Abstract
Dynamic MRI has been widely used to track the motion of the tongue and measure its internal deformation during speech and swallowing. Accurate segmentation of the tongue is a prerequisite step to define the target boundary and constrain the tracking to tissue points within the tongue. Segmentation of 2D slices or 3D volumes is challenging because of the large number of slices and time frames involved in the segmentation, as well as the incorporation of numerous local deformations that occur throughout the tongue during motion. In this paper, we propose a semi-automatic approach to segment 3D dynamic MRI of the tongue. The algorithm steps include seeding a few slices at one time frame, propagating seeds to the same slices at different time frames using deformable registration, and random walker segmentation based on these seed positions. This method was validated on the tongue of five normal subjects carrying out the same speech task with multi-slice 2D dynamic cine-MR images obtained at three orthogonal orientations and 26 time frames. The resulting semi-automatic segmentations of a total of 130 volumes showed an average dice similarity coefficient (DSC) score of 0.92 with less segmented volume variability between time frames than in manual segmentations.
Keywords: Tongue, motion, dynamic MRI, segmentation, random walker, deformable registration, super-resolution reconstruction
1. Introduction
The tongue is a crucial part of the oral cavity. Its behavior is critical for the production of speech, where its deformation shapes the vocal tract to produce sounds. It is also essential for eating, where it contains and propels the bolus during chewing and swallowing. Finally, the tongue is vital for breathing where every inhalation is accompanied by muscle activity designed to prevent the tongue from being pulled backward and closing off the airway [1].
Tongue malformation arising from natural malformation such as macroglos-sia or from surgery such as glossectomy to remove tongue cancer, creates situations that affect quality of life and may be life-threatening. The incidence of oral cancer has increased in the last four decades due to association to the human papillomatosis virus (HPV). Surgical ablation (glossectomy) and chemo-radiotherapy are the most common methods for treating tongue cancer patients [2]. It is crucial to understand the relationship between the anatomical structure of the tongue, its function, and the tumor for diagnosis, surgical planning, treatment quality assessment as well as scientific studies [3, 4]. However, the tongue is very difficult to study due to its location within the oral cavity, which makes measurement challenging. Characterization of tongue motion is also challenging because the tongue does not rely on rigid structures such as bones and cartilage, instead activates multiple muscles in a complex manner to produce a wide range of fast and precise movements [5]. Currently, there is no tool to directly characterize tongue motion and function with respect to the surgical approach and reconstruction procedure or chemo-radiation treatment in these patients [6]. The first step in these analyses is an algorithm to accurately and repeatably extract the tongue (or structure of interest) for use in further analysis.
Magnetic resonance imaging (MRI) is a widely used imaging modality for structural and functional analysis of the tongue and the vocal tract as it is nonionizing and shows excellent soft tissue contrast [3, 7–13]. In particular, fast MR imaging with tagging [14, 15] enables fast measurement and quantitative analysis of tongue motion during a specific speech or swallowing task [16, 17]. Since the major roles played by the tongue involve motion and the tongue is composed entirely of soft tissue, tagged-MRI is a natural method of exploring tongue behavior. There have been numerous attempts to compute 3D motion using dynamic MRI with MR tagging, mostly for cardiac motion analysis [18–22]. Several well-established algorithms such as tag-line intersection tracking [23–25], whole tag-line tracking [18, 20, 26, 27], harmonic phase (HARP) tracking [19, 21], Gabor filter-robust point matching-deformable model approach [28], and incompressible deformation estimation algorithm (IDEA) [22] enabled a computation of 2D and 3D motion fields from tagged-MR data. Despite the compelling need to study tongue function and this available technology, analysis of tagged-MRI data from the tongue has proved to be problematic because of the lack of an automatic or semiautomatic tongue segmentation method. The tongue surface is difficult to measure in tagged-MRI. The air at the surface creates error, and the sides and bottom of the tongue are contiguous with other tissues, which makes the tongue segmentation even more challenging. To resolve these dilemmas and proceed with the analysis of tongue motion from tagged-MRI data, we use separately acquired cine-MRI, collected with the same spatial and temporal parameters, so that boundaries are not obscured. Segmentation of the tongue is carried out on the cine images and then applied to the tagged-MRI data.
In our workflow, a set of multi-slice 2D tagged- and cine-MR images are acquired at three orthogonal orientations (axial, coronal, sagittal). Two-dimensional motion fields at each orientation and slice are computed from the tagged-MRI by HARP tracking [16, 29]. The tongue boundary is segmented from the cine-MR images and overlaid on the tagged dataset. Finally, IDEA [21, 22] is used to reconstruct 3D motion fields from the 2D motion fields within the segmented tongue under an incompressibility constraint. The success of this approach critically depends on the segmentation of all 2D cine images at all time frames to produce 3D segmented masks. This imposes significant burden to the user because there are a large number of cine images to be segmented (approximately 800 (2D) images per study in our case). In addition, it is often difficult to segment the periphery of the tongue on 2D images due to the insufficient image contrast between the tongue and neighboring soft tissues, which may lead to inconsistent segmentations between different time frames and different orientations, affecting the IDEA computation. As a consequence, user interaction and verification are crucial during and after 2D and 3D segmentations, which causes the segmentation to be the bottleneck of the entire workflow.
Image segmentation problems have long been studied in the medical imaging field. The gold-standard approach for accurate and robust segmentation is considered to be an expert’s manual delineation. Yet manual segmentation of time-varying structures on a series of 3D volumes is extremely time-consuming due to the large number of volumes. Manual segmentation is not only tedious, but also prone to inter- and intra-rater variability. Individual experimenters must determine landmarks based on image intensity differences and may not be as precise as automatic or semi-automatic segmentation algorithms in repeated measurements. There are numerous semi-or fully automatic algorithms available [30, 31] such as region growing approaches [32–35], classifiers or clustering approaches [36–38], model-based approaches [39–43], and atlas-based approaches [44–49]. Several methods were proposed to segment the tongue [50–52] and the vocal tract [53, 54]. These methods were applied to 2D MR images [50, 53, 54] or a static high-quality 3D MR images [51, 52]. Although these segmentation methods can be applied to the segmentation of time-varying volumes (i.e., motion) by repeatedly segmenting each volume at each time frame, they do not systematically process the entire set of volumes. Therefore, existing methods may not be efficient for our problem as they require the user interaction with individual volume if needed, and the user has to revisit the segmentation at each time frame to manually correct it if resulting segmentations are incorrect.
Our segmentation problem is challenging in several aspects: 1) There are a large number of images or volumes throughout the entire task cycle; in our experiments, there are 26 time frames per second and three image stacks, each with 7–14 slices. 2) Cine-MR images show relatively poor image contrast compared to conventional 3D high-resolution MR images, due to fast image acquisition. 3) The tongue may temporarily touch adjacent structures, such as the teeth or soft palate, at only a few time frames during the motion. This may lead automatic methods to incorrect segmentation of such boundaries. Therefore, user interactions, preferably minimal, are desired to guide the algorithm to correctly segment those regions or directly correct the segmentation results.
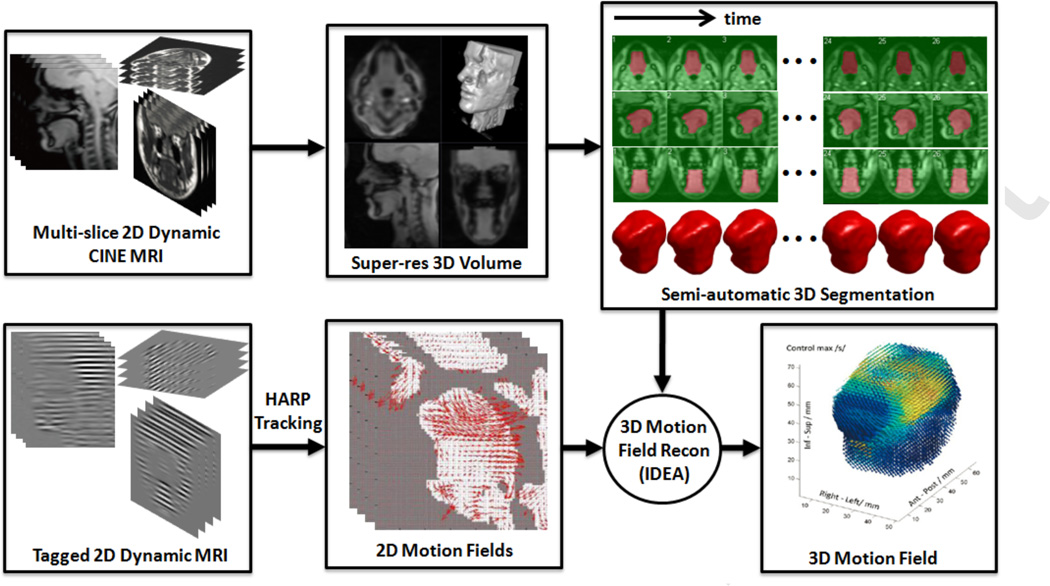
In this paper, we propose a semi-automatic segmentation method, which bridges the gap between the fast MR image acquisition and established 2D/3D motion analyses to complete the dynamic MRI-based tongue motion analysis workflow (see Fig. 1). The initial concept of our segmentation algorithm has been presented in [16, 55] with preliminary results. The proposed method computes a tongue mask at every time frame with minimal user input, thus significantly diminishing the segmentation burden for the user. Unlike our previously proposed method [16, 55], we directly segment a 3D super-resolution volume with isotropic voxel size that is reconstructed from 2D cine images at every time frame. The user has to input seeds on a few slices of the super-resolution volume at only one time frame, and seeds for the remaining time frames are automatically generated by 2D deformable registration and temporal stack segmentation. The super-resolution volume at every time frame is segmented by the random walker (RW) segmentation algorithm [56] using the generated seeds. The only manual interaction in the proposed method is the initial seeding on a few slices, which can be done in a few minutes. The successive segmentation will be automatically computed for all time frames. This method was validated on the tongues of five normal subjects carrying out the same speech task with multi-slice 2D dynamic cine-MR images obtained at three orthogonal orientations and 26 time frames.
