Abstract
The mature HIV-1 nucleocapsid protein (NCp7) is generated by sequential proteolytic cleavage of precursor proteins containing additional C-terminal peptides: NCp15 (NCp7-spacer peptide 2 (SP2)-p6); and NCp9 (NCp7-SP2). Here, we compare the nucleic acid chaperone activities of the three proteins, using reconstituted systems that model the annealing and elongation steps in tRNALys3-primed (-) strong-stop DNA synthesis and subsequent minus-strand transfer. The maximum levels of annealing are similar for all of the proteins, but there are important differences in their ability to facilitate reverse transcriptase (RT)-catalyzed DNA extension. Thus, at low concentrations, NCp9 has the greatest activity, but with increasing concentrations, DNA synthesis is significantly reduced. This finding reflects NCp9's strong nucleic acid binding affinity (associated with the highly basic SP2 domain) as well as its slow dissociation kinetics, which together limit the ability of RT to traverse the nucleic acid template. NCp15 has the poorest activity of the three proteins due to its acidic p6 domain. Indeed, mutants with alanine substitutions for the acidic residues in p6 have improved chaperone function. Collectively, these data can be correlated with the known biological properties of NCp9 and NCp15 mutant virions and help to explain why mature NC has evolved as the critical cofactor for efficient virus replication and long-term viral fitness.
Keywords: HIV-1, nucleocapsid precursor proteins, assembly intermediates, nucleic acid chaperone, reverse transcription, roadblock mechanism
1. Introduction
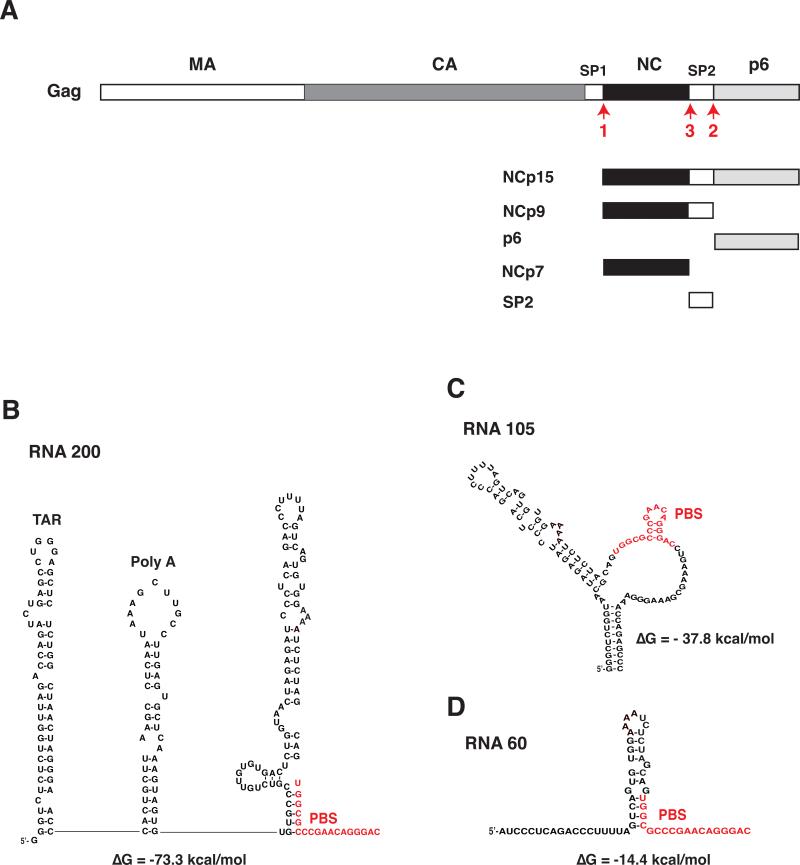
Human immunodeficiency type 1 (HIV-1) Gag (Bell and Lever, 2013) is a multidomain protein, which contains (from the N- to C-terminus): matrix (MA), capsid (CA), spacer peptide 1 (SP1), nucleocapsid (NC, also referred to as NCp7), SP2, and p6 (Fig. 1A) (Henderson et al., 1992; Mervis et al., 1988). During or shortly after budding of virus particles from the infected cell, maturation occurs and the viral protease (PR) cleaves Gag at specific sites in an ordered and sequential manner to generate the mature virus structural proteins (Adamson and Freed, 2007; Ganser-Pornillos et al., 2008; Lee et al., 2012; Swanstrom and Wills, 1997). The initial Gag cleavage event yields two products: (N-terminal) MA-CA-SP1 and (C-terminal) NCp7-SP2-p6, referred to as NCp15. Further PR cleavage of NCp15 releases NCp9 (NCp7-SP2) and p6 and in the final cleavage step, processing of NCp9 produces the mature NCp7 protein and SP2 (Fig. 1A).
Fig. 1.
Schematic representation of NC proteins and RNA templates used in this study. (A) Proteins produced by C-terminal cleavage of Gag. HIV-1 Gag is shown with each domain indicated by rectangles depicted as follows: MA, open; CA, dark gray; spacer peptide 1 (SP1), open; NCp7, closed; SP2, open; p6, light gray. The proteins derived from the Gag C-terminus, i.e., NCp15, NCp9, p6, NCp7, and SP2, are also shown. The red arrows labeled 1, 2, and 3 refer to the primary, secondary, and tertiary PR cleavages at the C-terminus of Gag (Swanstrom and Wills, 1997). (B), (C), and (D) Sequence of RNA templates and secondary structure, based on mFold analysis (Zuker, 2003): (B), RNA 200; (C), RNA 105; (D), RNA 60. Two major structural elements, i.e., the TAR and Poly A stem-loops, are present only in RNA 200. The other two templates contain varying amounts of sequence upstream of the PBS and in each case, the PBS is largely unpaired. RNA 105 also has unpaired bases downstream of the PBS in addition to a short 10-bp stem formed with bases upstream and downstream of the PBS. The PBS sequence in each template is highlighted in red. The predicted ΔG values are shown beneath the structures. The diagrams are not drawn to scale.
NCp7 is a small, basic nucleic acid binding protein containing two zinc-binding domains, i.e., zinc fingers (ZFs), each with the invariant CCHC motif, which are connected by a short basic linker peptide (Darlix et al., 2011; Darlix et al., 1995; Levin et al., 2005; Levin et al., 2010; Rein et al., 1998; Thomas and Gorelick, 2008). NCp7 and the NC domain in Gag are essential for multiple events in the virus life cycle including viral RNA dimerization and packaging, virus assembly, reverse transcription, and integration (reviewed in Darlix et al., 2011; Isel et al., 2010; Levin et al., 2005; Levin et al., 2010; Lyonnais et al., 2013; Mirambeau et al., 2010; Piekna-Przybylska and Bambara, 2011; Rein et al., 1998; Sleiman et al., 2012; Thomas and Gorelick, 2008). Importantly, NCp7 is a nucleic acid chaperone, i.e., it remodels nucleic acid structures to form the most thermodynamically stable conformations (Tsuchihashi and Brown, 1994) (reviewed in Darlix et al., 2011; Godet and Mély, 2010; Levin et al., 2005; Levin et al., 2010; Rein et al., 1998) (also see more recent refs. Hergott et al., 2013; Mitra et al., 2013; Wu et al., 2013; Wu et al., 2014). Effective chaperone activity depends on three properties: (i) aggregation of nucleic acids, which is important for annealing (associated with the basic residues); (ii) moderate duplex destabilizing activity (associated with the ZFs); and (iii) rapid on-off nucleic acid binding kinetics (Cruceanu et al., 2006a) (reviewed in Levin et al., 2005; Levin et al., 2010; Mirambeau et al., 2010; Wu et al., 2010a). This activity plays a critical role in ensuring specific and efficient reverse transcription and mediates primer placement, i.e., annealing of the tRNALys3 primer to the viral RNA genome, synthesis of (-) strong-stop DNA [(-) SSDNA]), and minus- and plus-strand transfer (Levin et al., 2005; Levin et al., 2010).
In addition to studies on the mature NC protein, the biological activity of the immediate NCp7 precursors, NCp15 and NCp9, has also been investigated. Under normal conditions, mature, infectious HIV-1 virions do not contain NCp15 and NCp9, which are transient intermediates in the virus assembly pathway (Henderson et al., 1992). In fact, blocking both C-terminal cleavage sites in Gag required for NCp15 processing, abolishes viral infectivity (Coren et al., 2007; de Marco et al., 2012) and results in assembly of virions with abnormal morphology (de Marco et al., 2012). If the cleavage site between NCp7 and SP2 is blocked giving rise to NCp9, there is some reduction (< 2-fold) in the number of particles that exhibit WT morphology (de Marco et al., 2012; Ohishi et al., 2011), but there is no clear consensus as to whether mutant virions are infectious, possibly due to differences in the constructs used or the different mutations used to maintain NCp9. For example, in one study it was reported that NCp9 mutants produced very little early viral DNA (<10%), implying that the virions were replication-negative (Ohishi et al., 2011), and in another, it was shown that blocking release of SP2 completely abolished replication (Kafaie et al., 2009). In contrast, other investigators have observed that NCp9 mutant virions were infectious in a single-cycle assay (Briggs and Kräusslich, 2011; Coren et al., 2007; Müller et al., 2009). However, in one study it was found that after four weeks in cell culture, normal processing was restored and the particles contained NCp7 instead of NCp9 (Coren et al., 2007). Studies on the behavior of NCp15 and NCp9 in several different experimental contexts, e.g., single-molecule DNA stretching (Cruceanu et al., 2006b), electron microscopic imaging of NC-DNA complexes (Mirambeau et al., 2007), biophysical and biochemical analysis of nucleic acid interactions (Wang et al., 2014), and formation of genomic RNA dimers (Jalalirad and Laughrea, 2010; Kafaie et al., 2009; Ohishi et al., 2011) showed differences between the two precursors and NCp7. However, until now, a detailed biochemical analysis comparing the nucleic acid chaperone activities of the three proteins in reverse transcription has not been reported.
