Abstract
Emerging evidence suggests that activation of adenosine monophosphate-activated protein kinase (AMPK), an energy gauge and redox sensor, controls the cell cycle and protects against DNA damage. However, the molecular mechanisms by which AMPKα isoform regulates DNA damage remain largely unknown. The aim of this study was to determine if AMPKα deletion contributes to cellular hyperproliferation by reducing p21WAF1/Cip1 (p21) expression thereby leading to accumulated DNA damage. The markers for DNA damage, cell cycle proteins, and apoptosis were monitored in cultured mouse embryonic fibroblasts (MEFs) isolated from wild type (WT, C57BL/6J), AMPKα1, or AMPKα2 homozygous deficient (AMPKα1−/−, AMPKα2−/−) mice by Western blot, flow cytometry, and cellular immunofluorescence staining. Deletion of AMPKα1, the predominant AMPKα isoform, but not AMPKα2 in immortalized MEFs led to spontaneous DNA double-strand breaks (DSB) which corresponded to repair protein p53-binding protein1 (53BP1) foci formation and subsequent apoptosis. Furthermore, AMPKα1 localizes to chromatin and AMPKα1 deletion down-regulates cyclin-dependent kinase inhibitor, p21, an important protein that plays a role in decreasing the incidence of spontaneous DSB via inhibition of cell proliferation. In addition, AMPKα1 null cells exhibited enhanced cell proliferation. Finally, p21 overexpression partially blocked the cellular hyperproliferation of AMPKα1-deleted MEFs via the inhibition of cyclin-dependent kinase 2 (CDK2). Taken together, our results suggest that AMPKα1 plays a fundamental role in controlling the cell cycle thereby affecting DNA damage and cellular apoptosis.
Keywords: AMPKα, p21, 53BP1, DNA damage, cell proliferation, apoptosis
1. Introduction
Several types of DNA damage, including oxidative damages, depurinations, single-strand breaks (SSB), and DNA double-strand breaks (DSB), occur naturally during DNA replication [1]. DSB are among the most deleterious lesions in the genome, as it can cause genomic rearrangements, chromosome breaks and translocations, leading to apoptosis, senescence, or tumorigenesis [2]. Most DNA damage can undergo DNA repair. For example, DSB elicit a cascade of protein recruitment to the chromatin surrounding DNA lesions that regulates DNA damage response signaling and repair [3]. Cells repair DSB by initiating either DNA nonhomologous end-joining (NHEJ), a mutation-prone pathway [4, 5], or homologous recombination (HR) [6]. DSB repair by HR is largely error-free, as it employs undamaged homologous sister chromatid DNA sequences as templates for repair [2]. NHEJ is the prevalent DSB repair pathway in higher eukaryotes [7]. Several molecules have been reported to be involved in DSB response and repair. For example, p53-binding protein 1 (53BP1, also known as TP53BP1) is a key effector of this DSB response [8, 9], as it promotes DNA damage repair by NHEJ [10–13].
Cyclin-dependent kinases (Cdks) play a pivotal role in the cell cycle. Among them, Cdk2 is essential for G1/S phase transition and S phase progression [14]. On the other hand, Cdk1 (formerly known as Cdc2) associated with Cyclin B is essential for regulating cell cycle entry and exit from mitosis [15, 16]. In response to DNA damage, numerous cell cycle signals are activated, which causes arrest in G1, thereby controlling progression through S phase and blocking entry into mitosis with damaged DNA. For example, p21WAF1/Cip1 (p21) binds to and suppresses Cdk2/cyclin E (CycE) complexes, thereby arresting cells at the G1/S checkpoint [17]. In addition, p21 plays a fundamental role in DNA damage response through inhibiting DNA synthesis via association and inhibition of proliferating cell nuclear antigen (PCNA) [18]. Recently, it was reported that p21 is critical in preventing excessive DNA damage accumulation in leukemia stem cells [19].
The well-known energy sensor, adenosine monophosphate-activated protein kinase (AMPK), consisting of a catalytic α subunit (α1 or α2) and regulatory β (β1 or β2) and γ (γ1, γ2, or γ3) units, also has a critical role in cell mitosis [20–22] and anti-oxidative stress [23]. Emerging data indicate that AMPK plays an important role in tumor suppression [24, 25]. It was recently reported that AMPK regulates UVB-induced DNA damage repair in skin tumor cells [26], as well as playing a role in NHEJ via the LKB1-AMPK signaling pathway [27]. However, the exact role of the two AMPKα isoforms and the mechanism by which AMPK might control the cell cycle and DNA damage remain elusive. In this study, we address the involvement of AMPKα1 in cellular hyperproliferation, DNA damage, and apoptosis by analyzing cell cycle proteins and DNA damage markers in AMPKα1−/− mouse embryo fibroblasts (MEFs). We demonstrate here, for the first time, that AMPKα1−/− MEFs exhibit hyperproliferation, high levels of DNA DSB markers, and consequent apoptosis, partially due to the p21 reduction. Importantly, p21 overexpression decreased the foci formation of DSB repair protein 53BP1 in AMPKα1−/− MEFs. These findings establish a new role for AMPKα1 in cell cycle and DNA damage, providing novel insights into the mechanism of tumor suppression mediated by AMPK.
2. Materials and methods
2.1. Materials and reagents
The following antibodies were obtained from Cell Signaling Technology (Beverly, MA): rabbit anti-AMPKα (2532), rabbit anti-phospho-AMPKα (Thr172) (2535), rabbit anti-phospho-histone H3 (Ser10) (9701), rabbit anti-phospho-p53 (Ser18) (9284), mouse anti-p53 (2524), rabbit anti-γ-H2AX (2577), anti-H2AX (2595), anti-phospho-Chk1 (Ser345) (2348), rabbit anti-53BP1 (4937), rabbit anti-cleaved caspase-3 (Asp175) (9664), rabbit anti-PARP (9542), rabbit anti-phospho-Cdk2 (Thr160) (2561), rabbit anti-Cdk2 (2546), rabbit anti-phospho-Cdk1(Thr161) (9114), and mouse anti-cyclin B1 (V152) (4135). The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): goat anti-AMPKα1 (sc-19128), goat anti-AMPKα2 (sc-19129), mouse anti-XRCC4 (sc-365118), mouse anti-p21 (sc-6246), mouse anti-GAPDH (sc-137179) and mouse anti-β-actin (sc-47778). Caspase-3 inhibitor Z-DEVD-FMK (Cat. # FMK004) and Caspase-9 inhibitor Z-LEHD-FMK (Cat. # FMK008) were purchased from R&D Systems, Inc. Other chemicals and organic solvents of the highest available grade were obtained from Sigma-Aldrich. Ampkα1−/− and Ampkα2−/− mice were described elsewhere [28, 29]. Mice were handled in accordance with study protocols approved by the Institutional Animal Care and Use Committee of the University of Oklahoma Health Sciences Center (Oklahoma City, OK).
