Abstract
Background & Aims
Distinguishing between eosinophilic esophagitis (EoE), gastroesophageal reflux disease (GERD), and proton pump inhibitor-responsive esophageal eosinophilia (PPI-REE) is challenging. We assessed whether immunohistochemical analysis of esophageal tissues for major basic protein (MBP), eotaxin3, and tryptase can be used in diagnosis of EoE and to differentiate EoE from PPI-REE.
Methods
We conducted a prospective study of 196 consecutive adults who underwent outpatient endoscopy at the University of North Carolina from 2009 through 2012. Incident cases of EoE were diagnosed per consensus guidelines. Patients with gastroesophageal reflux disease or dysphagia served as controls. PPI-REE was defined as a symptomatic and histologic response to a PPI. Immunohistochemistry was performed to quantify MBP, eotaxin3, and tryptase. The maximum density of epithelial staining was determined for each assay; levels were compared between EoE and control, and then EoE and PPI-REE groups, and receiver operator characteristic (ROC) curves were constructed.
Results
Esophageal tissues from patients with EoE (n=50) had a median 951 MBP-positive cells/mm2 whereas those from controls (n=123) had a median 2 MBP-positive cells/mm2 (P<.001). Samples from patients with EoE had a median 155 eotaxin3-positive cells/mm2 and those from controls (n=123) had 18 MBP-positive cells/mm2 (P<.001). Samples from patients with EoE had a median 249 tryptase-positive cells/mm2 and those from controls (n=123) had 11 tryptase-positive cells/mm2 (P<.001). Levels of MBP, eotaxin3, tryptase, and the combination of all 3 identified patients with EoE with area under the ROC curve values of 0.99, 0.94, 0.99, and 1.00. Analyses of only samples with eosinophil counts of 10–100 eos/hpf produced similar results. No marker distinguished EoE from PPI-REE. Esophageal tissues from patients with PPI-REE (n=23) had 987 MBP-positive cells/mm2 (P=.18, compared with controls), 160 eotaxin3-positive cells/mm2 (P=.33), and 243 tryptase-positive cells/mm2 (P=.28).
Conclusions
Esophageal tissues from patients with EoE have substantially higher levels of MBP, eotaxin3, and tryptase than controls, based in immunohistochemical analysis. Assays for the 3 markers identify patients with EoE 100% accuracy, but cannot distinguish EoE from PPI-REE.
Keywords: AUCROC, GERD, immunohistochemical assay, diagnostic, validation
Introduction
Eosinophilic esophagitis (EoE) is a chronic, allergen/immune-mediated disease defined by symptoms of esophageal dysfunction and an esophageal eosinophilic infiltrate of ≥15 eosinophils per high-power field (eos/hpf), in the absence of competing causes.1, 2 Distinguishing EoE from GERD is difficult because symptoms and high levels of esophageal eosinophilia can be common to both.3, 4 Additionally, proton pump-inhibitor-responsive esophageal eosinophilia (PPI-REE), a condition where esophageal eosinophilia and symptoms respond to a PPI, further complicates the diagnostic algorithm. PPI-REE is now recognized as the cause of esophageal eosinophilia in at least 30–40% of patients, and because clinical features are similar in EoE and PPI-REE, a PPI trial is required prior to diagnosing EoE.2, 5–7
We have previously demonstrated that staining esophageal biopsies for markers of inflammation and eosinophil activation can distinguish EoE from GERD.8, 9 Identifying epithelial mast cells with tryptase staining provided strong discrimination between the two conditions.8 Staining for major basic protein (MBP), an eosinophil granule protein, and eotaxin-3, a potent eosinophil chemokine, also had significant value for separating cases of EoE from those with GERD.9 However, these studies were retrospective and were conducted prior to the recognition of PPI-REE. No prospective data demonstrate the utility of these markers for the diagnosis of EoE. Additionally, it is unknown whether staining for inflammatory markers in PPI-REE patients prior to a PPI trial has utility for discriminating EoE and PPI-REE
The aims of this study were to assess the utility of MBP, eotaxin-3, and tryptase staining in the esophageal epithelium for diagnosis of EoE using EoE cases and non-EoE controls, and to determine whether the same set of stains could differentiate EoE cases from patients with PPI-REE prior to a PPI trial. We hypothesized that the stains would be discriminative in both situations.
