Abstract
Weight control through either dietary calorie restriction (DCR) or exercise has been associated with cancer prevention in animal models. However, the underlying mechanisms are not fully defined. Bioinformatics using genomics, proteomics, and lipidomics were employed to elucidate the molecular targets of weight control in a mouse skin cancer model. SENCAR mice were randomly assigned into 4 groups for 10 weeks: ad lib-fed sedentary control, ad lib-fed exercise (AE), exercise but pair-fed isocaloric amount of control (PE), and 20% DCR. Two hours after topical TPA treatment, skin epidermis was analyzed by Affymetrix for gene expression, DIGE for proteomics, and lipidomics for phospholipids. Body weights were significantly reduced in both DCR and PE but not AE mice versus the control. Among 39,000 transcripts, 411, 67, and 110 genes were significantly changed in DCR, PE, and AE, respectively. The expression of genes relevant to PI3K-Akt and Ras-MAPK signaling was effectively reduced by DCR and PE but not AE as measured through GenMAPP software. Proteomics analysis identified ~120 proteins, with 27 proteins significantly changed by DCR, including upregulated apolipoprotein A-1, a key antioxidant protein that decreases Ras-MAPK activity. Of the total 338 phospholipids analyzed by lipidomics, 57 decreased by PE including 5 phophatidylinositol species that serve as PI3K substrates. Although a full impact has not been determined yet, it appears the reduction of both Ras-MAPK and PI3K-Akt signaling pathways are cancer preventive targets that have been consistently demonstrated by three bioinformatics approaches.
Keywords: Bioinformatics, Weigh Control, Cancer Prevention, Dietary Calorie Restriction, Exercise
Introduction
Obesity has been identified as a risk factor for many chronic diseases including cancer. Since 1960, obesity rates have doubled in US men and nearly tripled in US women, with both groups at rates over 30% of the population.1,2 Obesity and overweightness contribute to cancer deaths in up to 14% for men and 20% for women over age 50, with higher rates of mortality for overweight cancer patients.3 With rising rates of obesity and chronic diseases, many studies have focused on various interventions to prevent obesity-related chronic diseases. In the early 20th century, dietary calorie restriction (DCR) was shown to prevent tumor growth, and it has become the most robust nutritional intervention for cancer prevention in animal models. As sedentary behavior has become endemic worldwide, exercise has also been identified as another lifestyle behavior that could reduce body weight and chronic disease risk. Weight control through DCR and/or exercise may be responsible, at least in animal models, for cancer prevention.
DCR is a weight control measure that decreases caloric intake of fats and carbohydrates while maintaining protein, fiber, and micronutrients. Typical studies have withheld calories between 20 and 40%.4 After a pilot study in 1909 by Moreschi, DCR was shown to prevent cancer in a variety of animal models and to be effective in reducing both chemically induced and spontaneous tumors.5 Additionally, increased energy expenditure through exercise is another form of weight control that may contribute to cancer prevention. Exercise has been strongly associated with cardiovascular health benefits, but the cancer preventive effect of exercise is less consistent. Exercise has demonstrated convincing evidence for cancer prevention in colorectal6 and breast cancer.7 Additionally, exercise may prevent prostate cancer progression as well as endometrial or lung cancer.8 Voluntary wheel running exercise provided cancer prevention by decreasing tumor size in a mouse skin cancer model.9,10 Some studies, however, have not shown a consistent protection by exercise.11–13 Furthermore, previous studies in our lab had mixed results of exercise for cancer prevention, with protection only seen with iso-caloric intake.14 Moore et al. demonstrated that a negative energy balance, as opposed to exercise alone, was responsible for inhibiting intestinal polyps in APCMin mice.15 Thus, to help differentiate between exercise and caloric restriction upon cancer signaling pathways, our study compared exercised mice with or without iso-caloric intake.
A variety of studies have been conducted to discover the biological mechanisms of weight control for cancer prevention. Many studies have focused on endocrine hormones and adipokines that are modified by increased adiposity. IGF-1 and leptin are two key factors that have been widely studied for cancer promotion. Increased IGF-1 levels from obesity have been associated with increased cancer risks. IGF-1 receptor has been shown to activate downstream pathways such as: MAPK, PI3K-Akt, and JAK/STAT which contribute to proliferation, antiapoptosis and gene transcription, respectively.16–17 Additionally, adipocyte-secreted adipokines such as leptin have been shown to promote cancer cell signaling.18–19
While traditional studies have been limited to examining a few select genes, proteins, or fat signaling phospholipids of interest to gauge the cancer preventive effect of weight control, recently developed -omics tools have enabled us to employ a more global approach to examine the etiology of cancer development. This study utilized a well-established mouse skin cancer model by 12-0-tetradecanoylphorbol-13-acetate (TPA) as a cancer promoter. Genomics, proteomics, and lipidomics are tools that are used to examine the interplay of profiling changes of gene, protein, and phospholipid expression on the development of cancer. The goal of this study was to implement genomic, proteomic and lipidomic methods to provide new information toward a mechanistic understanding of cancer prevention by weight control.
Materials and methods
Animals and treatment
Six-week old female SENCAR mice were purchased from NIH (Frederick, MD). Mice underwent a two-week training period to adjust to the new environment and treadmill exercise. Mice were housed individually at 24 ± 1 °C with a 12:12 light-dark cycle and given water ad libitum. Mice were divided into four treatment groups consisting of sedentary ad libitum-fed controls (control), ad-libitum-fed exercise (AE), pair-fed exercise (PE), and 20% DCR. Ad libitum controls and ad libitum exercise mice were allowed to freely access food, while the pair-fed exercise group was match-fed to the controls’ consumption. The basal AIN-93 and 20% DCR diets were made by Harland Teklad (Madison, WI). The 20% DCR diet that withheld calories from fat and carbohydrate is shown in Table 1. A speed adjustable rodent treadmill (Boston Gears, Boston, MA) was used for mice in exercise treatment groups. After two weeks training, the exercise groups ran on the treadmill at 13.4 m/min, 60 min per day and 5 days a week for 10 weeks. This exercise level has been rated as moderate intensity.20 Weekly body weights and food consumption measurements were taken.
Table 1.
