Abstract
Human pregnane X receptor (hPXR) regulates the expression of drug-metabolizing enzyme cytochrome P450 3A4 (CYP3A4) and drug transporters such as multidrug-resistance protein 1 (MDR1). PXR can be modulated by small molecules, including Federal Drug Administration (FDA)–approved drugs, thus altering drug metabolism and causing drug-drug interactions. To determine the role of FDA-approved drugs in PXR-mediated regulation of drug metabolism and clearance, we screened 1481 FDA-approved small-molecule drugs by using a luciferase reporter assay in HEK293T cells and identified the diuretic drug metolazone as an activator of PXR. Our data showed that metolazone activated hPXR-mediated expression of CYP3A4 and MDR1 in human hepatocytes and intestine cells and increased CYP3A4 promoter activity in various cell lines. Mammalian two-hybrid assays showed that hPXR recruits its co-activator SRC-1 upon metolazone binding in HepG2 cells, explaining the mechanism of hPXR activation. To understand the role of other commonly-used diuretics in PXR activation and the structure-activity relationship of metolazone, thiazide and non-thiazide diuretics drugs were also tested but only metolazone activates PXR. To understand the molecular mechanism, docking studies and mutational analysis were carried out and showed that metolazone binds in the ligand-binding pocket and interacts with mostly hydrophobic amino acid residues. This is the first report showing that metolazone activates PXR. Because activation of hPXR might cause drug-drug interactions, metolazone should be used with caution for drug treatment in patients undergoing combination therapy.
Keywords: PXR, metolazone, drug metabolism, CYP3A4, MDR1
1. Introduction
Nuclear receptors are ligand-activated DNA-binding transcription factors involved in a wide range of physiological processes [1, 2]. Pregnane X receptor (PXR) belongs to the nuclear receptor superfamily [3]. Like other nuclear receptors, PXR consists of a DNA binding domain (DBD), C-terminal ligand binding domain (LBD), and an activation function 2 domain (AF-2) [3]. PXR binds structurally diverse sets of ligands including clinical drugs, phytochemicals, dietary constituents, and some commonly used herbal medicines [3–7]. The crystal structures of the LBD of PXR showed that LBD is mostly hydrophobic, flexible, and larger than that of other nuclear receptors, thus explaining its promiscuity in binding molecules with different sizes and in different orientations [8–16]. PXR, heterodimerized with the retinoid X receptor (RXR), regulates gene expression by binding to the promoter region of its target genes [17]. Agonistic ligand binding causes PXR to recruit co-activators and induce the expression of its target genes, including those encoding the drug metabolism enzyme cytochrome P450 3A4 (CYP3A4) and the drug transporter multidrug-resistance protein 1 (MDR1), which are involved in metabolism and elimination of potentially harmful chemicals [18–21] and of more than 50% of clinical drugs [4, 22, 23]. PXR is mostly expressed in human livers and intestines, where CYP3A is also abundantly distributed [24, 25]. Therefore, induction of CYP3A4 resulting from the activation of PXR by clinical drugs might enhance the metabolism of other medications that are CYP3A4 substrates [3], causing unwanted drug-drug interaction, an important type of adverse drug events which might have life-threatening consequences [26].
Metolazone (MET) is a diuretic (saluretic, antihypertensive) drug that has been on the market for a number of years and mostly used for the congestive heart failure and high blood pressure [27–29]. MET is a quinazoline diuretic with similar properties to the thiazide diuretic and its action comes from the interference with the renal tubular mechanism of electrolyte reabsorption [30]. MET is a weak inhibitor of human carbonic anhydrase isoforms I and III, but very potent for isoforms VII, XII, and XIII [31]. The Role of MET in PXR-regulated drug metabolism is unknown.
In this study we focused on evaluating the potential of FDA-approved drugs to modulate hPXR because such interactions could alter drug metabolism and cause unintended drug-drug interactions. First, HEK293T cells transfected with PXR were used to screen 1481 FDA-approved small-molecule drugs by using a luciferase reporter assay. Our data showed that, MET activated human PXR (hPXR)-mediated CYP3A4 and MDR1 expression in human hepatocytes and intestine cells at both mRNA and protein levels. Competitive ligand binding assays were performed to confirm MET-PXR interaction and structure-activity relationships were studied. Interestingly, we have found that among all the diuretics tested only MET activates PXR. To understand the molecular mechanism underlying hPXR interactions with select drugs related to MET, docking studies and mutational analysis were carried out. This is the first report that identifies diuretic compound MET as an activator of hPXR.
2. Materials and Methods
2.1 Materials and chemicals
HepG2 liver carcinoma cells, HEK293T cells, LS180 and LS174T human intestinal epithelial cells (derived from human colorectal adenocarcinoma), were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cell culture reagents were obtained from Invitrogen (Carlsbad, CA). Anti-FLAG M2 antibody, anti–β-actin antibody, DMSO, and rifampicin were obtained from Sigma Aldrich (St. Louis, MO). Mouse monoclonal anti-CYP3A4 (K03) was previously described [32]; MDR1 and PXR (G-11) antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX). Charcoal/dextran-treated FBS was purchased from Hyclone (Logan, UT); blocking buffer and anti-mouse– and anti-rabbit–IR Dye secondary antibodies were from LI-COR Biosciences (Lincoln, NE). All other small molecules and analogs used in this study were purchased from Sigma Aldrich or Santa Cruz Biotechnology (Dallas, TX). All compounds were dissolved in DMSO as stock solution. Final DMSO concentration was either 0.28% (CYP3A4-luc and cytotoxicity assays) or 0.1% (for all other compound treatments, including two-hybrid assay, RT-PCR and Western-blotting).
2.2 Small-molecule screening in HEK293T cell–based luciferase assay
HEK293T cells (5000 cells/well; 25 μL/well) transiently transfected with FLAG-hPXR and CYP3A4-luciferase (CYP3A4-luc) were grown in 384-well, tissue culture–treated solid white plates (Corning) for 24 h prior to treatment with the drug compounds to be tested (70 nl; 10 mM in DMSO). Pintools were used to add compound to each well (final compound concentration: 28 μM; final DMSO concentration: 0.28%), and the plates were incubated for 24 h before the Dual-Glo luciferase assay (Promega) was performed and read on a EnVision microplate reader (PerkinElmer). A total of 1481 FDA-approved drug compounds obtained from various vendors (Sigma, AK Scientific, Inc., Chempacific, Toronto Research Chemicals, BIOMOL International, MP Biomedicals, BIOTREND Chemicals, LLC, Tocris Bioscience) were screened [33].
2.3 Cell culture, plasmids, and transfection
HepG2, LS180, and LS174T cells were maintained in modified Eagle’s minimal essential medium (ATCC) with 10% FBS, 2 mM L-glutamine, penicillin (100 units/mL), and streptomycin 100 μg/ml) at 37 °C in a humidified atmosphere containing 5% CO2. The pcDNA3-FLAG-hPXR (FLAG-hPXR) construct and the CYP3A4-luciferase (CYP3A4-luc) reporter were described previously [34]. The pcDNA3-FLAG-hPXR mutants were generated by Codex BioSolutions, Inc. (Gaithersburg, MD). Mutations were verified by performing nucleotide sequencing. Transfections were performed by using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer’s instructions. Human primary hepatocytes were obtained through the Liver Tissue and Cell Distribution System (Pittsburgh, PA), which is funded by NIH Contract #N01-DK-7-0004/HHSN267200700004C, and cultured in Williams’ medium E (Sigma) supplemented with primary hepatocyte maintenance supplements (Life Technologies).
2.4 Transient transfection and luciferase reporter gene assays
The methods for these procedures were described previously [20]. Briefly, the cells were transfected with Flag-hPXR, CMV-Renilla, and CYP3A4-luc plasmids by using FuGENE 6 (Roche Diagnostics). After 24 h, cells were seeded in 384-well plates (5000 cells/well) in phenol red–free medium containing 5% charcoal/dextran-treated FBS and incubated for another 24 h before treatment with compounds to be tested. Compounds were transferred by using pintools and incubated with the cells for 24 h before Dual-Glo Luciferase Assays (Promega, Madison, WI) were performed. Renilla luciferase activity was used to normalize the firefly luciferase activity. CYP3A4 promoter activity (percentage of activation; a.u.) was determined as described previously [34]. Rifampicin (7 μM) and DMSO were used as positive (100%, or a.u. = 100) and negative (0%, or a.u. = 0) controls, respectively in the dose-responsive evaluation of compound activity. Curve-fitting software (GraphPad Prism 4.0; GraphPad Software, La Jolla, CA) was used to generate the curves and to determine the EC50.
2.5 Cell viability assay
HepG2, LS180, and LS174T cells were transiently transfected with FLAG-hPXR and treated with compounds as described in Section 2.4 before the CellTiter-Glo assay (Promega) was used to measure cell viability. Briefly, CellTiter-Glo reagent was added to the wells and incubated at room temperature for 10 min in the dark. Luminescence was measured and recorded by using an Envision plate reader (PerkinElmer Life Sciences). DMSO was used as a negative control for cell viability. Values of the viability of compound-treated cells were expressed as a percentage of that of DMSO-treated cells.