Methods
Study design and case definitions
We conducted a prospective study at the University of North Carolina (UNC) from 2009–2012 enrolling consecutive adults (age 18–80) undergoing outpatient esophagogastroduodenoscopy (EGD). Subjects with either dysphagia or GERD symptoms (heartburn, regurgitation/vomiting, reflux, dyspepsia) or as an indication for endoscopy were enrolled from the two UNC GI procedure units. Exclusion criteria were: a known diagnosis of EoE; a non-EoE eosinophilic gastrointestinal disorder; anticoagulation; GI bleeding; esophageal varices; esophageal cancer; prior esophageal surgery; comorbidities or medical instability precluding enrollment; and inability to read or understand the consent form. Details of portions of this study design were previously reported,7 and it was a stated aim of this study to assess tissue biomarkers. Subjects provided informed consent, including consent for future use of stored specimens, prior to the endoscopy. This allowed all tissue analysis (see below) to be batch run for greatest consistency. The study was approved by the UNC IRB.
All cases of EoE were incident, and were defined as per consensus guidelines.1, 2 Specifically, they had at least one typical symptom of esophageal dysfunction (for example dysphagia, food impaction, or heartburn); ≥15 eos/hpf on esophageal biopsy after an 8 week PPI trial (20–40 mg twice daily of any of the available agents, selected and prescribed at the discretion of the clinician); and other causes of esophageal eosinophilia excluded.
Control subjects were those who underwent endoscopy for an indication of either dysphagia or GERD symptoms and did not meet clinical or histologic criteria for EoE. Controls could have other evidence of inflammation on biopsy (for example, mixed inflammation including eosinophils in the setting of erosive esophagitis). All controls were symptomatic, and there were no controls with eosinophilia but no symptoms.
Subjects with PPI-REE were defined as having at least one typical symptom of esophageal dysfunction; ≥15 eos/hpf on esophageal biopsy; improvement of esophageal eosinophilia to <15 eos/hpf after an 8 week PPI trial (same dosing as above); and improvement of symptoms by self-report at the time of the repeat endoscopy. For all study subjects, the inclusion/exclusion criteria and case definitions remained constant over the course of the entire study.
Clinical and histologic data
Demographics, symptoms, endoscopy indications and findings were recorded using a standardized case report form. During endoscopy, we obtained five research-protocol esophageal biopsies (two from the proximal, one from the mid, and two from the distal esophagus) to maximize EoE diagnostic sensitivity.10 Research-protocol gastric and duodenal biopsies were also collected to exclude concomitant eosinophilic gastroenteritis. At the discretion of the endoscopist, additional clinical biopsies were taken as indicated.
For histologic analysis, the study pathologists utilized our validated protocol to determine eosinophil counts.11 In brief, slides were masked to the study group, digitized, and viewed with Aperio ImageScope (Aperio Technologies, Vista, CA). After examination of five microscopy fields from each of the five biopsies, the maximum eosinophil density (eosinophils/mm2 [eos/mm2]) was determined. In order to compare the eosinophil densities to results from other studies, they were converted to eosinophil counts (eos/hpf) for an actual hpf size of 0.24 mm2, the most commonly reported field size in the literature and also the field size of the microscopes used at UNC.12
Immunohistochemistry
For immunohistochemistry (IHC), the following primary antibodies were used: anti-MBP (mouse, clone BMK 13, catalogue #MCA5751, 1:100 dilution, AbD Serotec, Kidlington, UK and Raleigh, NC); anti-eotaxin-3 (goat, #500-P156G, 1:100 dilution, PeproTech, Rocky Hill, NJ); and anti-mast cell tryptase (mouse, Clone AA1; #M7052, 1:3000 dilution; Dako, Carpinteria, CA). Sections (5 microns) were cut from formalin-fixed paraffin-embedded tissue corresponding to the location with the highest level of esophageal eosinophilia and were masked to study group. IHC staining was carried out using the Bond Autostainer (Leica Microsystems Inc., Norwell, MA). Sections were dewaxed (Bond Dewax solution, AR9222) and hydrated (Bond Wash solution, AR9590. Antigen retrieval was with pepsin (20 mins) for MBP, and with Bond-Epitope Retrieval solution (citrate, pH=6, AR9961) for eotaxin-3 and tryptase. Slides were incubated with the primary antibody, incubated with a peroxidase-labelled secondary antibody, stained with a diaminobenzidine chromogen, and counterstained with hematoxylin. Stained slides were dehydrated and coverslipped. Lung and tonsil were used for controls, as per manufacturer recommendations. Positive control slides were incubated with primary antibody to confirm the antibody was working as expected. Negative controls were stained with antibody diluent and the secondary antibody to assess background staining. These positive and negative controls used tissue expected to stain for the targeted antigen. Negative control tissue not expected to stain for the targeted antigen was used for stain optimization and control.
Stained slides were analyzed after being scanned, digitized, and viewed, similar to how eosinophil counts were determined and in accordance with our previously published protocols.8, 9, 13 The maximum density of positively staining cells (cells/mm2) in the esophageal epithelial layer for each antibody of interest was quantified in five microscopy fields using the Aperio Positive Pixel Count Algorithm, version 9.1.