2.2. Cell culture and transfection
Mouse embryonic fibroblasts (MEFs) were isolated from AMPKα1−/−, AMPKα2−/−, and WT embryos at 13.5-days post-coitus and cells were immortalized by the 3T3 protocol as described previously [30, 31]. Briefly, 13.5-day mouse embryo was decapitated, thoroughly minced, and trypsinized. The dissociated cells were re-suspended. To immortalize MEFs, cells were passaged continuously according to the 3T3 protocol (3 × 105 cells were plated per 60-mm dish every 3 days) until growth rates in culture stabilized. Cells were then cultured for an additional 15 passages (to about passage 35) and at that point were considered immortalized and used for experiments. MEFs were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS, L-Glutamine (2 mM) (Lonza, Walkersville, MD), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Life Technologies, Grand Island, NY). For cell synchronization in G0/G1 phase, MEFs were serum starved, and then re-incubated in complete medium for various times. MEFs were transiently infected with LacZ, p21, or AMPKα1 adenovirus (MOI = 50) for 24 or 48 h as previously reported [32].
2.3. Indirect immunofluorescence and microscopy
Cells were grown to exponential phase on poly-L-lysine-coated glass coverslips. Cell were fixed in 4% paraformaldehyde, permeabilized in 0.1% TritonX-100 and blocked with image-IT Fix or BSA (Invitrogen). Primary antibodies used were: mouse anti-γ-H2AX (1:100 v/v) or rabbit anti-γ-H2AX (1:100 v/v), rabbit anti-53BP1, and mouse anti-p21. DNA was stained with antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, CA). For indirect immunofluorescence, Alexa Fluor® 488 and 555 were used for detection of the protein. Confocal microscopy was performed using a Zeiss 710 confocal microscope (Oberkochen, Germany), with a 63× oil immersion lens. Image editing was performed in Adobe Systems Incorporated, San Jose, CA.
2.4. RNA extraction, cDNA synthesis, and real time PCR
Total mRNA was isolated and purified using the RNeasy mini kit from Qiagen (Valencia, CA) according to the manufacturer’s instructions. cDNA was synthesized from isolated mRNA using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA), as described previously [29] and by the manufacturer’s instructions. Real-time PCR was performed on a ABI PRISM 7700 sequence detection system (Applied Biosystems) with SYBR green PCR master mix (Applied Biosystems) and 1 μl of first-strand cDNA as template with specific primers for p21 (5′-CCTGGTGATGTCCGACCTGTT-3′, 5′-CCCCTTAGAAGTCCGGCGAG-3′) [33]. The levels of gene expression were determined relative to that of β-actin (5′-TGGGCCGCTCTAGGCACCA-3′, 5′-ACCGGAATCCCAAGTCCCC-3′).
2.5. Comet assay
Single cell DNA damage (double-stranded breaks) was analyzed by neutral comet assay using the Trevigen’s Comet Assay kit (4250-050-K, Gaithersburg, MD) according to the manufacturer’s instruction. Briefly, cells were suspended in 0.7% low melting point agarose and spread on glass slides precoated with 1% agarose. Slides were overlaid with coverslips that were removed after the gel solidified. The gel was treated with lysis solution (Trevigen) for 60 min at 4°C in the dark and electrophoresed at 1 V/cm for 20 min. Comet tails were stained and slides were captured by fluorescent microscope. Quantitative measurements of DNA damage were performed by using Comet Assay IV software (Perceptive Instruments).
2.6. Annexin V binding assay
Annexin V-FITC (Cat. # K101-100, BioVision, Milpitas, CA) was used to detect the phosphotidylserine exposure to the outer surface of cell membrane by following the manufacturer’s protocol. Briefly, cells were grown on cover slip in a 12 well plate with the desired treatment. Cells were washed with PBS and 1× binding buffer, then incubated with Annexin V-FITC (1:70 dilution) diluted in 1× binding buffer for 10 min. Then cells were washed twice with 1× binding buffer and the coverslips were mounted on microscope glass slides with Dako florescent mounting medium. Randomly selected fields were counted using a fluorescence microscope and quantitated using NIH Image software.
2.7. Flow cytometry analysis
Cells were serum starved for 24 hours, then cultured in regular culture medium for the indicated time. The treated cells were fixed in 80% ethanol and stained with 50 μg/ml propidium iodide (PI) in the presence of 10 μg of DNase-free RNase per ml [34]. Cell cycle profiles were determined by FACSDiVa (BD Bioscience, San Jose, CA) and data were analyzed using FCS Express V3 software. LacZ or p21 adenovirus-infected WT or AMPKα1−/− MEFs were serum-starved for 16 h, then incubated with regular culture medium plus 10 μM BrdU for 8 h. The cells were harvested and flow cytometric assay were performed by following the manufacturer’s protocol in BrdU Flow Kit (Cat. # 559619, BD Biosciences, San Jose, CA). In addition, exponentially growing WT or AMPKα1−/− MEFs were pulsed with BrdU (10 μM) for 40 min followed by washes with warmed serum-free medium twice and re-feeding with warmed and pre-gassed whole medium. Cells were fixed and stained using the FITC BrdU Flow Kit after 0, 40 min, 2, 4, and 6 h post pulse. The S and G2+M phase durations were calculated as previous report [35].
2.8. Protein extraction, immunoprecipitation, and immunoblotting
Whole cell extracts were prepared using cell lysis buffer (9803) from Cell Signaling Technology with protease and phosphatase inhibitor cocktails I and II (Cat. # BP-479 and BP-480, Boston BioProducts, MA). Protein samples (30–50 μg) were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and probed with different antibodies as previously described [36, 37]. Following incubation with the appropriate horseradish peroxidase-associated secondary antibodies (Cell Signaling Technology), signals were visualized with an enhanced chemiluminescence detection system (GE Healthcare) and quantified by densitometry. Equal loading of protein was verified by immunoblotting with anti-β-actin or -GAPDH antibody.
2.9. Subcellular fraction
Subcellular protein fractionation of cultured cells was performed as described in the instructions for the commercial kit (Cat. # 78840, Thermo Scientific, Rockford, IL).
2.10. Statistical analysis
Unless otherwise stated, data are presented as mean ± S.D. Differences between multiple means were evaluated by two-tailed Student’s t test or analysis of variance with post hoc Bonferroni corrections. A p value < 0.05 was considered statistically significant.