Figure 1.
Dynamic MRI-based tongue motion estimation workow
The remainder of this paper is organized as follows. In Section 2, we describe our dynamic MR image acquisition process. In Section 3, we describe the key methods and each step of our semi-automatic segmentation workflow which consists of 1) super-resolution volume reconstruction, 2) random walker segmentation, 3) temporal stack segmentation, and 4) super-resolution volume segmentation. Numerical results based on five normal subjects are presented in Section 4. In Section 5, we further discuss the advantage and future improvements as well as other potential applications. Finally, the paper concludes in Section 6.
2. Dynamic MR image acquisition
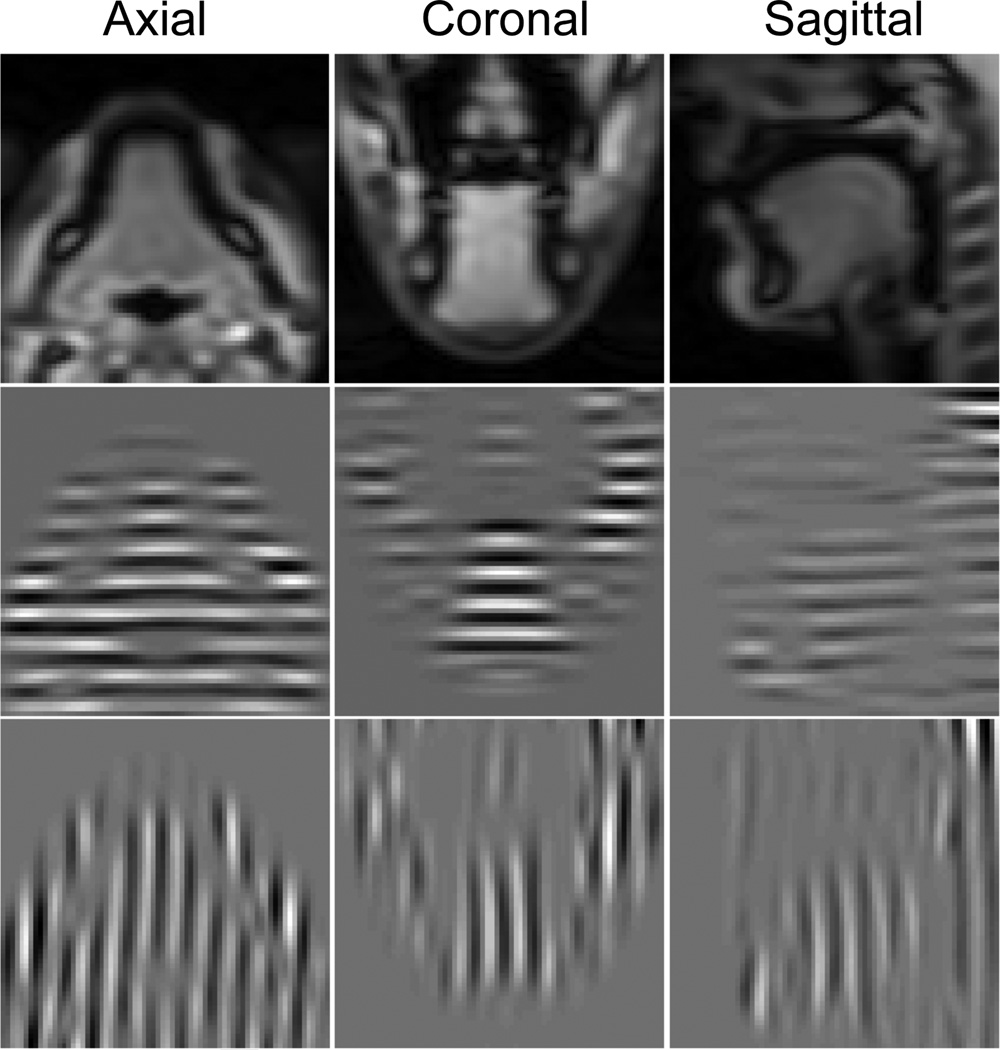
Our study uses T2-weighted multi-slice 2D dynamic cine- and tagged-MR images, acquired using a Siemens 3.0T Tim Trio system (Siemens Medical Solutions, Erlangen, Germany) at a frame rate of 26 frames per second. Both cine- and tagged-MR images were taken at exactly the same orientations using the same spatial and temporal parameters in the axial, coronal, and sagittal orientations while the subject repeated a speech task. The speech signal was simultaneously recorded during the image acquisition, and the MRI time frames for each slice were identified based on the speech phase. We used a fast MR image acquisition technique known as segmented k-space data acquisitions [57]. A set of k-space lines, i.e., partial Fourier information, were collected in a specified order but not constituting a complete coverage of k-space at each repetition. The complete k-space information was then assembled from the segmental repeated acquisitions in order to create an image by Fourier inversion. In our experiments, typically 9–12 axial, 9–14 coronal, and 7–9 sagittal images (depending on the subject’s tongue size) were acquired in each orientation over 26 time frames. The tagged-MR images contain horizontal and vertical tags. Each image is 128×128 pixels with a pixel size of 1.875×1.875 mm2, and both slice thickness and tag spacing are 6 mm. A user-chosen rectangular ROI was used to extract the tongue region on each slice for both the segmentation and the motion tracking. The choice of ROI does not change the segmentation results, but affects the computation time as the algorithm only segments the region inside the ROI. Fig. 2 shows an example of the cine- and tagged-MR images at three orientations.
Figure 2.
Cine-MR (top row) and horizontally (middle row) and vertically (bottom row) tagged-MR images at three orientations.
3. Methods
Fig. 1 shows our tongue motion estimation workflow. In this workflow, we first reconstruct super-resolution volumes from 2D cine images at all time frames. We then directly segment the 3D super-resolution volumes without requiring any 2D segmentation of the cine images. This approach enables the user to perform the segmentation using the 3D volumetric information of the target. The user needs to input seeds only on a few slices of the super-resolution volume at only one time frame. The user-given seeds are automatically propagated to the same slices at different time frames by 2D deformable registration. Instead of propagating seeds to all time frames, which is time-consuming, we do it for only a few selected time frames and segment a temporal stack of images across all time frames at the same slice location using random walker (RW) segmentation algorithm with the user-given and the propagated seeds. For time frames where no user-given seeds are available, seeds are automatically extracted from the segmentation of the temporal stack of images. Finally, the super-resolution volume at every time frame is automatically segmented by RW using the seeds provided by the user and the algorithm. The following sections describe each step of the proposed semi-automatic segmentation method.
3.1. Super-resolution volume reconstruction
In multi-slice 2D dynamic MR scans, through-plane (slice-selection direction) resolution is relatively poor compared to in-plane resolution, e.g., 6 mm versus 1.875 mm in this study. Low through-plane resolution limits the accuracy and robustness of volumetric image processing and analyses such as segmentation, registration, and 3D motion analysis. Instead of directly processing each multi-slice dataset that is insufficient for these tasks by itself, we derive a high-resolution, isotropic 3D volume from the three orthogonal 2D multi-slice image stacks using a super-resolution reconstruction technique developed in our group [58].
We first upsample each multi-slice image stack along the through-plane direction using a fifth-order B-spline interpolation to produce an isotropic volume. We then choose a target volume (sagittal in this study) and register the other two volumes (axial and coronal) to the target using mutual information as a similarity measure. Alignment by translation has been found to be sufficient to register these data sets. The image intensity mismatches between the three registered volumes are corrected by using a spline-based intensity regression method [58]. A super-resolution volume is then reconstructed by solving a maximum a posteriori (MAP) estimation problem:
| (1) |
where V is the super-resolution volume to be estimated, v1,v2,v3 are the processed three orthogonal volumes, p(v1,v2, v3|V) and p(V) represent the likelihood and the prior, respectively We assume that the image has additive white Gaussian noise, and use a Markov Random Field (MRF) prior to preserve the edge. We solve the MAP estimation problem in Eq. (1) by using a half-quadratic regularization technique that incorporates the region-based approach [59, 60]. The estimated super-resolution volume is 128×128x128 voxels with an isotropic voxel size of 1.875 mm. The super-resolution reconstruction is repeated for every time frame to yield a high-quality 4D MRI. The super-resolution volume not only provides a 3D volume with higher spatial resolution, but also reduces the blurring artifact caused by the misalignment between multi-slice images at three orientations. Therefore, direct segmentation of the 3D super-resolution volume yields an improved segmentation over that which can be achieved by segmenting the 2D images separately.
3.2. Random walker segmentation
Segmenting the tongue in all super-resolution volumes at all time frames is time-consuming as there are 26 volumes per study to segment in our case. As well, the insufficient image contrast between the tongue and adjacent soft tissues at the periphery of the tongue makes the segmentation task challenging. We use RW segmentation [56], which is a robust, graph-based, interactive semi-automatic algorithm, to find a globally optimal probabilistic multi-label image segmentation. RW is preferred as the user can interact to define proper boundaries in the region where image contrast between the target of interest and the surrounding structures is poor.
In the RW segmentation framework, a user specifies a small number of pixels with user-defined labels as seeds (in our case, on the tongue and the background). Each unlabeled pixel is assigned to the label with the greatest probability that a random walker starting at this pixel will reach one of the seeds with this label. In this framework, an image is considered as a graph that consists of a pair G = (V, E) with vertices (or nodes) v ∈ V and edges e ∈ E. An image pixel i corresponds to a node and is connected to the other node j by an edge eij We assign to each edge eij a Gaussian weighting function given by wij = exp{β(gi − gj)2} where gi indicates the image intensity at pixel i and β is a free parameter for which we used β = 30. It is known that the RW probabilities can be found by minimizing the combinatorial Dirichlet problem [56]
| (2) |
where D[x] is a combinatorial formulation of the Dirichlet integral, x is a real-valued vector defined over the set of nodes and L represents the combinatorial Laplacian matrix defined as in [18]. For the details of the algorithm, we refer readers to [56].