Statistical analysis
Statistical analysis was performed using Stata version 9 (Statacorp, College Station, TX). Bivariate analysis was performed with Chi-square for categorical variables, and t-tests, Wilcoxon Rank-sum, or Wilcoxon Signed-rank tests for continuous variables as appropriate. Receiver operating characteristic (ROC) curves were constructed and area under the curve (AUC) was calculated using EoE case status as defined by the consensus diagnostic guidelines as the gold standard.1, 2
For the first part of the analysis, ICH staining in the EoE cases (using samples from the post-PPI trial endoscopy) was compared to non-EoE controls. The AUC was calculated and compared for five different models predicting EoE case status: eosinophil count alone; MBP staining alone; eotaxin-3 staining alone; tryptase staining along; and a combined model containing MBP, eotaxin-3, tryptase staining, but no other clinical, endoscopic, or histologic features. For each stain we also determined the cut-point for cell density (cells/mm2) that maximized the sensitivity and specificity for EoE diagnosis, and calculated the sensitivity, specificity, and positive and negative likelihood ratios (LR). This analysis was repeated for pairs of stains (MBP/eotaxin-3; MBP/tryptase; and eotaxin-3/tryptase). Given that the majority of controls had no esophageal eosinophilia, we performed sensitivity analyses on subgroups of patients presenting diagnostic dilemmas. We first limited the study population to those with >10 but <100 eos/hpf, and then repeated the analysis with those with >10 but <30 eos/hpf. We also performed a subanalysis on the controls with definitive erosive reflux (symptoms of heartburn with erosive esophagitis). ROC curves were constructed for these sub-populations, and the AUC, sensitivity, specificity, and positive and negative LRs were calculated.
For the second part of the analysis, IHC staining in the EoE cases was compared to subjects with PPI-REE. ROC curves were constructed and AUCs were calculated. We compared the pre-PPI biopsies of PPI-REE patients to both the post-PPI biopsies of EoE patients and to the pre-PPI biopsies of EoE patients (in the subset of EoE patients for whom pre-PPI tissue was available). Additionally, we compared the pre-PPI biopsies of EoE patients to their matched post-PPI biopsies, to understand the impact of PPI therapy on these markers in EoE. Similarly, we compared the pre-PPI biopsies of PPI-REE patients to their matched post-PPI biopsies, to understand the impact of PPI therapy on these markers in PPI-REE.
Results
Characteristics of the study population
A total of 196 subjects had available tissue analyzed for this study, 50 with EoE, 23 with PPI-REE, and 123 non-EoE controls. EoE cases were a mean of 36 years, 62% were male, 94% were White, 95% had dysphagia, and the average maximum eosinophil count was 120 eos/hpf (Table 1). Subjects with PPI-REE were a mean of 48 years, 87% were male, 87% were white, and 96% had dysphagia. The pre-PPI average maximum eosinophil count was 65 eos/hpf, which decreased to 8 eos/hpf after PPI. The controls were a mean of 52 years, 41% were male, 76% were White, and 58% had dysphagia.
Table 1.
Baseline clinical characteristics of the study groups
| PPI-REE (n = 23) | EoE (n = 50) | Non-EoE controls (n = 123) | |
|---|---|---|---|
| Age at diagnosis (mean yrs ± SD) | 48.3 ± 11.1 | 35.9 ± 10.0 | 51.9 ± 15.3 |
| Male (n, %) | 20 (87) | 31 (62) | 51 (41) |
| White (n, %) | 20 (87) | 47 (94) | 94 (76) |
| Symptoms (n, %) | |||
| Dysphagia | 22 (96) | 36 (95) | 62 (58) |
| Heartburn | 2 (9) | 4 (8) | 23 (22) |
| Abdominal pain/dyspepsia | 0 (0) | 2 (4) | 20 (19)* |
| Nausea/vomiting/regurgitation | 1 (4) | 0 (0) | 4 (4)* |
| Atopic disorders | |||
| Asthma | 4 (24) | 15 (35) | 30 (31) |
| Atopic dermatitis | 2 (13) | 5 (12) | 9 (10) |
| Allergic rhinitis/sinusitis | 6 (35) | 31 (72) | 55 (57) |
| Food allergies | 1 (7) | 16 (37) | 26 (26) |
| EGD findings (n, %) | |||
| Normal | 1 (4) | 0 (0) | 31 (25) |
| Rings | 15 (65) | 45 (90) | 17 (14) |
| Stricture | 5 (22) | 18 (36) | 15 (12) |
| Narrowing | 2 (9) | 22 (44) | 5 (4) |
| Furrows | 14 (61) | 45 (90) | 10 (8) |
| Crêpe-paper | 1 (4) | 5 (10) | 0 (0) |
| White plaques/exudates | 8 (35) | 23 (46) | 8 (7) |
| Decreased vascularity | 0 (0) | 21 (42) | 3 (2) |
| Erosive esophagitis | 5 (22) | 0 (0) | 25 (20) |
| Schatzki’s ring | 5 (22) | 2 (4) | 9 (7) |
| Hiatal hernia | 7 (30) | 9 (18) | 49 (40) |
| Dilation performed | 8 (35) | 13 (26) | 29 (24) |
| Maximum eosinophil count (mean eos/hpf ± SD) | 65.2 ± 54.3 | 119.6 ± 101.6 | 3.5 ± 10.3 |
Of 20 controls with abdominal pain/dyspepsia, 3 had dysphagia and 2 had nausea or vomiting/regurgitation. Of 4 controls with nausea/vomiting/regurgitation, 1 had dysphagia.