3. Results
3.1. Subcellular localization of AMPKα isoforms and dynamic activation of AMPKα in MEFs
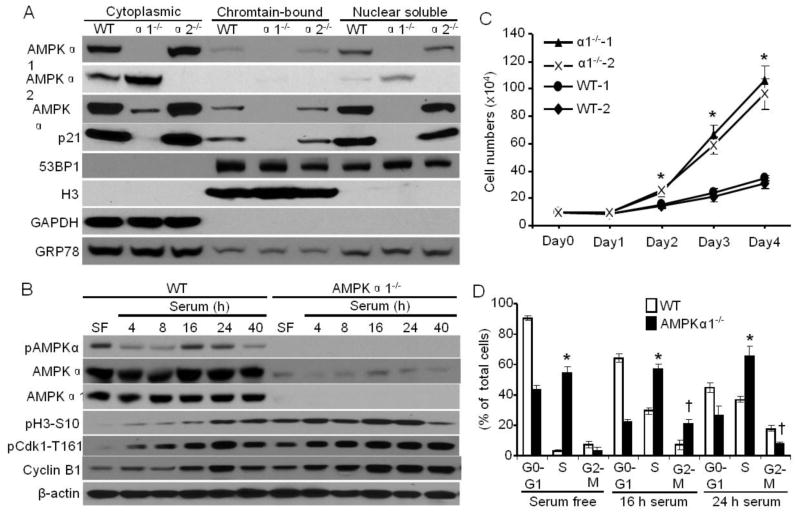
We examined the relative contribution of AMPKα1 and AMPKα2 isoform to total AMPKα in cytoplasmic and nuclear fractions. We performed subcellular fractionation followed by immunoblotting to assess the protein levels of AMPKα1, AMPKα2 and AMPKα in the cytoplasmic and nuclear fractions. As expected, AMPKα1 and AMPKα2 were not detected in either cytoplasmic or nuclear fractions of AMPKα1−/− or AMPKα2−/− MEFs, respectively (Fig. 1A). Since AMPKα1 is the predominant AMPKα isoform in MEFs [30], AMPKα1 deletion dramatically decreased the cytoplasmic, nuclear soluble, and chromatin-bound AMPKα (Fig. 1A), but increased AMPKα2 protein levels, which is in agreement with the result from Ras-transformed AMPKα1−/− MEFs [38]. AMPKα1 and AMPKα2 were predominantly found in the cytoplasmic region; however a small fraction of both were also located in the nuclear region. (Fig. 1A). Both AMPKα1 and total AMPKα were almost undetectable in the chromatin-bound and nuclear soluble fraction of AMPKα1−/− MEFs (Fig. 1A). Interestingly, AMPKα1, but not AMPKα2 was chromatin-bound (Fig. 1A).
Fig. 1.
AMPKα1 localizes to chromatin and is implicated in regulating the cell cycle. (A) MEFs were subcellularly fractionated by a commercially available kit from Thermo Scientific. AMPKα1, AMPKα2, AMPKα, p21, and 53BP1 were detected by Western blots. Histone H3 serves as a marker for chromatin. GAPDH serves as a cytoplasmic marker. GRP78 serves as a loading control for each fraction. Representative data from three independent experiments are shown. (B) AMPKα is activated during mitosis. WT and AMPKα1−/− MEFs were first serum-deprived for 24 hours, then cultured in regular culture medium for the indicated times. The cells were lysed and analyzed by Western blot using anti-pAMPKα-T172, -AMPKα, -AMPKα1, -Cyclin B1, -pCdk1-T161, or pH3-S10 antibody. Representative data from three independent experiments are shown. (C) WT and AMPKα1−/−-immortalized MEFs (two independent cell lines for each) were plated, and cells were counted at the indicated times. n=8, *p < 0.05 vs WT. (D) Flow cytometric analysis of cell cycle progression in WT and AMPKα1−/− MEFs serum starved for 24 hours or treated with serum for the indicated times. n=3, *p < 0.01 vs WT in S phase; † p < 0.01 vs WT in G2-M phase.
In WT MEFs, phosphorylated AMPKα on Thr-172 (pAMPKα-T172), an indicator of active AMPKα [39], was higher with serum deprivation, while AMPK activity was significantly inhibited during serum stimulation for 4–8 hours (Fig. 1B). pAMPKα in WT MEFs was increased during serum stimulation for 16 or 24 hours, then decreased, which was consistent with the upregulation of histone H3 phosphorylation at Ser10 (pH3-S10) (Fig. 1B), a typical marker of cell mitosis [40]. Our data is in line with previously published data demonstrating that AMPK activation is dynamic during the cell cycle of HeLa cancer cells [22] and HEK293 cells [41]. However, AMPKα1 deletion showed higher levels of pH3-S10 even under serum-free conditions, although pAMPKα was undetectable. Taken together, these results imply that AMPKα1 might act on chromatin and is required for normal cell cycle. Thus, loss of AMPKα1 might lead to a dysregulated cell cycle in MEFs.
3.2. AMPKα1 deletion enhances cell division in MEFs
It is reported that AMPK activation by glucose limitation suppresses cell-cycle progression via arresting the cell cycle at G0-G1 phase [42]. As depicted in Fig. 1C, AMPKα1 deletion significantly accelerated cell proliferation beginning at day 2 in culture compared with WT MEFs. As calculated, the mean population doubling time of AMPKα1−/− MEFs was around 18 h, while that of WT MEFs was about 35 h. Furthermore, flow cytometry data (Fig. 1D) demonstrated that AMPKα1 deletion had less cells in G0/G1 phase (about 40%) compared with WT MEFs (~ 92%) in response to serum starvation, however, AMPKα1−/− MEFs had more cells with S phase (about 55%) under either serum-free or serum-stimulated conditions. AMPKα1 deletion resulted in more cells in G2/M phase at 16 h after serum stimulation, however, fewer cells were in G2/M phase at 24 h of serum stimulation. Next, BrdU pulse-chase time course experiments demonstrated that AMPKα1−/− MEFs had shorter S traverse time (4.5 ± 1.1 h) than WT MEFs (11 ± 1.2 h). The G2/M phase traverse time (3.1 ± 0.4 h) in AMPKα1−/− MEFs was also shorter than that (8.8 ± 0.9 h) in WT MEFs. These results imply that AMPKα1−/− MEFs has reduced DNA replication time. AMPKα1 deletion enhances cell cycle progression from the G1 to S phase and G2 to M phase, which may be associated with the persistently high levels of Cyclin B1 and phosphorylated Cdk1 at T161 (Fig. 1B), an active form of Cdk1 [16, 43].