2.6 Mammalian two-hybrid assay
The CheckMate mammalian two-hybrid system (Promega) was performed as described previously [20] and consists of VP16-hPXR, Gal4-SRC-1, and a luciferase reporter (pG5-luc) co-transfected into HepG2 cells. The Gal4 vector (pBIND) also constitutively expresses Renilla luciferase, which was used as an internal transfection control. The Dual-Glo Luciferase Assay (Promega) was used to measure luciferase activity as an indicator of protein–protein interactions. The relative luciferase activity for pG5-luc was determined by normalizing firefly luciferase activity with Renilla luciferase activity.
2.7 Western blot analysis
All cell extracts were harvested in 1X RIPA buffer (Cell Signaling Technology, Inc.; Danvers, MA), and samples were centrifuged at 12,000g at 4°C for 25 min. The samples were then boiled in sample loading buffer (Invitrogen) containing SDS, and equal amounts of samples were resolved on a 4–12% SDS-PAGE gradient gel and then transferred onto a nitrocellulose membrane. Unbound sites on the membrane were blocked, and the membrane was incubated with the indicated antibodies overnight at 4°C. We used anti-CYP3A4, anti-MDR1, anti-PXR, anti-FLAG M2 (1:1000 dilutions) and anti-β-actin (1:5000 dilution) antibodies to detect CYP3A4, MDR1, PXR, FLAG-PXR and β-actin, respectively. All Western blot analyses were performed on the Odyssey Infrared Imaging system (LI-COR Biosciences; Lincoln, NE). The intensity of each protein band was quantified using ImageJ 1.48 software [35]. The intensity of each protein band was normalized to that of actin to generate the relative intensity, with the relative intensity of the DMSO treated sample set as “1”.
2.8 RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR) assays
Total RNA was isolated from LS180 cells, LS174T cells, and human primary hepatocytes by using Maxwell 16 LEV simplyRNA purification kits (Promega). Then, qRT-PCR was performed by using Taqman gene expression assays (Applied Biosystems, Carlsbad, CA) specific for CYP3A4, MDR1, and β-actin (ACTB), which was used as the reference gene according to the manufacturer’s protocol in an ABI 7900HT system (Applied Biosystems). The comparative Ct method was used for relative quantification for gene expression with the following formula: ΔCt = Ct (test gene) — Ct (ACTB); ΔΔCt (test gene) = ΔCt (test gene in treatment group) — ΔCt (test gene in vehicle or siPXR control group); the fold changes of mRNA = 2−ΔΔCt, which indicated the relative mRNA level of the corresponding transcript to that of the control samples. DMSO (solvent control) or siPXR and rifampicin were used as negative and positive control, respectively.
2.9 Small interfering RNA transfection
We knocked down hPXR expression by transiently transfecting small interfering RNA (siRNA) into cells by using the previously described protocol [36]. Briefly, LS180 cells stably expressing hPXR and CYP3A4-luc were cultured in 6-well plates and treated with a final concentration of 25 nM ON-Targetplus SMARTpool SiRNA targeting PXR (L-003415, Thermo Fisher Scientific, Waltham, MA) or Nontargeting Pool (D-001810, Thermo Fisher Scientific) by using Lipofectamine RNAiMAX (Life Technologies). After 72 h, the cells were washed, treated with indicated compounds for 48 h and then collected for qRT-PCR analysis.
2.10 Competitive ligand-binding assay
A LanthaScreen TR-FRET PXR competitive binding assay was conducted according to the manufacturer’s protocol as described previously [34]. Briefly, assays were performed in a volume of 20 μL in 384-well solid black plates (Corning) with 5 nM GST-hPXR ligand-binding domain, 40 nM fluorescent-labeled hPXR agonist (Fluomore PXR Green, Invitrogen), 5 nM terbium-labeled anti-GST antibody, and test compound at different concentrations. The reactions were incubated at 25°C for 60 min before the fluorescent emission of each well at 495 and 520 nm was measured by using a 340-nm excitation filter, 100-μs delay time, and 200-μs integration time on a PHERAStar plate reader (BMG Labtech, Durham, NC). The curve-fitting software GraphPad Prism 4.0 was used to generate the plot.
2.11 Molecular modeling and docking studies
Receptor files, ligands, and docking parameter files were prepared by using Molecular Operating Environment (MOE) [37]. The coordinates of the PXR-LBD-rifampicin X-ray crystal structure were taken from the Protein Data Bank (PDB: 1SKX) [38]. The crystal structure contained 2 PXR LBDs and 2 rifampicin molecules because PXR forms dimers in solution. One PXR-LBD-rifampicin complex was used for the docking studies. All hydrogen atoms and partial charges were added to the protein by using protonate 3D. The energy of the PXR-LBD-rifampicin complex molecule was minimized by using an energy minimization algorithm that uses the MMFF94x force field. The energy-minimized structure was used as the template for the docking studies. We chose the rifampicin binding site (active site) as our pharmacophore for docking studies. The placement of the small-molecule ligand was determined by the pharmacophore and rescored by using London DG. The placements of the ligands were refined again by force field. Finally, a three-dimensional pharmacophore model was generated by using MOE. Compounds were selected on the basis of scoring function (binding energy). The best conformation for each ligand was isolated on the basis of the S score, which measures interactions. The docking results were analyzed and the figures created in MOE.
2.12 Statistical analysis
Results are expressed as the mean ± standard deviation of at least 3 independent experiments (in at least triplicate for each experiment unless specified differently), and error bars indicate the standard deviation. Statistical analyses were performed using Student’s t-test. Differences were considered statistically significant for p ≤ 0.05 (*).
3. Results
3.1 Small-molecule screening of FDA-approved drugs identifies MET as a novel PXR activator
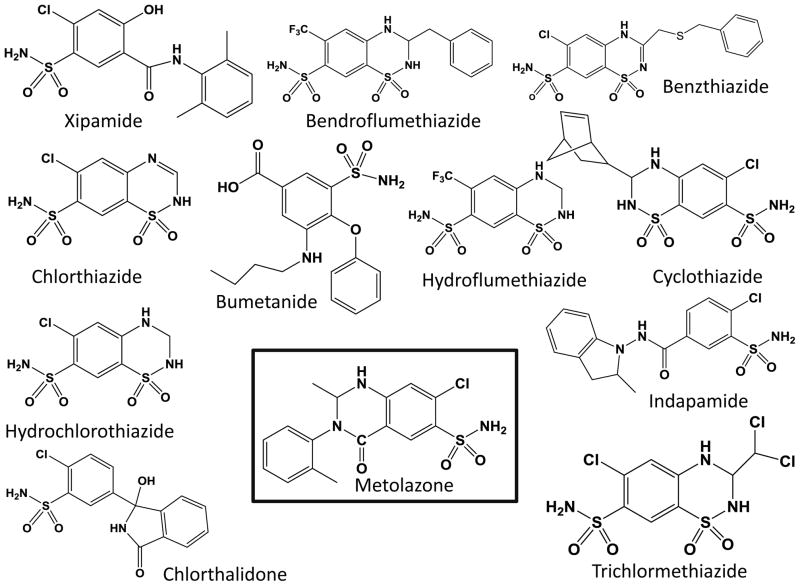
In total, 1481 FDA-approved drugs were screened in HEK293T cells transiently transfected with hPXR, CYP3A4-luc, and CMV-Renilla as described previously [20]. Our screen identified metolazone (MET) as an activator of PXR. We also found PXR activation by other drugs that have been previously published, including rifampicin [39], nimodipine [40], phenylbutazone [41], efavirenz [42, 43], montelukast [44], and diclofenac [45]. Analysis of the screening data showed that rifampicin and MET induced 193% and 182% activation of the CYP3A4-luc reporter (DMSO set as 0% and rifampicin at 5 μM set as 100%). Here, we have focused on PXR’s interaction with the diuretic drug MET. We also investigated other structurally and functionally related diuretics for their possible roles in PXR-mediated drug metabolism and clearance. Chemical structures of the compounds tested here are shown in Fig. 1.
Fig. 1. Structures of compounds selected for analysis.
Metolazone, identified as an activator of hPXR in the luciferase assay screening, is shown in the black box.