Experimental Diet Composition
| Diet Componentsa | Control Diet | 20% DCRb |
|---|---|---|
| Corn Oil | 5.0 | 3.7c |
| Casein | 20.0 | 20.0 |
| DL Methionine | 0.3 | 0.3 |
| Dextrose | 15.0 | 12.3d |
| Dextrin | 49.9 | 37.1d |
| Fiber | 5.0 | 5.0 |
| AIN-93 Mineral Mix | 3.5 | 3.5 |
| AIN-93 Vitamin Mix | 1.0 | 1.0 |
| Choline bitartrate | 0.25 | 0.25 |
| Total amount of food | 100.0 | 82.0 |
AIN-93 custom made diet by Harlan Teklad (Madison, WI)
DCR mice were fed 0.82 g of diet for every gram consumed by control mice
Dietary calorie restriction from fat source
Dietary calorie restriction from carbohydrate source
At the end of the experiment, the mice were topically treated with 6.4 nmol TPA in 200 µL acetone on the dorsal skin, which was shaved two days before treatment. The sham treatment was acetone without TPA. There is not much difference between the controls with or without acetone-treatment. Considering the huge bioinformatics data generated by omics tools, this study was focused on TPA- verse sham-treated data for cancer risk-associated results. Mice were sacrificed 2 hours after TPA treatment, and the dorsal skin samples were snap-frozen in liquid nitrogen and stored at −70 °C until further analyses.
Microarray Analysis
Microarray analysis was performed as described in our previous studies.14,21 Briefly, labeled cRNA was applied to an Affymetrix GeneChip Mouse Genome 430 2.0 Array containing 39,000 transcripts and 45,101 probe sets (Santa Clara, CA). The images were quantified by using GeneChip operating software 1.0 (GCOS 1.0; Affymetrix, Santa Clara, CA). The raw image readings were analyzed using Simpleaffy package from BioConductor at http://www.bioconductor.org. The data were normalized using either MAS or RMA algorithms. The genes that were differentially expressed between treatment groups were identified by using pair-wise comparison. Data were filtered by using 1.5-fold difference considered statistically significant at p<0.05.
Cytoscape v2.6.0 coupled with plug-in BiNGO v2.0 was used to map the predominant gene ontology categories of the differentially expressed genes.22 The GO annotations p-values were obtained by hypergeometric statistical test for cluster verse whole annotation. The test was adjusted by Benjamin and Hochberg false discovery rate, which is included in the BiNGO package. The dataset consisting of the significantly altered genes was loaded into GenMAPP2.0 (Gene Map Annotator and Pathway Profiler, www.genmapp.org) to analyze the effect of target gene expression on specific pathways. As reported previously,14, 22 RT-PCR on select genes was tested to confirm the microarray results (data not shown).
Proteomics Analysis
Mass spectrometry analysis for landmark identification: mouse skin tissues were homogenized, and the protein concentration was determined by utilizing Protein RC DC assay (Bio-Rad, Hercules, CA). The protein lysis was purified by ReadyPrep 2-D cleanup kit (Bio-Rad, Hercules, CA). SDS-PAGE was performed using a precast 8–20% gradient gel (Bio-Rad, Hercules, CA). The spots of interest were excised and subjected to in-gel digestion using proteomics grade trypsin (Sigma, St. Louis, MO). The digested peptides were analyzed on a MALDI TOF/TOF instrument (Bruker, MA) using α-cyano-4-hydroxycinnamic acid (Sigma, St. Louis, MO) as matrix. Peak annotation was carried out automatically using software Proteinscape supplied by the instrument manufacturer (Bruker, MA). The m/z-lists were submitted to MASCOT to search the NCBI protein sequence database.
For analysis of the 2-D gels: sample labeling with cyanine minimal dyes was carried out according to the manufacturer’s instructions (GE healthcare, Piscataway, NJ). Protein lysates (25 µg) were used for CyDye labeling and the ratio of protein to CyDye is 1 µg protein: 5 pmol CyDye. The internal standard was always labeled with Cy2, and the samples were labeled with Cy3 and Cy5 alternatively. Isoelectric focusing was carried out on a PROTEAN IEF Cell following manufacture’s instructions (Bio-Rad, Hercules, CA). SDS PAGE was conducted using a precast 8–20% gradient gel (Bio-Rad, Hercules, CA). After protein separation, the gels with Cydye labeled proteins were scanned using a Typhoon 9410 scanner (GE Healthcare, NJ) with a resolution of 50 µm. Spot detection was performed on the gel images using the DeCyder 6.5 software. Before the matching process, up to 20 landmarks were defined. After a match, the cycle of reviewing and confirming the matches and re-matching was repeated manually until there were no new level 1 mismatches. The differences between the two groups were analyzed by t-test, which is provided by Decyder 6.5. The gels containing non-labeled protein were stained with Coomassie blue for protein identification. Western blot analysis for select proteins was conducted to confirm the proteomics results (data not shown).
Lipidomics Analysis
Phospholipid analysis was performed as described in our previous publications.22, 23 In short, each frozen dorsal skin tissue was ground with liquid nitrogen. One gram of tissue was mixed with 2 ml solvent [chloroform/methanol (1:2) + 0.01% butylated hydroxytoluene], 1 ml of chloroform, and 1 ml of water. The mixture was centrifuged for 15 min at 1,000 rpm, and the lower layer was extracted. One ml of chloroform was added and the mixture was centrifuged for 15 min at 1,000 rpm followed by collection of the lower layer. The two lower layer extracts were combined for phospholipid analysis using an automated ESI/MS-MS.22, 23 Phospholipid analysis was able to determine phospholipid classes/subclasses such as Phosphatidic acid, PI, PC, lysoPC, alk(en)yl/acyl phosphocholine (ePC), PE, lysoPE, alk(en)yl/acyl phosphoethanolamine (ePE), phosphatidylserine (PS), alk(en)yl/acyl phosphoserine, sphingomyelin (SM), and ceramide PE. Phospholipid identification was based upon total mass/charge and fragment mass/charge consistent with the head group.
Statistical Analysis
Body weights were compared between treatment groups using one-way analysis of variance (ANOVA). Microarray and proteomics spots were considered statistically significant at 1.5-fold change using the student t-test with significance at p<0.05. Cytoscape v2.6.0 coupled with plug-in BiNGO v2.0 was used for mapping the predominant gene ontology categories and the significantly altered genes were loaded into GenMAPP (Gene Map Annotator and Pathway Profiler, www.genmapp.org) to analyze the specific pathways. We also provide information on protein descriptions and biological pathways using Affymetrix NetAffx web site, GeneSpring, and GenMapp relevant software platforms and databases. Lipidomics analysis was performed using one-way ANOVA and F test for significance, with pairwise comparison by the least significant difference method.