In the present study, we focus on the chaperone functions of NCp15, NCp9, and NCp7, using reconstituted systems that model authentic early reverse transcription events: (i) primer placement and synthesis of (-) SSDNA; and (ii) minus-strand transfer. Both of these systems provide a sensitive readout for chaperone activity, but in one case (primer placement and (-) SSDNA synthesis), the system is driven by RNA-RNA interactions, whereas in the other (strand transfer), by RNA-DNA interactions (Levin et al., 2005; Levin et al., 2010). We demonstrate that of the three NC proteins, NCp9 has the greatest activity in these assays at low protein concentrations, but at higher concentrations, reverse transcriptase (RT)-catalyzed elongation is inhibited. This inhibitory effect is consistent with NCp9's slow dissociation kinetics (Cruceanu et al., 2006b; Wang et al., 2014) and strong nucleic acid binding activity, which reflects the presence of its highly basic SP2 domain. In addition, we show that the presence of acidic residues in the p6 domain of NCp15 negatively impacts nucleic acid chaperone activity in DNA synthesis reactions. Collectively, our results help to explain why fully processed NCp7 has evolved as the critical cofactor for HIV-1 reverse transcription, replication, and optimal viral fitness.
2. Materials and methods
2.1. Materials
DNA and RNA oligonucleotides were obtained from Integrated DNA Technologies (IDT) (Coralville, IA). T4 polynucleotide kinase, proteinase K, SUPERaseIN, and Gel Loading Buffer II were purchased from Applied Biosystems (now Life Technologies) (Foster City, CA). [γ-33P]ATP, [α-33P]dCTP, and [α-33P]GTP were purchased from PerkinElmer (Shelton, CT). Purified tRNALys3 from human placenta was obtained from Bio S&T (Lachine, Québec, Canada). RNA transcripts were synthesized by using the Ambion MEGAshortscript T7 kit (Life Technologies). HIV-1 RT was obtained from Worthington (Lakewood, NJ). The SP2 peptide (FLGKIWPSHKGRPGNF) was purchased from GenScript (Piscataway, NJ). The sequences of the viral nucleic acid substrates and the RNA 200 and RNA 60 templates were derived from HIV-1 NL4-3 (GenBank Accession no. AF324493) (Adachi et al., 1986). The RNA 105 template was derived from the HIV-1 HXB2 sequence (GenBank Accession no. K03455), as described previously (Hargittai et al., 2001).
2.2. Recombinant NC proteins
The NC proteins (containing the HIV-1 NL4-3 sequence (see above, Section 2.1) were expressed in E. coli and were purified as described previously: (i) NCp7 (Carteau et al., 1999; Wu et al., 1996); and (ii) NCp9 and NCp15 (Cruceanu et al., 2006b). Mutations for expression of the mutant NCp15 proteins (Fig. 8A) are described in (Wang et al., 2014).
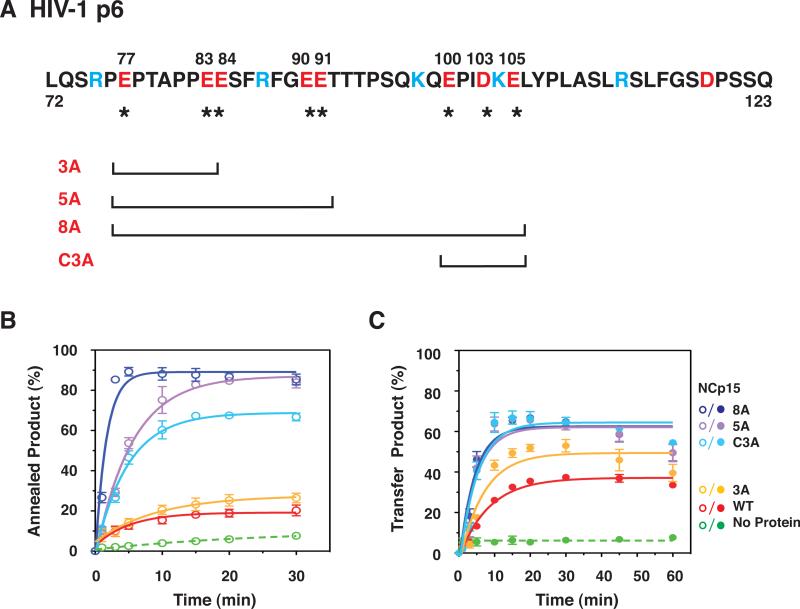
Fig. 8.
Effect of WT NCp15 and mutants with changes in the C-terminal p6 domain on the kinetics of minus-strand annealing and strand transfer. (A) Primary sequence of HIV-1 p6 and schematic representation of C-terminal NCp15 mutants. The acidic and basic residues are highlighted in red and blue, respectively. Acidic residues (E or D) that were changed to A are identified by their position number in the NCp15 sequence (above) and with an asterisk (underneath). The mutants are designated by the number of Ala substitutions. C3A refers to mutation of C-terminal residues 100, 103, and 105. The square brackets, one at each end, show the region that has the Ala substitutions for each mutant. (B) and (C) Kinetics of minus-strand annealing (B) and strand transfer (C) in reactions with WT NCp15 and mutants. In (B), the NC concentrations were all 0.2 μM; in (C), the concentrations were all 0.8 μM, with the exception of 8A, which was 0.4 μM. The data for all of the NC-containing reactions (solid lines) were fit to a single exponential equation. The curves are highlighted in color as follows: WT, red; 3A, yellow; C3A, light blue; 5A, lavender; and 8A, dark blue. The no protein controls (dashed lines) are highlighted in green. Symbols: Annealing, open; Minus-Strand Transfer, closed. The % annealed and transfer products were plotted against time of incubation.
Incubation of 32P-labeled RNA with increasing amounts of NCp9 or NCp15 had no effect on the integrity of the RNA, indicating that the protein preparations were free of RNase contamination (data not shown). Theoretical pI values were calculated with the Protparam web-based program (http://web.expasy.org/compute_pi/).
2.3. Synthesis of (-) SSDNA
The assay was conducted with two different primers: tRNALys3 and an 18-nt DNA oligonucleotide complementary to the primer binding site (PBS) sequence in viral RNA (DNA PBS) (Iwatani et al., 2003).
2.3.1. tRNALys3 primer
The RNA 200 template (Iwatani et al., 2003) and tRNALys3 primer were prepared by in vitro transcription with T7 RNA polymerase. The DNA clone used for synthesizing the tRNA was a generous gift from Christopher P. Jones and Karin Musier-Forsyth (The Ohio State University) (Hargittai et al., 2001). For assay of (-) SSDNA synthesis, 0.4 pmol of RNA 200 was annealed to 0.2 pmol of unlabeled tRNALys3 primer in a solution containing annealing buffer (50 mM Tris-HCl, pH 8.0, 75 mM KCl) and 0.5 units of SUPERaseIN at 37 °C for 15 min in the absence or presence of increasing concentrations of HIV-1 NCp7, NCp9, or NCp15. Following annealing, reaction buffer (50 mM Tris-HCl, pH 8.0, 75 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol [DTT]), 50 μM each of the four dNTPs, and 10 μCi of [α-33P]dCTP were added and the mixture was incubated further at 37 °C for 5 min. The reaction was initiated by addition of HIV-1 RT (0.2 pmol) and the mixture (final volume, 20 μl) was then incubated at 37 °C for 60 min. Reactions were terminated by addition of 1 μl of Proteinase K (20 mg/ml) and heating at 65 °C for 15 min, followed by addition of 8 μl of Gel Loading Buffer II. The samples were heated at 90 °C for 5 min prior to loading onto a denaturing 8% polyacrylamide/7 M urea gel. 33P-labeled DNA products, including (-) SSDNA, were quantified by using a Typhoon 9400 PhosphorImager (Molecular Dynamics) and ImageQuant software. To calculate the percentage of (-) SSDNA that was synthesized, the amount of (-) SSDNA product was divided by the sum of all products larger than the tRNALys3 primer and multiplied by 100.
2.3.2. DNA PBS primer
Three templates were used with this primer: RNA 200 (Iwatani et al., 2003); RNA 105, a T7 transcript made with a clone kindly provided by Christopher P. Jones and Karin Musier-Forsyth (Hargittai et al., 2001); and RNA 60, a synthetic oligonucleotide with the sequence 5’-AUC CCU CAG ACC CUU UUA GUC AGU GUG GAA AAU CUC UAG CAG UGG CGC CCG AAC AGG GAC. In this assay, 0.4 pmol of the indicated RNA template was annealed to 0.2 pmol of the DNA PBS primer, labeled with 33P at its 5’ end (Guo et al., 1995), in the absence or presence of HIV-1 NCp7 or NCp9 (as specified) in annealing buffer (50 mM Tris-HCl, pH 8.0, 75 mM KCl) at 37 °C for 15 min. The remainder of the assay was performed as described above (see Section 2.3.1), except that labeled dCTP was not added. To calculate the percentage of (-) SSDNA that was synthesized, the amount of (-) SSDNA was divided by the total amount of DNA and multiplied by 100.
2.4. Annealing assays
2.4.1. tRNALys3 annealing
tRNALys3 (0.3 pmol), internally labeled with 33P (transcription performed with [α-33P]GTP), was incubated with 0.6 pmol of RNA 200 in a final volume of 30 μl at 37 oC for 60 min. Reactions were carried out in the absence or presence of NC proteins in buffer containing 50 mM Tris-HCl (pH 8.0) and 75 mM KCl. After incubation, the reaction mixture was treated with SDS (final concentration, 1%) at 37 °C for 10 min and then extracted twice with phenol:chloroform (5:1 vol:vol, pH 4.5) to remove the protein. The annealed product was separated from unannealed tRNA by polyacrylamide gel electrophoresis (PAGE) in a 6% native gel (acrylamide:bis-acrylamide, 19:1). The amounts of unannealed and annealed tRNA were determined by PhosphorImager analysis. To calculate the percentage of annealed tRNA, the amount of annealed product was divided by the sum of annealed plus unannealed tRNA and multiplied by 100.