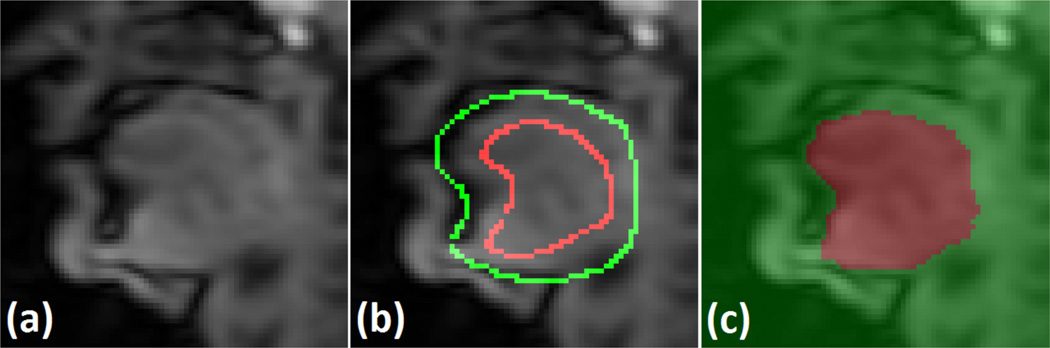
Fig. 3(a) shows an example sagittal image where the top-back of the tongue is touching the soft palate, showing no image contrast between these two structures. Fig. 3(b) shows an example of user-given seeds around the boundary of the tongue and the soft palate and Fig. 3(c) shows the resulting RW segmentation. This example demonstrates the capability of RW segmentation to accurately separate ambiguous regions with proper user interaction. In our method, RW is used not only for the segmentation of super-resolution volumes but also for the automatic seed generation by temporal stack segmentation. We now describe these two steps in the following sections.
Figure 3.
An example of RW segmentation of the tongue. (a) A sagittal image of the region where the tongue touches the soft palate showing very poor image contrast between these two structures. (b) A user-given seeds separating the tongue (red) and the background (green) including the soft palate. (c) RW segmentation of the tongue (red) and the background (green).
3.3. Temporal stack segmentation
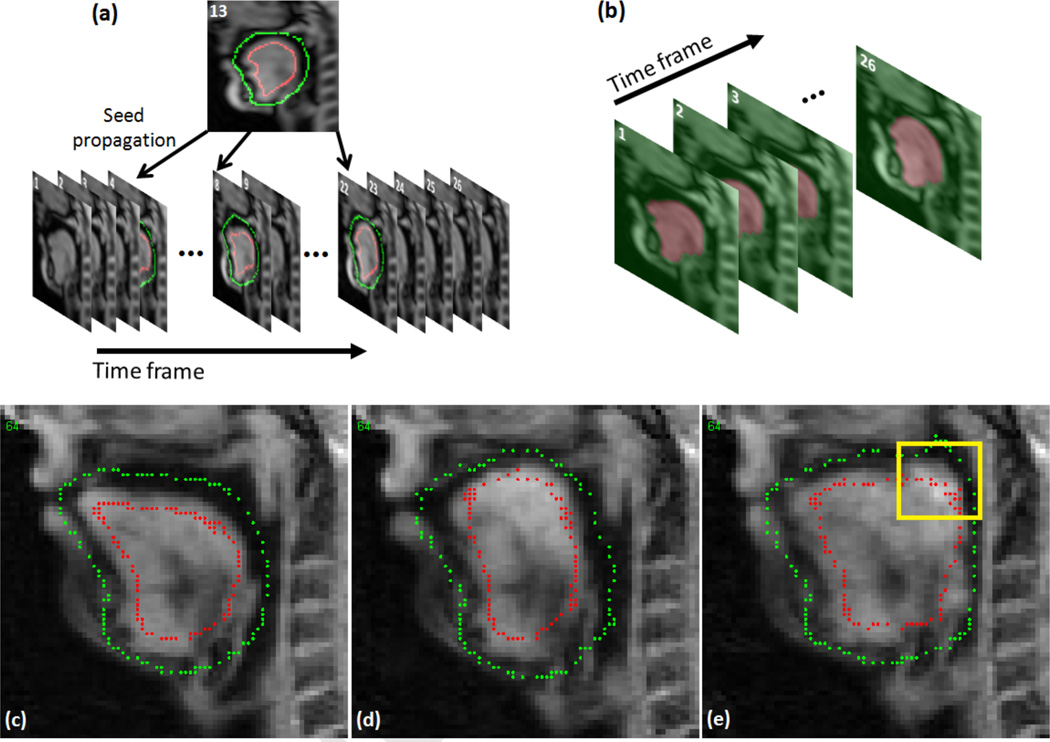
RW segmentation requires the user to input seeds only on a few slices of the target volume. However, it is laborious to segment all super-resolution volumes by manually inputting seeds due to the amount of data, i.e., 26 volumes per subject in our case. Therefore, we propose an approach to segment a temporal stack volume based on a small set of user-placed seeds at selected time frames from which seeds are automatically generated at all time frames. A temporal stack volume is a 3D volume that consists of a stack of 2D images at the same slice location and different time frames (see Fig. 4). For each user-chosen slice, we use time as the third dimension instead of through-plane direction to form a 3D temporal stack volume (2D target slice + time). The idea behind this is that the segmentation of temporal stack of images can be reliably computed by RW as images at the same slice location are smooth between adjacent time frames due to the fast image acquisition (26 frames per second). Seeds need to be input at only one time frame and then propagated to 3–4 other distributed time frames by 2D B-spline deformable registration [61] (see Fig. 4(a)). In case that the seeds are not properly propagated due to registration error, editing these incorrect seeds is trivial. Fig. 4(c)–(e) show an example of sagittal slice images with user-given seeds, properly propagated seeds, and incorrectly propagated seeds, respectively. In the case shown in Fig. 4(e), the tongue touched the soft palate and the seeds in the superior-posterior region of the tongue were moved to the soft palate (yellow box). However, the user can easily correct these incorrect seeds in the yellow box. The user-given and propagated seeds are then used to segment the 3D temporal stack volume using RW segmentation (Fig. 4(b)). The process is repeated for slices at different locations and orientations. Note that we only need to process several user-chosen slices (in this study, we only use 2–3 axial, 2–3 coronal, and 2–3 sagittal slices, a total of 6–9 slices) that are well-spread over the target volume. Since RW segmentation computes the probabilities of a random walker at each non-labeled pixel to reach the labeled pixels, i.e., seeds, to determine the segmented label on that pixel, it is desirable to spread the seeds over the volume rather than placing them only on specific regions or slices. These 3D temporal stack segmentations are then applied to automatic seed generation for the 3D super-resolution volume segmentations at all 26 time frames, of which process is described in the following section.
Figure 4.
An example of temporal stack segmentation at sagittal orientation. (a) Seed propagation from time frame 13 (user-given seeds) to other selected time frames (4, 8, 18, 22 in our case) by 2D deformable registration. (b) RW segmentation of temporal stack images using the user-given and propagated seeds in (a). (c) The user-given seeds on time frame 13. (d) An example of successful seed propagation. (e) An example of unsuccessful seed propagation.
3.4. Super-resolution volume segmentation
3D temporal stack segmentations of user-chosen slices (6–9 slices in our study) are used to generate seeds for the segmentation of 26 super-resolution volumes. For the time frames where no user-given seeds are available, seeds are extracted from the segmented 2D masks that are slices of the segmented temporal stack volume (see Fig. 4(b)). The segmented 2D mask M at each time frame is first eroded using a disk structuring element D to reliably extract seeds from the segmented 2D mask while eliminating potential errors from inaccurate segmentation near the boundary. For each label, eroded mask Me is computed by
| (3) |
where E is an Euclidean space, Ds is a translation of D by the vector s, i.e., Ds = {x + s\x ∈ D}, ∀s ∈ E. For each label l, boundary and the skeleton of the eroded mask are extracted. The union of the points on the boundary and the skeleton of the eroded mask becomes the extracted seeds for the 2D cine slice;
| (4) |
where i is the slice index and Nl is the number of labels. The super-resolution volume at each time frame is then segmented by RW using the user-given and automatically generated seeds that are available on 6–9 slices. Fig. 5 shows an example of automatically extracted seeds in axial, coronal, and sagittal slices from a 2D segmented mask at each orientation. Fig. 6 shows two examples of super-resolution volume segmentations performed on time frames 13 (seeds are provided by the user) and 1 (seeds are extracted from the temporal stack segmentations).