Results
Body Weight Effects
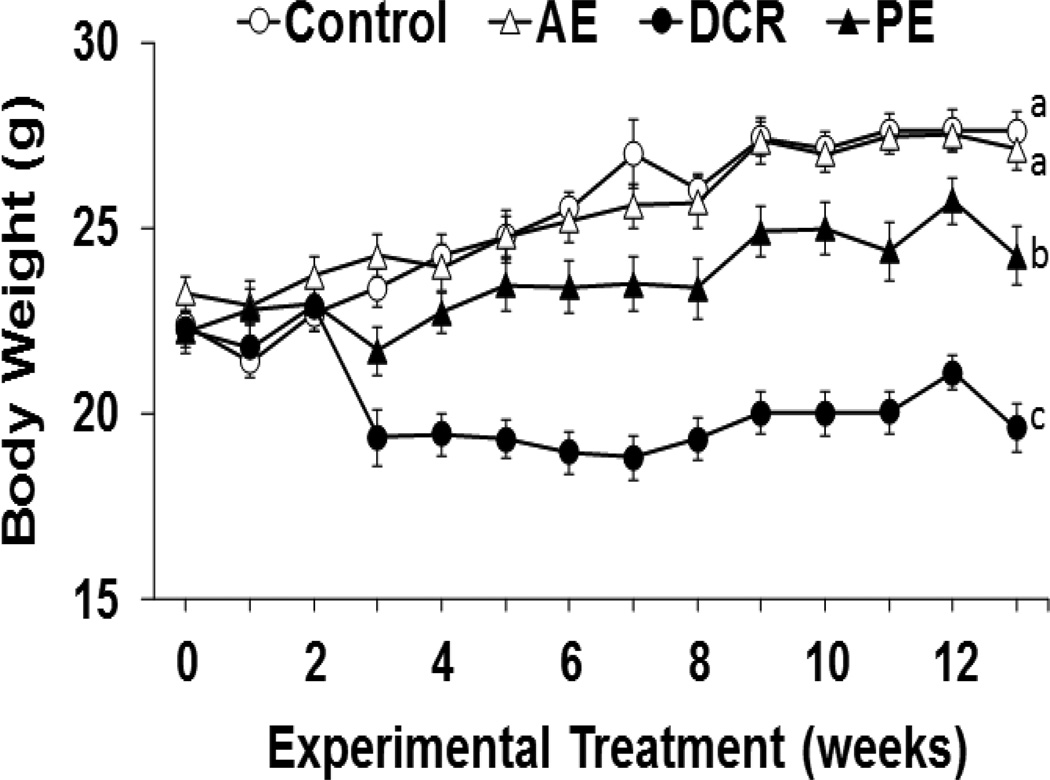
Body weight changes during the experiment are shown in Figure 1. The control group and AE groups demonstrated weight gains over the course of the study with an average body weight of 27 g. Both PE and DCR mice groups had significantly lower body weights of 22 and 19 g, respectively. The DCR treatment group had the most pronounced weight loss, followed by PE.
Figure 1.
Body Weight Effects. Data are shown for mice in the four groups: sedentary control (Control), ad-libitum exercise (AE), pair-fed exercise (PE), and dietary calorie restriction (DCR). Values are represented as mean ± SE, n=13–17. Means without a common letter differ, p<0.05.
Effect of Exercise on Food Intake
Table 2 shows food intake in all groups except for DCR mice that were fed 80% of the control and finished all the food daily without any variation for comparison. When compared with the sedentary control, PE but not AE had reduced food intake significantly in weeks 2, 3, 6, and 11. The overall food intake was also statistically reduced in PE verse the control.
Table 2.
Effects of exercise on food intake (g/week/mouse)*
| Control | AE | PE | |
|---|---|---|---|
| Week 1 | 25.6±2.2a | 24.6±2.6a | 24.3±2.7a |
| Week 2 | 29.5±2.4a | 28.4±2.7a | 24.5±1.3b |
| Week 3 | 31.5±4.7a | 28.9±3.4a | 26.2±2.0b |
| Week 4 | 28.6±3.5a | 27.6±5.8a | 26.4±2.1a |
| Week 5 | 27.7±3.1a | 26.9±6.2a | 26.5±1.6a |
| Week 6 | 35.2±5.4a | 33.2±2.9a | 26.5±1.2b |
| Week 7 | 29.4±3.4a | 28.3±3.3a | 27.0±0.6a |
| Week 8 | 29.8±4.2a | 28.3±3.8a | 27.8±1.3a |
| Week 9 | 29.5±3.4a | 28.9±3.8a | 27.9±1.6a |
| Week 10 | 31.3±4.1a | 30.9±4.5a | 30.5±0.7a |
| Week 11 | 35.8±6.5a | 35.4±3.4a | 27.4±2.2c |
| Week 12 | 27.6±3.8a | 26.8±5.1a | 27.1±1.4a |
Data are means ± SD, n = 15–18. Means in a row without a common letter differ, p < 0.05.
Effect of Weight Loss on Gene Expression Profile
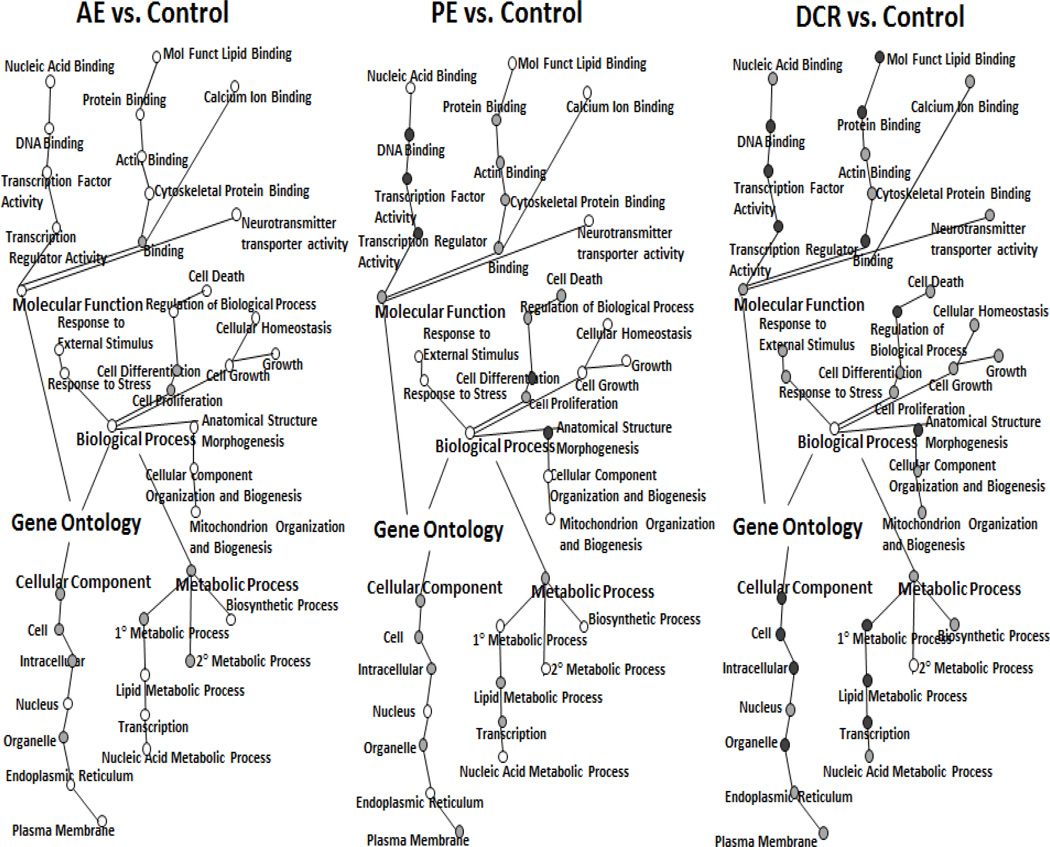
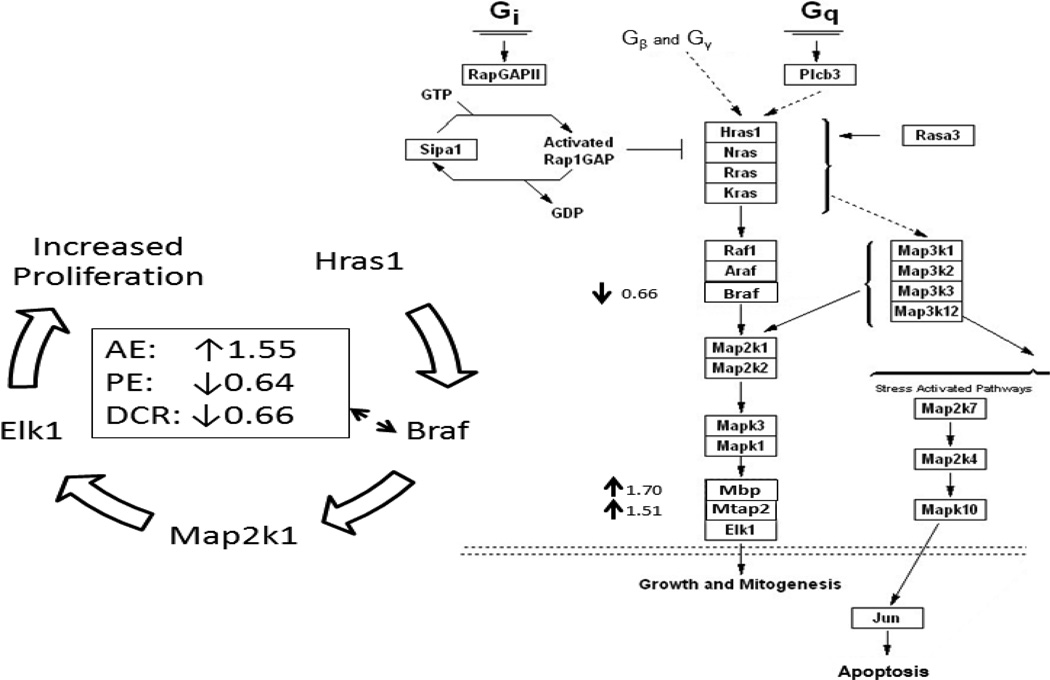
Among the 39,000 genes analyzed, 411 transcripts by DCR, 67 transcripts by PE, and 110 transcripts by AE were significantly changed compared with the control. The gene sets identified by microarray analysis that were significantly changed by weight control were further categorized using GO annotations. The GO categories were analyzed using BiNGO, a program which helps visualize functional themes. Figure 2 shows the visualization of significantly