2.4.2. Minus-strand annealing
A 128-nt DNA (DNA 128) representing (-) SSDNA (0.2 pmol), labeled at its 5’ end with 33P, was incubated at 37 °C with 0.2 pmol of acceptor RNA (RNA 148) (Guo et al., 1997; Heilman-Miller et al., 2004) in a final volume of 20 μl. Reactions were carried out in the absence or presence of the three NC proteins or NCp15 mutants in buffer containing 50 mM Tris-HCl (pH 8.0) and 75 mM KCl. To assay the kinetics of annealing, reaction mixtures were scaled up as needed and 10-μl aliquots were removed at the indicated times. Reactions were terminated as described above (see Section 2.3.1), except that following Proteinase K addition, the reaction mixtures were incubated at 37 °C for 15 min. Separation of the labeled DNA-RNA hybrid from unannealed (-) SSDNA by PAGE, analysis of the gel data, and calculation of the percentage of annealed DNA were performed as described previously (Wu et al., 2007).
2.5. Minus-strand transfer assay
The minus-strand transfer assay was performed as described previously (Wu et al., 2010b). Briefly, reaction mixtures contained reaction buffer (50 mM Tris-HCl (pH 8.0), 75 mM KCl, 1 mM DTT), DNA 128 labeled at its 5’ end with 33P, RNA 148, and HIV-1 RT, each at a final concentration of 10 nM, 0.5 units of SUPERaseIN, 100 μM each of the four dNTPs, and 1 mM MgCl2 in the absence or presence of NC proteins, as specified (final volume, 20 μl), and were incubated at 37 °C for 1 h. Termination of the reactions, PAGE, and PhosphorImager analysis were performed as described above (see Section 2.3.1). To assay the kinetics of strand transfer, reaction mixtures were scaled up as needed and 10-μl aliquots were removed at the indicated times. The percentage of strand transfer product formation was calculated by dividing the amount of transfer product by the total signal present in the gel lane and multiplying by 100 (Hergott et al., 2013).
2.6. Fluorescence anisotropy (FA) experiments
Equilibrium binding of HIV-1 NC proteins (NCp7, NCp9, NCp15, and NCp15 mutants) to a 20-nt ssDNA oligonucleotide containing bases complementary to the extreme 3’ terminus of the unique 3’ sequence (U3) (JL936, 5’-AlexaFluor 488-AGC TGC TTT TTG CCT GTA CT) and labeled at its 5’ end with a fluorescent tag, was measured using FA, as described in detail by Wu et al. (Wu et al., 2010b). The plot of anisotropy vs. protein concentration was fit using a one-site binding model (Iwatani et al., 2007) to obtain the apparent dissociation constant (Kd).
2.7. Aggregation assay
DNA 128 labeled at its 5’ end with 33P (0.2 pmol) was added to RNA 148 (0.2 pmol) in a final volume of 20 μl. Reactions were carried out in the absence or presence of NC proteins in buffer containing 50 mM Tris-HCl (pH 8.0) and 75 mM KCl at 37 °C for 60 min. Following incubation, reaction mixtures were centrifuged at 13,800 x g in a microcentrifuge (Sorvall LEGEND MICRO 17R) for 20 min at 4 °C. The supernatant (2 μl) was loaded onto a DEAE Filtermat (PerkinElmer) and the radioactivity was analyzed using a PhosphorImager and ImageQuant software. The percent radioactivity in the pellet was calculated as follows: the amount of radioactivity in the supernatant relative to the control sample (no NC; set to 100%) was determined and then subtracted from 100%. The data were plotted as a function of NC concentration.
In all of the assays described above (see Sections 2.3-2.7), the data represent the average of results obtained from three or more independent experiments. Error bars represent the standard deviation (SD). Note that the error bars for some of the data points were too small to see on the graphs.
3. Results
3.1. Activity of NCp7, NCp9, and NCp15 in an assay for tRNALys3-primed synthesis of (-) SSDNA
In this study, we set out to analyze the nucleic acid chaperone activities of NCp15 (NCp9-p6), NCp9 (NCp7-SP2), and NCp7 (Fig. 1A) in early reverse transcription events. In initial experiments, we measured (-) SSDNA synthesis (Fig. 2A), which consists of two reactions: (i) annealing of the 3’ 18 nt of tRNALys3 to the PBS in the viral RNA template (RNA 200, Fig. 1B); and (ii) RT-catalyzed elongation of annealed tRNALys3, yielding (-) SSDNA covalently attached to the tRNA primer. NC-mediated destabilization of the highly structured tRNA and RNA template is required for a positive read-out in this assay (Levin et al., 2010). Note that RNA 200 contains the repeat (R) region (including the 59-nt transactivation response element (TAR) stem-loop and the 5’ portion of the poly A stem-loop), U5 (including the 3’ portion of the poly A stem-loop followed by a stem-U-rich loop), and the PBS (Fig. 1B).
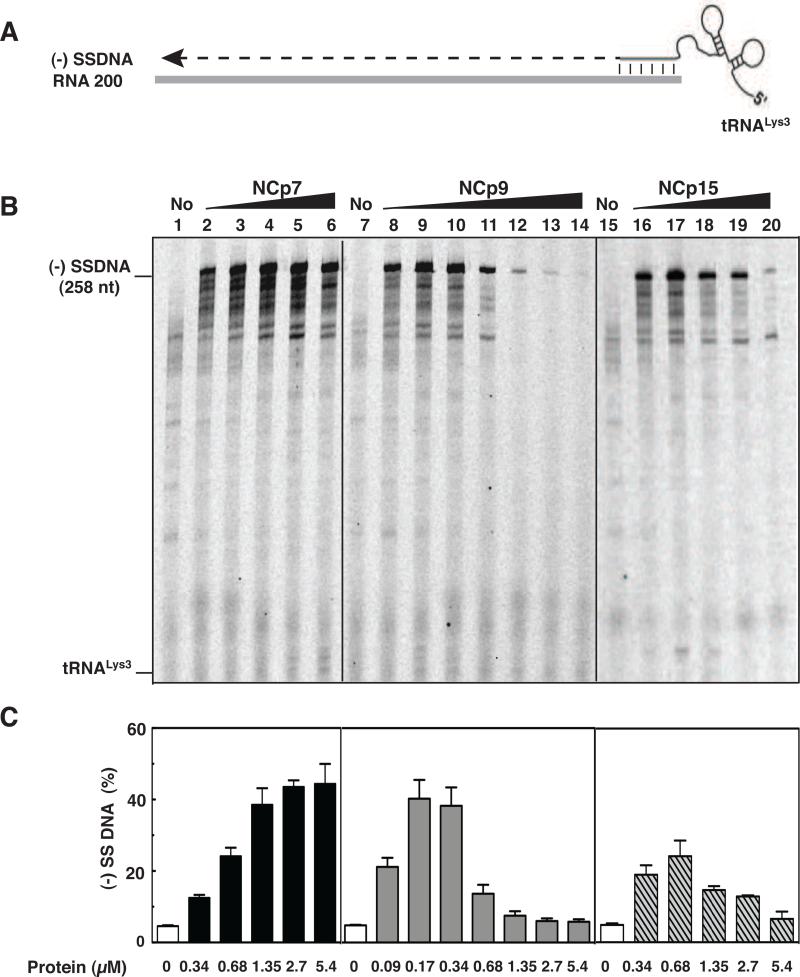
Fig. 2.
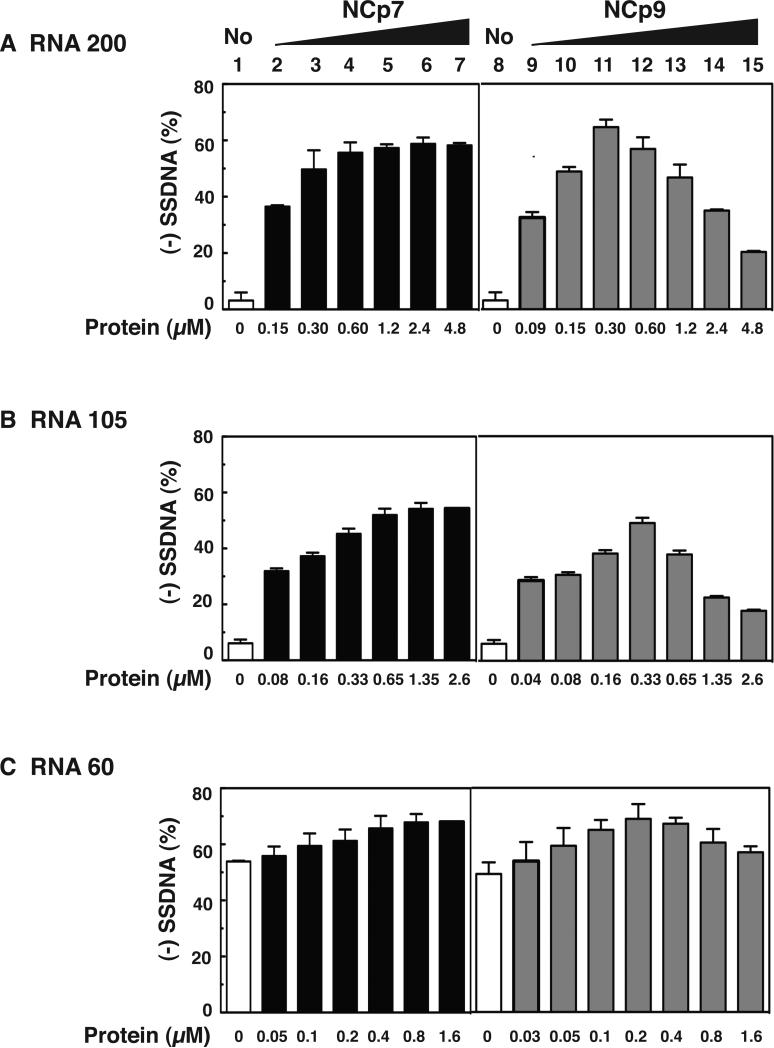
Effect of HIV-1 NC proteins on (-) SSDNA synthesis. (A) Reconstituted system used for assay of (-) SSDNA synthesis. The diagram shows annealing (vertical lines) of the 18 nt at the 3’ end of tRNALys3 to the complementary 18-nt PBS in RNA 200 (gray rectangle), which serves as the template for RT-catalyzed synthesis of (-) SSDNA (dashed line). The diagram is not drawn to scale. (B) Gel analysis. Unlabeled tRNALys3 was annealed to RNA 200 in the absence (lanes 1, 7, and 15) or presence of increasing concentrations of NCp7 (lanes 2 to 6), NCp9 (lanes 8 to 14), and NCp15 (lanes 16 to 20) and was extended by HIV-1 RT. The DNA products were separated by PAGE in a 6% denaturing gel. The position of marker tRNALys3 is shown on the left. (C) Bar graphs showing the percentage (%) of (-) SSDNA product synthesized as a function of NC protein concentration. Symbols: no protein, open bars; NCp7, closed bars; NCp9, gray bars; NCp15, hatched bars.