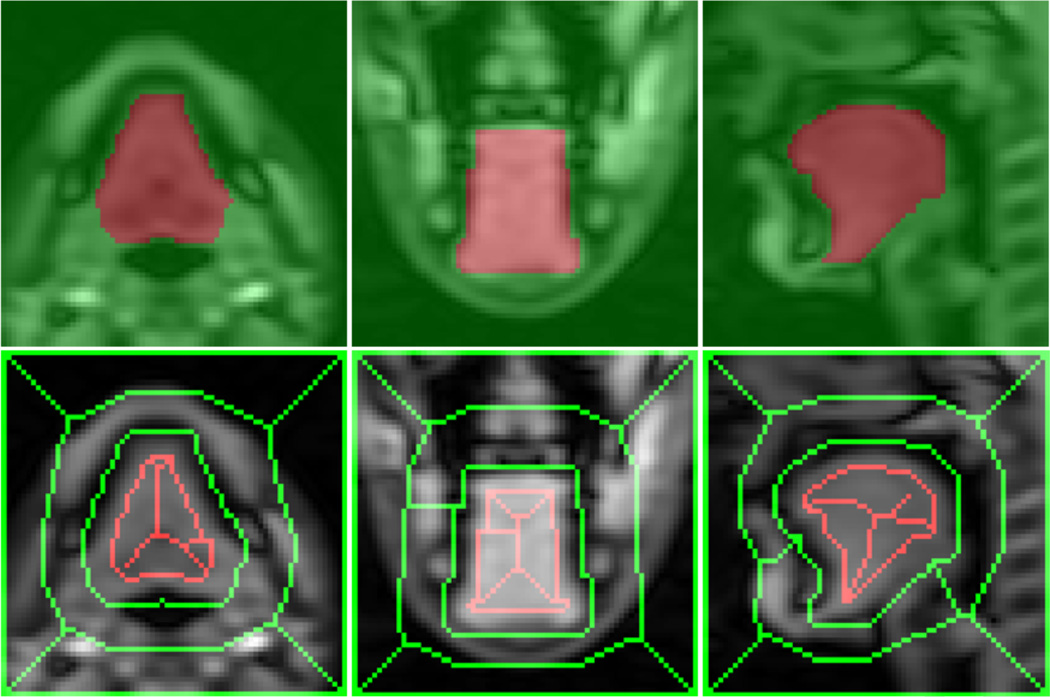
Figure 5.
Example of extracted seeds from the temporal stack of image segmentations. Bottom images show the seeds for the tongue (red) and the background (green) at three orientations extracted from the segmented masks on the top.
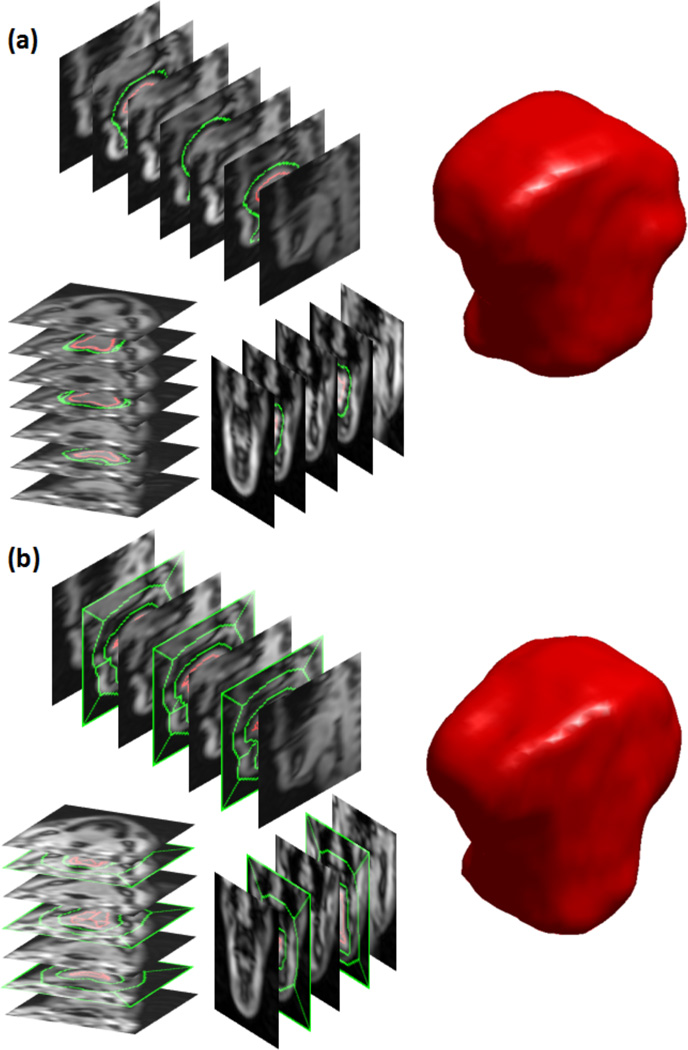
Figure 6.
Example of super-resolution volume segmentations. Seeds were provided at 8 slices (3 axial, 2 coronal, 3 sagittal slices) in the super-resolution volume to segment the tongue at each time frame. (a) User-given seeds at time frame 13, and the segmented 3D tongue from the super-resolution volume. (b) Automatically extracted seeds from the temporal stack segmentations at time frame 1, and the segmented 3D tongue from the super-resolution volume.
4. Experiments and results
We evaluated the proposed semi-automatic segmentation methods on MR images of five normal volunteers. Each subject performed the same speech task, repeating the word “asouk”, while multi-slice 2D dynamic cine- and tagged-MR images were acquired as described in Section 2. We chose this word for the following reasons. It takes less than a second to say, which is the temporal limit of the tagged MRI data collection. It begins with a “schwa” which is the neutral vowel, because the tongue is in the middle of the oral cavity and the shape of the vocal tract is close to a uniform cross sectional tube. Once the word “souk” begins there is little to no jaw motion, so tongue deformation is the primary mechanism for shaping the vocal tract. The motion from “s” to “k” is fairly unidirectionally backward and upward.
An isotropic super-resolution volume of 128 × 128 × 128 voxels with a voxel size of 1.875×1.875×1.875 mm3 was reconstructed at every time frame, yielding a total of 26 super-resolution volumes for each subject. A user-chosen ROI of approximately 70×70×70 voxels (ROI size varied case by case) was used for segmenting and tracking the tongue region. The user provided seeds on 2–3 axial, 2–3 coronal, and 2–3 sagittal slices only at time frame 13 (middle of 26 time frames), and the seeds were propagated to time frames 4, 8, 18, 22 by 2D B-spline deformable registration [61]. We specifically chose these time frames (4, 8, 13, 18, 22) to spread seeds out across 26 time frames, i.e., seeded every 4–5th time frame. For each slice with user-given seeds, 26 time frames were stacked to form a 3D temporal stack volume of ~ 70 × 70 × 26 voxels. The temporal stack volume was segmented by RW using the seeds available at 5 time frames (4, 8, 13, 18, 22). For the time frames where no seeds were provided (or propagated) by the user, seeds were automatically generated from the segmented mask of the temporal stack volume using the method described in Section 3.4. Note that this temporal stack segmentation was repeated only on 6–9 selected slices. 3D super-resolution volumes at all time frames were then automatically segmented by RW using the seeds provided by the user, propagated by the deformable registration, and generated from the temporal stack segmentations. Fig. 6 shows two example segmented surfaces of Subject 1 computed by RW at two time frames with user-provided (frame 13) and automatically generated (frame 1) seeds.
4.1. Segmentation quality and inter-rater variability
To evaluate the semi-automatic segmentation quality, a trained scientist (SA1) manually segmented all 130 super-resolution volumes (1 volume/time frame × 26 time frames × 5 subjects). For semi-automatic segmentation, three trained scientists (SA1–SA3), including the one who performed the manual segmentation (SA1), input seeds on slices of their own choices. Each user consistently input seeds on 3 axial, 3 coronal, and 3 sagittal slices at time frame 13. Dice similarity coefficients (DSCs) between the semi-automatic and the manual segmentations were computed at every time frame, and the averaged DSCs over 26 time frames are shown in Table 1. The segmentations differed mostly on the back of the tongue where we forced the exclusion of muscles extending outside the tongue. The variation among segmentations in this region was expected as there was no obvious image contrast between the muscles within and outside the tongue. Even with this ambiguity, the DSCs between different raters are very similar and the overall DSC for all three raters was 0.92.
Table 1.
Evaluation of the segmentation quality. Averaged DSCs between the semiautomatic (3 raters, SA1–SA3) and the manual segmentations for 26 time frames are computed.
| Subject | Number of slices with user-given seeds (axi,cor,sag) |
Average DSC |
||
|---|---|---|---|---|
| SA1 | SA2 | SA3 | ||
| S1 | 3, 3, 3 | 0.90 | 0.90 | 0.89 |
| S2 | 3, 3, 3 | 0.93 | 0.92 | 0.92 |
| S3 | 3, 3, 3 | 0.94 | 0.94 | 0.94 |
| S4 | 3, 3, 3 | 0.94 | 0.94 | 0.93 |
| S5 | 3, 3, 3 | 0.91 | 0.90 | 0.91 |
| Overall | 0.93 | 0.92 | 0.92 | |
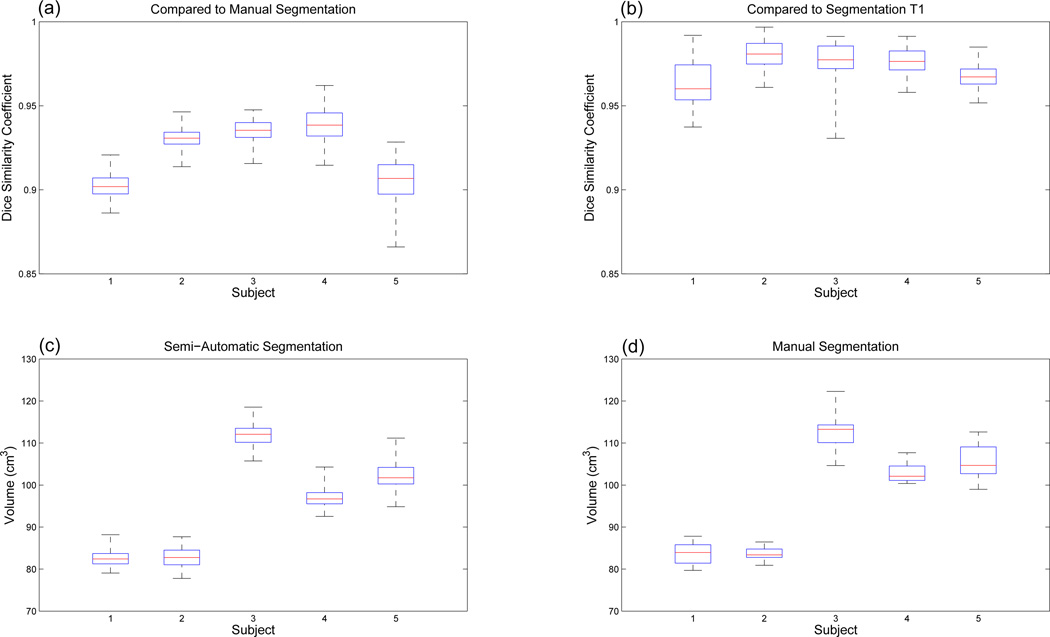
We also compared the volume variation of the segmented tongue across 26 time frames in the manual and the semi-automatic segmentations. Since the tongue is known to be incompressible, the volume of the segmented tongue mask at every time frame should not vary [16, 22]. As summarized in Table 2, the tongue volume varied on average (in terms of standard deviation) by 2.8 cm3 in manual segmentation, which comprised 2.8% of the tongue volume. In semi-automatic segmentations, the tongue volume varied by 2.3 cm3 (SA1), 2.3 cm3 (SA2), and 2.2 cm3 (SA3), comprising 2.4%, 2.4%, and 2.2% of the tongue volume, respectively. Although DSCs between different raters are very similar (see Table 1), the segmented volume sizes show noticeable differences in some cases. For example, the segmented volume of Subject 1 (S1) by the rater 3 (SA3) is larger than those by the other raters (SA1 and SA2). These differences happen mostly on the back of the tongue where different raters applied slightly different criteria to exclude muscles extending outside the tongue (see Fig. 7). Therefore, these differences stem from the rater performance rather than the algorithm, which is the nature of manual or semi-automatic segmentation. However, note that the semi-automatically segmented volume variability across 26 time frames is smaller for most cases (lower standard deviation) compared to the manual segmentation. The next section will discuss variability within and between these methods, however, this result shows that our semi-automatic method yields more consistent tongue segmentation across all time frames than the manual segmentation.