changed gene network by Cytoscape. The shading of the node indicates the degree of statistical significance: black node > grey node > white node. There is an overall impact of gene change by DCR > PE > AE. In comparison of the two exercised groups, PE had stronger impact than AE in biological processes such as cell death and cell differentiation, more molecular function gene expression in area such as DNA and protein binding, and little change in cellular components. In comparison of two weight loss groups, DCR had more impact on gene expression than PE in biological events related to stress, cell growth and homeostasis, metabolism, and many organelle differences in the cellular components section. DCR had distinctly more genetic changes in nearly all the GO categories as opposed to AE. Furthermore, Figure 3 shows examples of the pathway analysis by GenMapp, indicating the Raf MAP-Kinase pathway was significantly down-regulated by 0.64-fold and 0.66-fold change in PE and DCR groups, respectively, but upregulated by 1.55-fold in AE.
Figure 2.
BiNGO software representation showing Gene Ontology (GO) for significantly changed gene expression in (A) AL+Exe versus Control, (B) PF+Exe versus Control, (C) DCR versus Control. The coloring of the node indicates the degree of a statistical difference in gene expression of treatment group versus control at black node > grey node > white node.
Figure 3.
The Hras-Braf MAP-Kinase pathway visualized using GenMAPP software. Right panel: representative gene expression in Hras-Braf MAP-Kinase pathway for DCR verse the control, indicating 0.66-fold down-expression of Braf but 1.70- and 1.51-fold over-expression of Mbp (myelin basic protein) and Mtab2 (methanol cobalamin methyltransferase 2), respectively. Left panel: summary overview of Braf expression in AE, PE, and DCR verse the control, representing 1.55-fold increase in AE, but 0.64- and 0.66-fold decrease in PE and DCR, respectively.
Effect of Weight Loss on Protein Expression Profile
Using proteomic techniques, we were able to identify approximately 120 proteins. Among which, 22 proteins were significantly changed by DCR treatment. Some of the proteins had multiple spots including albumin (2), carbonic anhydrase 3 (3), enolase 3:beta muscle (3), and ATP Synthase (H+ transporting, mitochondrial F1 complex) (2). Table 3 lists the 10 proteins that were up-regulated and 12 proteins that were down-regulated by DCR.
Table 3.
