefterov, 2007; Koldamova et al, 2005b). Furthermore, in vivo studies using genetically modified mouse models of Alzheimer’s disease have shown that ABCA1 deficiency significantly reduces levels of APOE and increases Aβ and amyloid deposition, while overexpression of ABCA1 ameliorates Aβ accumulation (Fitz et al, 2012; Wahrle et al, 2005; Wahrle et al, 2008). Genetic studies also implicate aberrant cholesterol metabolism and ABCA1 gene expression in Alzheimer’s disease pathogenesis, as several single-nucleotide polymorphisms (SNPs) in ABCA1 have been proposed to modify the risk of early- and late-onset Alzheimer’s disease (Rodriguez-Rodriguez et al, 2007; Sun et al, 2012).
miR-106b regulation of ABCA1 and Aβ metabolism
Given that many lines of evidence implicate ABCA1 in regulating Aβ levels, identification of the basic mechanisms that govern neuronal ABCA1 expression represent attractive therapies for controlling Alzheimer’s disease. Indeed, Kim et al. hypothesized that miRNAs could play a role in regulating ABCA1 expression and Aβ metabolism (Kim et al, 2012). In particular, they identified miR-106b-5p as a negative regulator of ABCA1 expression and ApoA1-mediated cholesterol efflux in neuronal cells. Importantly, over-expression of miR-106b-5p in Neuro2a cells significantly increased Aβ secretion by upregulating Aβ production and preventing Aβ clearance. This decrease in Aβ clearance was attributed to the miR-106b-5p-mediated downregulation of ABCA1 as the reduction of Aβ clearance in cells overexpressing miR-106b-5p was rescued by overexpression of an ABCA1 construct that lacked the 3′UTR sequence. Interestingly, miR-106b-5p has also been shown to decrease the expression of amyloid precursor protein (APP) in neuronal and non-neuronal cells. As both APP and ABCA1 have opposing roles for regulating Aβ levels, the function of miR-106b-5p in controlling Aβ accumulation needs to be studied in vivo. Nevertheless, these findings suggest an important role for miR-106b-5p in regulating Aβ and the pathogenesis of Alzheimer’s disease via the post-transcriptional regulation of ABCA1 and APP.
miR-758 regulation of ABCA1 and cellular cholesterol efflux
In addition to miR-106b, studies from our group have shown that miR-758-5p also post-transcriptionally regulates the expression of ABCA1 (Ramirez et al, 2011). miR-758-5p is widely expressed in a variety of tissues and is repressed in cholesterol-loaded macrophages. Manipulation of miR-758-5p expression in mouse and human cells was shown to regulate ABCA1 expression and cholesterol efflux to ApoA1. Interestingly, miR-758-5p is also highly expressed in brain tissue and human neuronal cell lines (Ramirez et al, 2011). Overexpression of miR-758-5p in human neuroglioma cells markedly decreased ABCA1 expression and cellular cholesterol efflux, suggesting that miR-758-5p expression may influence neuronal cholesterol homeostasis (Ramirez et al, 2011). Moreover, miR-758-5p also negatively regulates the expression of several genes involved in neurological functions, such as sodium-coupled neutral amino acid transporter 1 (SLC38A1), neurite outgrowth (neurite outgrowth (neurotrimin) (NTM), ephrin type-A recetpor 7 (EPHA7), and myelin transcription factor 1-like (MYTL1), suggesting that miR-758-5p plays a key role in controlling cholesterol metabolism and may have important implications for the regulation of Alzheimer’s disease development (Ramirez et al, 2011).
miR-33 regulation of cholesterol homeostasis
Over the past years, many studies have highlighted the importance of miR-33a/b-5p in controlling lipid homeostasis (Gerin et al, 2010; Horie et al, 2013; Marquart et al, 2010; Najafi-Shoushtari et al, 2010; Rayner et al, 2010). miR-33a-5p and miR-33b-5p are evolutionarily conserved, intronic miRNAs that work together with their host genes, SREBP2 and SREBP1, to regulate cholesterol and fatty acid metabolism (Davalos et al, 2011; Rayner et al, 2010). While miR-33-5p has been shown to directly target many genes involved in cholesterol trafficking, fatty acid oxidation, and glucose metabolism, one of the best-characterized targets of miR-33 is ABCA1. Manipulation of miR-33-5p levels affects ABCA1 expression in macrophages and hepatocytes and result in changes in cellular cholesterol efflux to APOA1. Importantly, inhibition of miR-33-5p in vivo promotes ABCA1-mediated cholesterol efflux from macrophages and stimulates the production of HDL (Gerin et al, 2010; Marquart et al, 2010; Najafi-Shoushtari et al, 2010; Rayner et al, 2010). Concomitantly, mice lacking miR-33 have increased hepatic and macrophage ABCA1 expression and plasma HDL levels, thus highlighting the physiological role of miR-33-5p in modulating cholesterol levels (Horie et al, 2013). Consistent with this, two separate studies have shown that pharmacological inhibition of miR-33-5p in non-human primates increases hepatic ABCA1 expression and circulating HDL cholesterol (Rayner et al, 2011; Rottiers et al, 2013).