Table 2.
Evaluation of the segmented tongue volumes and the volume variability across time frames. The manual and semi-automatic (3 raters, SA1–SA3) segmentations are compared, and the mean and standard deviation of the sizes of the 26 segmented volumes for each subject are shown.
| Subject | Segmented volume mean ± s td (cm3) |
|||
|---|---|---|---|---|
| Manual | SA1 | SA2 | SA3 | |
| S1 | 83.9 ± 2.4 | 84.9 ± 1.5 | 86.7 ± 1.5 | 97.4 ± 1.8 |
| S2 | 83.5 ± 1.5 | 82.0 ± 2.3 | 82.7 ± 2.5 | 87.9 ± 2.4 |
| S3 | 112.7 ± 4.2 | 111.1 ± 3.1 | 114.6 ± 2.4 | 115.5 ± 2.1 |
| S4 | 102.8 ± 2.1 | 99.2 ± 2.2 | 99.8 ± 2.0 | 95.6 ± 2.2 |
| S5 | 105.6 ± 3.9 | 103.7 ± 2.7 | 99.2 ± 3.0 | 104.5 ± 2.5 |
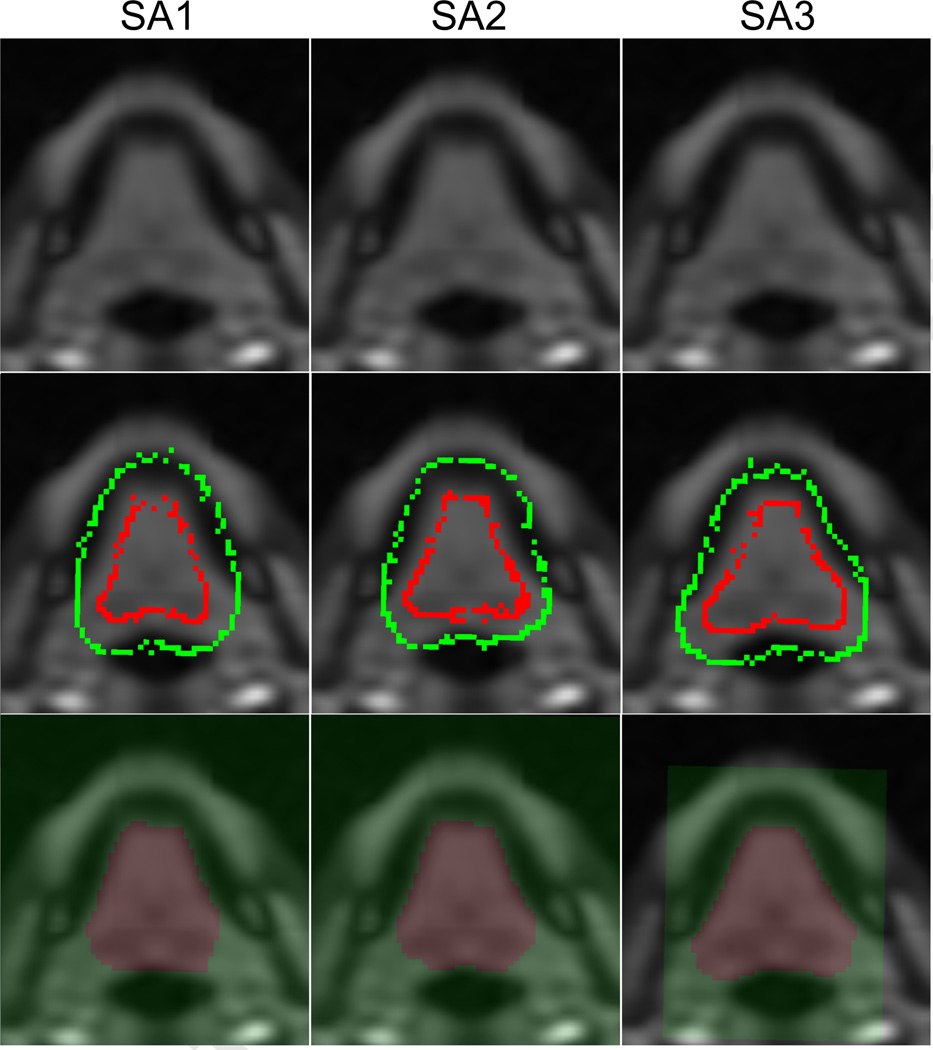
Figure 7.
Example of different seeding patterns in the back of the tongue region on Subject 1 (S1). (Top) Example axial slices on which seeds were input. SA1 and SA2 seeded on the axial slice 56 and SA3 seeded on slice 55. (Middle) User-given seeds for the tongue (red) and the background (green). (Bottom) Segmented tongue masks. SA3 used smaller ROI than SA1 and SA2. The tongue seeds (red) extend more in SA3 than the other two, resulting in larger segmented tongue (especially on the back).
4.2. Reproducibility: intra-rater variability
To evaluate the reproducibility of the segmentation, i.e., intra-rater variability, we repeated the semi-automatic segmentation eight times, Trials 1–8 (T1–T8). In this evaluation, the user (SA1) who performed the manual segmentation varied the number of slices between 2–3 at each orientation, and the location and combination of slices receiving input seeds. Fig. 8 shows the DSCs between the repeated semi-automatic segmentations and the manual segmentation as well as volume variability of the semi-automatic segmentations. For all 5 subjects and 8 repeated segmentations, DSC varied between 0.87 and 0.96 with an average DSC of 0.92, which is consistent with the results in Sec. 4.1. The overall volume variability for 8 repeats was lower than the manual segmentation except for only one case (S2). It was slightly higher than the manual segmentation for Subject 2 (S2), i.e., 2.3 versus 1.5 cm3 for which the manual segmentation was better than the other cases. Table 3 shows the results of the repeated segmentations for Subjects 4 and 5, which show the highest and the lowest averaged DSCs, respectively. The results indicate that the repeated semi-automatic segmentations are consistent with the same level of DSCs and the volume variabilities. Even for the most challenging case (S5) with the lowest averaged DSC and largest volume variability, the average DSC for all 8 repeated segmentations was 0.90 and the overall standard deviation of the segmented tongue volumes was lower than the single set of manual segmentations, i.e., 3.4 versus 3.9 cm3.
Figure 8.
DSC and volume box plots for the repeated semi-automatic segmentations and the manual segmentations. The red central mark is the median, the edges of the box are the 25th and 75th percentiles, and the whiskers extend to the most extreme data points. (a) DSCs between the repeated semi-automatic segmentations and the manual segmentations. (b) DSCs between the semi-automatic segmentations T1 and the other 7 sets of semi-automatic segmentations (T2-T8). (c) Volumes of the repeated semi-automatic segmentations. (d) Volumes of the manual segmentations.
Table 3.