Proteins that were up-regulated or down-regulated by Dietary Calorie Restriction (DCR) versus the Control utilizing a 2D-DlGE gel.a
| Proteins Up-Regulated by DCRb | Proteins Down-Regulated by DCRc |
|---|---|
| 6-phosphogluconolactonase | PDZ and LIM domain protein |
| trisephosphate isomerase | myosin light chain (phosphorytable) |
| kininogen 1 precursor | myosin A2 catalytic light chain |
| albumin | enolase 3: beta muscle |
| ornithine aminotransferase | gelsolin-like capping protein (capG) |
| carbonic anhydrase 3 | carbonic anhydrase 3 |
| apolipoprotein A-1 | ATP Synthase (H+ transporting, F1) |
| heat shock protein (cystallin related) | aldose reductase |
| Flavin reductase (NADPH-dependent) | UGP2 protein |
| peroxiredoxin 6 | phosphoglycerate kinase |
| aconitase 2 | |
| adenylate kinase isoenzyme 1 |
Proteins were scanned using a Typhoon 9410 scanner with resolution of 50 µm
Up-regulated proteins had fold-change of ≥1.50, p<0.05
Down-regulated proteins had fold-change of ≥1.50, p<0.05
Effect of Weight Loss on Phospholipid Expression Profile
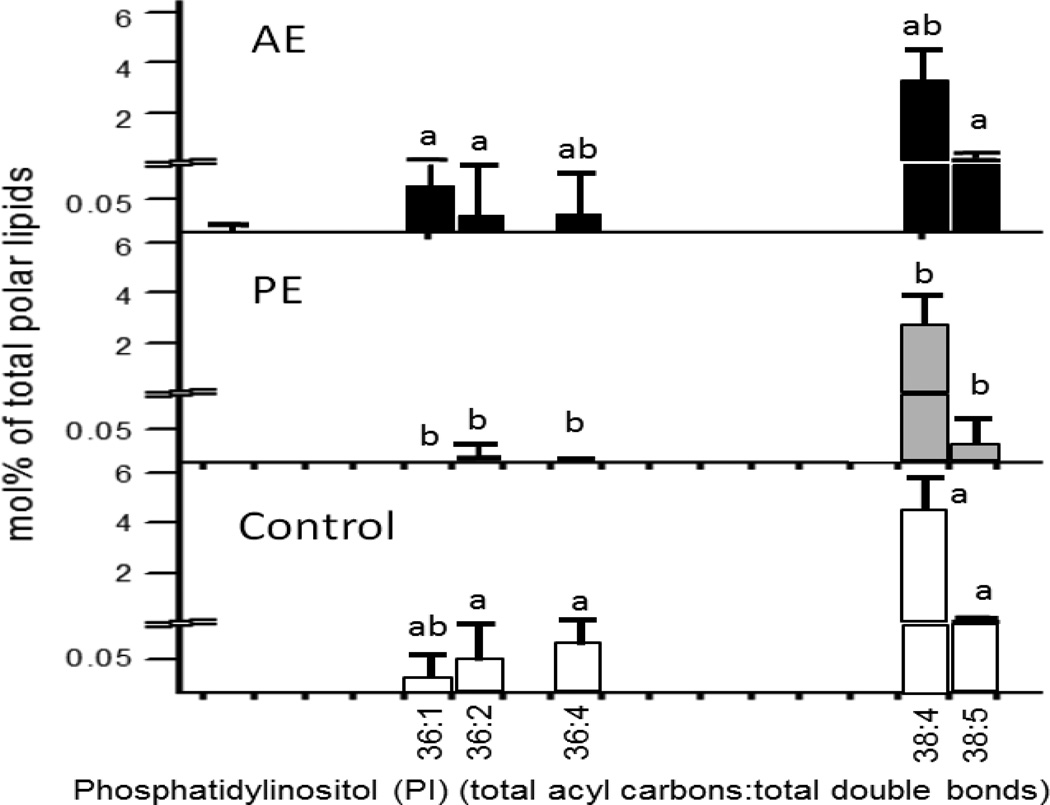
Among 338 phospholipid species analyzed, 57 species were significantly changed by exercise. Compared to sedentary controls, most phosphatidylinositol (PI), ether phosphatidylcholine (ePC), and some lysophosphatidylcholine (lysoPC) molecular species decreased significantly in exercise with pair feeding mice. It should be noted that five PI groups decreased in PE as opposed to AE. Figure 4 illustrates five PI species that were significantly decreased in PE but not AE.
Figure 4.
Molar percentage of phosphatidylinositol species between AE, PE, and control as analyzed by ESI/MS-MS. Values are shown as mean±SE, n=8–15. Means between treatment groups without a common letter differ, p<0.05.
Discussion
The current study demonstrated that both DCR and PE groups were effective in significantly lowering body weight as compared to control animals, with DCR having the most impact on body weight. Weight loss in PE group was not as pronounced as 20% DCR, possibly because the exercise was not strenuous enough to reach 20% calories burned in exercise.14 The body weight finding in this study confirms previous studies that have shown decreased food intake for a negative energy balance through PE and DCR may cause weight loss.14,21 It appears that negligible change of food intake in the AE group is responsible for the weight gain that was comparable to the sedentary control. Thus, DCR or negative food intake with exercise in PE group seems more effective than AE and control groups in reducing body weight.
It is usually expected that exercise should have prompted an elevation in food intake in compensation of energy expenditure. Some studies in humans have shown that moderate to high-intensity exercise leads to increased appetite and subsequent increase in food intake as well as body weight.24,25 However, our findings showed that food intake was reduced slightly in AE group and significantly in PE group compared to the controls. This is in agreement with a human study that found exercise decreased food intake in part through increased postprandial GLP-1, leading to a stronger satiety response.26 Furthermore, two studies conducted in rodent models also reported a decreased food consumption by a moderate exercise.27,28 The mechanisms by which exercise may reduce food intake have been associated with improved sensitivity to leptin resistance.29 In regard to the difference of food intake between AE and PE group, some mice in PE group could finish all the food, which means these mice were able to consume more if given free access to diet. This is probably one of the reasons that PE group had less food intake compared with AE group. In addition, fresh pellet diet was kept in freezer that usually contains about 10% of moisture. For the control group, mice were fed weekly and food intake was measured weekly, while PE sand AE mice were fed daily and food intake was weighted daily. It could be true there were more water loss in weekly-fed diet than that in daily-fed diet, which might account, at least in part, for an overestimation of food intake in the controls. Based on the current results, the decrease of body weight for exercise group was attributed mainly by less food intake rather than more energy expenditure. Future studies determining energy expenditure of each mouse and then incorporating it with food intake for body weight control are warranted.
The gene expression data indicate that caloric restriction obstructs cancer signaling pathways. The 2-hr period of TPA treatment was selected based upon a time-course study, in which AP-1:DNA binding and c-Jun mRNA were optimally induced at 2 hours after phorbol ester treatment.30 Our previous studies also confirmed the expression of tumor promotion-related genes was induced in response to 2-hr TPA after adjustment by the basal levels in acetone-treated TPA-free skin epidermis.14,21 However, it would be ideal if various time points including some late response genes were detected. The approaches to gene expression were distinguishable from our previous studies,14,21 since a new gene map annotator and pathway profiler was applied and since all the three bioinformatics approaches including microarray, proteomics, and lipidomics were simultaneously analyzed in this study. Out of the 39,000 transcripts measured, 559 genes were significantly different in the exercise treatment groups as compared to the controls. A total of 411 genes were changed by DCR, 67 genes by PE, and 110 genes by AE, illustrating the largest total number of genes changed occurred through DCR. To better grasp the specific molecular targets of DCR, BiNGO software was employed to analyze gene functions such as cellular components and regulations of biological processes like apoptosis, cell proliferation, and cell differentiation. DCR had the most change in functions related to cellular components and biological processes, while PE demonstrated moderate change in these functions versus AE. There was a progressive increase in gene change by treatment groups (DCR > PE > AE). While both DCR and PE modulated weight control, the genetic response shows a major distinction between DCR and PE. Many genes related to MAPK and PI3K signaling significantly decreased in both DCR and PE groups. DCR fostered more pronounced protection in genes related to pro-cancer pathways. For example, insulin-like growth factor binding protein 3, which is associated with IGF-1 activity, was reduced by 0.53-fold change in PE and 0.40-fold change in DCR. In looking for specific cellular pathways impacted by diet, we utilized GenMAPP software and found that DCR and PE both reduced the Braf/MAPK pathway involved in enhancing cellular proliferation, whereas this pathway was up-regulated by AE. While further studies will need to be conducted to elucidate the differences between DCR and PE gene ontology, our data clearly shows their effects on the Ras-MAPK pathway.
