Abstract
Background
Samples used for DNA isolation to be used for forensic odontology studies are often limited. The possibility to use tissue samples stored in formalin for a prolonged period, which contains nucleic acids of questionable quality, opens exciting possibilities for genetic and molecular biology studies useful in speciality of forensic odontology.
Aim
The present study defines substantial modification of existing protocols for total genomic isolation including mitochondrial DNA and proves the utility of such obtained mitochondrial DNA in microsatellite analyses.
Methods
50 dental tissue samples which were kept in neutral buffered formalin liquid bottles were taken for DNA isolation and subsequent analysis. For the isolation of total genomic DNA from tissue samples, a new protocol with substantial modifications from routine ones was adopted by us. Total genomic DNA from matched blood samples were extracted using standard phenol-chloroform extraction method.
Results
Polymerase Chain Reaction and Sequencing of such extracted DNA samples for mitochondrial D loop region were successful and the results were comparable with DNA extracted from normal sources of samples.
Conclusion
The present study reports for the first time that nucleic acids extracted from human dental tissue samples under prolonged formalin fixation times can be used for forensic odontology studies using the described methodology.
Keywords: Mitochondria, DNA, Forensic odontology, Isolation protocol
1. Introduction
The discipline of forensic odontology has grabbed recent attention across forensic experts and dental fraternity alike.1 Forensic odontology involves use of dental tissue remains in identification of humans. It has wide application from mass disasters to crime cases.2 During mass disaster, victim identification becomes difficult and then only the DNA profiling systems can reveal the true identity of a person.3 Similarly in criminal cases the identity of either the victim or criminal can be established through DNA identification.3 DNA analyses require expertise in molecular biology techniques.4 Hence, there have been calls for dentists in the discipline to get more expertise with the molecular biology methodologies. Sources for genomic DNA from dental field are very useful and but often are not taken properly leading to a loss of potentially useful data.
With the expansion of molecular biology techniques of nucleic acid analysis, knowledge about the utility of the preserved DNA is increasingly important.5 Fixed tissues are a huge resource of DNA in forensic odontology studies. These samples are often available at oral pathology centres or departments. Oral tissues during routine surgical procedures can also be easily obtained and put to future use. But, often, their collection and storage results in tissue fixation in formalin-containing solutions, ranging from few hours, days, months to years. Formalin treatment results in stiffening due to protein fixation at structural level and of DNA at molecular level, making DNA extraction difficult and its further use in molecular biology techniques unsuitable, though not impossible.
Use of mitochondrial microsatellites in forensic studies is well established.6 C tract region is one such region. The idea of extracting total genomic DNA including mitochondrial DNA (mtDNA) from tissue samples kept in formalin solution for long and using them in forensic odontology studies has not been tried to date.
2. Method & materials
50 dental tissue samples which were kept in neutral buffered formalin liquid bottles in the department of Oral & Maxillofacial Surgery were taken for DNA isolation and subsequent analysis. Due consent from concerned subjects were taken and ethical approval from institutional ethics committee was taken prior to the study. Total genomic DNA from matched blood samples were extracted using standard phenol–chloroform extraction method. From tissue samples the isolation of total genomic DNA, including mtDNA following protocol was adopted (List & Composition Reagents and Solutions used in the protocol are given in Table 1).
Table 1.
Reagents and solutions used in DNA isolation.
| List of reagents used | Solution preparationa | |
|---|---|---|
| Glycine (Merck Catalogue# 3570) Tris (Qualigens Catalogue# 10645) EDTA (Qualigens Catalogue# 118445000) Phenol (Sisco Research Laboratories Catalogue# 1649142) Chloroform (Qualigens Catalogue# 12307) Isoamyl alcohol (Sisco Research Laboratories Catalogue# 092945) 100% absolute Alcohol (Merck Emsure Catalogue# 100983) SDS (Life Technologies Catalog# 15525017) Proteinase K (Sigma–Aldrich Catalog#P2308) |
1X GTE buffer: | 100 mM glycine 10 mM Tris-HCL pH 8.0 1 mM EDTA |
| Cell Lysis Solution: | 10 mM Tris–HCl pH 8.0 0.1 mM EDTA pH 8.0 0.5% SDS |
|
| 24:1 v/v Chloroform and Isoamyl alcohol | ||
| 10 mg of Proteinase K in 1 ml of MiliQ (10 mg/ml) | ||
HINT: Reconstitute for the first time, aliquot it in 250 ul tubes, for uses, then store at −20 °C and reuse.