3.3. AMPKα1, not AMPKα2, deletion leads to increased DNA damage in MEFs
Since AMPK activation regulates UVB-induced DNA damage repair in skin tumor cells [26], we sought to identify whether AMPK regulates DNA damage signaling in MEFs. As shown in Fig. 2A, AMPKα1, not AMPKα2, deletion dramatically increased the protein level of serine 139-phosphorylated H2AX (γ-H2AX), an important and widely used molecular marker for DNA DSB [6, 44, 45]. Furthermore, compared to WT and AMPKa2−/−, AMPKα1 deletion significantly increased the basal levels of serine-18 phosphorylation of p53, an indicator of DNA damage associated with cellular apoptosis [30, 46], as well as serine-345 phosphorylation of Chk1, a general DSB sensor and response effector [47]. The percentage of cells with greater than 7 γ-H2AX foci per cell were markedly increased in AMPKα1−/− MEFs when compared with WT and AMPKα2−/− MEFs (Fig. 2B). To further verify the spontaneous accumulation of DNA-strand breaks in the absence of AMPKα1, the neutral comet assay was performed in both WT and AMPKα1−/− MEFs. In agreement with the data above, the comet assay demonstrated that AMPKα1 deletion exerts longer comet tails (Fig. 2C), which is closely associated with DNA damage [48]. These results suggest that AMPKα1 plays an important role in regulating DNA damage.
Fig. 2.
AMPKα1, not AMPKα2, deficiency leads to elevated DNA damage in MEFs. (A) (Upper) DNA damage signals in two independent cell lines for each genetic background were evaluated by Western blots with the indicated antibody. (Bottom) Quantification data from upper panel. n=6, *p < 0.05 vs WT. (B) Immunofluorescent staining of MEFs with indicated genotypes using nuclear (DAPI-blue) and DSB markers (γ-H2AX-red) (left). Scale bar = 50 μm. Quantitation for the percentages of cells with indicated number of γ-H2AX foci (right). WT MEFs (n=81); AMPKα1−/− MEFs (n=112); AMPKα2−/− MEFs (n=94). (C) Quantitation for comet tail assay measuring tail moment for WT and AMPKα1−/− MEFs. n=20, *p < 0.05 vs WT. Representative images are shown in the upper panel.
3.4. AMPKα1 deletion-elevated DNA damage contributes to the increased apoptosis
Since γ-H2AX induction and phosphorylation of p53 at S18 also exist in apoptotic cells [49], it is important to validate whether the observed increased γ-H2AX signal in AMPKα1-deleted MEFs is due to DNA damage or apoptosis. As shown in Fig. 3, AMPKα1 deletion dramatically enhanced cell apoptosis as demonstrated by increased poly(ADP-ribose) polymerase (PARP) cleavage (Fig. 3A and B) and Annexin V staining (Fig. 3C and D), which is consistent with a previous report [30]. The treatment with Caspase-3 specific inhibitor Z-DEVD-FMK [50] significantly inhibited PARP cleavage and Annexin V staining (Fig. 3C and D), but did not alter the γ-H2AX signaling (Fig. 3A). Moreover, the Caspase-9 specific inhibitor Z-LEHD-FMK [51] clearly suppressed apoptosis in AMPKα1−/− MEFs, indicated by the reduction of cleaved PARP and Caspase-3 (Fig. 3B) and Annexin V staining (Fig. 3C and D), however, the treatment had no effect on the elevated γ-H2AX (Fig. 3B). These data indicate that γ-H2AX induction in AMPKα1−/− MEFs is due to the increased DNA damage, which leads to the enhanced apoptosis.
Fig. 3.
Increased DNA damage contributes to the elevated apoptosis in AMPKα1−/− MEFs. (A) (Upper) Caspase-3 inhibitor (Cas-3i) Z-DEVD-FMK dramatically suppressed apoptosis in AMPKα1−/− MEFs, but had no effect on DNA damage signal as measured by γ-H2AX expression. (Bottom) Quantification of Western blot data. n=4, † p < 0.01 vs WT/Vehicle; # p < 0.01 vs α1−/−/Vehicle; *p < 0.001 vs WT/vehicle. (B) (Upper) Caspase-9 inhibitor (Cas-9i) Z-LEHD-FMK significantly inhibited apoptosis in AMPKα1−/− MEFs, but had no effect on DNA damage signal as measured by γ-H2AX expression. (Bottom) Quantification of Western blot data. n=3, *p < 0.001 vs WT/Vehicle; † p < 0.01 vs α1−/−/Vehicle. (C) Representative images showing phosphatidylserine externalization detected by Annexin V labeling in MEFs treated with Cas-3i and Cas-9i. Scale bar, 100 μm. (D) Quantification of Annexin V intensity. n=15, *p < 0.01 vs WT/Vehicle; †p < 0.05 vs α1−/−/Vehicle.
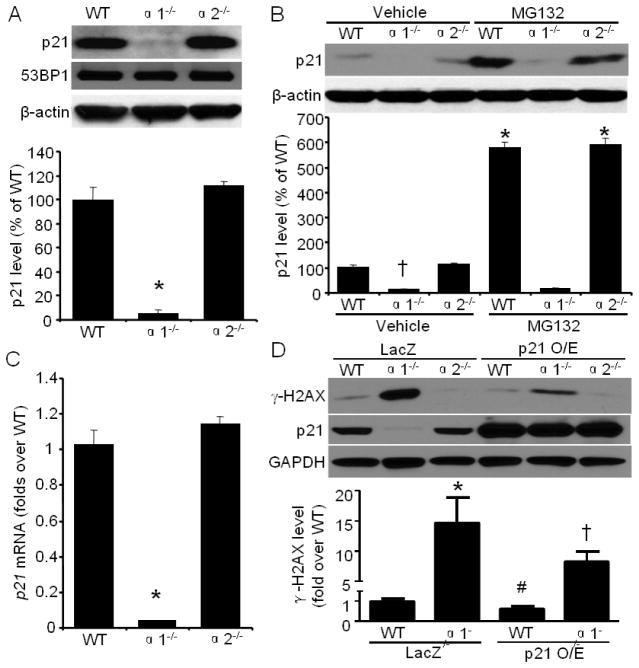
3.5. Elevated DNA damage in AMPKα1−/− MEFs is partially due to p21 reduction
One possibility for the increased DNA damage seen in AMPKa1−/− MEFs is the inability for the cell to activate DNA damage repair mechanisms. Since 53BP1 plays critical roles in the repair of damaged DNA, we investigated whether AMPKα1 deletion alters the protein levels of 53BP1. As depicted in Fig. 4A, AMPKα1 deletion did not change 53BP1 protein levels. Additionally, the amount of XRCC4, a DNA repair protein [52], is similar between WT and AMPKα1−/− MEFs (data not shown). Previous studies have indicated that p21 is involved in DNA lesion repair [18, 19]. Therefore, we tested whether AMPKα1 could modulate p21 expression. Indeed, AMPKα1 deletion dramatically reduced total, cytoplasmic, chromatin-bound, and nuclear soluble p21 protein levels compared with WT and AMPKα2 deletion (Fig. 4A and 1A). To examine whether p21 protein reduction is due to increased protein degradation, we treated cells with MG132, a potent inhibitor of the 26S proteasome [53]. MG132 treatment did not reverse the p21 reduction in AMPKα1−/− MEFs, whereas p21 protein levels in both WT and AMPKα2−/− MEFs were significantly increased (Fig. 4B), indicating the ubiquitin-proteasome system is not responsible for the p21 reduction in AMPKα1−/− MEFs. However, qRT-PCR assay demonstrated that p21 mRNA levels were markedly down-regulated by AMPKα1 deletion compared with WT and AMPKα2 deletion (Fig. 4C). Overexpression of p21 for 48 hours partially, but significantly reduced γ-H2AX signal in AMPKα1−/− MEFs (Fig. 4D). These results suggest that AMPKα1 deletion impairs p21 transcription hence leading to increased DNA damage which is partially rescued upon p21 overexpression.