Using 2D DIGE proteomics, we identified approximately 120 protein spots, which corresponded to 86 proteins within 2D gel. Among these proteins identified, we found that 22 proteins were significantly changed in DCR versus the control. The cellular functions of the proteins analyzed were common to pathways such as energy metabolism/glycolysis and cellular stress response. In analyzing the data, we identified proteins that may be key targets for cancer prevention and promotion such as Apolipoprotein A-1 (APOA1) which was up-regulated by DCR (Table 3). APOA1 has been studied extensively for its cancer protective properties through reducing inflammation.31 Over-expression of APOA1 mimetic peptides was associated with increased survival rates and inhibition of size and number of tumors in a mouse ovarian cancer model. APOA1 may foster cancer protection via binding lysophosphatidic acid (LPA), a proinflammatory lipoprotein that leads to cellular proliferation through the Ras-Rho GTPase crosstalk for cancer promotion.31,32 The inhibitory effect of APOA1 on Ras-MAPK pathway provides a possible link between the cancer prevention observed in microarray and proteomics data. Additionally, oxidized phospholipids contribute to pro-inflammation,33 and it is postulated that APOA1 may reduce this inflammatory response and exert pro-cancer effect through reducing MAPK activity.31 In contrast, gelsolin-like capping protein (capG), is an oncogenic protein down-regulated by DCR. Bahassi et al. demonstrated that capG is important for tumor cell motility and cell proliferation, therefore down-regulation of capG by the AP-1 transcription factor complex may contribute to cancer prevention.34 Increased cell motility in cancer is correlated with increased Ras-MAPK activity,32 as a result the down-regulation of capG may aid in the cancer preventive effects of DCR. Overall, it appears that DCR decreases cellular pathways related to cancer promotion including Ras-MAPK and PI3K-Akt signaling pathways, reduction of inflammation, and modulation of phospholipids by DCR for cancer prevention.22
Cancer prevention was further demonstrated through reduction of PI phospholipid species via PE treatment. Our lipidomics data indicated that weight control through PE reduced 5 PI species as opposed to the AE and control groups. The predominant form of PI in mouse tissue is PI 38:4.22,35 PI species can be substrates of PI3K involved in cellular signaling pathways related to cancer promotion via downstream PI3K-Akt signaling. This decrease in PI phospholipids is consistent with previous data which indicates that PIs are decreased in exercised groups.14 PE had the most pronounced reduction in PI substrates compared to AE or the control. Our lab has also shown that exercise-induced reduction in PI phospholipids leads to protection from further downstream cancer promoting events through PI3K-related signaling.22 We did not detect phospholipid profile in DCR group. When compared with AE, PE had shown a reduced food intake. The results of phospholipid change in PE may be, at least in part, suggested an impact of calorie restriction.
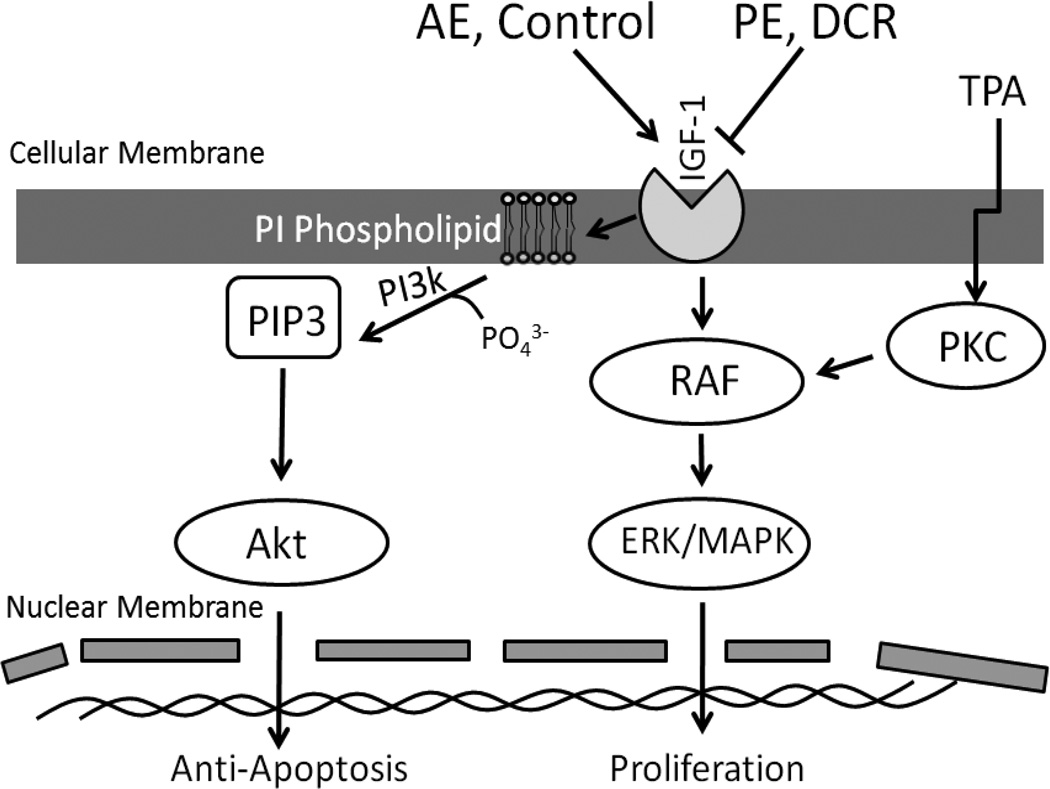
A major hormone decreased as a result of PI signaling following DCR or PE is IGF-1.36,37 Our lab has shown that IGF-1 restoration within PE mice reversed the reduction of PI phospholipids and PI-associated PI3K.23 Hence, it appears that IGF-1 is a key growth factor that promotes cancer development in overweight conditions, in part through promoting PI phosphorylation by PI3K and downstream Akt activity which contribute to anti-apoptosis.38,39 Hence, increased IGF-1-Akt anti-apoptosis, or inhibition of apoptosis, may lead to cancer promotion through turning off protective nuclear apoptosis genes. Through demonstrating that PIs were increased in the phospholipid membrane, our study helps to better understand the mechanism for IGF-1 and increased PI3K-Akt activity. Morimura et al. demonstrated that IGF-1 promotes colocalization of IGF-1 receptor and PIP3, which is a PI phosphorylated by PI3K.39 This colocalization of IGF-1 receptor and PIs could explain how PIs could amplify the signal for anti-apoptosis from IGF-1. Reduced IGF-1 levels could lead to decreased localization of PIs and subsequent reduction of downstream PI3K signaling. Figure 5 illustrates the possible mechanism through which weight control could lead to reduced IGF-1 levels, PI3K activity, and anti-apoptosis. Thus, our phospholipidomics data further illustrates the importance of the PI3K-Akt pathway in cancer prevention following weight control.