2.1. Protocol
-
1.
Cut thin slices of Formalin-fixed non-paraffin-embedded tissue of about 10 mg with a sterile scalpel blade.
-
2.
Briefly blot excess formalin from tissue.
-
3.
Place tissue in 0.5 mL of the 1× GTE buffer in a 2-mL centrifuge tube. This buffer acts a binding agent for excess formalin.
-
4.
Wash tissue six times, once every 12-h period for a total of 72 h. Additional washes may be needed until tissue becomes softened in texture.
-
5.
Air-dry or place tissue at 55 °C until dry. Do not over dry tissue.
-
6.
Cut tissue with sterile razor blade into 3–4 small pieces.
-
7.
Place the pieces of tissue into 1.5 ml microcentrifuge tube containing 300 ul Cell Lysis Solution. Incubate the mixture at 55 °C in a water bath for 1 h.
-
8.
Add 10 ul Proteinase K (10 mg/ml) to the mixture at intervals of 24 h for total period of 48 h. Incubate the mixture at 55 °C for 48 h. If the tissue is still not digested after 48 h, add further 10 ul of Proteinase K at 10 mg/ml for another 24 h.
-
9.
Once all tissue is in solution, cool sample to room temperature. Add equal volume (300 ul) of equilibrated phenol (pH 8.0) to the mixture. Invert tube several times mixing it well for 10 min
-
10.
Centrifuge for 10 min at 12,000 ×g to separate the layers.
-
11.
Carefully remove the upper aqueous layer discarding the lower phenol layer and add to a new tube containing equal volumes i.e 150 ul each of phenol and 24:1 chloroform:isoamyl alcohol. Be careful not to remove the protein-containing interface. Invert tube several times mixing it well for 10 min.
-
12.
Centrifuge for 10 min at 12,000 ×g to separate the layers.
-
13.
Transfer the supernatant i.e. upper aqueous layer to a fresh tube containing equal volume (300 ul) of 24:1 chloroform:isoamyl alcohol avoiding the interface material. Invert tube several times mixing it well for 10 min.
-
14.
Centrifuge for 10 min at 12,000 ×g to separate the layers.
-
15.
Remove the upper aqueous layer and place in a new 1.5 mL Eppendorf tube. Discard the lower chloroform:isoamyl alcohol layer.
-
16.
Add 2 volumes (600 ul) of absolute alcohol and invert gently a few times. The DNA precipitates as a thread like structure.
-
17.
Incubate the tube at −20 °C overnight to allow complete DNA precipitation.
-
18.
Centrifuge at high speed (>10,000 ×g) for 30 min at room temperature. Immediately following centrifugation, decant the liquid from the tube carefully. The DNA will have precipitated into a pellet at the bottom of the tube and may not be visible
-
19.
Give two washes with 70% alcohol and air-dry the pellet.
-
20.
Dissolve the DNA in 200 ul TE buffer for 4–6 h at 50 °C.
2.2. Mitochondrial DNA C tract amplification and sequencing
The mtDNA C tract amplification was done using a set of primer [Table 2] yielding a 109 bp fragment for all blood and tissue samples. Bidirectional direct capillary was done using BigDye® Direct Cycle Sequencing, Applied Biosystems, USA using manufacturer's recommended protocol. Both Forward and Reverse primers were used to analyse and confirm the sequences in the region of interest with the sequences.
Table 2.
Details of PCR primers & conditions.
| Primer | Sequence | Amplification conditions |
|---|---|---|
| Forward | 5′ ACAATTGAATGTCTGCACAGCCACTT 3′ | 94 °C for 5 min, 35 cycles of 94 °C for 45 s, 58 °C for 45 s and 72 °C for 45 s, followed by 72 °C for 10 min |
| Reverse | 5′ GGCAGAGATGTGTTTAAGTGCTG 3′ |
2.3. Statistical analyses
Mitochondrial DNA sequences from tissue and blood samples were compared and difference if any was scored. Student ‘t’ test was used to see significance of any such difference.