3.2 Functional characterization of MET in PXR-mediated pathways
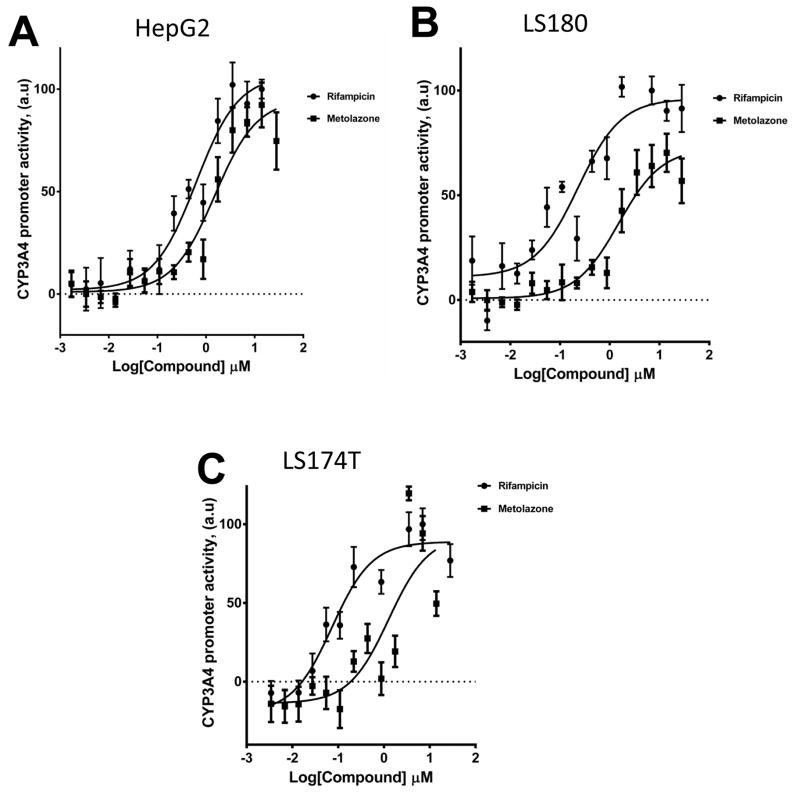
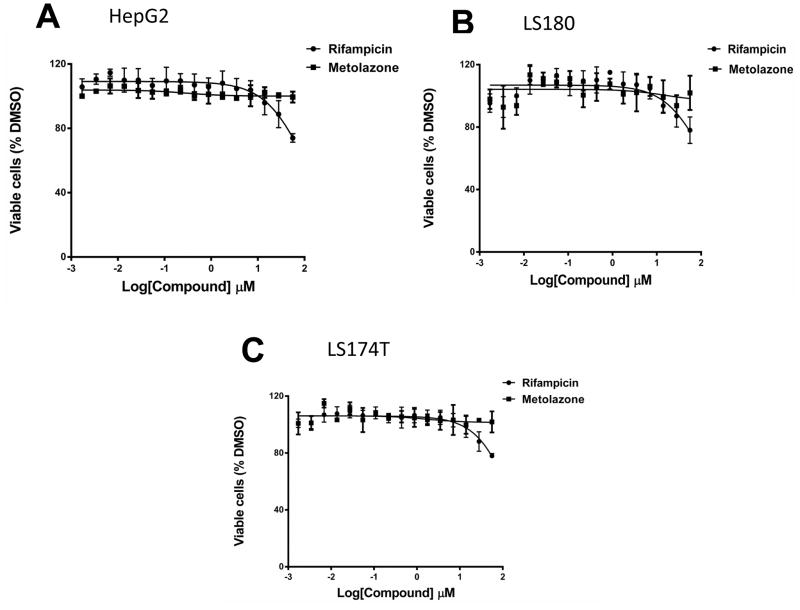
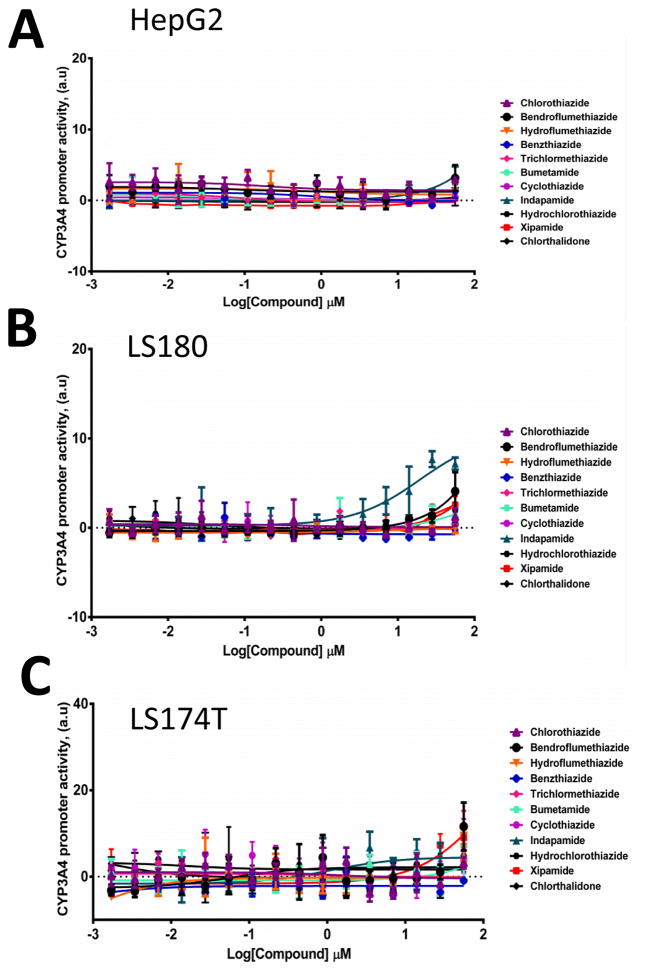
We used HepG2 human hepatocellular carcinoma cells and LS180 and LS174T human intestine cell lines to evaluate the agonistic or antagonistic activity of MET and other diuretics for the hPXR-regulated CYP3A4 promoter. MET was an activator of PXR activity (Fig. 2), with an estimated EC50 value of 0.7 μM to 1.5 μM (Table 1), and was not significantly cytotoxic even at high concentration (Fig. 3). Other diuretics did not noticeably activate PXR (Fig. 4).
Fig. 2. MET activates the PXR-regulated CYP3A4 promoter.
(A) HepG2, (B) LS180, and (C) LS174T cells transiently transfected with hPXR, CYP3A4-luc, and CMV-Renilla were treated for 24 h with indicated concentrations of rifampicin or MET prior to luciferase assay.
Table 1.
Rifampicin and MET activities in CYP3A4 promoter luciferase reporter assays
| Compounds | HepG2 (EC50, μM) | LS 180 (EC50, μM | LS174T (EC50, μM) |
|---|---|---|---|
| Rifampicin | 0.62 | 0.22 | 0.11 |
| Metolazone | 1.50 | 1.5 | 0.70 |
Fig. 3. Cytotoxicity of MET in (A) HepG2, (B) LS180, and (C) LS 174T cells.
Cells were treated with increasing concentrations (1.7 nM to 56 μM) of MET.
Fig. 4. MET-related diuretic compounds do not significantly activate the PXR-regulated CYP3A4 promoter.
(A) HepG2, (B) LS180, and (C) LS174T cells transiently transfected with hPXR, CYP3A4-luc, and CMV-Renilla were treated for 24 h with indicated concentrations of rifampicin or indicated diuretics prior to luciferase assay.
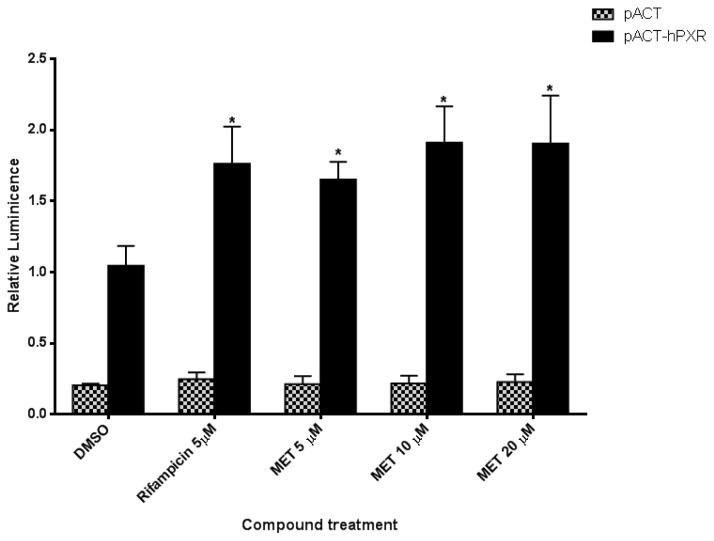
To understand the mechanism responsible for MET activation of hPXR, we performed a mammalian two-hybrid assay in which the interaction between hPXR and steroid receptor coactivator-1 (SRC-1) that is prompted by a PXR agonist such as rifampicin causes a change in specific reporter activity [46]. As shown in Fig. 5, similar to rifampicin, MET substantially increased the interaction between hPXR and SRC-1, which is consistent with the CYP3A4 promoter luciferase reporter assay data in which both rifampicin and MET activate CYP3A4 promoter (Fig. 2). These data suggest that MET enhanced the interaction of hPXR with SRC-1 to increase the activity of hPXR.