Utility of MBP, eotaxin-3, and tryptase for distinguishing EoE cases from controls
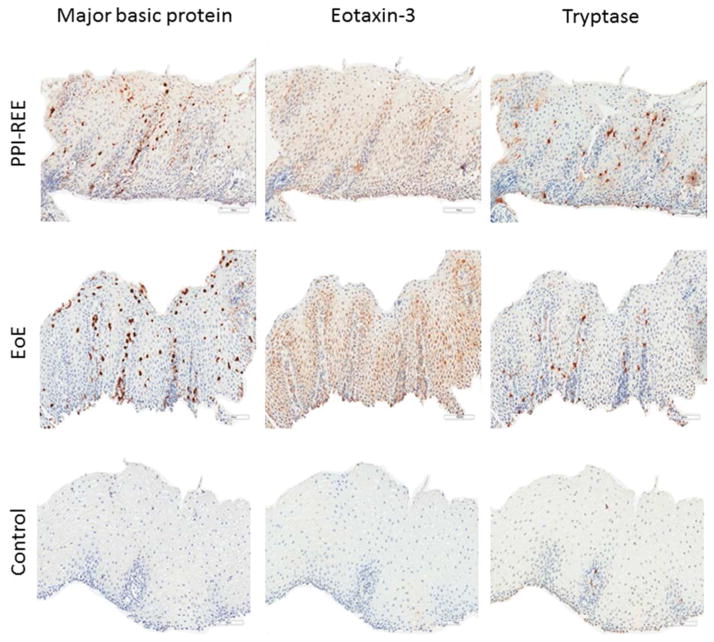
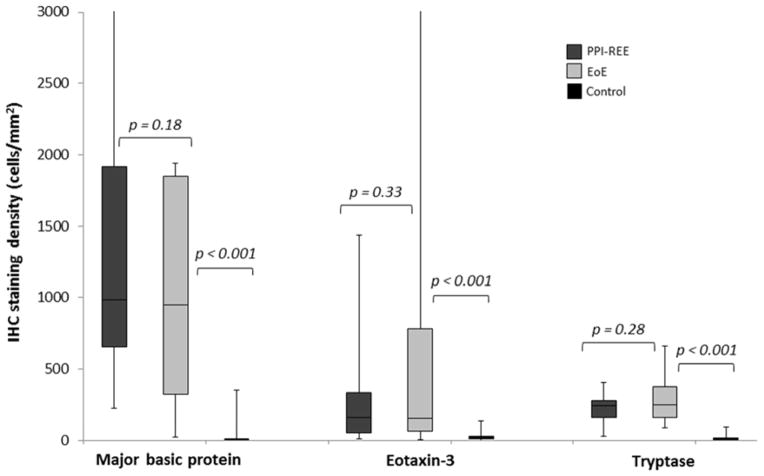
Staining for MBP, eotaxin-3, and tryptase was qualitatively different between EoE cases and controls (Figure 1). Quantitatively, staining levels were markedly higher in those with EoE compared to the controls (Figure 2). For MBP, EoE patients had a median of 951 cells/mm2 (IQR: 322-1849) and controls had 2 (0.4–6) (p<0.001). For eotaxin-3, the values were 155 (63-783) and 18 (10–31), respectively (p<0.001). For tryptase, the values were 249 (163–378) and 11 (7–19), respectively (p<0.001).
Figure 1.
Representative examples of immunohistochemistry staining. MBP, eotaxin-3, and tryptase are demonstrated for a PPI-REE, EoE, and GERD subject.
Figure 2.
Box and whiskers plots of staining in the PPI-REE, EoE, and control groups. The median value for each group is noted with the line in the box, the boxes represent the values from the 25th percentile to the 75th percentile, and the ends of the whiskers represent the maximum and minimum values.
