Abstract
The transcription factor STAT5 mediates prolactin signaling and controls functional development of mammary tissue during pregnancy. This study has identified the miR-193b locus, also encoding miRNAs 365-1 and 6365, as a STAT5 target in mammary epithelium. While the locus was characterized by active histone marks in mammary tissue, STAT5 binding and expression during pregnancy, it was silent in most non-mammary cells. Inactivation of the miR-193b locus in mice resulted in elevated mammary stem/progenitor cell activity as judged by limiting dilution transplantation experiments of primary mammary epithelial cells. Colonies formed by mutant cells were larger and contained more Ki-67 positive cells. Differentiation of mammary epithelium lacking the miR-193b locus was accelerated during puberty and pregnancy, which coincided with the loss of Cav3 and elevated levels of Elf5. Normal colony development was partially obtained upon ectopically expressing Cav3 or upon siRNA-mediated reduction of Elf5 in miR-193b-null primary mammary epithelial cells. This study reveals a previously unknown link between the mammary-defining transcription factor STAT5 and a microRNA cluster in controlling mammary epithelial differentiation and the activity of mammary stem and progenitor cells.
Keywords: Mir-193b, Mammary, Development, Micro Rnas, STAT5, Stem Cells
Introduction
Cytokines control a large panel of biological functions, many of them mediated by the JAK-STAT signal transduction pathway. The transcription factors Signal Transducer and Activator of Transcription 5A and 5B (collectively referred to as STAT5) are executors of such cytokines as prolactin, growth hormone, erythropoietin, and interleukins (Hennighausen and Robinson, 2008). In mammary tissue STAT5 is required for the establishment and maintenance of alveolar progenitors during puberty (Yamaji et al., 2009) as well as for alveolar proliferation and differentiation during pregnancy (Cui et al., 2004; Liu et al., 1997; Miyoshi et al., 2001). STAT5-regulated genes encode transcription factors, membrane proteins, enzymes and milk proteins (Hennighausen and Robinson, 2005). Concentration-dependent and stage-specific STAT5 binding to respective regulatory elements ensures the temporally coordinated activation of genetic programs (Kang et al., 2014; Yamaji et al., 2013).
In addition to controlling cell differentiation, STAT5 also impacts the biology of stem cells. STAT5-deficient HSCs are unable to compete with wild type cells in bone marrow transplantation assays and conditional ablation of STAT5 from adult HSCs in the mouse causes loss of quiescence and a rapid depletion of the long term repopulating stem cell pool (Wang et al., 2009). In contrast, transplantation experiments with primary mammary epithelial cells have not provided evidence for a role of STAT5 in mammary stem cells (Yamaji et al., 2009).
Cytokines, and their executive transcription factors, are considered modulators of biological programs rather than master regulators. MicroRNAs have also been implicated in fine-tuning physiological parameters. Evidence is building that cytokines, through STAT transcription factors, also control the expression of specific micro RNA genes. STAT3 and STAT5 have been implicated in the activation of the loci encoding miR-21, miR-15/16 (Li et al., 2010) and miR-17~92 (Feuermann et al., 2012). Although numerous studies have alluded to roles of miRNAs in breast cancer cell lines, their contribution to the development and function of mammary epithelium in mice is likely more subtle (Feuermann et al., 2014; Feuermann et al., 2012). Based on in vitro studies using cultured primary cells a role of miR-146b in alveolar differentiation has been proposed (Elsarraj et al., 2012). While it is assumed that miRNAs in general target cell autonomously the stromal presence of miR-212 was reported to be required for mammary epithelial development in mice (Ucar et al., 2010). However, a role for miR-212 in mammary development and function has been disputed (Kayo et al., 2014; Remenyi et al., 2013).
This study focused on the identification and biological understanding of miRNA loci in mammary tissue that are under control of the cytokine-induced transcription factor STAT5. Several years ago our lab has identified miR-193b as a cytokine-inducible miRNA in primary mammary and hematopoietic cells and we investigated its role in the development and physiology of brown adipose tissue (Feuermann et al., 2013; Sun et al., 2011). Now these studies were extended to mammary tissue and ChIP-seq analyses were used to identify additional miRNA loci under STAT5 control in mammary tissue. In addition we have used mouse genetics to explore the function of the miR-193b-365-1 locus in mammary epithelium and stem cells.
Results
The miR-193b locus is under STAT5 control
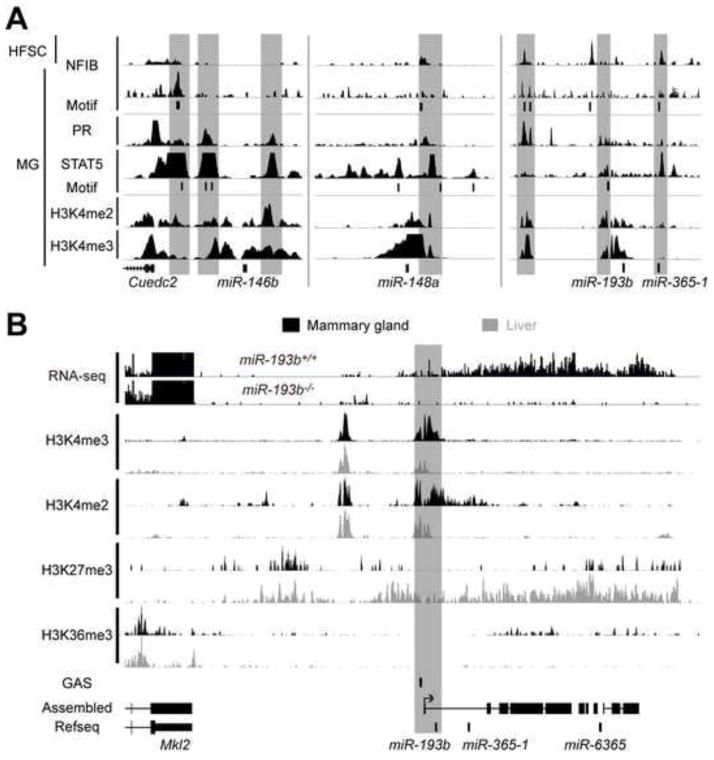