3. Results
Total genomic DNA from tissue samples kept in formalin solution for long periods (all > 1 Year) was isolated and visualized on 1% Agarose gel to check the quality and the quantity was confirmed by nanodrop readings with 260/280 and 260/230 values (Fig. 1). All the samples yielded DNA suitable for the running PCR (average 200 ng/ul from approximately 10 mg tissue sections; A260:A280 = 1.55) (Fig. 2). Upon mtDNA C tract amplification of all DNA samples, PCR products were of good quality as visualized by electrophoresis on 2% Agarose gels in 1× TBE (89 mM Tris–Borate, 89 mM boric acid, 2 mM EDTA) (Fig. 2). Further mtDNA C tract sequencing for 109 bp PCR products yielded good quality sequencing results which could be read well and compared with sequences obtained from corresponding blood samples. 37/50 paired samples perfectly matched (p > 0.05) (Figs. 3 and 4).
Fig. 1.
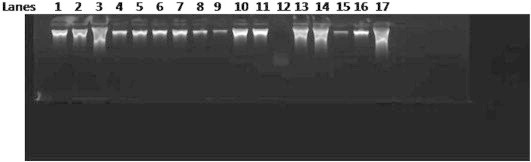
1% Agarose gel picture showing total genomic DNA.
Fig. 2.
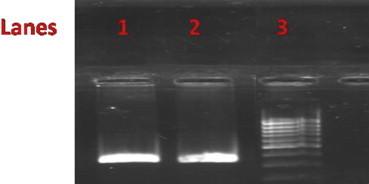
Lanes 1, 2 show mtDNA C tract PCR product while lane 3 shows 100 bp ladder.
Fig. 3.
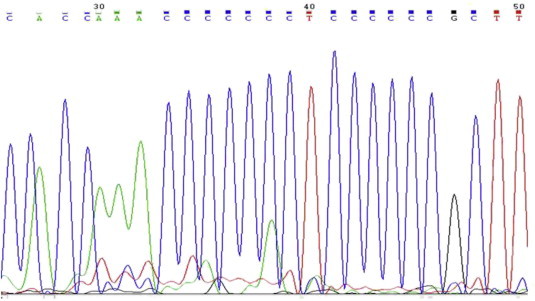
Tissue sample mitochondrial C tract sequence (7CT6C).
Fig. 4.
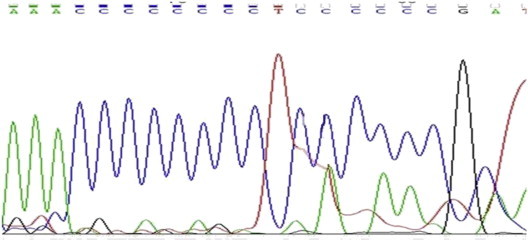
Blood sample mitochondrial C tract sequence (7CT6C).
4. Discussion
Formaldehyde as a 10% neutral buffered formalin is the most widely used universal fixative because it preserves a wide range of tissues and tissue components. Formalin preserved samples are quite common in dental practise. Routine day to day teeth extractions often yield tissues that are lacerated from the site and are waste hence thrown away. Preservation of such tissues in a dental office can put them to future use in forensic odontology analyses. However use of formalin is often frowned upon by molecular biologists. Formalin treatment results in cross-linking of biomolecules7,8 which not only complicates isolation of nucleic acid but also introduces polymerase “blocks” during PCR and this is directly proportional to the time spent in formalin. Few methods exists for the recovery of DNA from archival tissues9–13 and only 3 studies have analysed the ability to extract and study nucleic acids from samples after prolonged fixation and storage times.14–16 These all studies were limited by the fact for they started DNA extraction from paraffinized tissue. Therefore till date no study has been done to isolate DNA directly from tissues stored in formalin solution and checking its suitability for downstream applications such as PCR and sequencing.