To compare the chaperone activities of NCp7, NCp9, and NCp15, we performed the assay in the presence of increasing concentrations of each protein, separated the products by PAGE (Fig. 2B), and quantified the gel data by PhosphorImager analysis (Fig. 2C). At low protein concentrations (e.g., 0.17 μM), NCp9 clearly had the greatest activity. In fact, the level of (-) SSDNA synthesized with 0.17 or 0.34 μM NCp9 (~40 % yield of full-length (-) SSDNA) was only achieved in NCp7 reactions containing ≥1.35 μM protein. However, as the protein concentration was increased, NCp7 dramatically stimulated synthesis of (-) SSDNA (Fig. 2B and 2C, compare lanes 2 to 6 with lane 1), whereas the activities of NCp9 and NCp15 were ultimately reduced to base-line levels (compare lanes 8 to 14 with lane 7 [NCp9] and lanes 16 to 20 with lane 15 [NCp15]). This effect was particularly striking with NCp9. The maximum level of synthesis achieved in NCp15 reactions (0.68 μM protein, lane 17) was 2-fold lower than the highest levels observed with NCp9 and NCp7. Taken together, these results show that the protein concentration-dependence of NCp7's chaperone activity in (-) SSDNA synthesis reactions differs from the activities of NCp9 and NCp15.
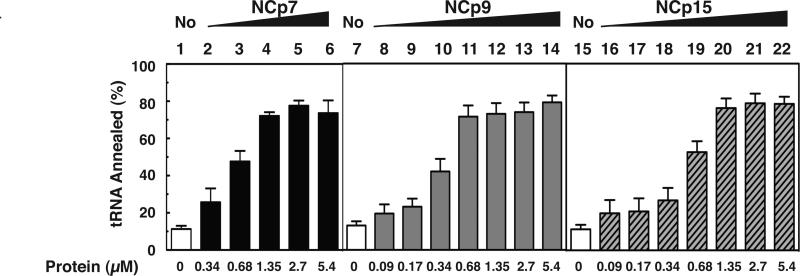
To determine whether it was the annealing or elongation step that was affected by high concentrations of NCp9 and NCp15, we assayed annealing alone as a function of NC concentration (Fig. 3 and Suppl. Fig. 1). The maximum extent of annealing was ~80% for all three NCs, but was achieved at a 2-fold lower concentration with NCp9 (0.68 μM) than with NCp7 and NCp15 (1.35 μM, respectively). Thus, the three NC proteins have similar annealing activities. Importantly, the data also imply that the observed reduction of (-) SSDNA synthesis (Fig. 2) was due to a decrease in RT-catalyzed DNA extension in reactions with NCp9 and NCp15 and not to a defect in annealing.
Fig. 3.
Effect of HIV-1 NC proteins on annealing of tRNALys3 to RNA 200. Annealing was performed with tRNALys3, uniformly labeled with 33P. Bar graphs show the % tRNA annealed as a function of NC protein concentration. Symbols: no protein, open bars; NCp7, closed bars; NCp9, gray bars; NCp15, hatched bars. The results are based on analysis of 6% native polyacrylamide gels, which separate annealed and unannealed tRNA Lys3. A representative gel is shown in Suppl. Fig. 1.
3.2. Effect of different primers and RNA templates of varying length on activities of NCp7 and NCp9 in assays of (-) SSDNA synthesis
To better understand the difference between the activities of NCp7 and NCp9, we systematically investigated the effects of varying the primers and templates used for the (-) SSDNA synthesis assay. In the reactions described thus far, the tRNALys3 primer was a T7 RNA transcript containing unmodified bases. However, substitution of human placenta tRNALys3 (having modified bases) in reactions with NCp9 did not change the outcome of the assay (Suppl. Fig. 2). Thus, the reduction in (-) SSDNA synthesis seen at high NCp9 concentrations was not related to the presence or absence of modified bases in the tRNA.
To determine whether the course of (-) SSDNA synthesis would be affected if an oligonucleotide primer were used, reactions were incubated with an 18-nt DNA PBS primer and the RNA 200 template in the presence of increasing concentrations of NCp7 and NCp9 (Fig. 4A). (-) SSDNA synthesis primed by the DNA oligonucleotide was efficient in the presence of NCp7 as reported previously (Ghosh et al., 1997; Iwatani et al., 2003) and the high level of product (~60%) remained fairly constant with concentrations ranging from 0.60 to 4.8 μM (lanes 5 to 7, respectively). With NCp9, the amount of (-) SSDNA reached the maximum level at 0.30 and 0.60 μM protein (lanes 11 and 12), but was significantly reduced at higher concentrations of NCp9 (compare lanes 11 and 12 with lanes 13 to 15). Interestingly, compared with the tRNALys3 reactions (Fig. 2), the maximum activity achieved with the DNA primer was higher (~65% vs. ~40%), but the reduction of activity with high NCp9 concentrations was ~2-fold less. These results reflect the fact that tRNALys3, but not the DNA PBS primer, must be destabilized by NC chaperone activity and indicate that the more stringent the NC requirement, the greater the inhibitory effect with NCp9. Note that in reactions with NCp15, reduction of activity with increasing protein concentration showed the same trend as that seen with NCp9, except that the maximum level of activity was lower (~43%) (data not shown). This finding is consistent with the data of Fig. 2.
Fig. 4.
Activity of NCp7 and NCp9 in assays for (-) SSDNA synthesis using three different RNA templates and a DNA PBS primer. Templates: (A) RNA 200; (B) RNA 105; (C) RNA 60. Bar graphs show the % (-) SSDNA that was synthesized in the absence (1 and 8, open bars) or presence of increasing concentrations of NCp7 (2 to 7, closed bars) and NCp9 (9 to 15, gray bars).
The effect of RNA template structure on (-) SSDNA synthesis was evaluated in reactions with the DNA PBS primer and NCp7 or NCp9, by testing two templates that are shorter than RNA 200: RNA 105 (Fig. 1C) and RNA 60 (Fig. 1D). RNA 105 is missing the TAR and poly A stem-loops that are present in RNA 200 (Fig. 1B). With this template (Fig. 4B), the maximum amount of product (~50%) was reached at a concentration of 0.65 μM NCp7 (Fig. 4B, lane 5) and remained constant when higher concentrations of protein were present. With NCp9, maximum synthesis of (-) SSDNA (~50%) occurred at 0.33 μM (Fig. 4B, lane 12) and was reduced as the concentration of protein was increased. These data mirror the results obtained with RNA 200 (Fig. 4A), except that the efficiency of (-) SSDNA synthesis was somewhat greater with RNA 200 than with RNA 105. Note that to copy the 5’ end of RNA 105, a stable 10-base pair (bp) stem containing 7 G-C bp (Fig. 1C) has to be transiently destabilized by NC. Since local structure is a major determinant of chaperone activity (Hargittai et al., 2004; Heilman-Miller et al., 2004; Wu et al., 2007), the 10-bp structural element could affect the efficiency of template utilization.
The RNA 60 template has a relatively weak secondary structure (ΔG, -14.4 kcal/mol) compared with the other two templates (Fig. 1D). It consists of three elements: a short stem-loop with two bulges and a 5-nt loop; and 18- and 14-nt ss regions at its 5’ and 3’ ends, respectively. In this case, NCp7 and NCp9 had very little effect on (-) SSDNA synthesis, although a low level of stimulatory activity with NCp7 (Fig. 4C, compare lane 1 with lanes 5 to 7) and some reduction in synthesis at high concentrations of NCp9 (Fig. 4C, compare lane 8 with lanes 13 to 15) were seen. These data reflect the minimal requirement for NC helix destabilizing activity in the DNA PBS-RNA 60 reactions.
Collectively, the results demonstrate that in reactions that require NC chaperone activity, the reduction in DNA synthesis observed with high concentrations of NCp9 is modulated by the structure of the RNA template and the primer.
3.3. Kinetics of minus-strand annealing and transfer
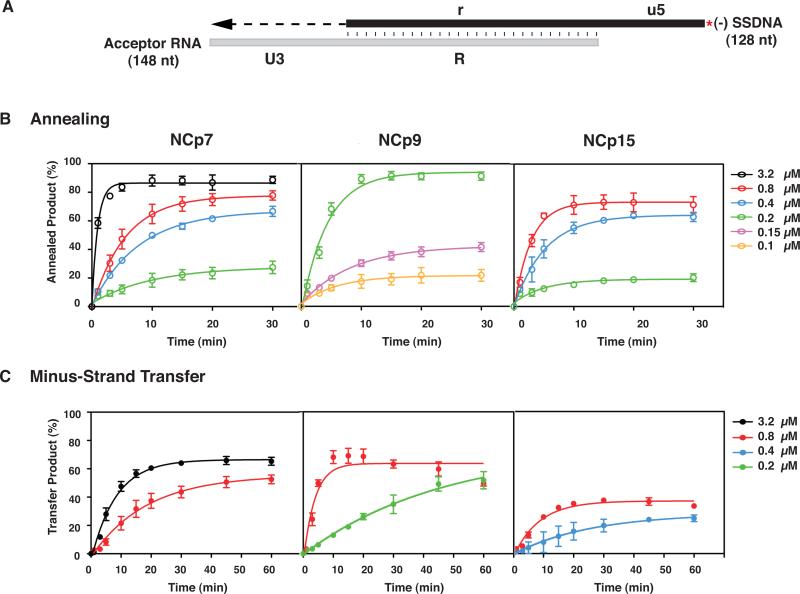
To further quantify differences in the nucleic acid chaperone activities of the NC proteins, we performed experiments with our reconstituted minus-strand transfer assay system (Heilman-Miller et al., 2004; Wu et al., 2010b). In this system, two reactions take place: (i) annealing of the complementary TAR-containing repeat (R) regions in the RNA 148 acceptor and (-) SSDNA (i.e., DNA 128) substrates; and (ii) RT-catalyzed elongation of annealed (-) SSDNA, using the 54-nt U3 sequence in the acceptor as the template (Fig. 5A). The kinetics of minus-strand annealing (Fig. 5B) and minus-strand transfer (i.e., annealing plus (-) SSDNA extension) (Fig. 5C) were analyzed. The concentrations chosen for assay were based on end-point dose response experiments (Fig. 7 and data not shown).