MicroRNA loci under STAT5 control were identified using ChIP-seq data sets for STAT5, progesterone receptor (PR), NFIB, H3K4me3 and H3K4me2 (Chang et al., 2013; Kang et al., 2014; Lain et al., 2013; Lemay et al., 2013; Rijnkels et al., 2013; Yamaji et al., 2013) and RNA-seq from wild type and Stat5-null mammary tissue. Histone marks characteristic of active promoters (H3K4me3) and enhancers (H3K4me2) and binding of transcription factors serve as indicators of transcribed loci. The loci encoding miR-193b, miR-146b and miR-148a fulfilled the defining criteria, i.e., binding of STAT5 and PR to putative regulatory sequences, the presence of H3K4me3 and H3K4me2 marks and STAT5-dependent transcription (Figure 1A). While the miR-146a locus is also a bona fide STAT3 target in breast cancer cells (Xiang et al., 2014) and miR-146b controls mammary cell differentiation in vitro (Elsarraj et al., 2013), no pertinent information exists on the roles of the miR-148a and miR-193b loci in mammary development. MiR-193b is unique in that it is expressed significantly only in a few cell types, mammary epithelium, brown adipose tissue (Feuermann et al., 2013; Sun et al., 2011) and hematopoietic progenitor cells (Kuchen et al., 2010). The miR-193b locus is transcribed as poly (A+) transcripts and also encodes miR-365-1 and miR-6365 (Figure 1B). H3K4me3 and H3K4me2 marks were identified over putative promoter and regulatory sequences in mammary epithelium. H3K27me3 levels spanning the locus were low and H3K36me3 marks covered the transcribed region, in agreement with an active gene locus (Figure 1B). In contrast, in liver tissue the locus was not bound by STAT5, displayed high H3K27me3 levels and there was a paucity of H3K36me3 marks (Figure 1B). STAT5-dependent expression of the miR-193b locus, as well as other miRNA loci, was interrogated with RNA-seq in mammary tissue at day 6 of pregnancy (p6) (Supplementary Table 1). Wild type mammary tissue, carrying both Stat5a and both Stat5b alleles, was compared with mutant tissue carrying only one Stat5b allele, which amounts to approximately 10% of total STAT5 levels (Yamaji et al., 2013). In contrast to a complete absence of STAT5, which inhibits alveologenesis, these low levels of STAT5 are sufficient for the formation of immature alveoli (Yamaji et al., 2013). Levels of miR-193b in control and mutant tissues were 5.6 and 1.7 FPKM, respectively (Supplementary Table 1), demonstrating that this locus is actively controlled by STAT5.
Figure 1. Features of the miR-193b locus.
(A) Integrative analysis of available ChIP-seq data (Chang et al., 2013; Kang et al., 2014; Lain et al., 2013; Lemay et al., 2013; Rijnkels et al., 2013; Yamaji et al., 2013) revealed regulatory regions (gray bars) at the miR-146b, miR-148a and miR-193b loci. HFSC, hair follicle stem cells; PR, prolactin receptor; MG, mammary gland; NFIB motif, TGGCC and STAT5 motif, TTCnnnGAA. (B) Analysis of RNA-seq and available histone ChIP-seq data reveals an active miR-193b locus in mammary tissue while it is silent in liver. Grey bar indicates putative transcription start site of the miR-193b transcript as described previously (Feuermann et al., 2013). MiR-193b was the first STAT5-regulated miRNA identified in this lab and the respective locus was inactivated in the mouse prior to the identification of the STAT5-controlled miR-146 and miR-148a.
Increased stem and progenitor cell activity in the absence of the miR-193b locus
MiR-193b was the first STAT5-regulated miRNA identified and the respective locus was inactivated in the mouse prior to the identification of the STAT5-controlled miR-146 and miR-148a. The miR-193b locus has been inactivated through the removal of a ~4 kb fragment spanning promoter and regulatory sequences and the entire coding region of miR-193b (Feuermann et al., 2013). RNA-seq analyses from control mammary tissue at day 13 of pregnancy (p13) demonstrated the presence of transcripts spanning the entire locus (Figure 1B). No transcripts were seen in miR-193b−/− mammary tissue, validating that this locus has been fully inactivated.
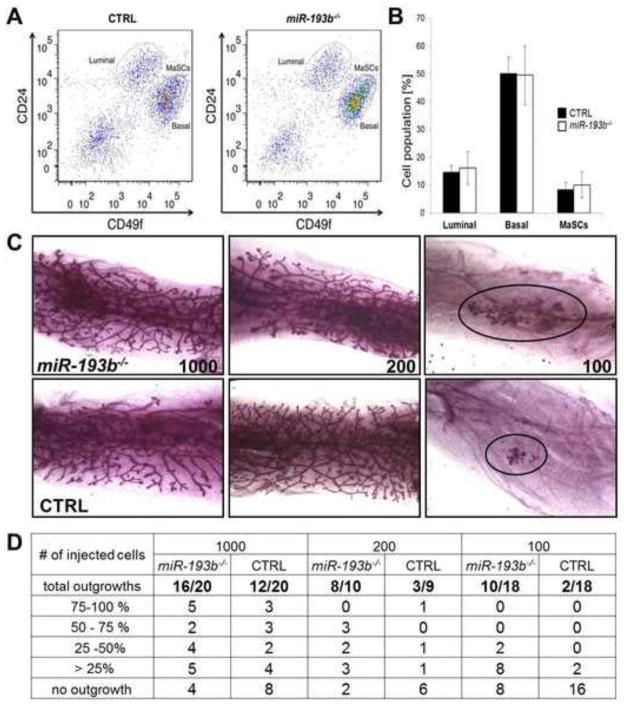
FACS analyses did not reveal overt differences between primary mammary epithelial cells (MECs) isolated from wild type and miR-193b−/− tissue of nulliparous mice at 12 weeks of age (Luminal; 16.26 ± 5.93 vs 14.64 ± 2.46 %, Basal; 49.52 ± 10.58 vs 50.15 ± 5.92 %, MaSCs; 10.07 ± 4.60 vs 8.43 ± 2.45 % in miR-193b−/− and control, respectively) (Figure 2A and B). Although the different cell populations, based on surface markers, were equivalent in wild type and mutant mice, limiting dilution transplantation experiments were used to determine their functional capacity. Decreasing numbers of primary mammary epithelial MECs were transplanted into cleared fat pads of recipient mice and ductal outgrowth was monitored after four weeks (Figure 2C). Upon implanting 1000 cells, 60% of wild type and 80% of mutant transplants formed a ductal tree. Upon transplanting 200 cells, only 33% of the wild type transplants but 80% of the mutants formed ducts. When 100 wild type cells were transplanted into each of 18 hosts, ductal outgrowth, albeit very small, was observed in only two mice. In contrast, more extended ductal outgrowth was observed in 10 out of the 18 hosts that had been injected with 100 mutant cells (Figure 2C and D). ELDA analysis (Hu and Smyth, 2009) estimated the repopulating frequency for wild type cells at 1/950 (1/580 to 1/1570). For mutant cells the frequency was 1/300 (1/200–1/500) with a p value of 0.0004. The divergent results of these two assays (FACS versus transplantation) suggest that the presence of specific epitopes might characterize specific cell populations but not their complete biological activity.