It is currently not impossible to reverse the effects of DNA fragmentation17–19 and in our opinion, the key step of successful DNA extraction is the one at which there is reversal of the DNA–protein cross-links and the neutralization of the excess formalin in the sample. Formaldehyde reacts reversibly with water to form methylene glycol. Tissues are rapidly penetrated by the methylene glycol20 but it has no role in fixation. The actual covalent chemical fixation depends on the fraction of the formaldehyde forming bonds with the tissue components and this formaldehyde comes from dissociation of methylene glycol. Interestingly, the equilibrium of this reaction strongly favours methylene glycol. Hence, formaldehyde though penetrates tissues rapidly as methylene glycol but fixes slowly as carbonyl formaldehyde. In our method we exploited this mechanism by using GTE (Glycine, Tris, EDTA) buffer which is glycine rich and alkaline in nature. Glycine reacts with formaldehyde to form either N-Methyleneglycine or Dimethylglycine and shifts the equilibrium towards formaldehyde thereby depleting the store of methylene glycol, the source of formaldehyde, in the tissue. Alkalinity of the buffer further aids in reversing the cross-linking of DNA and proteins. EDTA neutralizes DNAse activity preventing further DNA degradation. In this paper, we therefore report a substantial modification of methods for DNA extraction, fit for molecular biology work in forensic odontology studies, from long duration formalin stored tissues.
Subsequently the method involves heat treatment and proteinase K action21 which digests the denatured proteins and protein–protein cross-links followed by organic purification of the nucleic acids (Protocol). The method results in similar total yields of DNA per sample (as measured in ug/mL) compared to other extraction methods such as commercial kits marketed with the specific aim of processing formalin-fixed or paraffin-embedded (FFPE) material.
Use of mtDNA has been advocated as it possesses high copy number, maternal inheritance, and high degree of sequence variability.6,22 Moreover, nuclear DNA tends to degrade over long period of time and hence mtDNA offers an alternative. Mitochondrial microsatellite studies have long been done in forensic samples due to their polymorphic nature.23,24 In investigations involving missing persons, comparing the mtDNA profile of unidentified remains with the profile of a potential maternal relative is utilized. Nowadays, mtDNA samples from an individual are taken and compared either with sequence from crime site for identification or to available sequences on internet to confirm ethnicity or phylogeny. Our results provided an advancement in the field of forensic odontology by showing that DNA can be reliably amplified from tissues that have been maintained in formalin for considerable period of time (>1 Year). The sequencing results of 37 tissue samples matched with corresponding blood samples (p > 0.05). For rest of samples, difference can be attributed to the fact that C tract region is highly polymorphic site and resides in mitochondria which is in itself highly prone to mutation from external and internal stimuli. Thus there may be slight variability but this was not statistically in our study significant. Therefore, this simple, cost effective and non-laborious method should facilitate the molecular analysis of a large number of specimens fixed for long periods of time in retrospective studies.
5. Conclusion
In summary, we report for the first time that nucleic acids extracted from human dental tissue samples under prolonged formalin fixation times can be used for forensic odontology studies using the described methodology. The quality of DNA fragments is sufficient to study polymorphisms involving deletion/insertion or single nucleotide mutations.
Conflicts of interest
All authors have none to declare.
Acknowledgements
Rahul Pandey and Divya Mehrotra are thankful to Indian Council of Medical Research for research grant.
References
- 1.Datta P., Sood S., Rastogi P., Bhargava K., Bhargava D., Yadav M. DNA profiling in forensic dentistry. J Indian Acad Forensic Med. 2012;34(2):156–158. [Google Scholar]
- 2.Sarode S.C., Zarkar G.A., Kulkarni M.A., Desai R. Role of forensic odontology in the world's major mass disasters: facts and figures. SADJ. 2009;64:388–390. 392–393. [PubMed] [Google Scholar]
- 3.Sweet D., DiZinno J.A. Personal identification through dental evidence-tooth fragments to DNA. J Calif Dent Assoc. 1996;24:35–42. [PubMed] [Google Scholar]