Given the crucial role of miR-33-5p in altering processes that contribute to atherosclerosis and metabolic syndrome, the aforementioned studies have focused on studying the effects of miR-33 manipulation in cell types such as hepatocytes and macrophages. Surprisingly, thus far no group has investigated the role of miR-33-5p in controlling cholesterol metabolism in the brain, despite evidence that miR-33-5p is highly enriched in this organ. As ABCA1 plays an important role in regulating Aβ clearance and miR-33-5p strongly represses this transporter in various cell types, studying the post-transcriptional regulation of ABCA1 by miR-33-5p and its implications for Aβ accumulation in neuronal cells is warranted. Investigations analyzing the effect of miR-3-5p3 manipulation in mouse and non-human primate models, in terms of a potential treatment for Alzheimer’s disease, promises to be an exciting area of research in the future.
miRNAs THAT REGULATE CHOLESTEROL EFFLUX AND UPTAKE INDEPENDENTLY OF TARGETING ABCA1
miRNAs that regulate cellular cholesterol efflux
In addition to miRNAs that directly target the ABC transporters, several recent reports have uncovered novel miRNAs that regulate cellular cholesterol efflux by targeting the LXR transcription factors. In this regard, it has been shown that miR-1, miR-206 and miR-613 suppress LXR-induced lipogenesis and cholesterol efflux in macrophages and adipocytes (Ou et al, 2011; Zhong et al, 2013). Both miRNAs are expressed also in the brain but their physiological role in controlling brain cholesterol metabolism remains to be elucidated.
miRNAs that regulate cellular cholesterol uptake in neurons
The uptake of APOE-lipoproteins by neurons is mediated by the low-density lipoprotein receptor (LDLR), LDL-receptor-related protein (LRP) and APOE receptor 2 (ApoER2) (Boyles et al, 1989; Herz, 2001). To date, only a few miRNAs have been shown to control the expression of LRP1 in non-neurological cells. Song and colleagues found that miR-205-5p inhibits directly LRP-1 in tumor cell lines, such as U87 and SK-LU-1 (Kajihara et al, 2014). Moreover, APOE expression is also regulated at the post-transcriptional level by miRNAs. miR-1908-5p, miR-199a-5p and miR-199a-3p inhibit APOE expression (Pencheva et al, 2012). Even though these reports strongly suggest that both LRP-1 and APOE expression is regulated by miRNAs, it is still unknown whether these miRNAs are expressed in the brain and if they are relevant in controlling the expression of both genes in the central nervous system (CNS).
CONCLUSIONS
Work over the last decade has identified miRNAs as critical regulators in almost all biological processes. Dysregulation of miRNA expression has been associated with many human diseases including dyslipidemia, diabetes, cancer and neurological disorders. There is a growing interest in developing novel therapies by targeting the expression of miRNAs in vivo. In this regard, several pharmaceutical companies have recently developed pre-clinical trials that aim to assess the efficacy of antagonizing specific miRNAs. Some of these examples correspond to miR-33-5p for treating atherosclerotic vascular disease and metabolic syndrome (Rayner et al, 2011; Rottiers et al, 2013), miR-122-5p for treating HCV (Lanford et al, 2010), and miR-21-5p for treating renal and lung fibrosis (Kumarswamy et al, 2011). We can also speculate that delivery of anti-miR-33 oligonucleotides might increase ABCA1 in the brain, thus ameliorating Alzheimer’s disease. However targeting miRNAs in the CNS proves to be more challenging, as it first has to be evaluated whether antisense oligonucleotides can pass through the BBB. Another important hurdle is how specific and effective these experimental drugs could be in the brain. Further experimental work aimed to identify how a single miRNA controls the expression of a network of genes involved in a particular process in a specific tissue is therefore critical before this therapy can be translated into humans.
Figure 1. Contribution of miRNAs to the regulation of cholesterol uptake and efflux in the CNS.
miRNAs that were confirmed to target genes in neuronal cells are shown in bold. Other miRNAs listed were confirmed to target genes in non-neuronal cells.
HIGHLIGHTS.