Figure 2. Increased stem cell activity in the absence of the miR-193b locus.
(A) FACS analysis of mammary epithelium from 8–12 week-old nulliparous mice (wild type on left). (B) Graph showing MEC population from three wild type and three mutant mice. (C) Representative images of wholemounts of repopulated fat pads injected with the indicated numbers of MECs. (D) Ratio of the number of repopulated fat pads per number of fat pads injected with 1000, 200 and 100 MECs, respectively.
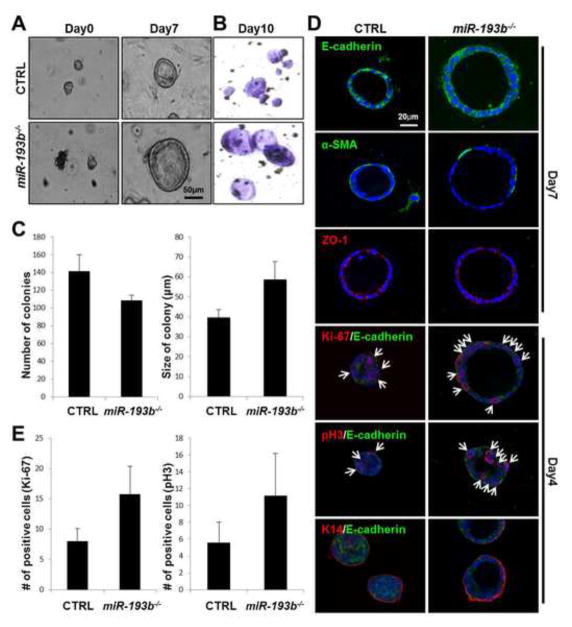
Next we gauged the proliferative capacity of alveolar cells in a defined in vitro setting by mammary colony formation assays. Primary MECs isolated from 8–12 week old miR-193b−/− and wild type nulliparous females were cultured and the formation of colonies was analyzed over a period of 10 days (Figure 3A and B). Reproducibly miR-193b−/− MECs had an approximately 20% reduced capacity to form colonies (Figure 3C). However, the size of colonies established from mutant MECs (58.8 ± 8.8μm) was approximately 50% bigger than that of controls (39.5 ± 4.2μm) (Figure 3A–C). Immunofluorescence staining for Cdh1 (E-cadherin) and Acta2 (a-SMA) verified the presence of a single epithelial layer and attached basal cells that were further characterized by the presence of Keratin 14 (K14) (Figure 3D). Correctly located TJP1 (ZO-1) demonstrated the presence of polarized epithelium.
Figure 3. Enhanced progenitor cell activity in the absence of miR-193b.
(A) Pictures were taken at days 0 and 7 after placing primary MECs in a colony formation culture system (Scale bar: 50 μm). (B) Representative pictures of colonies stained with crystal violet at day 10. (C) Graphs with the number and size of colonies. Three biological replicates were analyzed. (D) Confocal images of colonies stained for E-cadherin (Green), α-SMA (Green) and ZO-1 (Red) at day 7 and Ki-67 (Red), pH3 (Red) and K14 (Red) at day 4 with DAPI (Blue) as a counterstain (Scale bar: 20 μm). (E) Graphs with the number of positive cells stained with Ki-67 and pH3.
Whole mount nuclear staining with DAPI revealed an increased number of epithelial cells in miR-193b−/− colonies (Figure 3D). To compare the extent of proliferation in the presence and absence of miR-193b, colonies were stained with two different proliferation markers, Ki-67 and phosphorylated histoneH3 (pH3) after 4 days in culture (Figure 3D). MiR-193b−/− colonies were enlarged and had more cells positive for Ki-67 (miR-193b−/−: 15.80 ± 4.55, control: 8.00 ± 2.10) and pH3 (miR-193b−/−: 11.20 ± 5.02, control: 5.60 ± 2.41) (Figure 3E). These findings show increased proliferation of preferentially luminal cells in the absence of the miR-193b locus.