- 4.Saxena S., Sharma P., Gupta N. Experimental studies of forensic odontology to aid in the identification process. J Forensic Dent Sci. 2010;2(2):69–76. doi: 10.4103/0975-1475.81285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kavitha R. Molecular techniques in forensic dentistry. J Forensic Odontol. 2008;1:13–17. [Google Scholar]
- 6.Koyama H., Iwasa M., Ohtani S. Personal identification from human remains by mitochondrial DNA sequencing. Am J Forensic Med Pathol. 2002;23:272–276. doi: 10.1097/00000433-200209000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Tokuda Y., Nakamura T., Satonaka K. Fundamental study on the mechanism of DNA degradation in tissues fixed in formaldehyde. J Clin Pathol. 1990;43:748–751. doi: 10.1136/jcp.43.9.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlando V., Strutt H., Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11(2):205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 9.Bramwell N.H., Burns B.F. The effects of fixative type and fixation time on the quantity and quality of extractable DNA for hybridization studies on lymphoid tissue. Exp Hematol. 1988;16:730–732. [PubMed] [Google Scholar]
- 10.Serth J., Kuczyk M.A., Paeslack U., Lichtinghagen R., Jonas U. Quantitation of DNA extracted after micropreparation of cells from frozen and formalin-fixed tissue sections. Am J Pathol. 2000;156:1189–1196. doi: 10.1016/S0002-9440(10)64989-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagi N., Satonaka K., Horio M., Shimogaki H., Tokuda Y., Maeda S. The role of DNase and EDTA on DNA degradation in formaldehyde fixed tissues. Biotech Histochem. 1996;71:123–129. doi: 10.3109/10520299609117148. [DOI] [PubMed] [Google Scholar]
- 12.Douglas M.P., Rogers S.O. DNA damage caused by common cytological fixatives. Mutat Res. 1998;401:77–88. doi: 10.1016/s0027-5107(97)00314-x. [DOI] [PubMed] [Google Scholar]
- 13.Inadome Y., Noguchi M. Selection of higher molecular weight genomic DNA for molecular diagnosis from formalin-fixed material. Diagn Mol Pathol. 2003;12(4):231–236. doi: 10.1097/00019606-200312000-00007. [DOI] [PubMed] [Google Scholar]
- 14.Ferruelo A., El-Assar M., Lorente J.A. Transcriptional profiling and genotyping of degraded nucleic acids from autopsy tissue samples after prolonged formalin fixation times. Int J Clin Exp Pathol. 2011;4(2):156–161. [PMC free article] [PubMed] [Google Scholar]
- 15.Santos M.C., Saito C.P., Line S.R. Extraction of genomic DNA from paraffin-embedded tissue sections of human fetuses fixed and stored in formalin for long periods. Pathol Res Pract. 2008;204(9):633–636. doi: 10.1016/j.prp.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Savioz A., Blouin J.L., Guidi S., Antonarakis S.E., Bouras C. A method for the extraction of genomic DNA from human brain tissue fixed and stored in formalin for many years. Acta Neuropathol. 1997;93(4):408–413. doi: 10.1007/s004010050632. [DOI] [PubMed] [Google Scholar]
- 17.Srinivasan M., Sedmak D., Jewell S. Effect of fixatives and tissue processing on the Content and Integrity of nucleic acids. Am J Pathol. 2002;161(6):1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz-Cano S.J., Brady S.P. DNA extraction from formalin-fixed, paraffin embedded tissues: protein digestion as a limiting step for retrieval of high-quality DNA. Diagn Mol Pathol. 1997;6:342–346. doi: 10.1097/00019606-199712000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Gavrilov A., Razin S.V. Formaldehyde fixation of cells does not greatly reduce the ability to amplify cellular DNA. Anal Biochem. 2009;390(1):94–96. doi: 10.1016/j.ab.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 20.Start R.D., Layton C.M., Cross S.S., Smith J.H. Reassessment of the rate of fixative diffusion. J Clin Pathol. 1992;45:1120–1121. doi: 10.1136/jcp.45.12.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpi F.M., Di Pietro F., Vincenzetti S., Mignini F., Napolioni V. Human DNA extraction methods: patents and applications. Recent Pat DNA Gene Seq. 2011;5(1):1–7. doi: 10.2174/187221511794839264. [DOI] [PubMed] [Google Scholar]
- 22.da Silva R.H.A., Sales-Peres A., de Oliveira R.N., de Oliveira F.T., Sales-Peres S.H. Use of DNA technology in forensic dentistry. J Appl Oral Sci. 2007;15:156–161. doi: 10.1590/S1678-77572007000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranganathan K., Rooban T., Lakshminarayanan V. Forensic odontology: a review. J Forensic Odontol. 2008;1:4–12. [Google Scholar]
- 24.Nuzzolese E., Di Vella G. Future project concerning mass disaster management: a forensic odontology prospectus. Int Dent J. 2007;57:261–266. doi: 10.1111/j.1875-595x.2007.tb00130.x. [DOI] [PubMed] [Google Scholar]