Evaluation of the reproducibility of the semi-automatic segmentation. DSCs between the manual and eight semi-automatic segmentations, and between the first semiautomatic segmentation (T1) and the other seven segmentations (T2–T8) were computed. Individual segmented tongue volumes were also measured. The best and the worst cases are presented in this table.
| Sbj | Trial | Number of slices with user seeds (axi,cor,sag) |
DSC (vs manual) |
DSC (vs T1) |
Segmented volume (cm3) (mean ± std) |
|---|---|---|---|---|---|
| T1 | 3, 3, 3 | 0.94 | - | 99.2 ± 2.2 | |
| T2 | 3, 2, 3 | 0.94 | 0.99 | 98.3 ± 1.8 | |
| T3 | 3, 3, 2 | 0.94 | 0.98 | 97.3 ± 1.5 | |
| T4 | 3, 2, 2 | 0.94 | 0.98 | 96.7 ± 1.5 | |
| S4 | T5 | 2, 3, 3 | 0.94 | 0.97 | 96.7 ± 1.5 |
| T6 | 2, 2, 3 | 0.93 | 0.97 | 96.0 ± 1.9 | |
| T7 | 2, 3, 2 | 0.93 | 0.97 | 95.4 ± 1.7 | |
| T8 | 2, 2, 2 | 0.94 | 0.97 | 96.3 ± 1.8 | |
| Overall | 0.94 | 0.98 | 97.0 ± 2.1 | ||
| T1 | 3, 3, 3 | 0.91 | - | 103.7 ± 2.7 | |
| T2 | 3, 2, 3 | 0.90 | 0.97 | 102.8 ± 2.5 | |
| T3 | 3, 3, 2 | 0.91 | 0.97 | 103.3 ± 3.1 | |
| T4 | 3, 2, 2 | 0.90 | 0.96 | 104.0 ± 2.9 | |
| S5 | T5 | 2, 3, 3 | 0.90 | 0.96 | 102.2 ± 2.8 |
| T6 | 2, 2, 3 | 0.91 | 0.97 | 99.3 ± 2.6 | |
| T7 | 2, 3, 2 | 0.90 | 0.98 | 103.5 ± 2.5 | |
| T8 | 2, 2, 2 | 0.90 | 0.96 | 98.2 ± 2.6 | |
| Overall | 0.90 | 0.97 | 102.1 ± 3.4 | ||
It was observed that the manually segmented volumes are slightly larger than the semi-automatically segmented volumes. This happened because the semi-automatic segmentation algorithm and the human rater drew the boundary with slightly different criteria. Remember that the super-resolution volume was created by merging 3 sets of multi-slice images (at 3 different orientations). The resulting super-resolution volume was therefore blurry due to slight misalignment between the registered volumes as well as the low resolution of the original cine images (1.875 × 1.875 × 6 mm3 with slice thickness of 6 mm). The semi-automatic segmentation tends to separate two regions with different image intensities by choosing the mid-intensity as the boundary while the human rater tends to push the boundary to an extreme, i.e., close to one region. For example, on the tongue-air boundary, the semiautomatic algorithm chose the boundary in the middle gray region between the bright tongue and the black air, but the human rater included all blurred gray intensity as the tongue, which makes the manually segmented tongue larger than the semi-automatic segmentation. Note that this observation is solely based on the intra-rater variability study where we compared the repeated semi-automatic segmentations with the manual segmentation done by the same rater (SA1). However, this observation implies that there could be minor systematic difference between the manual and the semi-automatic segmentations if the rater does not select the mid-intensity between regions as the boundary during the manual segmentation.
For the same reason, we noticed that, even in the semi-automatic segmentations, the segmented tongue was slightly larger when the user input seeds on more slices, e.g., T1 compared to the others (T2–T8) in Table 3. This is because the user input seeds close to the boundary of the tongue while automatically extracted seeds were slightly inside due to the erosion operation. In this case, the segmentation at the time frame where user input (or propagated) seeds showed slightly larger segmented tongue compared to the other time frames with automatically extracted seeds, which resulted in increased volume variability. Therefore, cases with more slices with user-given seeds yielded slightly larger volume variability, e.g., T1. However, this caused a small variation in the resulting segmentations – only ~0.5 cm3 increase in standard deviation.
We also computed DSCs between the first segmentation (T1, with 3 axial, 3 coronal, and 3 sagittal slices with user-given seeds) and the other seven segmentations (T2–T8) to further evaluate the consistency of the segmentations (as seen in Table 3). In this case, DSC varied between 0.93 and 1.00 (after rounding to the nearest 1/100th) with an average DSC of 0.97. Fig. 8(b) shows these DSC plots for all 5 subjects and Table 3 shows DSCs of individual repeats for Subjects 4 and 5. These results demonstrate that the semi-automatic segmentation is reproducible with different choice of slices and different user-given seeds.
4.3. Computation time
The proposed semi-automatic segmentation was implemented on Matlab and ran on a PC with Intel Xeon CPU and 12 GB memory. Manual segmentation takes 20–30 minutes on average for each volume, which requires approximately 10 hours (20–30 minutes/volume × 26 volumes) for one subject. For the semi-automatic segmentation, seeding on a set of selected slices and propagating seeds to 4 different time frames take 2–3 minutes. The segmentation of a single temporal stack volume takes 2–3 seconds, which requires less than 30 seconds for segmenting temporal stack volumes for the selected 6–9 slices. The successive super-resolution volume segmentations for all 26 time frames take 2–5 minutes (5–10 seconds per each volume) depending on the ROI size. Since the super-resolution volume segmentation is automatic, the user only needs to interact with the software for the initial 3– 4 minutes. Overall, the whole semi-automatic segmentation process requires ~10 minutes at most, which implies that we can reduce the segmentation time by a factor of 60. Considering the actual user-interaction time, the user can reduce effort (in terms of time) by over 150 times without sacrificing performance.
4.4. Tongue motion analysis
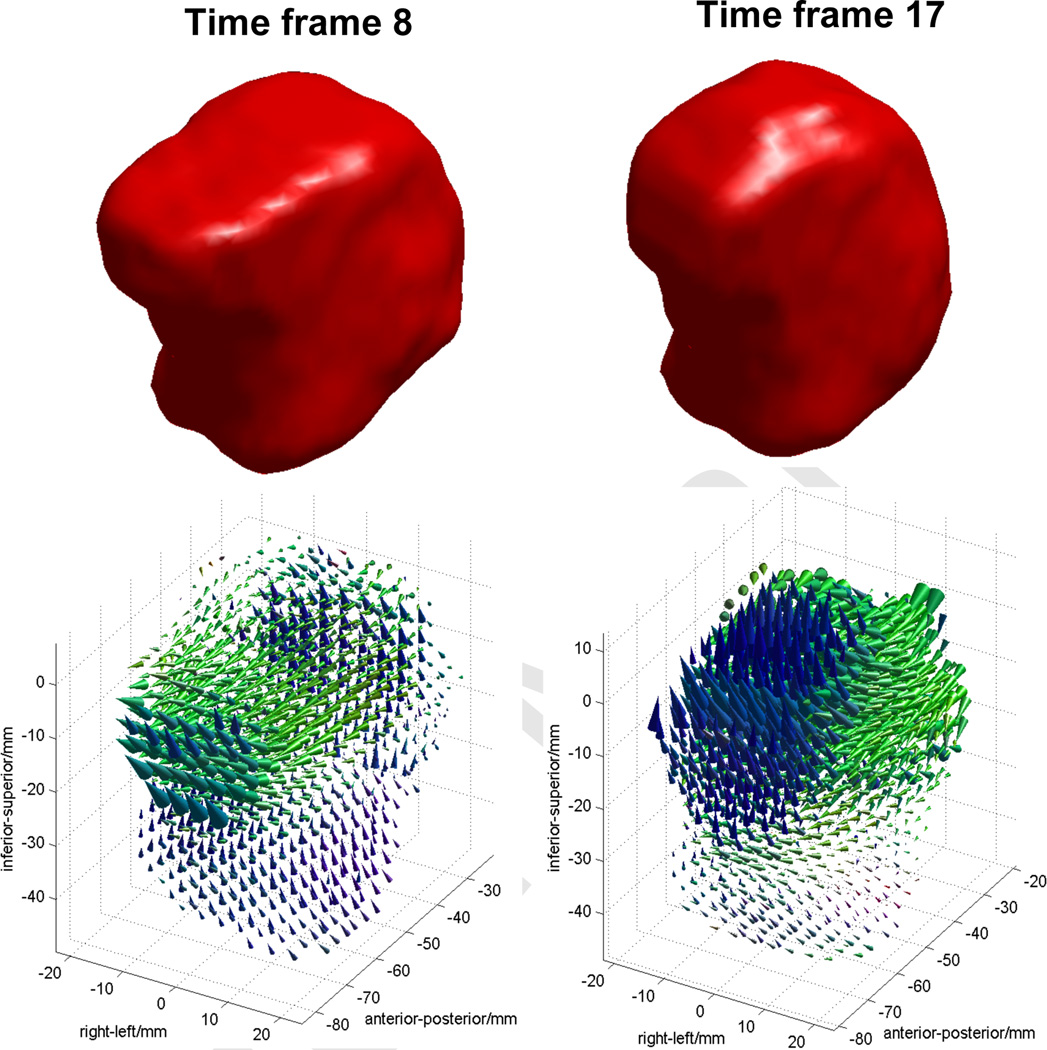
The proposed segmentation methods were originally developed for motion analysis of the tongue based on 4D MRI. To demonstrate the utility of the proposed methods in 3D tongue motion analysis, we have processed the tagged-MR images and the computed super-resolution volumes to compute 3D motion fields of the tongue for Subject 1 (S1). The 2D motion fields were first computed by the HARP tracking [19, 21] on the tagged-MR images. The super-resolution volumes were processed by the proposed semiautomatic segmentation to produce segmented 3D tongue masks for all 26 time frames. The 3D motion fields were then computed by IDEA [22] using the 2D motion fields and the segmented 3D tongue masks. Fig. 9 shows an example of the 3D tongue motion that visualizes 3D displacement vectors at individual voxels from the initial sound “a” to the sound “s” (time frame 8) and “k” (time frame 17) during the speech task of “asouk”. Each colored cone represents the direction (cone direction) and the amplitude (cone height) of the 3D displacement at each voxel position. The base of the cone represents the position of each voxel at the initial “a” sound and the tip corresponds to the moved position of the same voxel at the “s” or “k” sound. Cones were colored as red for the right-left motion, blue for the up-down motion, and green for the anterior-posterior motion for better visualization. The 3D displacement vectors imply that the tip of the tongue moves slightly forward (to the anterior direction) and also upward between the speech sounds “a” and “s”. From “a” to “k”, the top of the tongue moves upward while the back of the tongue moves forward. Further analysis on all normal and patient data sets are under investigation, and will be reported in a future publication.
Figure 9.