Precocious differentiation of mammary epithelium in the absence of the miR-193b locus
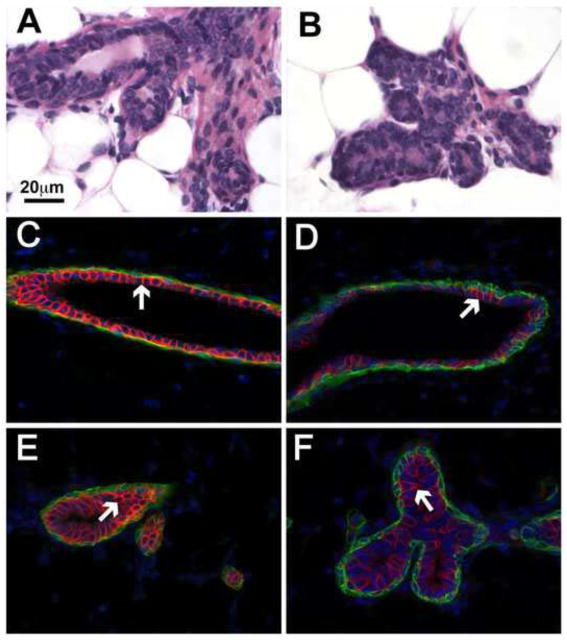
Proliferation and differentiation of mammary epithelium during pregnancy are tightly coupled and regulated processes. Mammary tissue from nulliparous mice (Figure 4) was analyzed to determine a potential contribution of miR-193b in epithelial differentiation. Histologically, alveolar and ductal structures from non-parous wild type and mutant mice were indistinguishable (Figure 4A and B). However, reduced expression of Slc12a2 (Nkcc1) in luminal cells from mutant mice was a harbinger of an accelerated differentiation program in the absence of the miR-193b locus (Figure 4C–F). At p13, the histological appearance of emerging alveoli in miR-193b−/− mice supported the presence of a precocious differentiation program (Figure 5). The number of cross-sectioned alveoli/mm2 was determined in 10 randomly selected fields from 3 mice in each group and a significant increase was observed in the absence of miR-193b (148/mm2 ± 1.8 in controls vs 198/mm2 ± 4.4 in miR-193b−/−: p = 0.0002). While alveoli in control mice displayed the typical undifferentiated appearance, signs of secretory differentiation such as fat globules were already visible in mutant epithelia.
Figure 4. Reduced NKCC1 in ductal epithelium in the absence of the miR-193b locus.
H&E staining of histological sections from mammary tissue from control (A) and miR-193b mutant (B) nulliparous mice. Immunofluorescence for NKCC1 (red) and a-SMA (green) in control (C, E) and mutant (D, F) tissues. Arrows point to luminal cells that express NKCC1.
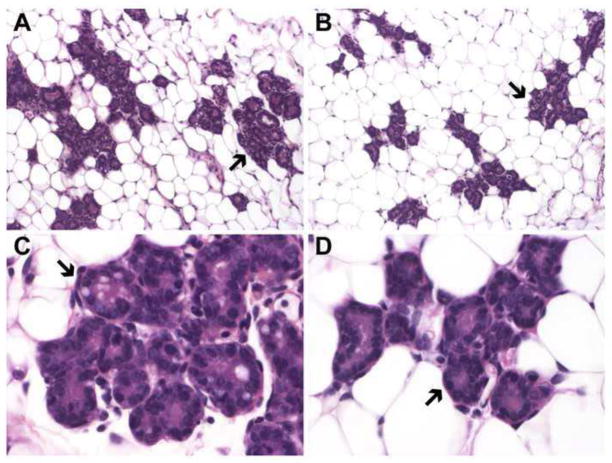
Figure 5. Precocious differentiation of mammary epithelial activity in the absence of the miR-193b locus.
Low (A and B) and high (C and D) magnification images of hematoxylin and eosin stained sections of p13 mammary tissues show increased alveolar development and accelerated differentiation in mutant (A and C) compared to controls (B and D). Arrows point to alveoli.
RNA-seq analyses were performed to evaluate the degree of differentiation of normal and mutant mammary epithelium at p13. Clustering analysis based on the expression of all genes revealed a marked difference between normal and mutant mammary epithelium (Figure 6A). Expression of approximately 450 genes was induced significantly in the absence of the miR-193b locus and expression of approximately 500 genes was reduced by more than 50% (Figure 6, Supplementary Table 2). One half of the upregulated genes were genuine STAT5 targets, mainly associated with the process of lactation, including transporter membrane proteins and milk proteins, such as Wap and Csn1s2b (Figure 6C). Several genes know to control alveolar differentiation were deregulated in the absence of miR-193b. Cav3 was expressed at 9 FPKM in control tissue and was reduced by 90% in the absence of miR-193b. Elf5 was expressed at 234 FPKM in controls and levels increased 1.8-fold in mutant tissue (Supplementary Table 2). Expression of Cidea, a transcriptional co-regulator controlling mammary function (Wang et al., 2012) was elevated four-fold in mutant tissue. Moreover, expression of several dual specific phosphatases was reduced in mutant tissue.
Figure 6. Identification of differentially expressed genes in the absence of miR-193b.
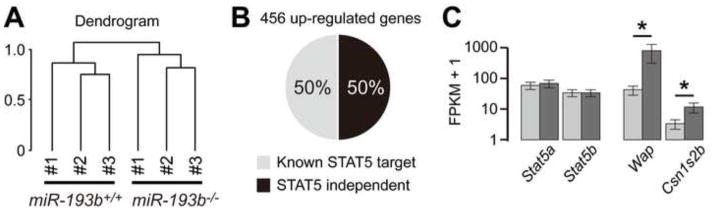
(A) Clustering analysis of RNA-seq result reveals a marked difference of gene expression between wild-type and mutant mammary tissues obtained at p13. Three biological replicates were used. (B) Percentage of STAT5-dependent genes among 456 induced genes is shown. (C) Expression levels of Stat5a, Stat5b, Wap and Csn1s2b are shown. *FDR-adjusted p value < 0.05.