Dysregulation of cholesterol metabolism in the brain has been associated to neurodegenerative disorders
ABCA1 expression regulates ApoE lipidation and Aβ clearance in the brain
miRNAs regulate gene expression at post-transcriptional level.
Therapeutic inhibition of miR-33 increases ABCA1 expression
Acknowledgments
The Fernandez-Hernando Lab is supported by grants from the National Institutes of Health (R01HL107953 and R01HL106063) and The Foundation Leducq. We apologize to those whose work could not be cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Blass JP. Cerebrometabolic abnormalities in Alzheimer’s disease. Neurological research. 2003;25:556–566. doi: 10.1179/016164103101201995. [DOI] [PubMed] [Google Scholar]
- Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. The Journal of biological chemistry. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles JK, Zoellner CD, Anderson LJ, Kosik LM, Pitas RE, Weisgraber KH, Hui DY, Mahley RW, Gebicke-Haerter PJ, Ignatius MJ, et al. A role for apolipoprotein E, apolipoprotein AI, and low density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. The Journal of clinical investigation. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht WJ, Harris FM, Chang S, Tesseur I, Yu GQ, Xu Q, Dee Fish J, Wyss-Coray T, Buttini M, Mucke L, Mahley RW, Huang Y. Neuron-specific apolipoprotein e4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:2527–2534. doi: 10.1523/JNEUROSCI.4315-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Vujic Spasic M, Altamura S, Elmen J, Lindow M, Kiss J, Stolte J, Sparla R, D’Alessandro LA, Klingmuller U, Fleming RE, Longerich T, Grone HJ, Benes V, Kauppinen S, Hentze MW, Muckenthaler MU. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. The Journal of clinical investigation. 2011;121:1386–1396. doi: 10.1172/JCI44883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Danik M, Champagne D, Petit-Turcotte C, Beffert U, Poirier J. Brain lipoprotein metabolism and its relation to neurodegenerative disease. Critical reviews in neurobiology. 1999;13:357–407. doi: 10.1615/critrevneurobiol.v13.i4.20. [DOI] [PubMed] [Google Scholar]
- Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim T, Tarling E, Kim T, Civelek M, Baldan A, Esau C, Edwards P. MicroRNA-144 Regulates Hepatic ABCA1 and Plasma HDL Following Activation of the Nuclear Receptor FXR. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Current opinion in lipidology. 2001;12:105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. Journal of lipid research. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- Dupree JL, Pomicter AD. Myelin, DIGs, and membrane rafts in the central nervous system. Prostaglandins & other lipid mediators. 2010;91:118–129. doi: 10.1016/j.prostaglandins.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. The Journal of biological chemistry. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T, Liu M, Li X, Tang H. miR-20a promotes proliferation and invasion by targeting APP in human ovarian cancer cells. Acta biochimica et biophysica Sinica. 2010;42:318–324. doi: 10.1093/abbs/gmq026. [DOI] [PubMed] [Google Scholar]
- Fernandez-Hernando C, Ramirez CM, Goedeke L, Suarez Y. MicroRNAs in metabolic disease. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:178–185. doi: 10.1161/ATVBAHA.112.300144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Hernando C, Suarez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Current opinion in lipidology. 2011;22:86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews Genetics. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fitz NF, Cronican AA, Saleem M, Fauq AH, Chapman R, Lefterov I, Koldamova R. Abca1 deficiency affects Alzheimer’s disease-like phenotype in human ApoE4 but not in ApoE3-targeted replacement mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:13125–13136. doi: 10.1523/JNEUROSCI.1937-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. The Journal of biological chemistry. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedeke L, Fernandez-Hernando C. Regulation of cholesterol homeostasis. Cellular and molecular life sciences: CMLS. 2012;69:915–930. doi: 10.1007/s00018-011-0857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramirez CM, Mattison JA, de Cabo R, Suarez Y, Fernandez-Hernando C. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol Cell Biol. 2013;33:2339–2352. doi: 10.1128/MCB.01714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ. Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Frontiers in cellular neuroscience. 2013;7:178. doi: 10.3389/fncel.2013.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Campenot RB, Vance DE, Vance JE. Glial lipoproteins stimulate axon growth of central nervous system neurons in compartmented cultures. The Journal of biological chemistry. 2004;279:14009–14015. doi: 10.1074/jbc.M313828200. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Campenot RB, Vance DE, Vance JE. Apolipoprotein E-containing lipoproteins protect neurons from apoptosis via a signaling pathway involving low-density lipoprotein receptor-related protein-1. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:1933–1941. doi: 10.1523/JNEUROSCI.5471-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J. The LDL receptor gene family: (un)expected signal transducers in the brain. Neuron. 2001;29:571–581. doi: 10.1016/s0896-6273(01)00234-3. [DOI] [PubMed] [Google Scholar]