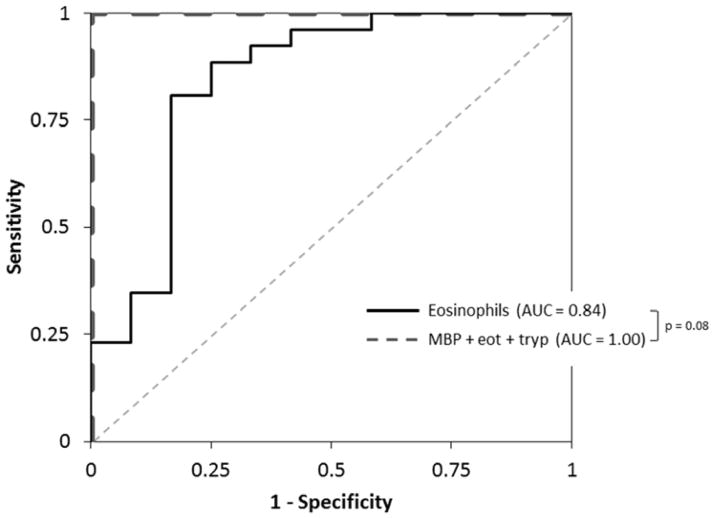
Analysis of operating characteristics showed these stains had excellent utility for diagnosis of EoE. The area under the ROC curves (AUC) for distinguishing EoE from controls were 0.99, 0.94, and 0.99, for MBP, eotaxin-3, and tryptase, respectively (Table 2). The AUC using the eosinophil count alone was 0.992, and the AUC for the combination of all three stains was 1.00. The sensitivity and specificity of using the three stains in combination for diagnosis of EoE in the absence of eosinophil count or other clinical information were 100% and 100%, respectively. When analyzing sets of two stains, for the pair of MBP/eotaxin-3, the AUC was 0.98, with a sensitive and specificity of 97% and 94%, respectively. For the pair of MBP/tryptase, the AUC was 1.00 with sensitivity and specificity of 100% and 100%. For the pair of eotaxin-3/tryptase, the AUC was 0.99, with sensitivity and specificity of 100% and 99%.
Table 2.
Operating characteristics of MBP, eotaxin-3, and tryptase IHC staining for diagnosis of EoE
| Eosinophil count (15 eos/hpf)† | MBP (99 positive cells/mm2)‡ | Eotaxin-3 (47 positive cells/mm2)‡ | Tryptase (89 positive cells/mm2)‡ | MBP, eotaxin-3, and tryptase (combination)# | |
|---|---|---|---|---|---|
| For all subjects | |||||
| ROC AUC | 0.992 | 0.989 | 0.937 | 0.999 | 1.000 |
| Sensitivity (%) | 100 | 96 | 88 | 100 | 100 |
| Specificity (%) | 94 | 94 | 89 | 99 | 100 |
| Positive LR | 17 | 16 | 8 | 100 | ∞ |
| Negative LR | 0 | 0.04 | 0.13 | 0 | 0 |
| For subjects with counts > 10 but < 100 eos/hpf (n = 38) | |||||
| ROC AUC | 0.843 | 0.891 | 0.902 | 0.996 | 1.000 |
| Sensitivity (%) | 100 | 96 | 78 | 100 | 100 |
| Specificity (%) | 42 | 55 | 92 | 91 | 100 |
| Positive LR | 1.7 | 2.2 | 9.8 | 11 | ∞ |
| Negative LR | 0 | 0.07 | 0.24 | 0 | 0 |
15 eos/hpf equals 63 eos/mm2 assuming a hpf size of 0.24 mm2.
These are the MBP, eotaxin-3, and tryptase staining densities that maximize sensitivity and specificity.
This uses the same cell staining density as the values for the individual stains.
When analysis was restricted to cases (n=26) and controls (n=12) with eosinophil counts >10 and <100 eos/hpf, similar results held (Table 2). The AUCs were 0.89, 0.90, and 0.99 for MBP, eotaxin-3, and tryptase, respectively. The AUC for the eosinophil count alone was 0.843, while the AUC for the combination of all three stains was 1.00 (Figure 3). In this analysis, seven of 12 controls (58%) would have been misclassified as EoE using the eosinophil count alone, while none would be misclassified using the three stains. For the combination of three stains, the sensitivity and specificity were again 100% and 100%. The data were also similar when the analysis was further restricted to cases (n = 6) and controls (n = 10) with eosinophil counts >10 and <30 eos/hpf, and to the comparison of all cases to controls with erosive reflux (n =13). For both of these sub-groups, the AUC was 1.00 for the combination of all three stains (data not shown).