Fig. 5. Mammalian two-hybrid assay confirms that compounds affect the interaction of wild-type hPXR with SRC-1.
Mammalian two-hybrid assays were performed in HepG2 cells transiently cotransfected with plasmids encoding Gal4-SRC-1 and the reporter gene pG5-luc, together with either pACT-hPXR or empty vector pACT as indicated. The cells were treated with DMSO; 5 μM rifampicin; or 5 μM, 10 μM, or 20 μM MET. Luciferase assays were performed 24 h after the compound treatment. The relative luminescence for pG5-luc was determined by normalizing firefly luciferase activity to Renilla luciferase activity. The values represent the means of five independent experiments, and the bars denote the SD (*p < 0.05; in the t-test comparisons were made between compound-treated and DMSO-treated samples in which pACT-hPXR was transfected).
3.3 MET induced CYP3A4 and MDR1 expression in human hepatocytes and intestine cells
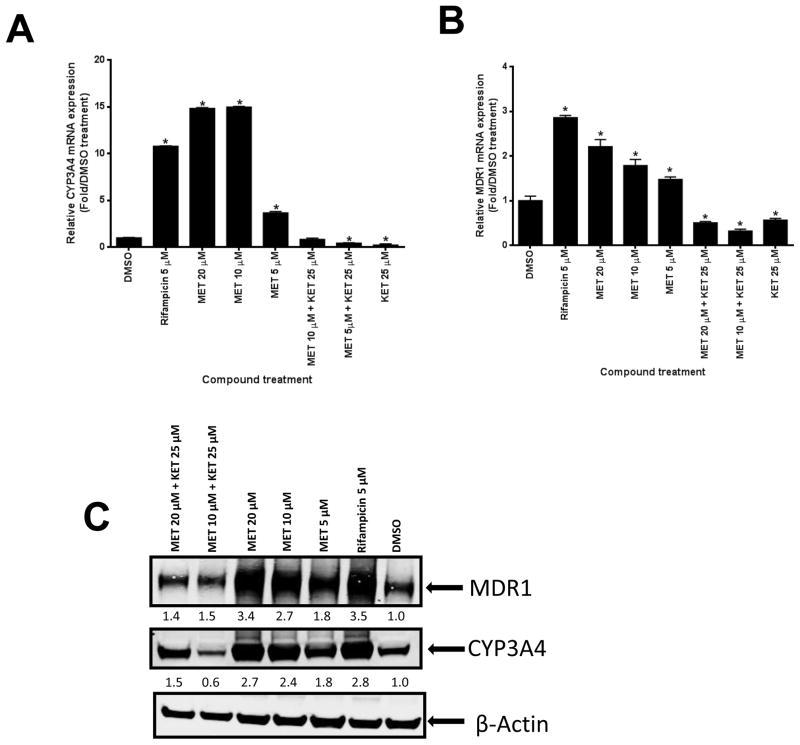
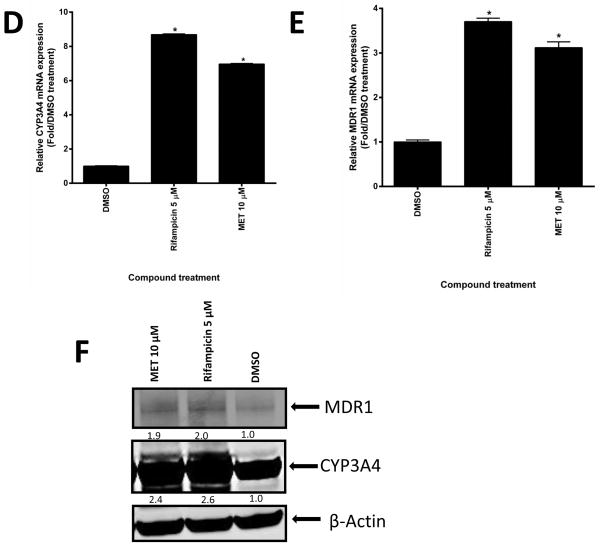
Induction of CYP3A4 and MDR1 expression was first assessed in human hepatocytes. As shown in Fig. 6A–C, MET induced the expression of CYP3A4 and MDR1 mRNA and protein. As expected, the hPXR inhibitor ketoconazole (KET) substantially reduced MET-induced CYP3A4 and MDR1 expression at both the mRNA and protein levels. The induction of CYP3A4 and MDR1 expression by MET was confirmed in human primary hepatocytes from a different donor (Fig. 6D–F). Overall, our data suggested that MET induced CYP3A4 and MDR1 expression in human primary hepatocytes.
Fig. 6. MET induced mRNA and protein expression of CYP3A4 and MDR1 in human hepatocytes.
(A–C: donor 1) (A) Human CYP3A4 and (B) MDR1 mRNA expression were analyzed by real-time PCR assays of human hepatocytes after 48-h treatment with different compounds as indicated (*p < 0.05; in the t-test, comparisons were made between compound-treated and DMSO-treated samples). (C) CYP3A4 and MDR1 protein levels in human hepatocytes were determined by Western blotting using anti-CYP3A4 and anti-MDR1 sequentially. The numbers below the protein bands indicate the relative intensity of the protein bands, with the DMSO treated sample set as “1”. (D–F: donor 2) (D) Human CYP3A4 and (E) MDR1 mRNA expression were analyzed as described in (A) and (B) (*p < 0.05; in the t-test, comparisons were made between compound-treated and DMSO-treated samples). (F) CYP3A4 and MDR1 were detected using anti-CYP3A4 and anti-MDR1 simultaneously, and quantified as described in (C). Anti-β-actin was finally used to detect β-actin in both (C) and (F).
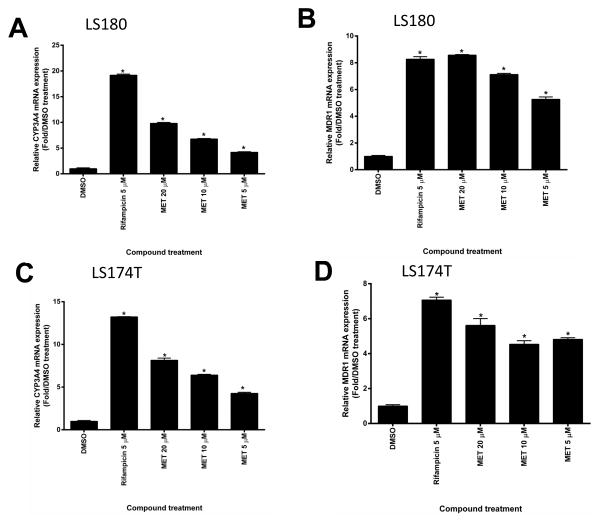
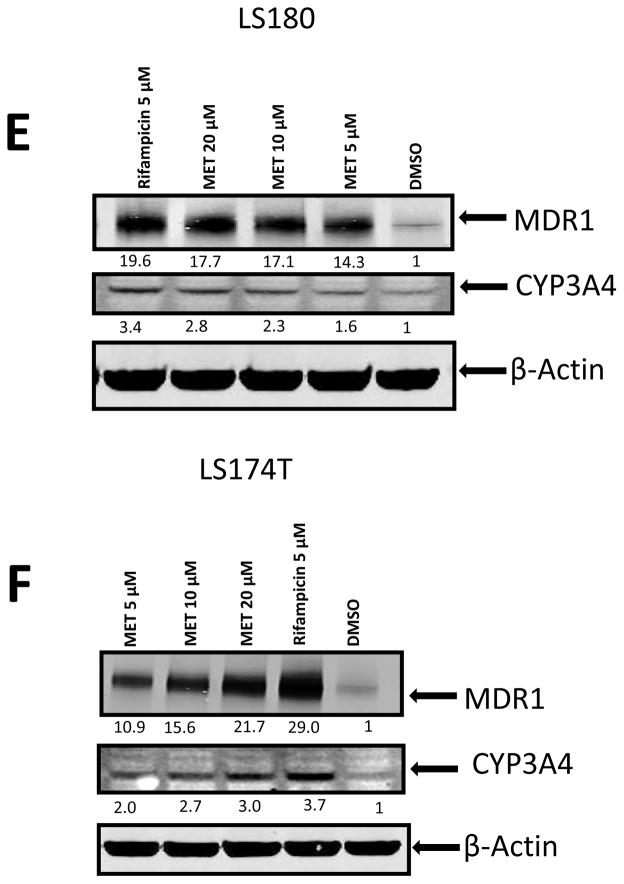
We also tested the effects of MET in human intestine cells LS174T and LS180 (Fig. 7). We chose to use these cell lines because they express endogenous hPXR which is ligand inducible and have been previously used to study the function of hPXR [4]. Similar to its induction effect in human primary hepatocytes, MET induced the expression of both CYP3A4 and MDR1 mRNA in LS180 (Fig. 7A and B) and LS174T (Fig. 7C and D) human intestine cells. Our data also showed that MET induced CYP3A4 and MDR1 protein expression consistent with the mRNA expression profile in both LS 180 cells (Fig. 7E) and LS 174T cells (Fig. 7F).