- Horie T, Nishino T, Baba O, Kuwabara Y, Nakao T, Nishiga M, Usami S, Izuhara M, Sowa N, Yahagi N, Shimano H, Matsumura S, Inoue K, Marusawa H, Nakamura T, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat Commun. 2013;4:2883. doi: 10.1038/ncomms3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. The Journal of clinical investigation. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Liu XQ, Wyss-Coray T, Brecht WJ, Sanan DA, Mahley RW. Apolipoprotein E fragments present in Alzheimer’s disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8838–8843. doi: 10.1073/pnas.151254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob-Roetne R, Jacobsen H. Alzheimer’s disease: from pathology to therapeutic approaches. Angewandte Chemie. 2009;48:3030–3059. doi: 10.1002/anie.200802808. [DOI] [PubMed] [Google Scholar]
- Kajihara I, Jinnin M, Harada M, Makino K, Honda N, Makino T, Igata T, Masuguchi S, Fukushima S, Ihn H. miR-205 down-regulation promotes proliferation of dermatofibrosarcoma protuberans tumor cells by regulating LRP-1 and ERK phosphorylation. Archives of dermatological research. 2014 doi: 10.1007/s00403-014-1452-z. [DOI] [PubMed] [Google Scholar]
- Kang MH, Zhang LH, Wijesekara N, de Haan W, Butland S, Bhattacharjee A, Hayden MR. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:2724–2732. doi: 10.1161/ATVBAHA.113.302004. [DOI] [PubMed] [Google Scholar]
- Karasinska JM, Hayden MR. Cholesterol metabolism in Huntington disease. Nature reviews Neurology. 2011;7:561–572. doi: 10.1038/nrneurol.2011.132. [DOI] [PubMed] [Google Scholar]
- Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C, Kim J. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Experimental neurology. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WS, Rahmanto AS, Kamili A, Rye KA, Guillemin GJ, Gelissen IC, Jessup W, Hill AF, Garner B. Role of ABCG1 and ABCA1 in regulation of neuronal cholesterol efflux to apolipoprotein E discs and suppression of amyloid-beta peptide generation. The Journal of biological chemistry. 2007;282:2851–2861. doi: 10.1074/jbc.M607831200. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Fitz NF, Lefterov I. The role of ATP-binding cassette transporter A1 in Alzheimer’s disease and neurodegeneration. Biochimica et biophysica acta. 2010;1801:824–830. doi: 10.1016/j.bbalip.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koldamova R, Lefterov I. Role of LXR and ABCA1 in the pathogenesis of Alzheimer’s disease - implications for a new therapeutic approach. Current Alzheimer research. 2007;4:171–178. doi: 10.2174/156720507780362227. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. The Journal of biological chemistry. 2005a;280:43224–43235. doi: 10.1074/jbc.M504513200. [DOI] [PubMed] [Google Scholar]
- Koldamova RP, Lefterov IM, Staufenbiel M, Wolfe D, Huang S, Glorioso JC, Walter M, Roth MG, Lazo JS. The liver X receptor ligand T0901317 decreases amyloid beta production in vitro and in a mouse model of Alzheimer’s disease. The Journal of biological chemistry. 2005b;280:4079–4088. doi: 10.1074/jbc.M411420200. [DOI] [PubMed] [Google Scholar]
- Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA biology. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. The FEBS journal. 2009;276:2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Tang Y, Zhou S, Toh BH, McLean C, Li H. Cholesterol involvement in the pathogenesis of neurodegenerative diseases. Molecular and cellular neurosciences. 2010;43:33–42. doi: 10.1016/j.mcn.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Liu W, Liu C, Zhu J, Shu P, Yin B, Gong Y, Qiang B, Yuan J, Peng X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiology of aging. 2012;33:522–534. doi: 10.1016/j.neurobiolaging.2010.04.034. [DOI] [PubMed] [Google Scholar]