An example of 3D displacement vectors of the tongue computed by the semiautomatic segmentations and HARP-IDEA. Time frame 8 (left column) shows the tongue motion from “a” to “s”, and time frame 17 (right column) shows the motion from “a” to “k” sound during a speech task of “asouk”.
5. Discussion
In our experiments, we used expert’s manual segmentation as the gold standard to evaluate the performance of the semi-automatic segmentation. However, manual segmentation of the tongue is challenging and may show variability due to the lack of sufficient features in the boundary of the tongue and the adjacent soft tissues. Additionally, the tongue boundary is uncertain in case that the tongue touches the neighboring soft tissues such as soft palate during the speech. Harandi et al. [51, 52] repeated manual segmentation of the tongue on a high-resolution MRI, and reported that the volume overlap index between different manual segmentations was 91% (equivalently DSC of 0.91). We expect slightly lower volume overlap in our case as we used cine-MRI of which image quality is poorer than high-resolution MRI. Although there exists uncertainty in manual segmentation, the variability of the expert’s manual segmentation of the tongue is not high and is considered as reasonable gold standard [51, 52]. Therefore, DSC of around 0.91 or higher is considered acceptable in our experiments. Our method achieved an average DSC of 0.92 when comparing to manual segmentation, and an average DSC of 0.97 between the repeated semi-automatic segmentations, which is more consistent than the manual segmentation of the high-resolution MRI.
In Section 4, we have demonstrated the accuracy and effectiveness of our semi-automatic segmentation on five tongue motion analysis cases. Here we would like to discuss a few issues that a user may experience during the semi-automatic segmentation process as well as other potential applications.
In order to minimize the user’s effort in seeding, we propagate the user-given seeds from one time frame to others by 2D deformable registration. Seed propagation by deformable registration, however, is sometimes erroneous due to the insufficient image contrast between the tongue boundary and other neighboring structures. In our experiments, the erroneous seed propagation happened usually at a local region where the tongue touched the soft palate or the user forced to separate muscles extending outside of the tongue. These seed misplacements were manually corrected before the temporal stack segmentations. Although this requires additional user-interactions, manual correction of seeds adds only a few seconds per image. Other segmentation methods would suffer from such errors on ambiguous boundaries with very poor image contrast, in which case the user has to correct the final segmentations. Note that correcting seeds in a few 2D slices is much easier than correcting final segmentations of 3D volumes.
To further reduce the user’s effort, we also extract seeds from the segmented temporal stack of images for time frames where no user-given seeds are available. We typically used skeletons and edges of the segmented mask to produce user-given seed-like patterns, and let the user easily modify them if needed. However, one can use the segmented mask itself as a set of seeds after image erosion to eliminate errors on the boundary. This will reduce the size of the unlabeled regions, thus further reducing the segmentation time.
In our experiments, we used only two labels, i.e., tongue and background, to segment the whole tongue for its tissue motion during speech. Since RW segmentation supports multi-label segmentation, more than two structures can be simultaneously segmented in the same manner. Even if multiple time-varying structures need to be segmented, the user only needs to add more seeds with different labels on a few chosen 2D slices, which requires very little additional effort compared to the 2-label segmentation. Therefore, the overall computation time does not change much, which makes the proposed method even more attractive than manual or other segmentation methods. For example, the vocal tract can be simultaneously segmented along with the tongue in our data sets. Its shape and volume variation during speech is of great interest to speech scientists for quantitative modeling and analysis of sound production. The application of the proposed method may revolutionize scientific grounds using MRI for the vocal tract as gold standard in speech, but also in emerging field in swallowing and obstructive sleep apnea.
Segmented structures can also be analyzed by other methods such as principal component analysis after registering them across all time frames. Such a method will enable the analysis of the target shape variation and determination of the major modes of variation, and also be useful for the data sets without tagged-MRI or for the regions where MR tagging is challenging or infeasible, e.g., the vocal tract. Other image-based motion analysis methods can also benefit from our segmentation methods. One direct application would be the cardiac motion analysis that can be computed by HARP and IDEA [19, 21, 22], which requires segmentation of the heart across multiple time frames. Another potential application is tumor and organ motion analysis in radiation therapy. For example, respiration-induced tumor and organ motion in lung and pancreatic cancer cases is of great concern in radiation therapy. 4D CT and/or 4D MRI are typically used to analyze the motion and determine the treatment margin. In these 4D imaging techniques, a single respiratory cycle is divided into ~10 phases, yielding ~10 volumes representing the patient anatomy at different breathing phases. To compute the radiation therapy margin accounting for the motion and analyze the variation in shape and location, both the tumor and the critical structures must be segmented at each phase. Therefore, this segmentation process can be greatly improved (in terms of computation time) by the proposed semi-automatic approaches.
6. Conclusions
In this paper, we proposed a semi-automatic segmentation method for 3D motion analysis of the tongue with dynamic MRI. The proposed method requires a small amount of user-interactions only at initial stages to guide the algorithm. A few temporal stack volume segmentations followed by 3D super-resolution volume segmentations using RW over all time frames enable an accurate and robust automatic segmentation of time-varying structures. Overall, the proposed method significantly reduces the segmentation burden while keeping a more consistent segmentation quality compared to the manual segmentation. Although it was applied to the segmentation of the tongue, it can be extended to the segmentation of any time-varying objects such as the heart or tumors or organs experiencing motion due to breathing.
Acknowledgements
This work was supported by NIH/NCI under Grant 1R01CA133015 and by NIH/NIDCD under Grant K99/R00DC9279.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sauerland EK, Mitchell SP. Electromyographic activity of intrinsic and extrinsic muscles of the human tongue. Tex. Rep. Biol. Med. 1975;33(3):444–455. [PubMed] [Google Scholar]
- 2.Shah JP, Gil Z. Current concepts in management of oral cancer -Surgery. Oral Oncology. 2009;45(4–5):394–401. doi: 10.1016/j.oraloncology.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone M, Davis E, Douglas A, Aiver M, Gullapalli R, Levine W, Lundberg A. Modeling tongue surface contours from cine-MRI images. J. speech, Lang., Hear. Res. 2001;44(5):1026–1040. doi: 10.1044/1092-4388(2001/081). [DOI] [PubMed] [Google Scholar]
- 4.Wilhelms-Tricarico R. Physiological modeling of speech production: Methods for modeling soft-tissue articulators. J. Acoust. Soc. Amer. 1995;97:3085–3098. doi: 10.1121/1.411871. [DOI] [PubMed] [Google Scholar]
- 5.Kier WM, Smith KK. Tongues, tentacles and trunks: the biomechanics of movement in muscular-hydrostats. Zool. J. Linnean Soc. 1985;83:307–324. [Google Scholar]
- 6.Bokhari WA, Wang SJ. Tongue reconstruction: recent advances. Curr. Opin. Otolaryngol Head Neck Surg. 2007;15(4):202–207. doi: 10.1097/MOO.0b013e3281fbd406. [DOI] [PubMed] [Google Scholar]
- 7.Narayanan S, Byrd D, Kaun A. Geometry, kinematics, and acoustics of tamil liquid consonants. J. Acoust. Soc. Amer. 1999;106:1993–2007. doi: 10.1121/1.427946. [DOI] [PubMed] [Google Scholar]
- 8.Narayanan S, Alwan A, Haker K. An articulatory study of fricative consonants using magnetic resonance imaging. J. Acoust. Soc. Amer. 1995;98:1325–1347. [Google Scholar]
- 9.Bresch E, Kim Y, Nayak K, Byrd D, Narayanan S. Seeing speech: Capturing vocal tract shaping using real-time magnetic resonance imaging. IEEE Signal Process. Mag. 2008;25(3):123–132. [Google Scholar]
- 10.Lakshminarayanan A, Lee S, McCutcheon M. MR imaging of the vocal tract during vowel production. J. Magn. Reson. Imaging. 1991;1(1):71–76. doi: 10.1002/jmri.1880010109. [DOI] [PubMed] [Google Scholar]
- 11.Stone M, Liu X, Chen H, Prince J. A preliminary application of principal components and cluster analysis to internal tongue deformation patterns. Comput. Methods Biomech. Biomed. Eng. 2010;13(4):493–503. doi: 10.1080/10255842.2010.484809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Napadow V, Chen Q, Wedeen V, Gilbert R. Intramural mechanics of the human tongue in association with physiological deformations. J. Biomech. 1999;32(1):1–12. doi: 10.1016/s0021-9290(98)00109-2. [DOI] [PubMed] [Google Scholar]
- 13.Takano S, Honda K. An MRI analysis of the extrinsic tongue muscles during vowel production. Speech Commun. 2007;49(1):49–58. [Google Scholar]
- 14.Zerhouni EA, Parish DM, Rogers WJ, Yang A, Shapiro EP. Radiology. 1988;169:59–63. doi: 10.1148/radiology.169.1.3420283. [DOI] [PubMed] [Google Scholar]