Fig. 5.
Kinetics of minus-strand annealing and transfer in the presence of NCp7, NCp9, and NCp15. (A) Reconstituted system used for the minus-strand transfer assay. The diagram shows the acceptor RNA with a portion of U3 (54 nt) and the R sequence from the 3’ end of the HIV-1 genome and (-) SSDNA with a portion of u5 and the r sequence, complementary to the sequence at the 5’ end of the viral genome (Guo et al., 1997; Heilman-Miller et al., 2004). Annealing of the R regions in RNA 148 and 33P-labeled DNA 128 is depicted by vertical lines. The red asterisk indicates that the (-) SSDNA is labeled at its 5’ end with 33P. The U3 sequence serves as the template for RT-catalyzed extension of annealed (-) SSDNA. The final DNA transfer product is 182 nt. The diagram is not drawn to scale. (B) Annealing and (C) Minus-Strand Transfer. Reactions were incubated at 37 °C with different concentrations of NCp7, NCp9, or NCp15 for up to 30 (B) or 60 (C) min and analyzed as described in Materials and Methods. The % annealed product (B) and % minus-strand transfer product (C) were plotted against time of incubation. The data were fit to a single exponential equation. Symbols: Annealing, open; Minus-Strand Transfer, closed. Protein concentrations are indicated by the following colors: 0.1 μM, yellow, 0.15 μM, lavender; 0.2 μM, green, 0.4 μM, blue; 0.8 μM, red and 3.2 μM, black. Note that in this series of experiments, the maximum values for the minus NC controls were less than 7% (annealing) and 3% (minus-strand transfer).
Fig. 7.
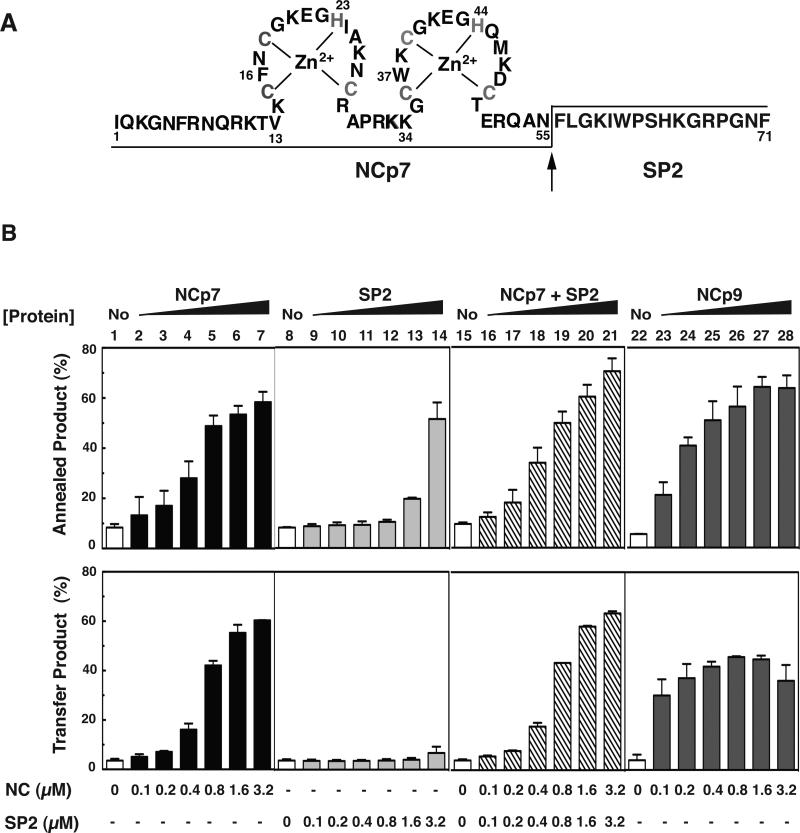
Minus-strand annealing and transfer activity of NCp7 in the absence or presence of HIV-1 SP2 added in trans. (A) Primary sequence of NCp9. The zinc coordinating residues (Cys and His) are shown in gray. The arrow indicates the PR cleavage site. The final products NCp7 (55 residues) and SP2 (16 residues) are separated by lines placed at the bottom or the top of the sequence. (B) Bar graphs. 33P-labeled DNA 128 was incubated with RNA 148 for 60 min in the absence (No, open bars, lanes 1, 8, 15, and 22) or presence of increasing concentrations of NCp7 as follows: NCp7, closed bars, lanes 2 to 7; SP2, light gray bars; lanes 9 to 14; NCp7+SP2 in trans, hatched bars, lanes 16 to 21; NCp9, dark gray bars, lanes 23 to 28). The % annealed product (upper series of bar graphs) or % transfer product (lower series of bar graphs) was plotted as a function of protein concentration. Note that in each NCp7+SP2 reaction, the two proteins were present in equal concentrations.
NCp7 stimulated annealing as a function of increasing protein concentration and time and the maximum level of annealed product (~90%) was reached at 10 min with 3.2 μM protein (Fig. 5B). By contrast, NCp9 gave very similar results at 10 min, but with a 16-fold lower protein concentration (0.2 μM). With 0.8 μM NCp7 and NCp15, the maximum amount of annealed product (~78 % and ~73%, respectively) was reached at 20 to 30 min and 10 min, respectively. At lower concentrations of NCp7 and NCp15 (i.e., 0.4 and 0.2 μM), the end point values were very similar.
The rates of annealing were calculated for reactions containing NCp7 and NCp15, each at 0.8 μM, and 0.2 μM NCp9 (Table 1, middle column). The values obtained with NCp7 and NCp15 were similar, with only a 2-fold higher rate observed with NCp15. The rate with NCp9 was similar to the rates for the other two proteins, but at a 4-fold lower concentration. To account for these differences, we used FA to measure the nucleic acid binding affinity of each NC protein for a 20-nt ssDNA oligonucleotide (complementary to the first 20 nt in U3) (Table 2, middle column). Consistent with the data of Fig. 5B and Table 1, the Kd values for NCp7 and NCp15 showed little difference (176±12.7 nM and 151±5.1 nM, respectively), whereas the Kd value for NCp9 (45±1.7 nM) was ~3- to 4-fold lower than the values obtained for NCp7 and NCp15. Taken together, these data show that of the three proteins, NCp9 has the highest affinity for binding ssDNA and the greatest activity in the annealing assay, due to the higher cationic density provided by its SP2 domain.
Table 1.
Rates of annealing or strand transfer (kobs values)a
| Protein | Annealing (min−1) | Strand transfer (min−1) |
|---|---|---|
| NCp7 (0.8 μM) | 0.18 ±0.008b | 0.047 ±0.015 |
| NCp15 (0.8 μM) | 0.35 ±0.029 | 0.093 ±0.019 |
| NCp9 (0.8 μM) | n.d.c,d | 0.22 ±0.055 |
| NCp9 (0.2 μM) | 0.23 ±0.032 | nd.e |
Rates were determined by fitting the data from Figure 5B (annealing) and 5C (strand transfer) to a single exponential equation. The data represent the average of results obtained from at least three independent experiments.
The error determinations represent the SD.
n.d., not determined.
Rate was too fast to measure accurately at concentrations > 0.2 μM.
Rate was too slow to measure accurately at 0.2 μM.
Table 2.
Apparent dissociation constants for binding of NC proteins to 20-nt ssDNA and theoretical isoelectric points (pI)
| Protein/Peptide | (nM) | pI |
|---|---|---|
| NCp7 | 176 ± 12.7a | 9.93 |
| NCp15 | 151 ± 5.1 | 9.59 |
| 3A | 151 ± 12.6 | 9.90 |
| C3A | 73 ± 15.5 | 9.90 |
| 5A | 65 ± 9.6 | 10.10 |
| 8A | 50 ± 8.9 | 10.44 |
| NCp9 | 45 ± 1.7 | 10.19 |
| SP2 | n.d.b | 11.17 |
| p6 | n.d. | 4.48 |
The error determinations represent the SD.
n.d., not determined.
We also assayed the kinetics of minus-strand transfer (Fig. 5C). At 0.8 μM, maximum activity was reached by NCp7 at 60 min (53%), by NCp15 at 30 min (37%), and by NCp9 at 10 min (70%); however, with NCp9, the amount of transfer product decreased to ~50% at 60 min. (The reason for this is not known, but is possibly related to NCp9's strong aggregating activity (see below, Section 3.4 and Fig. 6 and 7), which may remove the nucleoprotein complex from solution). Interestingly, with 3.2 μM NCp7, maximum strand transfer occurred by 30 min (65%), comparable to the maximum value for NCp9 at 0.8 μM. At a lower concentration (0.2 μM) of NCp9, the transfer product increased with time and by 45 min, a stable value of ~50% was reached. In contrast, the reaction with 0.4 μM NCp15 was inefficient and the maximum amount of transfer product was only 25%.
Fig. 6.
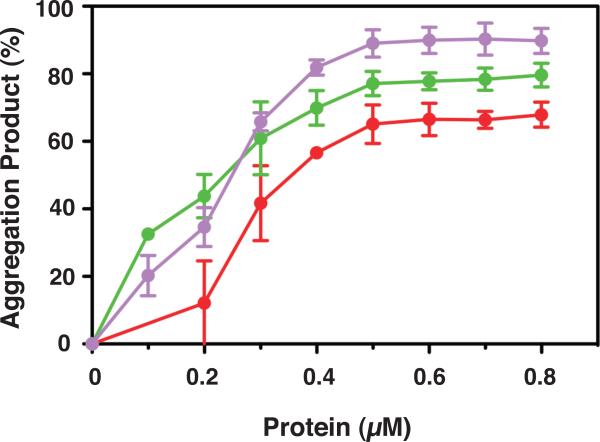
Effect of NC proteins on nucleic acid aggregation. 33P-labeled DNA 128 was incubated with RNA 148 for 60 min at 37 °C in the absence or presence of increasing concentrations of NCp7, NCp9, and mutant 8A, under the conditions used for annealing in minus-stand transfer. The % aggregated nucleic acid present in the reaction mixture following incubation was plotted against protein concentration. The curves are highlighted in color as follows: NCp7, red; NCp9, green; mutant 8A, lavender.