Figure 3.
Receiver operating characteristic (ROC) curves for diagnosis of EoE. The dotted gray line represents a test that performs no better than chance (area under the curve [AUC] of 0.5). The solid black line is the eosinophil count alone and the dashed dark gray line is for the combination of all three stains.
Utility of MBP, eotaxin-3, and tryptase for distinguishing EoE cases from PPI-REE cases
In contrast to the analysis between cases of EoE and controls, there were not clear qualitative differences between EoE patients and PPI-REE (Figure 1). Quantitatively, staining levels for all three markers were also similar (Figure 2). Compared with the EoE staining results noted above, PPI-REE patients had a median of 987 cells/mm2 (IQR: 655–1917) for MBP (p=0.18), 160 (53–334) for eotaxin-3 (p=0.33), and 243 (163–280) for tryptase (p=0.28).
Analysis of the operating characteristics showed these stains did not distinguish between confirmed EoE cases and PPI-REE subjects before they had a PPI trial. The AUCs were 0.60, 0.58, 0.58, and 0.72 for MBP, eotaxin-3, tryptase, and the combination of all three stains, respectively.
A total of 21 baseline (pre-PPI) EoE samples were available for analysis, and there were no significant differences in staining patterns between EoE and PPI-REE. The baseline median cell counts for the EoE group prior to PPI treatment were 1581 cells/mm2 (IQR: 991–2387) for MBP, 266 (162–1092) for eotaxin-3, and 244 (159–348) for tryptase (p=0.37, 0.06, and 0.70, respectively, compared to the PPI-REE subjects, and p=0.35, 0.69, and 0.91, respectively compared to paired post-PPI EoE samples). The stains showed no utility for distinguishing EoE and PPI-REE, with AUCs of 0.58, 0.69, 0.54, and 0.67 for MBP, eotaxin-3, tryptase, and the combination of all three stains.
The analysis of the post-PPI samples in the PPI-REE group showed marked decreases in the levels of staining after the PPI trial. The post-PPI treatment median cell counts were 22 cells/mm2 (IQR 10–67) for MBP, 21 (11–33) for eotaxin-3, and 56 (28–116) for tryptase (p<0.001, p=0.001, and p<0.001, respectively, compared to the paired pre-PPI PPI-REE samples).
Discussion
We conducted a prospective study to determine whether staining the esophageal epithelium for markers previously demonstrated to be associated with EoE, MBP, eotaxin-3, and tryptase, would distinguish EoE cases from non-EoE controls with GERD or dysphagia. We additionally sought to define whether these stains could differentiate EoE from PPI-REE. Our results confirmed our hypothesis that these stains distinguished EoE from controls. In fact, when the three stains were used in combination, and in the absence of any clinical, endoscopic, or histologic data, they predicted EoE case status with perfect accuracy. Moreover, when we limited the analysis to the subpopulation with intermediate levels of esophageal eosinophilia, the group in which there is most often a diagnostic dilemma, the combination of stains also performed perfectly.
In contrast, we found that these stains did not have utility for separating EoE from PPI-REE, as both sets of subjects had similarly high staining levels. While this was counter to our hypothesis, it does raise the intriguing question of whether EoE and PPI-REE may share some common pathogenic basis.
The three tissue biomarkers used in this study were selected based on their role in the pathogenesis of EoE, that they were unlikely to be increased in GERD, and our previous work with these makers.8, 9, 14–22 Specifically, tissue cytokine levels and expression of eotaxin-3 are elevated in EoE.14–17 MBP has been used to localize eosinophils and characterize degranulation,20, 22 and MBP staining and degranulation is increased in EoE as compared to GERD.9, 19, 21, 22 Mast cell-specific genes have been shown to be upregulated in EoE,16, 23 and higher levels of mast cells have been reported in EoE.8, 17, 19, 21, 24, 25
Because the esophageal eosinophil count alone does not distinguish EoE and GERD,1, 3, 4 it would be desirable to have a set of predictors that are more specific for EoE. IHC is an attractive option as it is commercially available, relatively inexpensive, and, with good adherence to standardized protocols, reproducible between labs. In two related previous studies, we found that MBP, eotaxin-3, and tryptase held promise for diagnosis of EoE.8, 9 The results from the present study demonstrate in a prospective cohort the combined utility of these markers for the diagnosis of EoE. There is particular clinical utility in patients with intermediate eosinophil counts, and in our study 7 of 12 controls (58%) in this intermediate range of 10–100 eos/hpf would have been misclassified using the eosinophil count alone, whereas none would have been misclassified using the three stains. To optimize the usefulness of this panel, we have created an on-line tool that allows the clinician or pathologist to enter the results for each of the three stains used in this study and to calculate the probability of an EoE diagnosis (https://gicenter.med.unc.edu/cedas/eoe_calculator.html). In addition, using two of the three stains (ie MBP/tryptase or eotaxin-3/tryptase), yields comparable results to staining for all three markers, and could potentially reduce cost associated with this test.