Fig. 7. MET induced mRNA and protein expression of CYP3A4 and MDR1 in LS180 and LS174T intestinal cells.
Real-time PCR analysis of (A) CYP3A4 and (B) MDR1 mRNA expression in LS180 cells and (C) CYP3A4 and (D) MDR1 mRNA expression in LS174T cells after 48-h treatment with different compounds as indicated (*p < 0.05; in the t-test comparisons were made between compound-treated and DMSO-treated samples). Western blot showing CYP3A4 and MDR1 protein levels in (E) LS180 and (F) LS174T cells upon MET treatment. The experiments were repeated two times and the data show one representative experiment. The numbers below the protein bands indicate the relative intensity of the protein bands, with the DMSO treated sample set as “1”. In both (E) and (F), anti-CYP3A4 and anti-MDR1 were used to detect CYP3A4 and MDR1 sequentially. Anti-β-actin was finally used to detect β-actin.
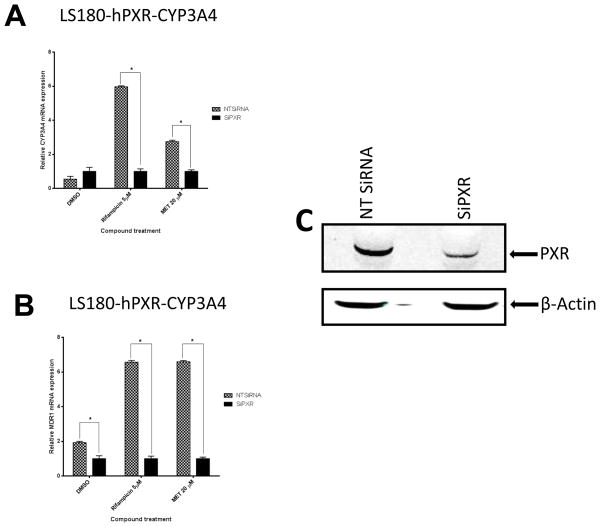
To determine whether MET’s induction of hPXR target gene expression was hPXR-dependent, we performed hPXR knockdown experiments. In order to clearly observe the effect of hPXR knockdown, we need a cellular system that has high levels of hPXR and hPXR-inducible target genes CYP3A4 and MDR1. As previously described LS180-hPXR-CYP3A4-luc cells express ectopic hPXR and have higher levels of CYP3A4 and MDR 1 in response to hPXR agonists [4]. In treated LS180-hPXR-CYP3A4-luc cells, when hPXR expression was knocked down by using siRNA, the induction of PXR target genes CYP3A4 and MDR1 mRNA levels were substantially diminished compared to non-targeting control siRNA upon indicated compound treatment (Fig. 8A–B). The efficiency of hPXR knockdown (>80%) in LS180-hPXR-CYP3A4-luc cells is shown in Fig. 8C. Therefore, our data suggests that MET induction of CYP3A4 and MDR1 is hPXR-dependent.
Fig. 8. Knockdown of hPXR expression abolished MET-induced CYP3A4 and MDR1 expression in LS180-hPXR-CYP3A4-luc cells.
Human (A) CYP3A4 and (B) MDR1 mRNA expression levels were determined in LS180-hPXR-CYP3A4-luc cells transiently transfected with hPXR siRNA (siPXR) or non-target control siRNA (NTsiRNA) before 48-h treatments with the indicated compounds. (*p < 0.05; in the t-test comparisons were made between siPXR-treated and NTsiRNA-treated samples for each treatment group). (C) Protein expression profile of hPXR from one representative sample in LS180-hPXR-CYP3A4-luc cells upon siRNA knockdown of hPXR expression. Anti-PXR and anti-β-actin were used to detect PXR and β-actin, respectively.
3.4 Structure-activity relationship and selectivity of MET
To investigate the biological role of diuretics on hPXR-regulated pathways and gain insight into the SAR, several thiazide and non-thiazide diuretics were tested (Fig. 1). We found that none of the diuretics except MET substantially activated PXR-regulated CYP3A4 promoter activity in both liver cells and intestine cells (Fig. 2 and 4), suggesting that only MET might bind to and activate hPXR. Analysis of related structures showed an ortho methyl phenyl ring occupying a groove where the phenyl ring is stabilized by hydrophobic residues of PXR’s LBD. However, compounds such as chlorthiazide and hydrochlorothiazide that lack the phenyl ring are inactive and contain a sulfone instead of carbonyl (-CO) on a ring structure. Hydrochlorothiazide and chlorthiazide have molecular architecture similar to that of MET but are missing the functional groups present in MET, further suggesting a tight SAR.
3.5 Molecular modeling and ligand binding studies in vitro explain the binding and mechanism of interaction of MET with PXR’s LBD
Molecular modeling data show that MET binds in the LBD of PXR with the highest binding score of −8.9 kcal/mol. Docking studies show that MET might have more than one orientation in the LBD because it is comparatively small in size. The best 2 orientations of MET binding are shown in Fig. 9A and B, and the binding energies are −8.9 and −8.1 kcal/mol. Molecular docking data also showed that the phenyl ring attached to ring nitrogen does not change its position substantially, but the rest of the molecule does. Additionally, the cyclothiazide molecule has a binding pattern similar to that of MET, but cyclothiazide does not activate PXR, indicating that the phenyl ring is important for the interaction with PXR. The flexibility of the phenyl ring comes from an extra –CH2 group between the ring carbon and phenyl, making the molecule inactive as shown in bendroflumethiazide. Docking data also showed that, unlike MET, cyclothiazide does not form any hydrogen bond with the polar amino acid residues of PXR. Therefore, our molecular modeling data explain the tight SAR observed for MET.
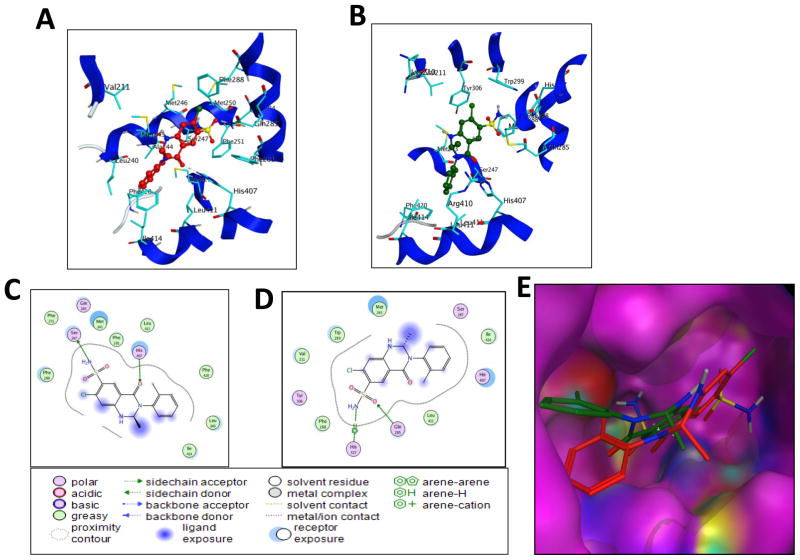
Fig. 9. Binding orientations of MET at the PXR LBD binding site.
(A) Docking mode three-dimensional (3D) interaction scheme of predicted binding orientations of MET (shown in red) at the PXR LBD binding site (Orientation 1). (B) Docking mode 3D interaction scheme of another predicted binding orientation of MET (shown in green) at the PXR LBD binding site (Orientation 2). (C) Docking mode two-dimensional (2D) interaction scheme of predicted binding pose of MET at the PXR LBD binding site (Orientation 1). (D) Docking mode 2D interaction schemes of predicted binding pose of MET at the PXR LBD binding site (Orientation 2). (E) Surface representation of the PXR LBD bound to MET in a different orientation. MET orientations are shown in stick with green and red color.