The rates of minus-strand transfer were calculated from the experimental data for reactions containing 0.8 μM protein (Table 1, last column). Of the three proteins, NCp9 had the highest kobs value (0.22 ±0.055 min−1), ~4.5-fold higher than the rate for NCp7 and 2.3-fold higher than the rate for NCp15. Although the rates for strand transfer were lower than the rates for annealing alone (Table 1, compare values in middle and last columns), the rank order was the same in both assays, i.e., NCp9>NCp15~NCp7.
3.4. Aggregation activities of NCp7 and NCp9
NC proteins are able to aggregate nucleic acids, which is important for facilitating annealing reactions (Anthony and Destefano, 2007; Le Cam et al., 1998; Mirambeau et al., 2007; Mirambeau et al., 2006; Stoylov et al., 1997). To determine whether the aggregation activities of the individual NC proteins could be correlated with their behavior in assays for annealing and nucleic acid binding affinity, we measured the percentage of (-) SSDNA that is aggregated as a function of increasing concentrations of NC proteins in minus-strand annealing reactions (Fig. 6). Both NCp7 and NCp9 promoted aggregation in a dose-dependent manner, but NCp9 displayed the highest aggregation activity. At 0.2 μM, the aggregation product represented ~45% of the total DNA in the reaction and at concentrations ≥ 0.5 μM, the aggregated fraction reached ~80%. In contrast, NCp7 showed lower aggregation activity, which increased more gradually as the protein concentration was raised from 0.2 to 0.8 μM. At 0.8 μM, the amount of aggregated nucleic acid reached close to 70%, the same value observed for NCp9 at only 0.4 μM, The highly efficient aggregation activity of NCp9 is consistent with the annealing and nucleic acid binding data presented above and presumably reflects the strong basic character of NCp9 (pI, 10.19 [Table 2, last column]). Note that data for NCp15 are not presented here, since NCp15 does not form authentic aggregates upon sedimentation (see below, Section 4) (Mirambeau et al., 2007; Wang et al., 2014).
3.5. Contribution of SP2 to NCp9 chaperone activity
NCp9 is even more basic than NCp7 because its C-terminus consists of the short 16-amino acid SP2 peptide (Fig. 1A and 7A), having a calculated pI value of 11.17 (Table 2, last column). To determine whether SP2 contributes to NCp9 nucleic acid chaperone activity, we investigated the effect of a synthetic SP2 peptide on minus-strand annealing and strand transfer. We performed both assays with either SP2 alone or with NCp7+SP2 (addition of SP2 in trans) and compared the results with the activities of NCp7 alone and NCp9 (Fig. 7B).
As shown in Fig. 7B (upper panel), SP2 alone stimulated annealing to a significant extent (annealed product, ~50%), but only at the highest concentration tested (lane 14, 3.2 μM); at the same concentration of NCp7, the % annealed product reached a value of 60% (lane 7). Moreover, when 0.1 to 0.4 μM SP2 was added in trans to a reaction with NCp7 (ratio of NCp7:SP2 in each reaction, 1:1), the activity was the same as that of NCp7, but lower than that of NCp9 (compare lanes 16, 17, 18 with lanes 1, 2, 3 and 23, 24, 25). Reactions containing high concentrations of NCp7+SP2 or NCp9 resulted in similar levels of annealing (compare lanes 19 to 21 with lanes 26 to 28). At a concentration of 3.2 μM, a higher level of annealing was observed with NCp7+SP2 (lane 21) than with NCp7 alone (lane 7).
In contrast to these results, SP2 had no effect on minus-strand transfer (Fig. 7B, lower panel, lanes 8 to 14) and it did not increase the activity of NCp7 when added in trans (compare lanes 1 to 7 with lanes 15 to 21). These observations presumably reflect the fact that SP2 lacks the requisite helix destabilizing activity needed to transiently unwind the TAR structures. At low concentrations (0.1 to 0.4 μM), NCp9 had a greater stimulatory effect on strand transfer than either NCp7 or NCp7+SP2 (compare lanes 23 to 25 with lanes 2 to 4 and 16 to 18). However, at ≥ 1.6 μM, the amount of transfer product was lower with NCp9 than with NCp7 alone or NCp7+SP2 (compare lanes 27 and 28 with lanes 6 and 7 and lanes 20 and 21). When the concentrations of NCp7 and NCp9 were increased even higher to 4.8 or 6.4 μM, NCp7 activity was unchanged, but the reduction in NCp9 activity was more striking than that seen in the Fig. 7 assay (Suppl. Fig. 3).
3.6. Nucleic acid chaperone activities of NCp15 WT and mutant proteins
The C-terminal p6 domain of NCp15 is acidic (pI, 4.48 [Table 2, last column]) due to the presence of 7 Glu and 2 Asp residues (Fig. 8A). We hypothesized that this domain might be responsible for the fact that NCp15 nucleic acid chaperone activity is weaker than that of NCp9 and NCp7, especially in assays that involve annealing plus elongation. To approach this question, we made a series of mutants in which the acidic residues in p6 were changed to Ala (Fig. 8A) and then compared the activities of WT and mutant proteins in time-course assays for minus-strand annealing (Fig. 8B) and strand transfer (Fig. 8C). Annealing reactions all contained 0.2 μM protein so that differences in activity could be highlighted. With WT NCp15, the maximum amount of annealed product (~20%) was reached after incubation for 30 min; with mutant 3A, annealing reached a maximum value of ~30%. Mutants with a greater number of Ala substitutions (5A and 8A) as well as mutant C3A (Ala substitutions in three acidic C-terminal residues) all showed greatly improved activity compared to WT, with mutant 8A having the greatest activity (~90%) after only 5 min of incubation. Maximum annealing with mutants C3A and 5A was reached by 15 min and was ~65% and 80%, respectively.
The rates of annealing were calculated for the reactions with mutants C3A, 5A, and 8A (Table 3) and the rank order was 8A>C3A~5A. Interestingly, the rate for mutant 8A was 3-fold higher than that of NCp9 at the same concentration (Table 1, middle column). The rates for the WT and 3A reactions were too slow to be calculated accurately. However, the trend is clear: reducing the acidic character of NCp15 improved the rate and extent of annealing and in general, the greater the number of Ala substitutions, the greater the activity. Moreover, a decrease in Kd values follows the same general trend (Table 2 [middle column]), indicating that nucleic acid binding affinity becomes more efficient as the protein becomes less acidic. For example, mutant 8A (pI, 10.44, Table 2, last column), which has only two acidic residues in p6, has a Kd value of 50±8.9 nM; this is essentially the same value as that obtained for NCp9 (45±1.7 nM) (Table 2). Interestingly, mutant 8A also exhibited strong aggregating activity, even higher than that of NCp9, and reached a maximum value of ~90% at concentrations ≥ 0.5 μM (Fig. 6).
Table 3.
Rates of annealing or strand transfer (kobs values)a for WT NCp15 and mutants
| Protein | Annealing (min−1) | Strand Transfer (min−1) |
|---|---|---|
| NCp15 | n.d.b,c | 0.093±0.019d |
| 3A | n.d.b,c | 0.11±0.035 |
| C3A | 0.23±0.034 | 0.17±0.049 |
| 5A | 0.19±0.024 | 0.18±0.054 |
| 8A | 0.64±0.15 | 0.20±0.048e |
Rates were determined by fitting the data from Figure 8B (annealing) and 8C (strand transfer) to a single exponential equation. The data represent the average of results obtained from at least three independent experiments.
n.d., not determined.
Rate was too slow to measure accurately.
The error determinations represent the SD.
Rate was determined by using 0.4 μM protein.
To measure the kinetics of strand transfer, all samples were assayed at a concentration of 0.8 μM, with the exception of mutant 8A, which was assayed at 0.4 μM (Fig. 8C). At concentrations > 0.4 μM, 8A activity was reduced. The maximum strand transfer level was reached at 20 min by WT (36%) and mutant 3A (52%) and at 10 min by C3A, 5A, and 8A (each ~65%). Reduction of the plateau values by 60 min was observed with each of the mutants, reminiscent of the behavior of NCp9 (Fig. 5C). The rates of strand transfer for WT and 3A were ~2-fold lower than those for C3A and 5A. The 8A rate was the same as the rates for C3A and 5A, but at a 2-fold lower protein concentration (Table 3, last column).
Thus, taken together, the data presented in Fig. 8, Table 2, and Table 3 strongly support our prediction that the acidic residues in the p6 domain of NCp15 negatively modulate the efficiency of nucleic acid binding, annealing, and RT-catalyzed DNA elongation in minus-strand transfer.
4. Discussion
The goal of this study was to investigate the nucleic acid chaperone activities of the NCp15 and NCp9 precursor proteins in early steps of reverse transcription and to gain new insights into the rationale for selection of processed NCp7 as the HIV-1 NC protein required for long-term viral fitness and replication. Our approach was to use well-characterized reconstituted systems that model (-) SSDNA synthesis and minus-strand transfer, since these reactions are crucial for successful virus replication and also provide a sensitive measure of chaperone function. The results demonstrate that while both NCp9 and NCp15 have strong annealing activity, they are unable to sustain efficient RT-catalyzed DNA elongation.