Figure 5.
Overview of the molecular pathways through which weight control may impact for cancer prevention.
Our current data demonstrate the cellular signaling pathways of Ras-MAPK and PI3K-Akt as key cancer preventive targets based on three independent bioinformatics approaches. A limitation of our study was that the proteomics analysis did not detect many protein kinases, as we may have expected. A more recently developed technique of phosphoproteomics may aid in identifying more kinases related to signaling pathways.
Taken together, this study identified PI3K-Akt and Ras-MAPK as two major pathways related to weight control and cancer prevention seen through all three bioinformatics approaches. Microarray data showed that the Ras-MAPK pathway was down-regulated in DCR and PE, but increased in AE. Our proteomics data showed that APOA1 and capG are proteins that are modified for cancer prevention by Ras-MAPK. APOA1 leads to a decrease in proinflammatory response that may be helpful for cancer prevention through modulating Ras-MAPK. CapG was also a protein reduced by DCR that is indicative of reduced Ras-MAPK and PI3K-Akt activity. Finally, lipidomics data showed reduced levels of PI species with isocaloric exercise (PE), suggesting how weight control can reduce the PI3K-Akt pathway and its downstream effects on apoptosis. The three areas of bioinformatics utilized give us a more global overview for the protective effect of weight control through both isocaloric exercise and calorie restriction on cancer prevention. It seems that weight control helps to prevent against cancer through reduction in hormones from excess body fat such as IGF-1 and/or leptin and their further downstream molecular targets as concluded in Figure 5.
ACKNOWLEDGEMENTS
This work was supported in part by NIH/NCI CA167678, NIH/INBRE RR16475, and Kansas State University Foundation #J-33950 for Nutrition and Cancer. This is a journal contribution #13-226-J of the Kansas Agricultural Experiment Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: ZZ, LL, RB and WW participated in the experimental design. JS, YJ, XS, JC, MY, JX and JT performed the experimental procedure and/or data analysis. JS, YJ, BK and WW wrote the manuscript. All authors reviewed and edited the manuscript.
REFERENCES
- 1.Calle EE, Kaaks R. Overweigh, Obesity, and Cancer: Epidemiological Evidence and Proposed Mechanisms. Nat Rev Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 2.Flegal KM, Carroll MD, Ogen CL, Curtin LR. Prevalence and Trends in Obesity Among US Adults, 1999–2008. J Am Med Assoc. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 3.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, Obesity, and Mortality from Cancer in a Prospectively Studied Cohort of U.S. Adults. N Engl J Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 4.Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Kritchevsky D. Caloric Restriction and Experimental Carcinogenesis. Toxicological Sciences. 1999;52:13–16. doi: 10.1093/toxsci/52.2.13. [DOI] [PubMed] [Google Scholar]
- 6.Basterfield L, Reul J, Mathers JC. Impact of Physical Activity on Intestinal Cancer Development in Mice. The Journal of Nutrition. 2005;135:3002S–3008S. doi: 10.1093/jn/135.12.3002S. [DOI] [PubMed] [Google Scholar]
- 7.Jiang W, Zhu Z, Thompson HJ. Effects of Physical Activity and Restricted Energy Intake on Chemically Induced Mammary Carcinogenesis. Cancer Prevention Research. 2009;2:338–344. doi: 10.1158/1940-6207.CAPR-08-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedenreich CM, Orenstein MR. Physical activity and cancer prevention: etiologic evidence and biological mechanisms. J Nutr. 2002;132:3456S–3464S. doi: 10.1093/jn/132.11.3456S. [DOI] [PubMed] [Google Scholar]
- 9.Conney AH, Lou YR, Nghiem P, Bernard JJ, Wagner GC, Lu YP. Inhibition of UVB-induced nonmelanoma skin cancer: a path from tea to caffeine to exercise to decreased tissue fat. Top Curr Chem. 2013;329:61–72. doi: 10.1007/128_2012_336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Cui XX, Huang MT, Liu Y, Wagner GC, Lin Y, Shih WJ, Lee MJ, Yang CS, Conney AH. Inhibition of progression of androgen-dependent prostate LNCaP tumors to androgen independence in SCID mice by oral caffeine and voluntary exercise. Nutr Cancer. 2012;64:1029–1037. doi: 10.1080/01635581.2012.716899. [DOI] [PubMed] [Google Scholar]
- 11.Gillette CA, Zhu Z, Westerlind KC, Melby CL, Wolfe P, Thompson HJ. Energy availability and mammary carcinogenesis: effects of calorie restriction and exercise. Carcinogenesis. 1997;18:1183–1188. doi: 10.1093/carcin/18.6.1183. [DOI] [PubMed] [Google Scholar]
- 12.Thompson HJ, Ronan AM, Tiaco KA, Tagliaferro AR. Effect of type and amount of dietary fat on the enhancement of rat mammary tumorigenesis by exercise. Cancer Res. 1989;49:1904–1908. [PubMed] [Google Scholar]
- 13.Thompson HJ, Westerlind K, Snedden JR, Briggs S, Singh M. Inhibition of mammary carcinogenesis by treadmill exercise. J Natl Cancer Inst. 1995;87:453–455. doi: 10.1093/jnci/87.6.453. [DOI] [PubMed] [Google Scholar]
- 14.Xie L, Jian Y, Ouyang P, Chen J, Doan H, Herndon B, Sylvester JE, Zhang K, Molteni A, Reichle M, Zhang R, Haub MD, Baybutt RC, Wang W. Effects of Dietary Calorie Restriction or Exercise on the PI3K and Ras Signaling Pathways in the Skin of Mice. J Biol Chem. 2007;282:28025–28035. doi: 10.1074/jbc.M604857200. [DOI] [PubMed] [Google Scholar]
- 15.Moore T, Carbajal S, Beltran L, Perkins SN, Yakar S, Leroith D, Hursting SD, Digiovanni J. Reduced susceptibility to two-stage skin carcinogenesis in mice with low circulating insulin-like growth factor I levels. Cancer Res. 2008;68:3680–3688. doi: 10.1158/0008-5472.CAN-07-6271. [DOI] [PubMed] [Google Scholar]
- 16.Gallagher EJ, LeRoith D. Minireview: IGF-1, Insulin, and Cancer. Endocrinology. 2011;152:2546–2551. doi: 10.1210/en.2011-0231. [DOI] [PubMed] [Google Scholar]