- Long JM, Lahiri DK. MicroRNA-101 downregulates Alzheimer’s amyloid-beta precursor protein levels in human cell cultures and is differentially expressed. Biochemical and biophysical research communications. 2011;404:889–895. doi: 10.1016/j.bbrc.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciotta S, Meregalli M, Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Frontiers in cellular neuroscience. 2013;7:265. doi: 10.3389/fncel.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch DH, Nagler K, Schumacher S, Goritz C, Muller EC, Otto A, Pfrieger FW. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- Maulik M, Westaway D, Jhamandas JH, Kar S. Role of cholesterol in APP metabolism and its significance in Alzheimer’s disease pathogenesis. Molecular neurobiology. 2013;47:37–63. doi: 10.1007/s12035-012-8337-y. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyk MJ, Irons MB. Smith-Lemli-Opitz syndrome: phenotype, natural history, and epidemiology. American journal of medical genetics Part C, Seminars in medical genetics. 2012;160C:250–262. doi: 10.1002/ajmg.c.31343. [DOI] [PubMed] [Google Scholar]
- Ou Z, Wada T, Gramignoli R, Li S, Strom SC, Huang M, Xie W. MicroRNA hsa-miR-613 targets the human LXRalpha gene and mediates a feedback loop of LXRalpha autoregulation. Molecular endocrinology. 2011;25:584–596. doi: 10.1210/me.2010-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel AC, Zavadil J, Castrillo A, Jungsu K, Suarez Y, Fernandez-Hernando C. Control of Cholesterol Metabolism and Plasma HDL Levels by miRNA-144. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, Thibodeau SN, Osborne D. Preclinical evidence of Alzheimer’s disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. The New England journal of medicine. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez E, Mateo I, Llorca J, Sanchez-Quintana C, Infante J, Garcia-Gorostiaga I, Sanchez-Juan P, Berciano J, Combarros O. Association of genetic variants of ABCA1 with Alzheimer’s disease risk. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2007;144B:964–968. doi: 10.1002/ajmg.b.30552. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, Hansen HF, Whittaker S, Henry S, Brookes R, Najafi-Shoushtari SH, Chung RT, Whetstine JR, Gerszten RE, Kauppinen S, Naar AM. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saher G, Simons M. Cholesterol and myelin biogenesis. Sub-cellular biochemistry. 2010;51:489–508. doi: 10.1007/978-90-481-8622-8_18. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Gotz J. Decoding the non-coding RNAs in Alzheimer’s disease. Cellular and molecular life sciences: CMLS. 2012;69:3543–3559. doi: 10.1007/s00018-012-1125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder F, Jefferson JR, Kier AB, Knittel J, Scallen TJ, Wood WG, Hapala I. Membrane cholesterol dynamics: cholesterol domains and kinetic pools. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine. 1991;196:235–252. doi: 10.3181/00379727-196-43185. [DOI] [PubMed] [Google Scholar]
- Sodhi RK, Singh N. Liver X receptors: emerging therapeutic targets for Alzheimer’s disease. Pharmacological research: the official journal of the Italian Pharmacological Society. 2013;72:45–51. doi: 10.1016/j.phrs.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YM, Li HL, Guo QH, Wu P, Hong Z, Lu CZ, Wu ZY. The polymorphism of the ATP-binding cassette transporter 1 gene modulates Alzheimer disease risk in Chinese Han ethnic population. The American journal of geriatric psychiatry: official journal of the American Association for Geriatric Psychiatry. 2012;20:603–611. doi: 10.1097/JGP.0b013e3182423b6a. [DOI] [PubMed] [Google Scholar]
- Valenza M, Rigamonti D, Goffredo D, Zuccato C, Fenu S, Jamot L, Strand A, Tarditi A, Woodman B, Racchi M, Mariotti C, Di Donato S, Corsini A, Bates G, Pruss R, Olson JM, Sipione S, Tartari M, Cattaneo E. Dysfunction of the cholesterol biosynthetic pathway in Huntington’s disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:9932–9939. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE. Lipid imbalance in the neurological disorder, Niemann-Pick C disease. FEBS letters. 2006;580:5518–5524. doi: 10.1016/j.febslet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Vance JE. Dysregulation of cholesterol balance in the brain: contribution to neurodegenerative diseases. Disease models & mechanisms. 2012;5:746–755. doi: 10.1242/dmm.010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Barbato C, Ciotti M, Cogoni C, Ruberti F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. The Journal of biological chemistry. 2010;285:18344–18351. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, Holtzman DM. Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. The Journal of biological chemistry. 2005;280:43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, Paul SM, Holtzman DM. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. The Journal of clinical investigation. 2008;118:671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeagle PL. Modulation of membrane function by cholesterol. Biochimie. 1991;73:1303–1310. doi: 10.1016/0300-9084(91)90093-g. [DOI] [PubMed] [Google Scholar]
- Zhong D, Huang G, Zhang Y, Zeng Y, Xu Z, Zhao Y, He X, He F. MicroRNA-1 and microRNA-206 suppress LXRalpha-induced lipogenesis in hepatocytes. Cellular signalling. 2013;25:1429–1437. doi: 10.1016/j.cellsig.2013.03.003. [DOI] [PubMed] [Google Scholar]