In the assay for tRNALys3 annealing to a viral RNA template, NCp9 facilitated maximum annealing at a 2-fold lower concentration than NCp7 or NCp15, but the same level of activity was ultimately achieved by all three proteins (Fig. 3 and Suppl. Fig. 1). Similar results were obtained for annealing in the minus-strand transfer system (Fig. 5B). More significant differences in the activities of the three NC proteins were apparent in the complete systems, i.e., annealing plus elongation. When (-) SSDNA synthesis or minus-strand transfer was assayed at low concentrations, NCp9 had the highest activity, while NCp15 activity had the poorest (Fig. 2 and 5). However, as the NCp9 or NCp15 concentration was raised, the amount of product was reduced, eventually reaching almost base-line level (Fig. 2, 4, 5, Suppl. Fig. 1 and 3). In earlier work, Lener et al. reported a similar concentration effect in an assay for (-) SSDNA synthesis, but for NCp9 (referred to as NCp7(1-72)) as well as NCp7 (referred to as NCp7(1-55)) (Lener et al., 1998). In contrast, we consistently observe an increase in NCp7 activity as a function of increasing protein concentration (Fig. 2) (Jones, C.P., Goodrich, A., Wu, T., Levin, J.G., and Musier-Forsyth, K., manuscript in preparation). The reason for this discrepancy is not clear.
Our finding that under optimal conditions, NCp9 is a more efficient nucleic acid chaperone than either NCp7 or NCp15 in a number of different assays is consistent with the results of a recent parallel study (Wang et al., 2014). The data raise several questions: (i) Are there differences in the properties of the three NC proteins that would account for differences in their behavior in our reverse transcription assay systems? (ii) How do the activities of NCp9 and NCp15 explain the infectivity data obtained with processing mutants that contain either NCp9 or NCp15 instead of NCp7?
The aggregation data of Fig. 6 demonstrate that NCp9 has higher activity than NCp7 in the minus-strand annealing reaction. The short, highly basic SP2 domain is responsible for the enhanced activity of NCp9 in annealing and aggregation assays and even stimulates the annealing activity of NCp7 when present in trans at a high concentration (Fig. 7B, upper panel). Analysis by transmission electron microscopy showed that NCp9-DNA aggregates formed in reverse transcription reactions are less mobile than NCp7 aggregates, i.e., release of both double-stranded DNA and NCp9 that should occur after completion of DNA synthesis is more limited than with NCp7 (Mirambeau et al., 2007). This result is consistent with NCp9 data for DNA synthesis reactions (Fig. 2, 4, 5C, 7, Suppl. Fig. 2 and 3) and with single-molecule DNA stretching studies (Cruceanu et al., 2006b; Wang et al., 2014), which show that NCp9 as well as NCp15 dissociate more slowly than NCp7 from bound nucleic acid.
Interestingly, behavior similar to that exhibited by NCp9 and NCp15 has been encountered in two other experimental systems: RT-catalyzed DNA extension in the presence of high concentrations of Gag (Roldan et al., 2005; Wu et al., 2010b) and APOBEC3G (A3G) (Bishop et al., 2008; Iwatani et al., 2007). Both Gag (Cruceanu et al., 2006b) and A3G (Chaurasiya et al., 2014; Iwatani et al., 2007) exhibit even slower on-off nucleic acid binding kinetics than either of the two NC precursors. The slow dissociation rates of these proteins affect RT's ability to traverse the template and we have referred to this phenomenon as the “roadblock mechanism” (Levin et al., 2010; Wu et al., 2010b).
Aggregates formed by NCp15 also differ from those of NCp7. Electron microscopic imaging revealed that NCp15 coats ssDNA, behaving like a ss nucleic acid binding protein, but does not form the coaggregates (dense spheroid bodies) seen with NCp7 or NCp9 (Mirambeau et al., 2007; Mirambeau et al., 2006). Moreover, dynamic light scattering studies showed that whereas NCp7 and NCp9 coaggregates formed with RNA scatter light effectively, only weak scattering activity is observed with NCp15-RNA complexes and the size of the NCp15 “aggregates” is significantly smaller than NCp7 and NCp9 aggregates (NCp9>NCp7>>NCp15) (Wang et al., 2014).
How can we rationalize the poor chaperone activity of NCp15 in RT-catalyzed extension reactions? Its slow on-off nucleic acid binding kinetics and inability to form proper aggregates are certainly factors. However, ultimately the source of NCp15's unique properties derives from the fact that unlike NCp9 and NCp7, it contains the acidic p6 domain. In this respect, NCp15 resembles HTLV-1 NC, which has a basic N-terminal domain and an acidic C-terminal domain. Studies on the chaperone activity of the HTLV-1 protein support the proposal that the two domains interact with each other (Morcock et al., 2000; Qualley et al., 2010). Recently, several lines of evidence including NMR analysis were reported, strongly suggesting that the p6 domain of NCp15 is in a fold-back conformation that allows interaction with the basic ZFs and thereby reduces the net positive charge of the N-terminal NC domain (Wang et al., 2014).
In analogy to mutational analysis performed with HTLV-1 NC (Qualley et al., 2010), reducing the negative charge of the C-terminal p6 domain, in this case by changing acidic residues to Ala, improves chaperone activity. Indeed, the 8A mutant (Fig. 8A), which is more basic than NCp9, is actually somewhat more efficient than NCp9 in the annealing and aggregation assays, although the nucleic acid binding affinities are similar (Table 2). Interestingly, an 8A mutant virus is not infectious (Wang et al., 2014), indicating that simply replacing acidic residues in p6 with Ala is not sufficient to restore full biological activity. The p6 domain in Gag has a key role in virus budding (Göttlinger et al., 1991; Huang et al., 1995) as well as incorporation of Vpr into virus particles (Jenkins et al., 2001; Kondo et al., 1995; Lu et al., 1995; Paxton et al., 1993) and covalent linkage to NCp7-SP2 may interfere with these functions. Note too that the fold-back model (Wang et al., 2014) implies that residues closer to the C-terminus of p6 would be more important for interaction with the basic ZFs than more N-terminal residues. In turn, this would explain why the C3A mutation results in considerably more activity than the 3A mutation (Fig. 8).
Taken together, these findings demonstrate that the p6 domain negatively regulates the chaperone activity of NCp15 with the result that biological activity is profoundly affected. Thus, in cells infected with NCp15 mutant virus, there are distinct effects on reverse transcription (Coren et al., 2007), proviral DNA integration (Coren et al., 2007), production of mature genomic RNA dimers (Jalalirad and Laughrea, 2010; Kafaie et al., 2009), proper core morphology (de Marco et al., 2012), and infectivity (Coren et al., 2007; de Marco et al., 2012; Müller et al., 2009).
Unlike NCp15, NCp9 forms mature genomic RNA dimers as well as NCp7 (Jalalirad and Laughrea, 2010; Kafaie et al., 2009), although in one study, it was shown that the NCp9 dimers behave differently from WT (Ohishi et al., 2011). Interestingly, several groups have reported that the effect of blocking the NCp7-SP2 cleavage site has little effect on infectivity in a single-cycle assay (Coren et al., 2007; de Marco et al., 2012; Müller et al., 2009). Moreover, NCp9 mutant virions do not exhibit a transdominant effect in phenotypic mixing experiments (Müller et al., 2009) and deletion of SP2 has only a minor effect on infectivity (de Marco et al., 2012). Nevertheless, after four weeks of passage, NCp9 virions revert to WT (Coren et al., 2007). This suggests that NCp9's robust nucleic acid binding, aggregation, and chaperone activities observed in our study are not optimal for long-term viral replication, which clearly requires eventual cleavage of SP2 from NCp7. It is intriguing to note that when WT virus replication was assayed in the presence of low concentrations of a PR inhibitor (PI), processing at the NCp7-SP2 cleavage site was particularly affected (Müller et al., 2009). Virions contained both NCp7 and NCp9, a scenario that represents a more “natural” phenotypic mixing experiment. As the concentration of PI was increased, the amount of NCp9 was also increased, and there was a dramatic loss of infectivity; by 200 nM, infectivity was completely abolished. Although it is not clear why this loss of infectivity in a single round of infection was seen only with limiting concentrations of PR (Müller et al., 2009), these results further emphasize the crucial link between NC precursor processing and successful virus replication.
In summary, we have demonstrated that despite having efficient annealing and helix destabilizing activities, the immediate NC precursors, NCp15 and NCp9, are not as effective as mature NC in facilitating RT-catalyzed synthesis of (-) SSDNA and minus-strand transfer. These results reflect the activity imposed on the NC domain by the C-terminal peptides in each protein. Thus, the acidic domain of NCp15 negatively regulates nucleic acid chaperone activity and neutralization of the negative charge converts NCp15 to a protein with properties of NCp9. The highly basic SP2 domain in NCp9 is associated with unusually strong aggregating activity and nucleic acid binding affinity, which together with NCp9's slow on-off binding kinetics, lead to inhibition of RT movement along the nucleic acid template during DNA elongation (roadblock mechanism). Viewed collectively, our findings help to further explain why complete processing of the NC precursors is critical for long-term virus replication and fitness.
Supplementary Material
Acknowledgements
We thank Donald G. Johnson, William J. Bosche, and Catherine V. Hixon for preparing the expression constructs and recombinant proteins used in this study. We also thank Christopher P. Jones and Karin Musier-Forsyth for the RNA 105 and tRNALys3 clones. We are indebted to Mithun Mitra for advice on the aggregation and fluorescence anisotropy assays, to Wei Wang, Ioulia Rouzina, and Karin Musier-Forsyth for generously sharing their results prior to publication, and to all of these colleagues for stimulating and valuable discussion. This work was supported in part by the Intramural Research Program at the National Institutes of Health (Eunice Kennedy Shriver National Institute of Child Health and Human Development [T.W. and J.G.L.]). This project has also been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E with Leidos Biomedical Research Inc. (R.J.G.).