Fig. 4.
Increased DNA damage is partially due to p21 reduction in AMPKα1−/− MEFs. (A) (Upper) AMPKα1 deletion significantly down-regulated p21, but not 53BP1. (Bottom) Quantification of Western blot data. n=8, *p < 0.001 vs WT. (B) Proteasome inhibitor, MG132 (20 μM, 8 h) did not reverse p21 reduction in AMPKα1−/− MEFs. n=4, †p < 0.001 vs WT, *p < 0.001 vs WT/vehicle or α2−/−/vehicle, respectively. (C) Quantitative RT-PCR analysis of p21 expression in MEFs. β-actin was used as endogenous control. Values are mean ± SEM of four independent experiments, *p < 0.001 vs WT. (D) p21 overexpression partially reduced DNA damage in AMPKα1−/− MEFs. n=4, *p < 0.01 vs WT/LacZ; #p < 0.05 vs WT/LacZ; †p < 0.01 vs α1−/−/LacZ.
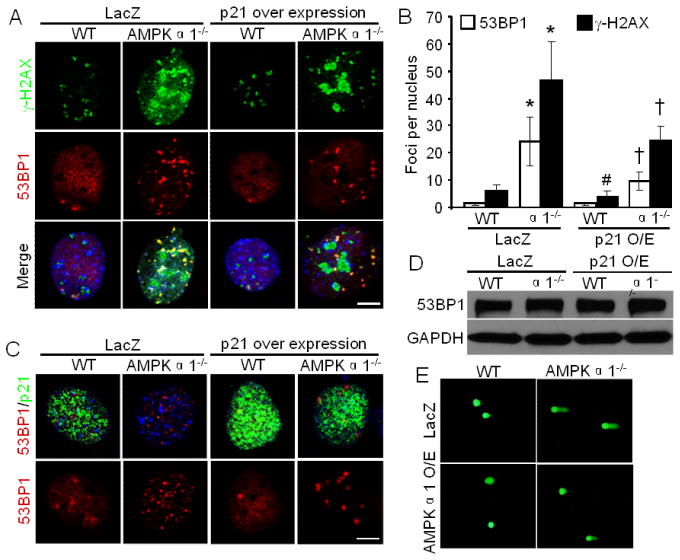
3.6. p21 inhibits 53BP1 foci formation
Next, we investigated whether p21 overexpression affects the foci formation of repair protein 53BP1. The overall 53BP1 protein level was similar in WT and AMPKα1−/− MEFs (Fig. 1A and 4A). Initial qualitative assessment indicated that there was a stronger γ-H2AX signal in AMPKα1−/− MEFs compared to WT MEFs (Fig. 2A) which was confirmed by immunofluorescence (Fig. 5A). No 53BP1 foci was observed in about 85% of WT MEFs, which may be due to less DNA damage (Fig. 5B and 2B). In contrast, 68% of AMPKα1−/− MEFs exhibited 53BP1 foci formation. p21 overexpression significantly decreased 53BP1 foci number in AMPKα1−/− MEFs, which is in line with the decreased foci number of γ-H2AX. p21did not show colocalization with 53BP1. Furthermore, p21 overexpression did not alter the total 53BP1 protein levels (Fig. 5D). In addition, AMPKα1 overexpression decreased DNA damage in AMPKα1−/− MEFs demonstrated by comet assay. These results imply that p21 overexpression inhibits 53BP1 foci formation highly associated with DNA damage [9] in AMPKα1-deleted MEFs.
Fig. 5.
53BP1 foci formation is inhibited by p21 expression in AMPKα1−/− MEFs. (A) p21 overexpression decreases 53BP1 foci formation in AMPKα1−/− MEFs. Representative images showing 53BP1 and γ-H2AX foci formation. Scale bar = 10 μm. (B) Quantitative analysis of 53BP1 and γ-H2AX foci per nucleus. n=20, *p < 0.01 vs WT/LacZ; †p < 0.01 vs α1−/−/LacZ for 53BP1 or γ-H2AX, respectively; #p < 0.05 vs WT/LacZ. (C) Representative images showing 53BP1 foci formation and p21 expression. Scale bar = 10 μm. (D) p21 overexpression does not alter total 53BP1 protein. (E) AMPKα1 overexpression attenuates DNA damage determined by comet assay.