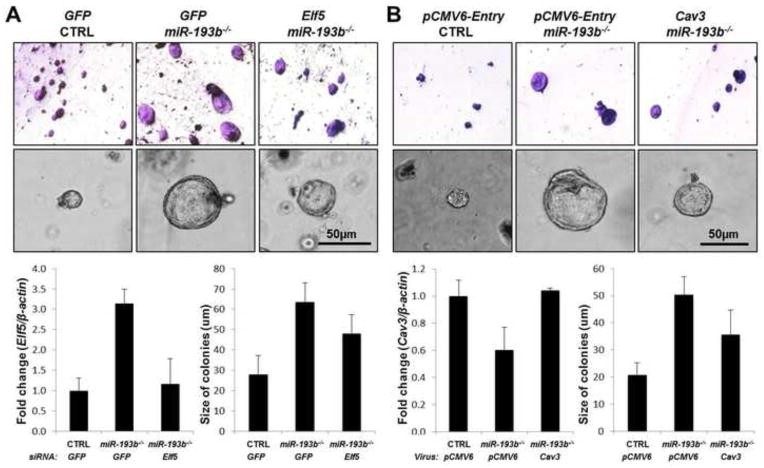
Precocious mammary epithelial differentiation has been observed upon loss of Caveolin-3 (Sotgia et al., 2009) or tyrosine phosphatase PTP1B (Milani et al., 2013), inappropriate activation of STAT5 (Dong et al., 2010; Vafaizadeh et al., 2010) or overexpression of ELF5 (Oakes et al., 2008). To determine whether deregulated expression of Cav3 and Elf5 in the absence of miR-193b could be causal to the observed biology, we tested their functional role in vitro (Figure 7). Short interfering RNAs (siRNAs) were used to reduce ELF5 levels in miR-193b−/−MECs and separately Cav3 was overexpressed. Subsequently these cells were cultured in matrigel for 6 days to evaluate the effect of altering the expression of these potential targets on colony behavior. Expression of Elf5 and Cav3 in control and mutant cells was established by real-time qPCR. Elf5 expression in miR-193b−/− cells (3.13 ± 0.36) was 3-fold higher compared to controls (1.00 ± 0.31). In the presence of Elf5-siRNAs, its expression was significantly reduced in miR-193b−/− (1.15 ± 0.63). Whole mount assay validated that the absence of miR-193b resulted in larger colonies as had already been shown in an earlier experiment (Figure 3). Knock-down of Elf5 reduced colony size (control: 28.18 ± 8.95, miR-193b−/−: 63.56 ± 9.49, miR-193b−/− treated with Elf5 siRNAs: 48.17 ± 9.14 μm). Cav3 expression was 40% lower in miR-193b−/− cells (0.60 ± 0.16) compared to controls (1.00 ± 0.11) and expression was restored in Cav3-overexpressing cells (1.04 ± 0.02). Overexpression of Cav3 in miR-193b−/− MECs (control: 20.83 ± 4.36, miR-193b−/−: 50.28 ± 6.70, miR-193b−/− overexpressing Cav3: 35.81 ± 8.87 μm) resulted in colonies with an intermediate size. These results indicate that miR-193b controls mammary epithelial proliferation and differentiation in part through regulating expression of Elf5 and Cav3.
Figure 7. Identification of functional roles of ELF5 and caveolin3 in miR-193b−/− cultures.
(A) Colonies generated by miR-193b−/− MECs treated with Elf5-siRNAs. (B) Colonies generated by miR-193b−/− MECs with restored Cav3 expression. Whole mount assay (top, original magnification; x5) and representative pictures of colonies (lower, scale bar: 50 μm) at day 6. Quantitative real-time qPCR analysis_ of Elf5 and Cav3 (left). Graph with the size of colonies (right).
Discussion
Since the miR-193b locus, encoding also miR365-1 and miR-6365, is under STAT5 control, its loss could affect functions attributed to STAT5. Notably, enhanced in vivo activity of MaSCs in the absence of the miR-193b locus resembled accelerated cycling of HSCs lacking STAT5 (Wang et al., 2009). However, loss of STAT5 in mammary stem cells has no overt consequences on their repopulation capacity (Yamaji et al., 2009) suggesting also STAT5-independent functions of the miR-193b locus. It is not clear whether any of the three miRNAs in this locus control MaSC activity by themselves or in combination with each other. While several studies have shown that specific genes, such as Lgr5 (Plaks et al., 2013), Fak (Luo et al., 2013) and Ezh2 (Pal et al., 2013) positively control MaSC activity (Fu et al., 2014), the miR-193b locus restricts MaSC activity in vivo and the proliferative capacity of progenitor cells.
Development and differentiation of mammary epithelium in non-parous mice and in early pregnancy is enhanced in the absence of the miR-193b locus, which coincides with larger sized colonies derived from primary MECs, indicative of a role in coordinating mammary alveolar differentiation. Execution of temporal differentiation programs can be controlled at different levels, mainly through the expression of positive and negative regulators and the activation of STAT5. Loss of the miR-193b locus resulted in elevated levels of the mammary transcription factor ELF5 and the reduction of Caveolin-3, suggesting that their respective deregulation was, at least in part, responsible for the altered biology of mutant tissue. This notion was supported as partial correction of Elf5 and Cav3 expression in miR-193b−/− cells alleviated the effects on colony growth in vitro. Consistent with these findings, the phenotypes of transgenic mice overexpressing ELF5 (Oakes et al., 2008) and those of Cav3 gene knock-out mice (Sotgia et al., 2009) mimic the precocious differentiation observed in the absence of the miR-193b locus. It is likely that precocious differentiation is the result of the deregulation of several genes, possibly also including those encoding dual specific phosphatases.
Differentiation of mammary epithelium is not only controlled cell intrinsically but also responds to cues from the embedding stroma. While this study demonstrated a cell-autonomous role for the miR-193b locus in the epithelial compartment, other miRNAs are likely to control mammary development through stromal cells in a paracrine fashion. Notably, the presence of miR-212 in the stromal compartment of mammary tissue is required for mammary epithelial development (Ucar et al., 2010) although these findings have been questioned (Kayo et al., 2014; Remenyi et al., 2013). A study (Llobet-Navas et al., 2014) described a key role of the miR-424 cluster in orchestrating remodeling of mammary tissue during involution, highlighting the contributions of miRNAs in modulating biological programs.
Extensive literature has been published on the potential role of miRNAs in breast cancer cell lines but there is a paucity of in vivo studies. Studies with primary cells have demonstrated that miR-146b, a known STAT3 target (Xiang et al., 2014), promotes alveolar progenitor cells in a cell line-based organ culture system (Elsarraj et al., 2013). STAT5 and PR are also binding to respective regulatory sequences in this locus further supporting the notion that miR-146b executes hormone function in mammary epithelium. MiR-22 and miR-205 are enriched in mammary progenitor cells (Ibarra et al., 2007) but an in vivo role has not been investigated. Based on ChIP-seq data from this lab and others (Lain et al., 2013) both these loci are recognized by STAT5 and PR supporting a role in the physiology of the mammary gland.