Our IHC panel was not able to distinguish EoE from PPI-REE, as both conditions had markedly elevated levels of MBP, eotaxin-3, and tryptase. To our knowledge, this is the first study to provide immunohistologic data for a group of PPI-REE subjects, and these data provocatively suggest that patients with PPI-REE may share some of the same pathogenic features as patients with EoE, including elevated levels of mast cells. This may explain the difficulty in distinguishing PPI-REE from EoE based on clinical, endoscopic, and routine histologic features alone.7 It also may provide a basis for the recent observation that selected cytokine levels may be similar.26 Moreover, it is interesting to note that after the PPI trial, staining levels markedly decreased, correlating with the reduction in esophageal eosinophilia. Further investigation is required to understand the exact relation between EoE and PPI-REE and to elucidate pathogenic mechanisms of each.
This study has several limitations. First, it was performed at a single center, so results may not be generalizable. However, features of the EoE, PPI-REE, and control populations here are comparable to other previously reported populations.6, 10, 27, 28 Second, this study enrolled adults, so it is unknown if the data could be applied to children. Our prior studies, however, did analyze samples from children and found similar results.8, 9 Third, the number of control subjects in our subanalysis with intermediate levels of esophageal eosinophilia was relatively small. Fourth, given the widespread use of empiric PPI therapy, only a subset of subjects with EoE had PPI-naïve biopsies available for evaluation. While this shortcoming is unavoidable in all clinical studies of this type, it is notable that the staining levels for the EoE cases were similar both before and after the PPI trial. A related point is that pH testing was not performed to attempt to investigate whether reflux might have been the etiology of PPI-REE in a subgroup of patients. However, pH testing has not been shown to differentiate PPI-REE from EoE or predict PPI response.5, 6 Finally, even with the quantitative image analysis strategy used here, IHC can still be a variable technique. We mitigated this limitation by using an autostainer and a strict image analysis protocol. However, for the diagnostic cut-points determined in this study to hold, clinicians and pathologists will need to use a protocol similar to the one presented here.
This study has substantial strengths as well. The study design was prospective, allowing full characterization of patients, standardized tissue acquisition and processing protocols, and minimizing the likelihood of misclassification. We also used a previously validated pathology protocol to determine the eosinophil counts, obtained biopsies to exclude concomitant eosinophilic gastrointestinal disorders, and conducted a sensitivity analysis in the subgroup of patients in which these stains would have the highest clinical utility. The importance of studies validating preliminary findings is increasingly being recognized, and this study was designed to do just that. Moreover, the techniques used are readily available. To assist with the incorporation of these techniques into clinical practice, we have created a freely accessible EoE predictive calculator (see link above).
In conclusion, this prospective study showed that patients with EoE have substantially higher levels of MBP, eotaxin-3, and tryptase staining in the esophageal epithelium compared with GERD and dysphagia controls. These results held for the subset of controls with esophageal eosinophilia, and the combination of the three stains perfectly predicts EoE case status. However, staining levels were similar in EoE and PPI-REE cases. While these markers currently do not have value for distinguishing these two conditions prior to the PPI trial, the data present some of the first immunopathologic profiling of PPI-REE and raise the question of whether EoE and PPI-REE may represent different clinical manifestations of a common underlying pathogenic mechanism.
Acknowledgments
We gratefully acknowledge Mervi M. Eeva, Bentley R. Midkiff, Nana Nikolasichvili Feinberg, and Ying Li in the UNC Translational Pathology Lab for their technical assistance.
Grant support: This study was conducted with support, in part, by a grant from the CURED Foundation, the AGA-June and Donald O. Castell, MD Esophageal Clinical Research Award, and NIH K23 DK090073. It also utilized the Histology Core of the UNC Center for Gastrointestinal Biology and Disease (NIH P30DK034987) and the UNC Translational Pathology Lab (NIH P30CA016086).
Abbreviations
- AUC
area under the curve
- EGD
esophagogastroduodenoscopy
- EoE
eosinophilic esophagitis
- eos/hpf
eosinophils per high-power field
- GERD
gastroesophageal reflux disease
- IHC
immunohistochemistry
- MBP
major basic protein
- PPI
proton-pump inhibitors
- PPI-REE
PPI-responsive esophageal eosinophilia
- ROC
receive operator characteristic
- UNC
University of North Carolina
Footnotes
Disclosures: None of the authors have competing interests related to this manuscript.