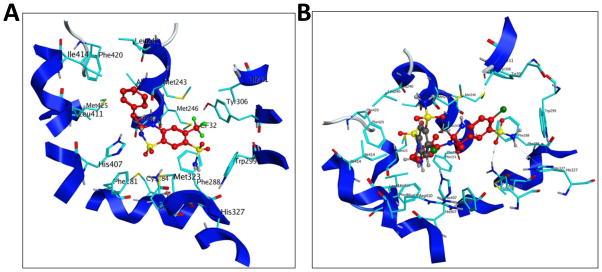
From the binding orientation, it is evident that MET contacts with mostly hydrophobic amino acid residues of PXR, with a couple of hydrogen bonds with polar amino acid residues for each orientation of MET. Crystallographic analysis of all the PXR-ligand structures previously published show a similar mode of binding. Our docking results showed that the following PXR residues are important for MET interactions; Phe 288, Trp 299, Met 246, Met 243, Lys 210, His 327, Ser 247, Val 211, Tyr 306, Gln 285, Met 323, Leu 239, Leu 240, His 407, Phe 281, Lys 210, Leu 324, Phe 251, Phe 281, and Phe 420 (Fig. 9). The amino acids of PXR’s LBD that form hydrogen bonds with MET are Gln 285, His 407, and Ser 247. Docking mode 3D interactions of binding sites of PXR’s LBD with MET orientations are shown in Fig. 9A and B, and docking mode two-dimensional (2D) interaction schemes of predicted binding poses of MET are shown in Fig. 9C and D. A comparison of the best 2 orientations of MET are shown in the binding site of the surface representation of PXR’s LBD (Fig. 9E). The docking structure of bendroflumethiazide with PXR’s LBD showed that the phenyl ring attached to the ring is upward, not sitting in the groove created by hydrophobic residues Arg410, Leu411, Ile414, His407, Met 243, Leu 240, and Phe 420 (Fig. 10A). A comparison of the docking mode of MET and the related compound hydrochlorothiazide showed that hydrochlorothiazide does not satisfy the requirement to fit in the LBD, as it mostly sits in the same space as the phenyl ring attached to the bicyclic ring, leaving the rest of the binding site open and resulting in no interaction with PXR (Fig. 10B).
Fig. 10. Binding mode of bendroflumethiazide and comparison of the binding modes of MET and thiazide diuretic hydrochlorothiazide to PXR’s LBD.
(A) Docking mode three-dimensional (3D) interaction scheme of predicted binding orientation of bendroflumethiazide (shown in red) at the PXR LBD binding site. (B) Comparison of the binding mode of MET (red) and hydrochlorothiazide (grey) to PXR’s LBD.
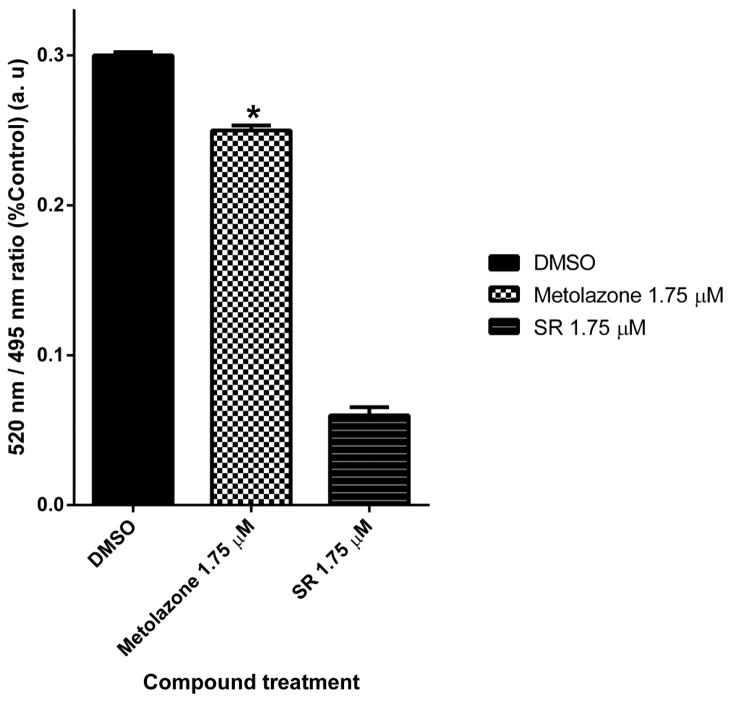
To test whether hPXR-LBD directly binds to MET, we used a cell-free competitive hPXR-LBD binding assay in which a fluorescein-labeled PXR ligand is used as a tracer. At higher concentrations, MET itself is fluorescent at the wavelength used for the assay. Therefore, it was not feasible to determine the IC50 values of MET-PXR interactions in this experiment. However, at the 1.75 μM concentration, it decreased tracer binding to hPXR-LBD substantially compared with vehicle control (Fig. 11), indicating that MET might directly bind to hPXR.
Fig. 11. Binding of MET to PXR’s LBD in a TR-FRET competitive binding assay.
SR12813 (SR) was used as a control. (*p < 0.05; in the t-test comparisons were made between Metolazone-treated and DMSO-treated samples).
3.6 MET modulates the activity of critical hPXR mutants differently
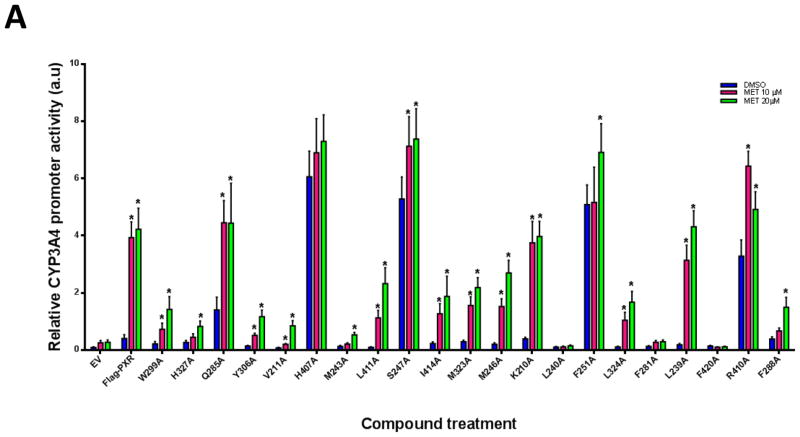
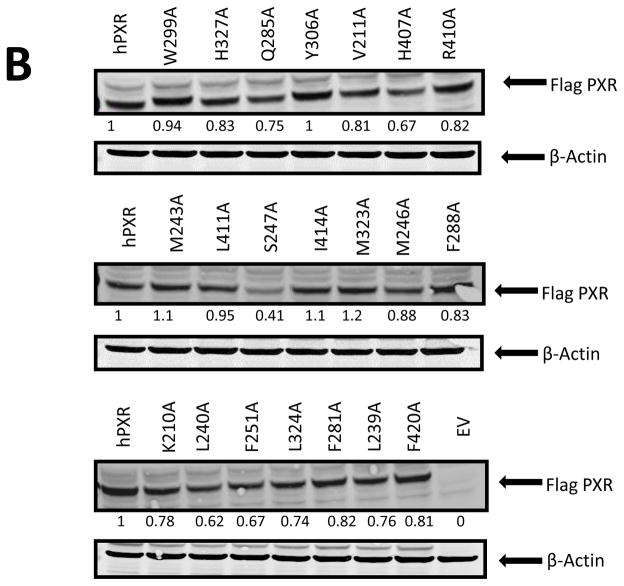
Molecular modeling studies showed that ligands can bind in different orientations and interact with various residues of PXR. To experimentally confirm the docking data and the interactions between MET and the identified PXR residues, we individually mutated critical PXR amino acids to alanine and then performed CYP3A4 promoter luciferase reporter assays in HepG2 cells. The following hPXR mutants were tested: W299A, H327A, Q285A, Y306A, V211A, H407A, M243A, S247A, L411A, I414A, M323A, M246A, K210A, L240A, F251A, L324A, F281A, L239A, F420A, R410A, and F288A. All the mutants were tested at various ligand concentrations (Fig. 12A). Our results showed that MET increased CYP3A4 promoter activity substantially for all PXR mutants except L240A, F281A, and F420A, which are loss-of-function mutants. The basal activities (the CYP3A4 promoter activity in the presence of DMSO) of H407A, S247A, F251A and R410A are substantially higher than that of the wild-type hPXR. While all 4 mutants (H407A, S247A, F251A and R410A) are induced by MET to a level similar to or higher than that of the wild-type hPXR, their folds of induction (MET-induced CYP3A4 promoter activity/DMSO-treated CYP3A4 promoter activity) are substantially lower than the wild-type hPXR. For example, in response to DMSO, 10 and 20 μM of MET, the CYP3A4 promoter activities are 0.40, 3.93, and 4.22 for wild-type hPXR, and 3.28, 6.43, and 4.92 for R410A, and the folds induction for 10 and 20 μM of MET are 9.83 and 10.55 for wild-type hPXR and 1.96 and 1.50 for R410A. The decrease in fold induction is more substantial for H407, S247A and F251A (analysis not shown). Q285A also has higher basal and lower net MET-induced activity. In addition, H327A, M243A, L411A, L239A, W299A, Y306A, V211A, I414A, M323A, M246A, L324A, and F288A showed either altered basal or MET-induced activities. However, when considered the lower basal activity, the activity induced by 20 μM of MET remains very similar to wild-type hPXR for the following mutants: W299A, Y306A, V211A, I414A, M323A, L324A, and M246A. Particularly, K210A behaves very similar to the wild-type hPXR in both basal and MET-induced activities. Although W299, Y306, V211, I414, M323, M246, L324A, and K210 were predicted from the docking studies to be critical for MET binding, mutation of these residues to alanine, particularly that of K210, did not noticeably alter their ability to be induced by MET. Western blot analysis in HepG2 cells showed that all the PXR mutants expressed comparable protein levels upon 20 μM MET treatment except S247A, which expressed a noticeably lower protein level (Fig. 12B). These results experimentally reveal the functional consequence when the residues predicted to be critical for MET binding are mutated, and provide additional insight into the direct interactions of MET with PXR LBD.