- 15.Axel L, Dougherty L. MR imaging of motion with spatial modulation of magnetization. Radiology. 1989;171:841–845. doi: 10.1148/radiology.171.3.2717762. [DOI] [PubMed] [Google Scholar]
- 16.Xing F, Lee J, Murano E, Stone M, Prince JL. Estimating 3D tongue motion with MR images; Monterey, CA. 46th Annual Asilomar Conference on Signals, Systems, and Computers.2012. [Google Scholar]
- 17.Xing F, Murano EZ, Lee J, Woo J, Stone M, Prince JL. MRI analysis of 3D normal and post-glossectomy tongue motion in speech. 10th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2013:816–819. doi: 10.1109/ISBI.2013.6556600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denny TS, Prince JL. Reconstruction of 3-D left ventricular motion from planar tagged cardiac MR images: An estimation theoretic approach. IEEE Trans. Med. Imag. 1995;14(4):625–635. doi: 10.1109/42.476104. [DOI] [PubMed] [Google Scholar]
- 19.Osman NF, Kerwin WS, McVeigh ER, Prince JL. Cardiac motion tracking using CINE harmonic phase (HARP) magnetic resonance imaging. Magn. Reson. Med. 1999;42(6):1048–1060. doi: 10.1002/(sici)1522-2594(199912)42:6<1048::aid-mrm9>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haber I, Metaxas DN, Axel L. Three-dimensional motion reconstruction and analysis of the right ventricle using tagged MRI. Med. Imag. Anal. 2000;4(4):335–355. doi: 10.1016/s1361-8415(00)00028-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Prince JL. Shortest path refinement for motion estimation from tagged MR images. IEEE Trans. Med. Imag. 2010;29(8):1560–1572. doi: 10.1109/TMI.2010.2045509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Abd-Elmoniem K, Stone M, Murano EZ, Zhuo J, Gullapalli R, Prince JL. Incompressible deformation estimation algorithm (IDEA) from tagged MR images. IEEE Trans. Med. Imag. 2011;31(2):326–340. doi: 10.1109/TMI.2011.2168825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amini AA, Chen Y, Curwen RW, Manu V, Sun J. Coupled B-snake grides and constrained thin-plate splines for analysis of 2-D tissue deformations from tagged MRI. IEEE Trans. Med. Imag. 1998;17(3):344–356. doi: 10.1109/42.712124. [DOI] [PubMed] [Google Scholar]
- 24.Kerwin WS, Prince JL. Cardiac material markers from tagged MR images. Med. Imag. Anal. 1998;2(4):339–353. doi: 10.1016/s1361-8415(98)80015-7. [DOI] [PubMed] [Google Scholar]
- 25.Young AA, Kraitchman DL, Dougherty L, Axel L. Tracking and finite element analysis of stripe deformation in magnetic resonance tagging. IEEE. Trans. Med. Imag. 1995;14(3):413–421. doi: 10.1109/42.414605. [DOI] [PubMed] [Google Scholar]
- 26.Guttman MA, Prince JL, McVeigh ER. Tag and contour detection in tagged MR images of the left ventricle. IEEE. Trans. Med. Imag. 1994;13(1):74–88. doi: 10.1109/42.276146. [DOI] [PubMed] [Google Scholar]
- 27.Guttman MA, Zerhouni EA, MvVeigh ER. Analysis of cardiac function from MR images. IEEE Comput. Graph. Appl. 1997;17(1):30–38. doi: 10.1109/38.576854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen T, Wang X, Chung S, Metaxas D, Axel L. Automated 3D motion tracking using Gabor filter bank, robust point matching, and de-formable models. IEEE. Trans. Med. Imag. 2009;29(1):1–11. doi: 10.1109/TMI.2009.2021041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parthasarathy V, Prince JL, Stone M, Murano E, Nessaiver M. Measuring tongue motion from tagged cine-MRI using harmonic phase (HARP) processing. J. Acoust. Soc. Am. 2007;121(1):491–504. doi: 10.1121/1.2363926. [DOI] [PubMed] [Google Scholar]
- 30.Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2000;2:315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- 31.Balfar MA, Ramli AR, Saripan MI, Mashohor S. Review of brain MRI image segmentation methods. Artif. Intell. Rev. 2010;33:261–274. [Google Scholar]
- 32.Adams R. Seeded region growing. IEEE. Trans. Pattern Anal. Mach. Intell. 1994;16(6):641–647. [Google Scholar]
- 33.Mangin JF, Frouin V, Regis IBJ, Krahe J, Lopez From 3D magnetic resonance images to structural representations of the cortex topography using topology preserving deformations. J. Math. Imaging Vis. 1995;5:297–318. [Google Scholar]
- 34.Gibbs P, Buckley DL, Blackband SJ, Horsman A. Tumour volume detection from MR images by morphological segmentation. Phys. Med. Biol. 1996;41:2437–2446. doi: 10.1088/0031-9155/41/11/014. [DOI] [PubMed] [Google Scholar]
- 35.Manousakas IN, Undrill PE, Cameron GG, Redpath TW. Split-and-merge segmentation of magnetic resonance medical images: performance evaluation and extension to three dimensions. Comput. Biomed. Res. 1998;31:393–412. doi: 10.1006/cbmr.1998.1489. [DOI] [PubMed] [Google Scholar]
- 36.Coleman GB, Andrews HC. Image segmentation by clustering. Proc. IEEE. 1979;5:773–785. [Google Scholar]
- 37.Liang Z, MacFall JR, Harrington DP. Parameter estimation and tissue segmentation from multispectral MR images. IEEE Trans. Med. Imag. 1994;13:441–449. doi: 10.1109/42.310875. [DOI] [PubMed] [Google Scholar]
- 38.Kapur T, Grimson WEL, Kikinis R, Wells WM. Enhanced spatial priors for segmentation of magnetic resonance imagery; 1st Int. Conf. Med. Image Comput. Comp. Assist. Interv; 1998. pp. 457–468. [Google Scholar]
- 39.Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3D model-based neuroanatomical segmentation. Human Brain Mapping. 1995;3(3):190–208. [Google Scholar]
- 40.Kelemen A, Szekely G, Gerig G. Elastic model-based segmentation of 3-D neuroradiological data sets. IEEE. Trans. Med. Imag. 1999;18(10):828–839. doi: 10.1109/42.811260. [DOI] [PubMed] [Google Scholar]
- 41.Li SZ. Markov Random Field Modeling in Image Analysis. 3rd edn. London: Springer-Verlag; 2009. [Google Scholar]
- 42.Heimann T, Delingette H. Biomedical Image Processing, Biological and Medical Physics, Biomedical Engineering, chap. 11. Model-based segmentation. Berlin, Heidelberg: Springer; 2011. pp. 279–303. [Google Scholar]
- 43.Bogovic JA, Prince JL, Bazin P-L. A multiple object geometric de-formable model for image segmentation. Comput. Vision Image Underst. 2013;117:145–157. doi: 10.1016/j.cviu.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gee JC, Reivich M, Bajcsy R. Elastically deforming a three-dimensional atlas to match anatomical brain images. J. Comput. As. 1993;17:225–236. doi: 10.1097/00004728-199303000-00011. [DOI] [PubMed] [Google Scholar]
- 45.Andreasen NC, Rajarethinam R, Cizadlo T, Arndt S, II VWS, Flashman LA, O’Leary DS, Ehrhardt JC, Yuh WTC. Automatic atlas-based volume estimation of human brain regions from MR images. J. Comput. Assist. Tomogr. 1996;20:98–106. doi: 10.1097/00004728-199601000-00018. [DOI] [PubMed] [Google Scholar]
- 46.Christensen GE, Joshi SC, Miller MI. Volumetric transformation of brain anatomy. IEEE. Tran. 1997;16:864–877. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- 47.Lorenzo-Valdes M, Sanchez-Ortiz GI, Mohiaddin R, Ruechkert D. Atlas-based segmentation and tracking of 3D cardiac MR images using non-rigid registration MICCAI 2002, Lecture Notes in Computer Science. 2002;2488:642–650. [Google Scholar]
- 48.Rohlfing T, Brandt R, Menzel R, Maurer CR. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neu. 2004;21:1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 49.Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage. 2009;46(3):726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 50.Peng T, Kerrien E, Berger MO. A shape-based framework to segmentation of tongue contours from MRI data; IEEE International Conference Acoustics Speech and Signal Processing (ICASSP); 2010. pp. 662–665. [Google Scholar]
- 51.Harandi NM, Abugharbieh R, Fels S. Minimally interactive MRI segmentation for subject-specific modelling of the tongue, Lecture Notes in Computational Vision and Biomechanics. 2014;13:53–64. [Google Scholar]
- 52.Harandi NM, Abugharbieh R, Fels S. 3D segmentation of the tongue in MRI: a minimally interactive model-based approach, Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization. 2014:1–11. [Google Scholar]
- 53.Bresh E, Narayanan S. Region segmentation in the frequency domain applied to upper airway real-time magnetic resonance images. IEEE Trans Med Imag. 2009;28(3):323–338. doi: 10.1109/TMI.2008.928920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eryildirim A, Berger MO. A guided approach for automatic segmentation and modeling of the vocal tract in MRI images; European Signal Processing Conference (EUSIPCO-2011).2011. [Google Scholar]
- 55.Lee J, Woo J, Xing F, Murano EZ, Stone M, Prince JL. Semiautomatic segmentation of the tongue for 3D motion analysis with dynamic MRI. 10th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2013:1465–1468. doi: 10.1109/ISBI.2013.6556811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grady L. Random walks for image segmentation. IEEE Trans Pattern Anal. Mach. Intell. 2006;28(11):1768–1783. doi: 10.1109/TPAMI.2006.233. [DOI] [PubMed] [Google Scholar]
- 57.McVeigh ER, Atalar E. Cardiac tagging with breath-hold cine MRI. Magn. Reson. Med. 1992;28:318–327. doi: 10.1002/mrm.1910280214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woo J, Murano EZ, Stone M, Prince JL. Reconstruction of high-resolution tongue volumes from MRI. IEEE Trans. Biomed. Eng. 2012;59(12):3511–3524. doi: 10.1109/TBME.2012.2218246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villain JIN, Goussard Y, Allain M. Three-dimensional edgepreserving image enhancement for computed tomography. IEEE Trans. Med. Imag. 2003;22(10):1275–1287. doi: 10.1109/TMI.2003.817767. [DOI] [PubMed] [Google Scholar]
- 60.Deterministic edge-preserving regularization in computed imaging. IEEE Trans. Imag. Process. 1997;6(2):298–311. doi: 10.1109/83.551699. [DOI] [PubMed] [Google Scholar]
- 61.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. elastix: A toolbox for intensity-based medical image registration. IEEE Trans. Med. Imag. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]