Although computational approaches have been used to predict miRNA targets, gene expression profiling from miRNA knock-out tissues have failed to reliably validate proposed targets (Feuermann et al., 2013; Feuermann et al., 2014; Feuermann et al., 2012). Similarly, the genes deregulated in mammary tissue lacking the miR-193b locus are not predicted targets of miR-193b, miR-365-1 and miR-6365. However, transcriptional analyses of mutant tissue led to the identification of transcripts, including Elf5 and Cav3, whose altered levels can account for the observed phenotypes as demonstrated in this study (Figure 8). Notably, ELF5 also controls the activity of mammary stem and progenitor cells (Oakes et al., 2008) and Caveolin-3 is a negative regulator of alveolar differentiation (Sotgia et al., 2009). This study, for the first time, demonstrates an intersection between miRNAs and cytokine-STAT5-mediated developmental programs in mammary epithelium. It remains an open question whether the miR-193b locus asserts its biology through targeting specific developmental genes or through a yet to be determined mechanism.
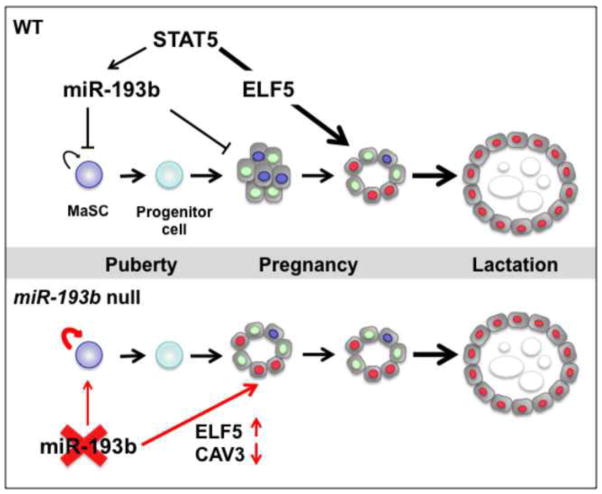
Figure 8.
Proposed model linking the miR-193b locus to the biology of mammary stem cells and differentiating epithelium. The miR-193b locus is under STAT5 control and negatively controls the activity of mammary stem and progenitor cells. This conclusion is based on mouse genetics, where loss of miR-193b resulted in enhanced stem and progenitor cell activity. Loss of miR-193b also resulted in precocious differentiation of mammary epithelium during pregnancy and a concomitant induction of Elf5 expression and reduced Cav3 expression. The functional contribution of ELF5 and caveolin3 in mammary alveolar differentiation was determined in in vivo loss-of-function and gain-of-function studies using colony formation assays in this research.
Materials and methods
Mice and transplantation of primary mammary epithelial cells
Mice with a targeted miR-193b locus have been described (Feuermann et al., 2013). Mammary tissue from nulliparous mice was chopped and digested at 37°C, shaking at 200rpm for 2 hrs in complete EpiCult-B medium (EpiCult-B medium with 5% fetal bovine serum, 10 ng/mL recombinant human epidermal growth factor, 10 ng/mL recombinant human basic fibroblast growth factor, 0.0004% heparin) supplemented with 300 U/mL collagenase 100 U/mL hyaluronidase. After lysis of red blood cells in NH4Cl, a single-cell suspension was obtained by sequential dissociation of the fragments with pre-warmed 0.25% trypsin-EDTA for 1–2 min, followed by pre-warmed 5 mg/mL dispase II plus 0.1 mg/mL DNase I (DNase; Sigma) for 2 min, and filtration through a 70-μm mesh. All reagents were from StemCell Technologies, Inc. unless otherwise specified. Defined numbers of isolated MECs were injected into the cleared fat pads of immunocompromised hosts (nu/nu athymic). Four weeks later the transplants were harvested and outgrowths were evaluated on carmine stained whole mounts. Stem cell frequencies were determined by ELDA (Hu and Smyth, 2009).
MECs preparation and FACS analysis
Mammary epithelial cells (MECs) were isolated using a standard protocol and non-epithelial cells were removed using the EasySep™ Mouse Mammary Stem Cell Enrichment Kit (STEMCELL TECHNOLOGY, #19757). MECs were stained with biotinylated anti-CD24-fluorescein isothiocyanate (FITC, BD Biosciences), anti-CD49f-R-phycoerythrin (R-PE, BD Biosciences) and 7-amino-actinomycin D (7AAD, BD Biosciences). FACS analysis was performed using FACSAria (BD Biosciences). Only live cells were gated from the 7AAD negative portion and FACS analysis of the CD49f and CD24 cell population in mammary epithelial cells yielded four different cell populations: negative (CD24−CD49f−), luminal (CD24hiCD49flo) and basal (CD24+CD49fhi) cells. The latter ones were divided into two different populations: myoepithelial (CD24loCD49fhi) and the stem cell-enriched (CD24midCD49fhi). Our analysis and interpretation of the data are based on defined epitope distribution used by others (Shackleton et al., 2006; Stingl et al., 2006) and our lab (Yamaji et al., 2009).
Colony formation assays
106 MECs from 8–14 week-old virgin mice were plated overnight on ultra-low attachment plates (Corning) with monolayer media (DMEM/F12, 5% FBS, 1% Pen/Strep, 1X Insulin-Transferrin-Selenium-X Supplement (ITS-X), 1 μg/ml hydrocortisone and 10 ng/ml EGF) at 37°C to allow formation of aggregates. 200 aggregates were plated in GFR-matrigel with naked media (DMEM/F12, 1% Pen/Strep and 1X ITS-X) and incubated at 37°C for 20 min to let the mixture gel. Aggregates in GFR-matrigel were cultured with EGF media (DMEM/F12, 1% Pen/Strep, 1X ITS-X, 10 ng/ml EGF and 0.0004% heparin), which was changed every 48hr.
RNA-seq analysis
RNA-seq was performed with total RNAs extracted from mammary gland tissues of wild type (miR-193b+/+) and mice lacking the miR-193b locus (miR-193b−/−) at day 13 of pregnancy (p13). Analysis was performed as described previously (Feuermann et al., 2013).