- 17.Novosyadlyy R, Lann DE, Vijayakumar A, et al. Insulin-Mediated Acceleration of Breast Cancer Development and Progression in a Nonobese Model of Type 2 Diabetes. Cancer Res. 2010;70:741–751. doi: 10.1158/0008-5472.CAN-09-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Tian J, Lv Y, et al. Leptin induces functional activation of cyclooxygenase-2 through JAK2/STAT3, MAPK/ERK, and PI3K/AKT pathways in human endometrial cancer. Cancer Sci. 2009;100:389–395. doi: 10.1111/j.1349-7006.2008.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luhn P, Dallal CM, Weiss J, Black A, Huang WY, Lacey JV, Hayes RB, Stanczyk FZ, Wentzensen N, Brinton LA. Circulating adipokine levels and endometrial cancer risk in the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. Cancer Epidemiol Biomarkers Prev. 2013;22:1304–1312. doi: 10.1158/1055-9965.EPI-13-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernando P, Bonen A, Hoffman-Goetz L. Predicting submaximal oxygen consumption during treadmill running in mice. Can J Physiol Pharmacol. 1993;48:854–857. doi: 10.1139/y93-128. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Xie L, Sylvester J, Wang J, Bai J, Baybutt R, Wang W. Different gene expression of skin tissues between mice with weight controlled by either calorie restriction or physical exercise. Exp Biol Med. 2007;232:473–480. [PubMed] [Google Scholar]
- 22.Ouyang P, Jiang Y, Doan HM, Xie L, Vasquez D, Welti R, Su X, Lu N, Herndon B, Yang SS, Jeannotte R, Wang W. Weight Loss via exercise with controlled dietary intake may affect phospholipid profile for cancer prevention in murine skin tissues. Cancer Prev Res. 2010;3:466–477. doi: 10.1158/1940-6207.CAPR-09-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Ma H, Su X, Chen J, Xu J, Standard J, Lin D, Wang W. IGF-1 Mediates Exercise-Induced Phospholipid Alteration in the Murine Skin Tissues. J Nutr Food Sci. 2012;S2:003. [Google Scholar]
- 24.Finlayson G, Caudwell, Gibbons C, Hopkins M, King N, Blundell J. Low fat loss response after medium-term supervised exercise in obese is associated with exercise-induced increase in food reward. Journal of Obesity. 2011:615624. doi: 10.1155/2011/615624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toti L, Bartalucci A, Ferrucci M, Fulceri F, Lazzeri G, Lenzi P, Soldani P, Gobbi P, La Torre A, Gesi M. High-intensity exercise training induces morphological and biochemical changes in skeletal muscles. Biol Sport. 2013;30:301–309. doi: 10.5604/20831862.1077557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martins C, Kulseng B, King NA, Holst JJ, Blundell JE. The effects of exercise-induced weight loss on appetite-related peptides and motivation to eat. J Clin Endocrinol Metab. 2010;95:1609–1616. doi: 10.1210/jc.2009-2082. [DOI] [PubMed] [Google Scholar]
- 27.Ebal E, Cavalie H, Michaux O, Lac G. Effect of a moderate exercise on the regulatory hormones of food intake in rats. Appetite. 2007;49:521–524. doi: 10.1016/j.appet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Lira FS1, Yamashita AS, Rosa JC, Tavares FL, Caperuto E, Carnevali LC, Jr, Pimentel GD, Santos RV, Batista ML, Jr, Laviano A, Rossi-Fanelli F, Seelaender M. Hypothalamic inflammation is reversed by endurance training in anorectic-cachectic rats. Nutr Metab. 2011;8:60. doi: 10.1186/1743-7075-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krawczeski Carhuatanta KA, Demuro G, Tschöp MH, Pfluger PT, Benoit SC, Obici S. Voluntary exercise improves high-fat diet-induced leptin resistance independent of adiposity. Endocrinology. 2011;152:2655–2664. doi: 10.1210/en.2010-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Przybyszewski J, Yaktine AL, Duysen E, Blackwood D, Wang W, Au A, Birt DF. Inhibition of phorbol ester-induced AP-1-DNA binding, c-Jun protein and c-jun mRNA by dietary energy restriction is reversed by adrenalectomy in SENCAR mouse epidermis. Carcinogenesis. 2001;22:1421–1427. doi: 10.1093/carcin/22.9.1421. [DOI] [PubMed] [Google Scholar]
- 31.Su F, Kozak KR, Imaizumi S, Gao F, Amneus MW, Grijalva V, Ng C, Wagner A, Hough G, Farias-Eisner G, Anantharamaiah GM, Van Lenten BJ, Navab M, Fogelman AM, Reddy ST, Farias-Eisner R. Apoliliprotein A-I (apoA-1) and apoA-1 mimetic peptides inhibit tumor development in a mouse model of ovarian cancer. PNAS. 2010;107:19997–20002. doi: 10.1073/pnas.1009010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahai E, Olson MF, Marshall CJ. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 2001;20:755–766. doi: 10.1093/emboj/20.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bochkov VN, Oskolkova OV, Birukov KG, Levonen AL, Binder CJ, Stöckl J. Generation and biological activities of oxidized phospholipids. Antioxid Redox Signal. 2010;12:1009–1059. doi: 10.1089/ars.2009.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahassi el M, Karyala S, Tomlinson CR, Sartor MA, Medvedovic M, Hennigan RF. Critical regulation of genes for tumor cell migration by AP-1. Clin Exp Metastasis. 2004;21:293–304. doi: 10.1023/b:clin.0000046132.46946.dd. [DOI] [PubMed] [Google Scholar]
- 35.Postle AD, Dombrowsky H, Clarke H, Pynn CJ, Koster G, Hunt AN. Mass spectroscopic analysis of phosphatidylinositol synthesis using 6-deuteriated-myo-inositol: comparison of the molecular specificities and acyl remodelling mechanisms in mouse tissues and cultured cells. Biochem Soc Trans. 2004;32:1057–1059. doi: 10.1042/BST0321057. [DOI] [PubMed] [Google Scholar]
- 36.Jiang Y, Wang W. Potential Mechanisms of Cancer Prevention By Weight Control. Biophys Reviews Lett. 2008;3:421–437. [Google Scholar]
- 37.King B, Jiang Y, Su X, Xu J, Xie L, Standard J, Wang W. Weight control, endocrine hormones and cancer prevention. Exp Biol Med. 2013;238:502–508. doi: 10.1177/1535370213480695. [DOI] [PubMed] [Google Scholar]
- 38.Xie L, Wang W. Weight control and cancer prevention: role of IGF1-mediated signaling pathways. Experimental Biology and Medicine. 2013;238:127–132. doi: 10.1177/1535370213477602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morimura S, Suzuki K, Takahashi K. Nonmuscle myosin IIA is required for lamellipodia formation through binding to WAVE2 and phosphatidylinositol 3,4,5-triphosphate. Biochem Biophys Res Commun. 2011;404:834–840. doi: 10.1016/j.bbrc.2010.12.069. [DOI] [PubMed] [Google Scholar]