Fig. 12. Comparison of CYP3A4 promoter activation by hPXR or hPXR mutants upon MET treatment.
(A) HepG2 cells transiently transfected with CYP3A4-luc and CMV-Renilla together with either empty vector (EV), wild-type hPXR (Flag-hPXR), or hPXR mutant were treated for 24 h with indicated concentrations of MET prior to luciferase assay. The relative luciferase units (a.u.) were determined by normalizing, with the Renilla luciferase used as the control (*p < 0.05; in the t-test comparisons were made between compound-treated and DMSO-treated samples for each construct, either wild-type or mutated hPXR). (B) The expression of wild-type and mutated Flag-hPXR after 20-μM MET treatment. Actin expression level was used to verify equal loading of lysates. The experiments were repeated two times and the data show one representative experiment. The numbers below the protein bands indicate the relative intensity of the protein bands, with the wild-type Flag-PXR sample set as “1”. Anti-FLAG M2 and anti-β-actin were used to detect Flag-PXR and β-actin, respectively.
4. Discussion
PXR is recognized for its role in regulating drug metabolism and disposition by regulating mostly CYP3A4 and MDR1 expression. Therefore, activation of PXR by small molecules including marketed drugs may cause drug-drug interactions. Drug-drug interactions or the generation of toxic levels of a drug metabolites result from enhanced drug metabolism might cause the failure of drug development and clinical therapy [47], even withdrawal of drugs from the market due to rare incidences [48]. Identification of currently used FDA approved drugs that modulate PXR and subsequently, understanding drug-drug interactions will improve therapeutic outcomes. To our knowledge, this is first report of MET that increases CYP3A4 and MDR1 expression by interacting with hPXR.
Xenobiotic sensor PXR has a promiscuous ligand binding site and can accommodate structurally diverse ligands. Our data showed that MET-mediated activation of PXR is very selective as both structurally and functionally related compounds were unable to induce PXR target genes. Other diuretics studied here did not substantially activate PXR, indicating that there is no cross-talk between PXR signaling and the pathways that are biologically modulated by diuretics. SAR and molecular modeling data also showed that bicyclic system with different functional groups and their positions are important for PXR recognition. It is worthy to note that -CO instead of sulfone (-SO2-) might also be crucial for PXR binding activity. All the compounds except MET have a sulfonamide group as part of the ring, which may render these compounds inactive. Although bendroflumethiazide and benzthiazide compounds have phenyl rings, these phenyl groups are separated by at least one methylene group, making them inactive. Distance separation between the bicyclic ring and phenyl group indicates the specificity of phenyl ring position to be on the nitrogen on the bicyclic ring. Xipamide is similar to MET in terms of functional groups. However, because xipamide’s -CO group is not locked in a bicyclic ring, which causes free rotation of the amide bond, the compound does not possess PXR-activating activity, indicating that a bicyclic system is required for PXR binding activity. Therefore, SAR study showed that functional groups and position of substitutions are very critical for binding and activation of PXR. For a better understanding of SAR, further work will be required using various MET analogs. Overall, phenyl ring, sulfone and -CO functional groups are crucial for PXR specific recognition and biological activity. Also, it was observed that phenyl group position should be on the bicyclic nitrogen. Together, our findings showed that although PXR has a promiscuous ligand binding pocket, it is highly selective to MET instead of other compounds that are either structurally or functionally related to MET. Given that mutation to alanine of several residues predicted to be critical for binding of MET to hPXR did not alter their ability to be induced by MET, it is possible that MET binds in different orientations in PXR LBD. Similar results were shown previously for SR12813 which binds at three different orientations in PXR LBD [8]. Our docking studies showed that Phe 420 and Leu 240 residues of hPXR are very critical for the interaction process with MET. Indeed, both F420A and L240A are complete loss-of-function mutants as seen in the mutagenesis study. Mutants of polar amino acid residues such as Q285A, H407A, S247A and charged amino acid residue R410A substantially enhance their basal activity and decrease their MET-inducibility accordingly. Understanding the structural basis of the interaction between MET and hPXR may enable more-informed drug design of compounds in the future.
It is already advised that MET should be used cautiously in patients with liver disease [27]. Our discovery that MET induces CYP3A4 and MDR1 through PXR may also have important clinical implications for MET’s use, as activation of PXR may cause drug-drug interactions. Similar undesired effects of drugs via PXR have been reported previously. For example, co-treatment of patients with St. John’s wort and other drugs caused failure of the therapeutic drugs, mainly due to activation of PXR by St. John’s wort, which causes CYP3A4 induction resulting in enhanced drug metabolism [49, 50]. Given that xenobiotics activate hPXR by post-translational modifications such as phosphorylation [34, 51], further investigation is needed to determine whether and how MET could indirectly activate hPXR in addition to the direct-binding mechanism we report here.
In summary, we identified the diuretic drug MET that induces mRNA and protein expression of CYP3A4 and MDR1 by activating PXR and may also recruit the PXR coactivator SRC-1. We also identified different functional groups of MET that are important for PXR activation. These results will help our understanding of MET’s role in drug metabolism. The discovery of MET as an activator of hPXR suggests that it should be used cautiously in patients undergoing combination therapy.
Acknowledgments
This work was supported by the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children’s Research Hospital, National Institutes of Health National Institute of General Medical Sciences [Grants GM086415 & GM110034], and National Institutes of Health National Cancer Institute [Grant P30-CA21765]. Human primary hepatocytes were obtained through the Liver Tissue and Cell Distribution System (Pittsburgh, PA), which is funded by NIH Contract #N01-DK-7-0004/HHSN267200700004C. We thank Jing Wu, Dr. Sergio Chai, Asli Goktug, Dr. Su Sien Ong, and Dr. Rajendra Tangallaphy for their help and many valuable suggestions and discussions, and we thank Dr. Cherise Guess for editing the manuscript.