Histology and immunostaining
Tissues were fixed in formalin and processed for paraffin embedding and hematoxylin and eosin staining by standard procedures. For immunostaining deparaffinized 5 μm sections were treated in a Biocare Decloaker. Anti-NKCC1 (1:1000; generous gift from Dr. J. Turner, NIDCR, NIH) and smooth muscle actin (1: 1000; Sigma) were applied after blocking with 3% goat serum. Secondary antibodies (1:400) were conjugated with Alexafluor 488 and 594, respectively. Images were captured with a Retiga EXi camera and processed with Image-Pro Plus software.
Colony formation cultures were fixed with 2% paraformaldehyde for 20 min at RT and permeabilized with 0.5% TritonX-100 in PBS for 10 min at 4°C. Rins them three times with 100 mM glycine in PBS and blocked with 10% FBS in IF buffer (NaN3 7.7mM. 0.1% BSA, 0.2% TritonX-100, 0.05% Tween20) for 1 hr at RT then incubate with the indicated antibodies diluted in blocking buffer overnight at 4°C. Antibodies used were: anti-E-cadherin (BD Biosciences, 610182), anti-aSMA (Sigma, A2547), anti-ZO-1 (Miyoshi et al., 2001), anti-Ki67 (abcam, ab16667), anti-pH3 (millipore, 06–570) and anti-K14 (Covance, PRB-155p). After washing three times with IF buffer, incubate with secondary antibodies (Alexa 594 goat anti-rabbit (Life technologies, A11037) and Alexa 488 anti-mouse (Life technologies, A11029) and DAPI (Sigma, D9564) were applied for 45min at RT. The preparations were analyzed with a LSM700 Zeiss confocal microscope.
siRNA treatment and transient transfection assay
For knockdown experiments, siRNA targeting the Elf5 gene (ON-TARGETplus Elf5 siRNA, J-044985-12) and GFP Duplex I siRNA (P-002048-01-20) as positive control were purchased from Dharmacon. siRNA transfection were conducted using Lipofectamine RNAiMAX (Invitrogen, 13778-075). For overexpression experiments, cDNA clone for Cav3 (MR226246) and pCMV6-Entry vector (PS100001) as positive control were purchase from OriGene. MECs were transfected using FuGene HD Transfection Reagent (Promega, E2311).
Supplementary Material
Highlights.
The miR-193b locus in mammary epithelium is under the control of the transcription factor STAT5
Temporal differentiation of mammary epithelial during pregnancy depends on miR-193b and is mediated by ELF5 and caveolin3
Mammary stem/progenitor cell activity is curtailed by miR-193b
Acknowledgments
This work was supported by the Intramural Research Program (IRP) of NIDDK, NIH. We thank Dr. Harold Smith (NIDDK Genomics core) for conducting NGS.
Footnotes
Contributions
KHY: design of experiments, colony formation assays, immunohistochemistry, data analysis, writing manuscript; KK: design of experiments, computational analysis of RNA-seq and ChIP-seq data, data analysis, writing manuscript; YF: identification of miR-193b as a STAT5 target in mammary tissue, design of experiments, RNA-seq experiments; SJJ: mouse experiments, histology; GWR: design of experiments; transplantations, histology, data analysis, writing manuscript; LH: design of experiments, data analysis, writing manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chang CY, Pasolli HA, Giannopoulou EG, Guasch G, Gronostajski RM, Elemento O, Fuchs E. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature. 2013;495:98–102. doi: 10.1038/nature11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–47. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Tong T, Reynado AM, Rosen JM, Huang S, Li Y. Genetic manipulation of individual somatic mammary cells in vivo reveals a master role of STAT5a in inducing alveolar fate commitment and lactogenesis even in the absence of ovarian hormones. Dev Biol. 2010;346:196–203. doi: 10.1016/j.ydbio.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsarraj HS, Hong Y, Valdez K, Carletti M, Salah SM, Raimo M, Taverna D, Prochasson P, Bharadwaj U, Tweardy DJ, Christenson LK, Behbod F. A novel role of microRNA146b in promoting mammary alveolar progenitor cell maintenance. J Cell Sci. 2013;126:2446–58. doi: 10.1242/jcs.119214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsarraj HS, Stecklein SR, Valdez K, Behbod F. Emerging functions of microRNA-146a/b in development and breast cancer: microRNA-146a/b in development and breast cancer. J Mammary Gland Biol Neoplasia. 2012;17:79–87. doi: 10.1007/s10911-012-9240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuermann Y, Kang K, Gavrilova 0, Haetscher N, Jin Jang S, Hyun Yoo K, Jiang C, Gonzalez FJ, Robinson GW, Hennighausen L. MiR-193b and miR-365-1 are not required for the development and function of brown fat in the mouse. RNA Biol. 2013;10:1807–14. doi: 10.4161/rna.27239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuermann Y, Kang K, Shamay A, Robinson GW, Hennighausen L. MiR-21 Is under Control of STAT5 but Is Dispensable for Mammary Development and Lactation. PLoS One. 2014;9:e85123. doi: 10.1371/journal.pone.0085123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuermann Y, Robinson GW, Zhu BM, Kang K, Raviv N, Yamaji D, Hennighausen L. The miR-17/92 cluster is targeted by STAT5 but dispensable for mammary development. Genesis. 2012;50:665–71. doi: 10.1002/dvg.22023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu N, Lindeman GJ, Visvader JE. The mammary stem cell hierarchy. Curr Top Dev Biol. 2014;107:133–60. doi: 10.1016/B978-0-12-416022-4.00005-6. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6:715–25. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–21. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–43. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Yamaji D, Yoo KH, Robinson GW, Hennighausen L. Mammary-Specific Gene Activation Is Defined by Progressive Recruitment of STAT5 during Pregnancy and the Establishment of H3K4me3 Marks. Mol Cell Biol. 2014;34:464–73. doi: 10.1128/MCB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayo H, Kiga K, Fukuda-Yuzawa Y, Hedlund S, Murakami K, De La Rosa-Velazquez IA, Kimura T, Shimoda K, Tanabe M, Fukao T. miR-212 and miR-132 are dispensable for mouse mammary gland development. Nat Genet. 