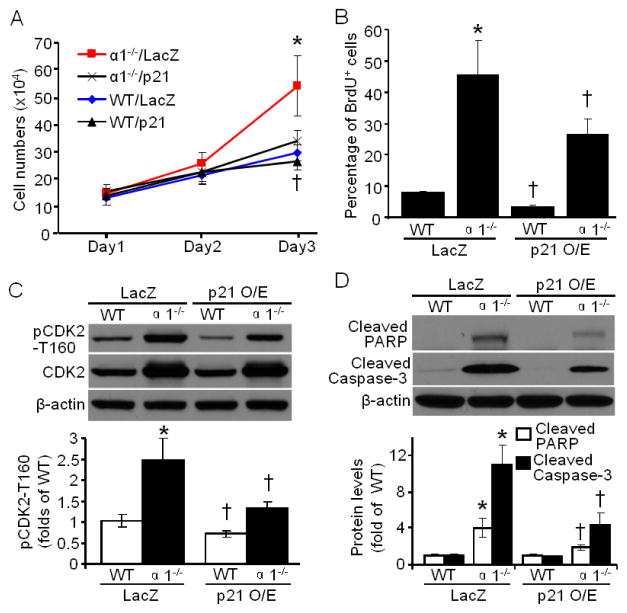
3.7. p21 overexpression partially abrogates the hyperproliferation and apoptosis in AMPKα1−/− MEFs
Given the role of p21 in DNA repair and cell cycle regulation, we analyzed whether p21 overexpression can normalize the phenotype of AMPKα1−/− MEFs by evaluating cellular proliferation. As depicted in Fig. 6A, p21 overexpression significantly attenuated cell proliferation of AMPKα1−/− MEFs, while only mildly inhibiting the proliferation of WT MEFs. After synchronizing the cells with serum-free medium for 16 h, LacZ or p21 adenovirus-infected MEFs were labelled with BrdU for 8 h. The flow cytometric results indicated that deletion of AMPKα1 increased the BrdU-positive cells by 6 times as compared with WT (Fig. 6B), implying that there is more cells in S-phase for AMPKα1−/− MEFs. Importantly, p21 overexpression significantly decreased BrdU-positive cells in AMPKα1−/− MEFs by 46%, as compared to LacZ infection (Fig. 6B). Furthermore, the CDK2 phosphorylation at T160, an active form of CDK2 [54], in AMPKα1−/− MEFs, was markedly ablated by p21 overexpression (Fig. 6C). In addition, the apoptotic signal in AMPKα1−/− MEFs was profoundly blunted by p21 overexpression (Fig. 6D).
Fig. 6.
p21 overexpression attenuates cellular hyperproliferation and apoptosis in AMPKα1−/− MEFs. (A) p21 overexpression blocked the hyperproliferation of AMPKα1−/− MEFs. n=10, *p < 0.01 vs α1−/−/p21; † p < 0.05 vs WT/LacZ. (B) p21 overexpression decreased the percentage of BrdU positive cells in AMPKα1−/− and WT MEFs. n=4, *p < 0.01 vs WT/LacZ; †p < 0.01 vs WT/LacZ, α1−/−/LacZ, respectively. (C) (Upper) p21 overexpression significantly inhibited phosphorylation of CDK2 at T160 (pCDK2-T160), while had no effect on total CDK2 protein level. (Bottom) Quantification of Western blot data. n=3, *p < 0.001 vs WT/LacZ; † p < 0.01 vs WT/LacZ, or α1−/−/LacZ, respectively. (D) p21 overexpression suppressed apoptotic signal (PARP and Caspase-3 cleavage) in AMPKα1−/− MEFs. n= 3, *p < 0.001 vs WT/LacZ; † p < 0.01 vs α1−/−/LacZ.
4. Discussion
In the present study, we have demonstrated that AMPKα1, but not AMPKα2 deletion, mediates aberrant cell proliferation, spontaneous DNA DSB damage, and apoptosis in MEFs. The mechanism underlying this process is partly due to p21 reduction. The enhanced cell proliferation in AMPKα1−/− MEFs is due to the G1 to S transition and impaired G2/M arrest resulting from activation of Cdk2 and Cdk1 by p21 reduction. p21 overexpression decreases 53BP1 foci formation in AMPKα1-deleted MEFs. These findings indicate that AMPKα1 is a pivotal regulator for cell cycle, DNA DSB and resultant apoptosis.
A previous report indicated that AMPKα is associated with the efficient repair of UVB-induced DNA damage in SKH-1 mouse skin, via the regulation of xeroderma pigmentosum group C (XPC) [26], a crucial initiator of global genome nucleotide excision repair [55]. Here, we have, for the first time, demonstrated that AMPKα1 isoform deletion down-regulated p21 and transient overexpression of p21 partially reduced this DNA damage (Fig. 4D and Fig. 5A), suggesting that p21 may play an integral part in DNA damage and repair. Recently, it is reported that p21 inhibits DNA damage through interaction with PCNA [18]. These results are consistent with the findings in leukemia stem cells [19]. Furthermore, a critical and previously unrecognized role of p21 as a gatekeeper of AMPKα1 deletion-induced DNA damage was demonstrated. p21 overexpression decreased 53BP1 foci formation in AMPKα1-deficient cells via an unknown mechanism. How 53BP1 is recruited to DSB sites has been recently studied. Histone modifications are involved in 53BP1 recruitment to DSB sites [10]. Our data imply that p21 may be an additional 53BP1 effector. In addition, p21 reduction in AMPKα1-deleted MEFs is unlikely p53-dependent, a widely reported molecular mechanism [56–59], since p53 is up-regulated by AMPKα1 deletion (Fig. 2A). Hence, the mechanism involved in p21 reduction by AMPKα1 deletion or inhibition warrants further investigation.
Increasing evidence indicates that AMPK controls the cell cycle [29, 60] and mitosis [20–22] via distinct mechanisms. For example, constitutive expression of AMPK-related kinase NUAK1 leads to gross aneuploidies in WI-38 human fibroblast [61]. Additionally, p21 is required for proper cell cycle control and consequent chromosome stability in Trp53 515C/515C mice (encoding p53R172P, the corresponding murine p53 mutant) which is deficient for apoptosis but retains a partial cell cycle arrest function [62]. Unrepaired DNA damage is a trigger for cellular apoptosis and/or enhanced cell proliferation [63], and AMPKα1 deletion enhances MEFs apoptotic signaling [30]. Here, AMPKα1 deletion increased Cdk2 protein levels and decreased p21, as well as stimulated cell proliferation. p21 overexpression partially inhibited Cdk2 phosphorylation at T160 and consequently hindered cell hyperproliferation and the resultant DNA damage. In addition, p21 overexpression partially suppressed apoptosis in AMPKα1−/− MEFs, which is due to the increased DNA damage.
Loss of AMPKα1 culminates in elevated DNA damage, which when prolonged can lead to accumulation of a lethal amount of damaged DNA or mutated DNA, thereby resulting in apoptosis or hyperproliferation associated with cancer development. As AMPK is an important modulator of cell metabolism, it will be interesting to examine whether AMPKα1 regulates DNA damage, in part via modulating fuel switching.
In summary, our studies reveal an important role for AMPKα1 in cell biology and connect two hallmarks of tumor cells: hyperproliferation and DNA damage [64–66], which may be due to p21 reduction. Given the importance of AMPK in the cell cycle, these findings hold profound implications for understanding the molecular mechanisms by which AMPK functions as a promising tumor suppressor or senescence blocker.
Highlights.