Abbreviations
- A3G
APOBEC3G
- bp
base pair
- CA
capsid
- DTT
dithiothreitol
- FA
fluorescence anisotropy
- HIV-1
human immunodeficiency virus type 1
- HTLV-1
human T-cell leukemia virus type 1
- MA
matrix
- NC
nucleocapsid
- nt
nucleotides
- PAGE
polyacrylamide electrophoresis
- PBS
primer binding site
- PR
protease
- PI
protease inhibitor
- R
repeat
- RT
reverse transcriptase
- SP
spacer peptide
- SD
standard deviation
- [-] SSDNA
(-) strong-stop DNA
- TAR
transactivation response element
- U3
unique 3’ sequence
- U5
unique 5’ sequence
- WT
wild type
- ZFs
zinc fingers
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- Anthony RM, Destefano JJ. In vitro synthesis of long DNA products in reactions with HIV-RT and nucleocapsid protein. J. Mol. Biol. 2007;365:310–324. doi: 10.1016/j.jmb.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell NM, Lever AML. HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol. 2013;21:136–144. doi: 10.1016/j.tim.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Verma M, Kim E-Y, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JAG, Kräusslich HG. The molecular architecture of HIV. J. Mol. Biol. 2011;410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DSB, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat Chem. 2014;6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coren LV, Thomas JA, Chertova E, Sowder RC, II, Gagliardi TD, Gorelick RJ, Ott DE. Mutational analysis of the C-terminal Gag cleavage sites in human immunodeficiency virus type 1. J. Virol. 2007;81:10047–10054. doi: 10.1128/JVI.02496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J. Mol. Biol. 2006a;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, Rein A, Rouzina I, Williams MC. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006b;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J-L, Godet J, Ivanyi-Nagy R, Fossé P, Mauffret O, Mély Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- de Marco A, Heuser A-M, Glass B, Kräusslich HG, Müller B, Briggs JAG. Role of the SP2 domain and its proteolytic cleavage in HIV-1 structural maturation and infectivity. J. Virol. 2012;86:13708–13716. doi: 10.1128/JVI.01704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Williams J, Powell MD, Levin JG, Le Grice SFJ. Mutating a conserved motif of the HIV-1 reverse transcriptase palm subdomain alters primer utilization. Biochemistry. 1997;36:5758–5768. doi: 10.1021/bi963045e. [DOI] [PubMed] [Google Scholar]
- Godet J, Mély Y. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biol. 2010;7:687–699. doi: 10.4161/rna.7.6.13616. [DOI] [PubMed] [Google Scholar]
- Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U. S. A. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Henderson LE, Bess J, Kane B, Levin JG. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wu W, Yuan ZY, Post K, Crouch RJ, Levin JG. Defects in primer-template binding, processive DNA synthesis, and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to Escherichia coli RNase H. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- Hargittai MRS, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- Hargittai MRS, Mangla AT, Gorelick RJ, Musier-Forsyth K. HIV-1 nucleocapsid protein zinc finger structures induce tRNALys,3 structural changes but are not critical for primer/template annealing. J. Mol. Biol. 2001;312:985–997. doi: 10.1006/jmbi.2001.5021. [DOI] [PubMed] [Google Scholar]
- Heilman-Miller SL, Wu T, Levin JG. Alteration of nucleic acid structure and stability modulates the efficiency of minus-strand transfer mediated by the HIV-1 nucleocapsid protein. J. Biol. Chem. 2004;279:44154–44165. doi: 10.1074/jbc.M401646200. [DOI] [PubMed] [Google Scholar]
- Henderson LE, Bowers MA, Sowder II RC, Serabyn SA, Johnson DG, Bess JW, Jr., Arthur LO, Bryant DK, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergott CB, Mitra M, Guo J, Wu T, Miller JT, Iwatani Y, Gorelick RJ, Levin JG. Zinc finger function of HIV-1 nucleocapsid protein is required for removal of 5'-terminal genomic RNA fragments: a paradigm for RNA removal reactions in HIV-1 reverse transcription. Virus Res. 2013;171:346–356. doi: 10.1016/j.virusres.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isel C, Ehresmann C, Marquet R. Initiation of HIV reverse transcription. Viruses. 2010;2:213–243. doi: 10.3390/v2010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Chan DSB, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Rosen AE, Guo J, Musier-Forsyth K, Levin JG. Efficient initiation of HIV-1 reverse transcription in vitro. Requirement for RNA sequences downstream of the primer binding site abrogated by nucleocapsid protein-dependent primer-template interactions. J. Biol. Chem. 2003;278:14185–14195. doi: 10.1074/jbc.M211618200. [DOI] [PubMed] [Google Scholar]
- Jalalirad M, Laughrea M. Formation of immature and mature genomic RNA dimers in wild-type and protease-inactive HIV-1: Differential roles of the Gag polyprotein, nucleocapsid proteins NCp15, NCp9, NCp7, and the dimerization initiation site. Virology. 2010;407:225–236. doi: 10.1016/j.virol.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Jenkins Y, Pornillos O, Rich RL, Myszka DG, Sundquist WI, Malim MH. Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6Gag. J. Virol. 2001;75:10537–10542. doi: 10.1128/JVI.75.21.10537-10542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafaie J, Dolatshahi M, Ajamian L, Song R, Mouland AJ, Rouiller I, Laughrea M. Role of capsid sequence and immature nucleocapsid proteins p9 and p15 in human immunodeficiency virus type 1 genomic RNA dimerization. Virology. 2009;385:233–244. doi: 10.1016/j.virol.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Kondo E, Mammano F, Cohen EA, Göttlinger HG. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J. Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam E, Coulaud D, Delain E, Petitjean P, Roques BP, Gérard D, Stoylova E, Vuilleumier C, Stoylov SP, Mély Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lee SK, Potempa M, Swanstrom R. The choreography of HIV-1 proteolytic processing and virion assembly. J. Biol. Chem. 2012;287:40867–40874. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lener D, Tanchou V, Roques BP, Le Grice SFJ, Darlix J-L. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y-L, Bennett RP, Wills JW, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J. Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyonnais S, Gorelick RJ, Heniche-Boukhalfa F, Bouaziz S, Parissi V, Mouscadet J-F, Restle T, Gatell JM, Le Cam E, Mirambeau G. A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: From nucleocapsid-condensed RNA to Vpr-bridged DNA. Virus Res. 2013;171:287–303. doi: 10.1016/j.virusres.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mervis RJ, Ahmad N, Lillehoj EP, Raum MG, Salazar FHR, Chan HW, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J. Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Borde I, Reboud-Ravaux M, Restle T, Gorelick RJ, Le Cam E. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS ONE. 2007;2:e669. doi: 10.1371/journal.pone.0000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, Le Cam E. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Gorelick RJ. Features, processing states and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010;7:724–734. doi: 10.4161/rna.7.6.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M, Wang W, Vo M-N, Rouzina I, Barany G, Musier-Forsyth K. The N-terminal zinc finger and flanking basic domains represent the minimal region of the human immunodeficiency virus type-1 nucleocapsid protein for targeting chaperone function. Biochemistry. 2013;52:8226–8236. doi: 10.1021/bi401250a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcock DR, Kane BP, Casas-Finet JR. Fluorescence and nucleic acid binding properties of the human T-cell leukemia virus-type 1 nucleocapsid protein. Biochim. Biophys. Acta. 2000;1481:381–394. doi: 10.1016/s0167-4838(00)00181-3. [DOI] [PubMed] [Google Scholar]
- Müller B, Anders M, Akiyama H, Welsch S, Glass B, Nikovics K, Clavel F, Tervo H-M, Keppler OT, Kräusslich H-G. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J. Biol. Chem. 2009;284:29692–29703. doi: 10.1074/jbc.M109.027144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi M, Nakano T, Sakuragi S, Shioda T, Sano K, Sakuragi J-I. The relationship between HIV-1 genome RNA dimerization, virion maturation and infectivity. Nucleic Acids Res. 2011;39:3404–3417. doi: 10.1093/nar/gkq1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton W, Connor RI, Landau NR. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Bambara RA. Requirements for efficient minus strand strong-stop DNA transfer in human immunodeficiency virus 1. RNA Biol. 2011;8:230–236. doi: 10.4161/rna.8.2.14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualley DF, Stewart-Maynard KM, Wang F, Mitra M, Gorelick RJ, Rouzina I, Williams MC, Musier-Forsyth K. C-terminal domain modulates the nucleic acid chaperone activity of human T-cell leukemia virus type 1 nucleocapsid protein via an electrostatic mechanism. J. Biol. Chem. 2010;285:295–307. doi: 10.1074/jbc.M109.051334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- Roldan A, Warren OU, Russell RS, Liang C, Wainberg MA. A HIV-1 minimal Gag protein is superior to nucleocapsid at in vitro tRNA Lys3 annealing and exhibits multimerization-induced inhibition of reverse transcription. J. Biol. Chem. 2005;280:17488–17496. doi: 10.1074/jbc.M501310200. [DOI] [PubMed] [Google Scholar]
- Sleiman D, Goldschmidt V, Barraud P, Marquet R, Paillart J-C, Tisné C. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res. 2012;169:324–339. doi: 10.1016/j.virusres.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Stoylov SP, Vuilleumier C, Stoylova E, de Rocquigny H, Roques BP, Gérard D, Mély Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N. Y.: 1997. pp. 263–334. [PubMed] [Google Scholar]
- Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Naiyer N, Mitra M, Li J, Williams MC, Rouzina I, Gorelick RJ, Wu Z, Musier-Forsyth K. Distinct nucleic acid interaction properties of HIV-1 nucleocapsid protein precursor NCp15 explain reduced viral infectivity. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku335. doi: 10.1093/nar/gku335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein's nucleic acid chaperone activity and retroviral replication. Virus Res. 2013;171:263–277. doi: 10.1016/j.virusres.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, Naufer MN, McCauley MJ, Gorelick RJ, Rouzina I, Musier-Forsyth K, Williams MC. Differential contribution of basic residues to HIV-1 nucleocapsid protein's nucleic acid chaperone function and retroviral replication. Nucleic Acids Res. 2014;42:2525–2537. doi: 10.1093/nar/gkt1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Rouzina I, Williams MC. Single-molecule stretching studies of RNA chaperones. RNA Biol. 2010a;7:712–723. doi: 10.4161/rna.7.6.13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Datta SAK, Mitra M, Gorelick RJ, Rein A, Levin JG. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: biological implications. Virology. 2010b;405:556–567. doi: 10.1016/j.virol.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Heilman-Miller SL, Levin JG. Effects of nucleic acid local structure and magnesium ions on minus-strand transfer mediated by the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2007;35:3974–3987. doi: 10.1093/nar/gkm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Henderson LE, Copeland TD, Gorelick RJ, Bosche WJ, Rein A, Levin JG. Human immunodeficiency virus type 1 nucleocapsid protein reduces pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.