Footnotes
Conflict of interest
The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 2.Evans RM. The nuclear receptor superfamily: a Rosetta Stone for physiology. Mol Endocrinol. 2005;19:1429–38. doi: 10.1210/me.2005-0046. [DOI] [PubMed] [Google Scholar]
- 3.Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259–66. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y-M, Lin W, Chai SC, Wu J, Ong SS, Schuetz EG, et al. Piperine activates human pregnane X receptor to induce the expression of cytochrome P450 3A4 and multidrug resistance protein 1. Toxicol Appl Pharmacol. 2013;272:96–107. doi: 10.1016/j.taap.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang TK. Activation of pregnane X receptor (PXR) and constitutive androstane receptor (CAR) by herbal medicines. AAPS J. 2009;11:590–601. doi: 10.1208/s12248-009-9135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.di Masi A, De Marinis E, Ascenzi P, Marino M. Nuclear receptors CAR and PXR: molecular, functional, and biomedical aspects. Mol Aspects Med. 2009;30:297–343. doi: 10.1016/j.mam.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Staudinger JL, Ding X, Lichti K. Pregnane X receptor and natural products: beyond drug-drug interactions. Expert Opin Drug Metab Toxicol. 2006;2:847–57. doi: 10.1517/17425255.2.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–33. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- 9.Watkins RE, Noble SM, Redinbo MR. Structural insights into the promiscuity and function of the human pregnane X receptor. Curr Opin Drug Discov Devel. 2002;5:150–8. [PubMed] [Google Scholar]
- 10.Watkins RE, Maglich JM, Moore LB, Wisely GB, Noble SM, Davis-Searles PR, et al. 2.1 A crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry. 2003;42:1430–8. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- 11.Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J Mol Biol. 2003;331:815–28. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 12.Xue Y, Moore LB, Orans J, Peng L, Bencharit S, Kliewer SA, et al. Crystal structure of the pregnane X receptor-estradiol complex provides insights into endobiotic recognition. Mol Endocrinol. 2007;21:1028–38. doi: 10.1210/me.2006-0323. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee M, Robbins D, Chen T. Modulation of xenobiotic receptors by steroids. Molecules. 2013;18:7389–406. doi: 10.3390/molecules18077389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani S, Huang H, Sundarababu S, Liu W, Kalpana G, Smith AB, et al. Activation of the steroid and xenobiotic receptor (Human pregnane X receptor) by nontaxane microtubule-stabilizing agents. Clin Cancer Res. 2005;11:6359–69. doi: 10.1158/1078-0432.CCR-05-0252. [DOI] [PubMed] [Google Scholar]
- 15.Venkatesh M, Wang H, Cayer J, Leroux M, Salvail D, Das B, et al. In vivo and in vitro characterization of a first-in-class novel azole analog that targets pregnane X receptor activation. Mol Pharmacol. 2011;80:124–35. doi: 10.1124/mol.111.071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekins S, Erickson JA. A pharmacophore for human pregnane X receptor ligands. Drug Metab Dispos. 2002;30:96–9. doi: 10.1124/dmd.30.1.96. [DOI] [PubMed] [Google Scholar]
- 17.Chen T. Nuclear receptor drug discovery. Curr Opin Chem Biol. 2008;12:418–26. doi: 10.1016/j.cbpa.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Ihunnah CA, Jiang M, Xie W. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim Biophys Acta. 2011;1812:956–63. doi: 10.1016/j.bbadis.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krasowski MD, Ni A, Hagey LR, Ekins S. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR. Mol Cell Endocrinol. 2011;334:39–48. doi: 10.1016/j.mce.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee M, Chen T. Differential regulation of CYP3A4 promoter activity by a new class of natural product derivatives binding to pregnane X receptor. Biochem Pharmacol. 2013;86:824–35. doi: 10.1016/j.bcp.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie W, Uppal H, Saini SP, Mu Y, Little JM, Radominska-Pandya A, et al. Orphan nuclear receptor-mediated xenobiotic regulation in drug metabolism. Drug Discov Today. 2004;9:442–9. doi: 10.1016/S1359-6446(04)03061-2. [DOI] [PubMed] [Google Scholar]
- 22.Veith H, Southall N, Huang R, James T, Fayne D, Artemenko N, et al. Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries. Nat Biotechnol. 2009;27:1050–5. doi: 10.1038/nbt.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Tang Y, Guo C, Wang J, Boral D, Nie D. Nuclear receptors in the multidrug resistance through the regulation of drug-metabolizing enzymes and drug transporters. Biochem Pharmacol. 2012;83:1112–26. doi: 10.1016/j.bcp.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrighton SA, Schuetz EG, Thummel KE, Shen DD, Korzekwa KR, Watkins PB. The human CYP3A subfamily: practical considerations. Drug Metab Rev. 2000;32:339–61. doi: 10.1081/dmr-100102338. [DOI] [PubMed] [Google Scholar]
- 26.Juurlink DN, Mamdani M, Kopp A, Laupacis A, Redelmeier DA. Drug-drug interactions among elderly patients hospitalized for drug toxicity. Jama-J Am Med Assoc. 2003;289:1652–8. doi: 10.1001/jama.289.13.1652. [DOI] [PubMed] [Google Scholar]
- 27.Hillenbrand P, Sherlock S. Use of Metolazone in the Treatment of Ascites due to Liver Disease. Br Med J. 1971;4:266–70. doi: 10.1136/bmj.4.5782.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pridjian G, Pridjian C, Danchuk S, Ianosi-Irimie M, Vu HV, Puschett JB. Beneficial effects of metolazone in a rat model of preeclampsia. J Pharmacol Exp Ther. 2006;318:1027–32. doi: 10.1124/jpet.106.104901. [DOI] [PubMed] [Google Scholar]
- 29.Lazkani M, Ota KS. The role of outpatient intravenous diuretic therapy in a transitional care program for patients with heart failure: a case series. J Clin Med Res. 2012;4:434–8. doi: 10.4021/jocmr1106w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabrielli M, Gasbarrini A, Santoliquido A, Flore R, Gaetani E, Flex A, et al. Metolazone-induced cholestatic liver disease. Dig Liver Dis. 2001;33:501. doi: 10.1016/s1590-8658(01)80029-3. [DOI] [PubMed] [Google Scholar]
- 31.Temperini C, Cecchi A, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. Interaction of indapamide and related diuretics with 12 mammalian isozymes and X-ray crystallographic studies for the indapamide-isozyme II adduct. Bioorg Med Chem Lett. 2008;18:2567–73. doi: 10.1016/j.bmcl.2008.03.051. [DOI] [PubMed] [Google Scholar]
- 32.Schuetz EG, Schinkel AH, Relling MV, Schuetz JD. P-glycoprotein: a major determinant of rifampicin-inducible expression of cytochrome P4503A in mice and humans. Proc Natl Acad Sci U S A. 1996;93:4001–5. doi: 10.1073/pnas.93.9.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morfouace M, Shelat A, Jacus M, Freeman BB, 3rd, Turner D, Robinson S, et al. Pemetrexed and gemcitabine as combination therapy for the treatment of group3 medulloblastoma. Cancer Cell. 2014;25:516–29. doi: 10.1016/j.ccr.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin W, Wu J, Dong H, Bouck D, Zeng FY, Chen T. Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem. 2008;283:30650–7. doi: 10.1074/jbc.M806132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ong SS, Goktug AN, Elias A, Wu J, Saunders D, Chen TS. Stability of the human pregnane X receptor is regulated by E3 ligase UBR5 and serine/threonine kinase DYRK2. Biochem J. 2014;459:193–203. doi: 10.1042/BJ20130558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molecular Operating Environment (MOE) software M. Chemical Computing Group Inc; http://www.chemcomp/com. [Google Scholar]
- 38.Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, et al. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol. 2005;19:1125–34. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- 39.Bertilsson G, Heidrich J, Svensson K, -sman M, Jendeberg L, Sydow-Bäckman M, et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci U S A. 1998;95:12208–13. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan Y, Li L, Kim G, Ekins S, Wang H, Swaan PW. Identification and validation of novel human pregnane X receptor activators among prescribed drugs via ligand-based virtual screening. Drug Metab Dispos. 2011;39:337–44. doi: 10.1124/dmd.110.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khandelwal A, Krasowski MD, Reschly EJ, Sinz MW, Swaan PW, Ekins S. Machine learning methods and docking for predicting human pregnane X receptor activation. Chem Res Toxicol. 2008;21:1457–67. doi: 10.1021/tx800102e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma D, Lau AJ, Sherman MA, Chang TK. Agonism of human pregnane X receptor by rilpivirine and etravirine: comparison with first generation non-nucleoside reverse transcriptase inhibitors. Biochem Pharmacol. 2013;85:1700–11. doi: 10.1016/j.bcp.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Hariparsad N, Nallani SC, Sane RS, Buckley DJ, Buckley AR, Desai PB. Induction of CYP3A4 by efavirenz in primary human hepatocytes: comparison with rifampin and phenobarbital. J Clin Pharmacol. 2004;44:1273–81. doi: 10.1177/0091270004269142. [DOI] [PubMed] [Google Scholar]
- 44.Sinz M, Kim S, Zhu Z, Chen T, Anthony M, Dickinson K, et al. Evaluation of 170 xenobiotics as transactivators of human pregnane X receptor (hPXR) and correlation to known CYP3A4 drug interactions. Curr Drug Metab. 2006;7:375–88. doi: 10.2174/138920006776873535. [DOI] [PubMed] [Google Scholar]
- 45.Creusot N, Kinani S, Balaguer P, Tapie N, LeMenach K, Maillot-Maréchal E, et al. Evaluation of an hPXR reporter gene assay for the detection of aquatic emerging pollutants: screening of chemicals and application to water samples. Anal Bioanal Chem. 2010;396:569–83. doi: 10.1007/s00216-009-3310-y. [DOI] [PubMed] [Google Scholar]
- 46.Pondugula SR, Brimer-Cline C, Wu J, Schuetz EG, Tyagi RK, Chen T. A phosphomimetic mutation at threonine-57 abolishes transactivation activity and alters nuclear localization pattern of human pregnane x receptor. Drug Metab Dispos. 2009;37:719–30. doi: 10.1124/dmd.108.024695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuhr U. Induction of drug metabolising enzymes: pharmacokinetic and toxicological consequences in humans. Clin pharmacokinet. 2000;38:493–504. doi: 10.2165/00003088-200038060-00003. [DOI] [PubMed] [Google Scholar]
- 48.Watkins PB, Whitcomb RW. Hepatic dysfunction associated with troglitazone. N Engl J Med. 1998;338:916–7. doi: 10.1056/NEJM199803263381314. [DOI] [PubMed] [Google Scholar]
- 49.Moore LB, Goodwin B, Jones SA, Wisely GB, Serabjit-Singh CJ, Willson TM, et al. St. John’s wort induces hepatic drug metabolism through activation of the pregnane X receptor. Proc Natl Acad Sci U S A. 2000;97:7500–2. doi: 10.1073/pnas.130155097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izzo AA, Ernst E. Interactions between herbal medicines and prescribed drugs: an updated systematic review. Drugs. 2009;69:1777–98. doi: 10.2165/11317010-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 51.Pondugula SR, Tong AA, Wu J, Cui J, Chen T. Protein phosphatase 2Cbetal regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. Drug Metab Dispos. 2010;38:1411–6. doi: 10.1124/dmd.110.032128. [DOI] [PMC free article] [PubMed] [Google Scholar]