AMPKα1 deletion leads to cellular hyperproliferation
AMPK α1 deletion promotes DNA damage thereby leading to apoptosis
Deletion of AMPKα1 is associated with a reduction in p21
p21 overexpression partially decreases DNA damage
p21 overexpression partially inhibits cell proliferation via CDK2 inhibition
Acknowledgments
This work was supported by funding from the following agencies: National Institutes of Health RO1 (HL079584, HL080499, HL074399, HL089920, HL096032, HL105157, and HL110488), the Warren Chair in Diabetes Research of the University of Oklahoma Health Sciences Center (all to M.-H.Z.), Scientist Development Grant (11SDG5560036) from National Center of American Heart Association, Oklahoma Center for the Advancement of Science and Technology OCAST grant (HR12-061) (both to P.S.), and National Natural Science Foundation of China (81303109, to H.X.).
Abbreviations
- 53BP1
p53-binding protein 1
- AMPK
adenosine monophosphate-activated protein kinase
- DSB
double-strand breaks
- MEFs
mouse embryonic fibroblasts
- NHEJ
nonhomologous end-joining
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J. 2012;443:13–26. doi: 10.1042/BJ20112100. [DOI] [PubMed] [Google Scholar]
- 2.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual review of genetics. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS genetics. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strande N, Roberts SA, Oh S, Hendrickson EA, Ramsden DA. Specificity of the dRP/AP lyase of Ku promotes nonhomologous end joining (NHEJ) fidelity at damaged ends. J Biol Chem. 2012;287:13686–13693. doi: 10.1074/jbc.M111.329730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renkawitz J, Lademann CA, Kalocsay M, Jentsch S. Monitoring homology search during DNA double-strand break repair in vivo. Mol Cell. 2013;50:261–272. doi: 10.1016/j.molcel.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 10.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, Durocher D. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature structural & molecular biology. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, Wesemann DR, Lee JE, Tubbs A, Sleckman BP, Daniel JA, Ge K, Alt FW, Fernandez-Capetillo O, Nussenzweig MC, Nussenzweig A. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satyanarayana A, Hilton MB, Kaldis P. p21 Inhibits Cdk1 in the absence of Cdk2 to maintain the G1/S phase DNA damage checkpoint. Mol Biol Cell. 2008;19:65–77. doi: 10.1091/mbc.E07-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindqvist A, van Zon W, Karlsson Rosenthal C, Wolthuis RM. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS biology. 2007;5:e123. doi: 10.1371/journal.pbio.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deibler RW, Kirschner MW. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol Cell. 2010;37:753–767. doi: 10.1016/j.molcel.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 18.Perucca P, Cazzalini O, Mortusewicz O, Necchi D, Savio M, Nardo T, Stivala LA, Leonhardt H, Cardoso MC, Prosperi E. Spatiotemporal dynamics of p21CDKN1A protein recruitment to DNA-damage sites and interaction with proliferating cell nuclear antigen. Journal of cell science. 2006;119:1517–1527. doi: 10.1242/jcs.02868. [DOI] [PubMed] [Google Scholar]
- 19.Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, Ronchini C, Ronzoni S, Muradore I, Monestiroli S, Gobbi A, Alcalay M, Minucci S, Pelicci PG. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457:51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 20.Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, Villen J, Wang B, Kim SR, Sakamoto K, Gygi SP, Cantley LC, Yaffe MB, Shokat KM, Brunet A. Chemical genetic screen for AMPKalpha2 substrates uncovers a network of proteins involved in mitosis. Mol Cell. 2011;44:878–892. doi: 10.1016/j.molcel.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazquez-Martin A, Lopez-Bonet E, Oliveras-Ferraros C, Perez-Martinez MC, Bernado L, Menendez JA. Mitotic kinase dynamics of the active form of AMPK (phospho-AMPKalphaThr172) in human cancer cells. Cell Cycle. 2009;8:788–791. doi: 10.4161/cc.8.5.7787. [DOI] [PubMed] [Google Scholar]
- 22.Thaiparambil JT, Eggers CM, Marcus AI. AMPK regulates mitotic spindle orientation through phosphorylation of myosin regulatory light chain. Mol Cell Biol. 2012;32:3203–3217. doi: 10.1128/MCB.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song P, Zou MH. Regulation of NAD(P)H oxidases by AMPK in cardiovascular systems. Free Radic Biol Med. 2012;52:1607–1619. doi: 10.1016/j.freeradbiomed.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, Huang W, Tao R, Ibaragi S, Lan F, Ido Y, Wu X, Alekseyev YO, Lenburg ME, Hu GF, Luo Z. Inactivation of AMPK alters gene expression and promotes growth of prostate cancer cells. Oncogene. 2009;28:1993–2002. doi: 10.1038/onc.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Z, Zang M, Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth. Future oncology. 2010;6:457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu CL, Qiang L, Han W, Ming M, Viollet B, He YY. Role of AMPK in UVB-induced DNA damage repair and growth control. Oncogene. 2013;32:2682–2689. doi: 10.1038/onc.2012.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ui A, Ogiwara H, Nakajima S, Kanno S, Watanabe R, Harata M, Okayama H, Harris CC, Yokota J, Yasui A, Kohno T. Possible involvement of LKB1-AMPK signaling in non-homologous end joining. Oncogene. 2014;33:1640–1648. doi: 10.1038/onc.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viollet B, Andreelli F, Jorgensen SB, Perrin C, Geloen A, Flamez D, Mu J, Lenzner C, Baud O, Bennoun M, Gomas E, Nicolas G, Wojtaszewski JF, Kahn A, Carling D, Schuit FC, Birnbaum MJ, Richter EA, Burcelin R, Vaulont S. The AMP-activated protein kinase alpha2 catalytic subunit controls whole-body insulin sensitivity. J Clin Invest. 2003;111:91–98. doi: 10.1172/JCI16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song P, Wang S, He C, Liang B, Viollet B, Zou MH. AMPKalpha2 deletion exacerbates neointima formation by upregulating Skp2 in vascular smooth muscle cells. Circ Res. 2011;109:1230–1239. doi: 10.1161/CIRCRESAHA.111.250423. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Wang S, Song P, Zou MH. Inhibition of AMP-activated protein kinase alpha (AMPKalpha) by doxorubicin accentuates genotoxic stress and cell death in mouse embryonic fibroblasts and cardiomyocytes: role of p53 and SIRT1. J Biol Chem. 2012;287:8001–8012. doi: 10.1074/jbc.M111.315812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu MJ, Song P, Shirwany N, Liang B, Xing JJ, Viollet B, Wang X, Zhu Y, Zou MH. Impaired Expression of Uncoupling Protein 2 Causes Defective Postischemic Angiogenesis in Mice Deficient in AMP-Activated Protein Kinase alpha Subunits. Arterioscler Thromb Vasc Biol. 2011;31:1757–1765. doi: 10.1161/ATVBAHA.111.227991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward IM, Difilippantonio S, Minn K, Mueller MD, Molina JR, Yu X, Frisk CS, Ried T, Nussenzweig A, Chen J. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol Cell Biol. 2005;25:10079–10086. doi: 10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song P, Wei J, Wang HC. Distinct roles of the ERK pathway in modulating apoptosis of Ras-transformed and non-transformed cells induced by anticancer agent FR901228. FEBS Lett. 2005;579:90–94. doi: 10.1016/j.febslet.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 35.Terry NH, White RA. Flow cytometry after bromodeoxyuridine labeling to measure S and G2+M phase durations plus doubling times in vitro and in vivo. Nat Protoc. 2006;1:859–869. doi: 10.1038/nprot.2006.113. [DOI] [PubMed] [Google Scholar]