2014;46:802–4. doi: 10.1038/ng.2990. [DOI] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, Hu-Li J, Lu K, Dubois W, Kitamura Y, Charles N, Sun HW, Muljo S, Schwartzberg PL, Paul WE, O’Shea J, Rajewsky K, Casellas R. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–39. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lain AR, Creighton CJ, Conneely OM. Research resource: progesterone receptor targetome underlying mammary gland branching morphogenesis. Mol Endocrinol. 2013;27:1743–61. doi: 10.1210/me.2013-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay DG, Pollard KS, Martin WF, Freeman Zadrowski C, Hernandez J, Korf I, German JB, Rijnkels M. From genes to milk: genomic organization and epigenetic regulation of the mammary transcriptome. PLoS One. 2013;8:e75030. doi: 10.1371/journal.pone.0075030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Miskimen KL, Wang Z, Xie XY, Brenzovich J, Ryan JJ, Tse W, Moriggl R, Bunting KD. STAT5 requires the N-domain for suppression of miR15/16, induction of bcl-2, and survival signaling in myeloproliferative disease. Blood. 2010;115:1416–24. doi: 10.1182/blood-2009-07-234963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–86. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- Llobet-Navas D, Rodriguez-Barrueco R, Castro V, Ugalde AP, Sumazin P, Jacob-Sendler D, Demircan B, Castillo-Martin M, Putcha P, Marshall N, Villagrasa P, Chan J, Sanchez-Garcia F, Pe’er D, Rabadan R, Iavarone A, Cordon-Cardo C, Califano A, Lopez-Otin C, Ezhkova E, Silva JM. The miR-424(322)/503 cluster orchestrates remodeling of the epithelium in the involuting mammary gland. Genes Dev. 2014;28:765–82. doi: 10.1101/gad.237404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Zhao X, Chen S, Liu S, Wicha MS, Guan JL. Distinct FAK activities determine progenitor and mammary stem cell characteristics. Cancer Res. 2013;73:5591–602. doi: 10.1158/0008-5472.CAN-13-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani ES, Brinkhaus H, Dueggeli R, Klebba I, Mueller U, Stadler M, Kohler H, Smalley MJ, Bentires-Alj M. Protein tyrosine phosphatase 1B restrains mammary alveologenesis and secretory differentiation. Development. 2013;140:117–25. doi: 10.1242/dev.082941. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Shillingford JM, Smith GH, Grimm SL, Wagner KU, Oka T, Rosen JM, Robinson GW, Hennighausen L. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J Cell Biol. 2001;155:531–42. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes SR, Naylor MJ, Asselin-Labat ML, Blazek KD, Gardiner-Garden M, Hilton HN, Kazlauskas M, Pritchard MA, Chodosh LA, Pfeffer PL, Lindeman GJ, Visvader JE, Ormandy CJ. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008;22:581–6. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B, Bouras T, Shi W, Vaillant F, Sheridan JM, Fu N, Breslin K, Jiang K, Ritchie ME, Young M, Lindeman GJ, Smyth GK, Visvader JE. Global changes in the mammary epigenome are induced by hormonal cues and coordinated by Ezh2. Cell Rep. 2013;3:411–26. doi: 10.1016/j.celrep.2012.12.020. [DOI] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, Werb Z. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3:70–8. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi J, van den Bosch MW, Palygin O, Mistry RB, McKenzie C, Macdonald A, Hutvagner G, Arthur JS, Frenguelli BG, Pankratov Y. miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS One. 2013;8:e62509. doi: 10.1371/journal.pone.0062509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnkels M, Freeman-Zadrowski C, Hernandez J, Potluri V, Wang L, Li W, Lemay DG. Epigenetic modifications unlock the milk protein gene loci during mouse mammary gland development and differentiation. PLoS One. 2013;8:e53270. doi: 10.1371/journal.pone.0053270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Sotgia F, Casimiro MC, Bonuccelli G, Liu M, Whitaker-Menezes D, Er O, Daumer KM, Mercier I, Witkiewicz AK, Minetti C, Capozza F, Gormley M, Quong AA, Rui H, Frank PG, Milliman JN, Knudsen ES, Zhou J, Wang C, Pestell RG, Lisanti MP. Loss of caveolin-3 induces a lactogenic microenvironment that is protective against mammary tumor formation. Am j Pathol. 2009;174:613–29. doi: 10.2353/ajpath.2009.080653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mirl93b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–65. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar A, Vafaizadeh V, Jarry H, Fiedler J, Klemmt PA, Thum T, Groner B, Chowdhury K. miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nat Genet. 2010;42:1101–8. doi: 10.1038/ng.709. [DOI] [PubMed] [Google Scholar]
- Vafaizadeh V, Klemmt P, Brendel C, Weber K, Doebele C, Britt K, Grez M, Fehse B, Desrivieres S, Groner B. Mammary epithelial reconstitution with gene-modified stem cells assigns roles to Stat5 in luminal alveolar cell fate decisions, differentiation, involution, and mammary tumor formation. Stem Cells. 2010;28:928–38. doi: 10.1002/stem.407. [DOI] [PubMed] [Google Scholar]
- Wang W, Lv N, Zhang S, Shui G, Qian H, Zhang J, Chen Y, Ye J, Xie Y, Shen Y, Wenk MR, Li P. Cidea is an essential transcriptional coactivator regulating mammary gland secretion of milk lipids. Nat Med. 2012;18:235–43. doi: 10.1038/nm.2614. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li G, Tse W, Bunting KD. Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement. Blood. 2009;113:4856–65. doi: 10.1182/blood-2008-09-181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Birkbak NJ, Vafaizadeh V, Walker SR, Yeh JE, Liu S, Kroll Y, Boldin M, Taganov K, Groner B, Richardson AL, Frank DA. STAT3 Induction of miR-146b Forms a Feedback Loop to Inhibit the NF-kappaB to IL-6 Signaling Axis and STAT3-Driven Cancer Phenotypes. Sci Signal. 2014;7:ra11. doi: 10.1126/scisignal.2004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji D, Kang K, Robinson GW, Hennighausen L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res. 2013;41:1622–36. doi: 10.1093/nar/gks1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, Hennighausen L. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–7. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.