- 36.Song P, Wu Y, Xu J, Xie Z, Dong Y, Zhang M, Zou MH. Reactive nitrogen species induced by hyperglycemia suppresses Akt signaling and triggers apoptosis by upregulating phosphatase PTEN (phosphatase and tensin homologue deleted on chromosome 10) in an LKB1-dependent manner. Circulation. 2007;116:1585–1595. doi: 10.1161/CIRCULATIONAHA.107.716498. [DOI] [PubMed] [Google Scholar]
- 37.Song P, Zhang M, Wang S, Xu J, Choi HC, Zou MH. Thromboxane A2 Receptor Activates a Rho-associated Kinase/LKB1/PTEN Pathway to Attenuate Endothelium Insulin Signaling. J Biol Chem. 2009;284:17120–17128. doi: 10.1074/jbc.M109.012583. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Phoenix KN, Devarakonda CV, Fox MM, Stevens LE, Claffey KP. AMPKalpha2 Suppresses Murine Embryonic Fibroblast Transformation and Tumorigenesis. Genes & cancer. 2012;3:51–62. doi: 10.1177/1947601912452883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang W, Zhu Z, Thompson HJ. Dietary energy restriction modulates the activity of AMP-activated protein kinase, Akt, and mammalian target of rapamycin in mammary carcinomas, mammary gland, and liver. Cancer Res. 2008;68:5492–5499. doi: 10.1158/0008-5472.CAN-07-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 41.Mao L, Li N, Guo Y, Xu X, Gao L, Xu Y, Zhou L, Liu W. AMPK phosphorylates GBF1 for mitotic Golgi disassembly. Journal of cell science. 2013;126:1498–1505. doi: 10.1242/jcs.121954. [DOI] [PubMed] [Google Scholar]
- 42.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 43.Coulonval K, Kooken H, Roger PP. Coupling of T161 and T14 phosphorylations protects cyclin B-CDK1 from premature activation. Mol Biol Cell. 2011;22:3971–3985. doi: 10.1091/mbc.E11-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S, Liu X, Bawa-Khalfe T, Lu LS, Lyu YL, Liu LF, Yeh ET. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat Med. 2012;18:1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
- 45.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 46.Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol. 2004;24:976–984. doi: 10.1128/MCB.24.3.976-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smits VA, Reaper PM, Jackson SP. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol. 2006;16:150–159. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 48.Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Molecular biotechnology. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Zeng L, Wang J, Chau JF, Lai KP, Jia D, Poonepalli A, Hande MP, Liu H, He G, He L, Li B. A positive role for c-Abl in Atm and Atr activation in DNA damage response. Cell death and differentiation. 2011;18:5–15. doi: 10.1038/cdd.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaudry D, Gonzalez BJ, Basille M, Pamantung TF, Fontaine M, Fournier A, Vaudry H. The neuroprotective effect of pituitary adenylate cyclase-activating polypeptide on cerebellar granule cells is mediated through inhibition of the CED3-related cysteine protease caspase-3/CPP32. Proc Natl Acad Sci U S A. 2000;97:13390–13395. doi: 10.1073/pnas.97.24.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozoren N, Kim K, Burns TF, Dicker DT, Moscioni AD, El-Deiry WS. The caspase 9 inhibitor Z-LEHD-FMK protects human liver cells while permitting death of cancer cells exposed to tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2000;60:6259–6265. [PubMed] [Google Scholar]
- 52.Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic acids research. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tawa NE, Jr, Odessey R, Goldberg AL. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J Clin Invest. 1997;100:197–203. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 56.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He G, Siddik ZH, Huang Z, Wang R, Koomen J, Kobayashi R, Khokhar AR, Kuang J. Induction of p21 by p53 following DNA damage inhibits both Cdk4 and Cdk2 activities. Oncogene. 2005;24:2929–2943. doi: 10.1038/sj.onc.1208474. [DOI] [PubMed] [Google Scholar]
- 58.Kim HS, Hwang JT, Yun H, Chi SG, Lee SJ, Kang I, Yoon KS, Choe WJ, Kim SS, Ha J. Inhibition of AMP-activated protein kinase sensitizes cancer cells to cisplatin-induced apoptosis via hyper-induction of p53. J Biol Chem. 2008;283:3731–3742. doi: 10.1074/jbc.M704432200. [DOI] [PubMed] [Google Scholar]
- 59.Nicol SM, Bray SE, Derek Black H, Lorimore SA, Wright EG, Lane DP, Meek DW, Coates PJ, Fuller-Pace FV. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene. 2013;32:3461–3469. doi: 10.1038/onc.2012.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Igata M, Motoshima H, Tsuruzoe K, Kojima K, Matsumura T, Kondo T, Taguchi T, Nakamaru K, Yano M, Kukidome D, Matsumoto K, Toyonaga T, Asano T, Nishikawa T, Araki E. Adenosine monophosphate-activated protein kinase suppresses vascular smooth muscle cell proliferation through the inhibition of cell cycle progression. Circ Res. 2005;97:837–844. doi: 10.1161/01.RES.0000185823.73556.06. [DOI] [PubMed] [Google Scholar]
- 61.Humbert N, Navaratnam N, Augert A, Da Costa M, Martien S, Wang J, Martinez D, Abbadie C, Carling D, de Launoit Y, Gil J, Bernard D. Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. EMBO J. 2010;29:376–386. doi: 10.1038/emboj.2009.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci U S A. 2006;103:19842–19847. doi: 10.1073/pnas.0606343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hiraku Y, Yamashita N, Nishiguchi M, Kawanishi S. Catechol estrogens induce oxidative DNA damage and estradiol enhances cell proliferation. Int J Cancer. 2001;92:333–337. doi: 10.1002/ijc.1193. [DOI] [PubMed] [Google Scholar]
- 64.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d’Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 65.Ogrunc M, Di Micco R, Liontos M, Bombardelli L, Mione M, Fumagalli M, Gorgoulis VG, d’Adda di Fagagna F. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell death and differentiation. 2014;21:998–1012. doi: 10.1038/cdd.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]