Author contributions (all authors approved the final draft):
Dellon: Project conception, study design, data acquisition and interpretation, manuscript drafting, critical revision
Speck: Data acquisition (slide review for eosinophil counts), critical revision
Woodward: Data acquisition (slide review for eosinophil counts), critical revision
Covey: Data acquisition (slide review for eosinophil counts), critical revision
Rusin: Data acquisition (slide review for eosinophil counts), critical revision
Gebhart: Patient recruitment, data acquisition and management, critical revision
Chen: Project conception, immunohistochemistry supervision, critical revision
Woosley: Pathology supervision and eosinophil count review; critical revision
Shaheen: Project conception, study design, supervision, data interpretation, critical revision
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, Burks AW, Chehade M, Collins MH, Dellon ES, Dohil R, Falk GW, Gonsalves N, Gupta SK, Katzka DA, Lucendo AJ, Markowitz JE, Noel RJ, Odze RD, Putnam PE, Richter JE, Romero Y, Ruchelli E, Sampson HA, Schoepfer A, Shaheen NJ, Sicherer SH, Spechler S, Spergel JM, Straumann A, Wershil BK, Rothenberg ME, Aceves SS. Eosinophilic esophagitis: Updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. e6. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 2.Dellon ES, Gonsalves N, Hirano I, Furuta GT, Liacouras C, Katzka DA. ACG Clinical Guideline: Evidence based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis. Am J Gastroenterol. 2013;108:679–92. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 3.Dellon ES, Gibbs WB, Fritchie KJ, Rubinas TC, Wilson LA, Woosley JT, Shaheen NJ. Clinical, endoscopic, and histologic findings distinguish eosinophilic esophagitis from gastroesophageal reflux disease. Clin Gastroenterol Hepatol. 2009;7:1305–1313. doi: 10.1016/j.cgh.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigo S, Abboud G, Oh D, Demeester SR, Hagen J, Lipham J, Demeester TR, Chandrasoma P. High intraepithelial eosinophil counts in esophageal squamous epithelium are not specific for eosinophilic esophagitis in adults. Am J Gastroenterol. 2008;103:435–42. doi: 10.1111/j.1572-0241.2007.01594.x. [DOI] [PubMed] [Google Scholar]
- 5.Dranove JE, Horn DS, Davis MA, Kernek KM, Gupta SK. Predictors of response to proton pump inhibitor therapy among children with significant esophageal eosinophilia. J Pediatr. 2009;154:96–100. doi: 10.1016/j.jpeds.2008.07.042. [DOI] [PubMed] [Google Scholar]
- 6.Molina-Infante J, Ferrando-Lamana L, Ripoll C, Hernandez-Alonso M, Mateos JM, Fernandez-Bermejo M, Duenas C, Fernandez-Gonzalez N, Quintana EM, Gonzalez-Nunez MA. Esophageal Eosinophilic Infiltration Responds to Proton Pump Inhibition in Most Adults. Clin Gastroenterol Hepatol. 2011;9:110–7. doi: 10.1016/j.cgh.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Dellon ES, Speck O, Woodward K, Gebhart JH, Madanick RD, Levinson S, Fritchie KJ, Woosley JT, Shaheen NJ. Clinical and Endoscopic Characteristics do Not Reliably Differentiate PPI-Responsive Esophageal Eosinophilia and Eosinophilic Esophagitis in Patients Undergoing Upper Endoscopy: A Prospective Cohort Study. Am J Gastroenterol. 2013;108:1854–60. doi: 10.1038/ajg.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dellon ES, Chen X, Miller CR, Fritchie KJ, Rubinas TC, Woosley JT, Shaheen NJ. Tryptase staining of mast cells may differentiate eosinophilic esophagitis from gastroesophageal reflux disease. Am J Gastroenterol. 2011;106:264–71. doi: 10.1038/ajg.2010.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dellon ES, Chen X, Miller CR, Woosley JT, Shaheen NJ. Diagnostic utility of major basic protein, eotaxin-3, and leukotriene enzyme staining in eosinophilic esophagitis. Am J Gastroenterol. 2012;107:1503–11. doi: 10.1038/ajg.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonsalves N, Policarpio-Nicolas M, Zhang Q, Rao MS, Hirano I. Histopathologic variability and endoscopic correlates in adults with eosinophilic esophagitis. Gastrointest Endosc. 2006;64:313–9. doi: 10.1016/j.gie.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 11.Dellon ES, Fritchie KJ, Rubinas TC, Woosley JT, Shaheen NJ. Inter- and intraobserver reliability and validation of a new method for determination of eosinophil counts in patients with esophageal eosinophilia. Dig Dis Sci. 2010;55:1940–9. doi: 10.1007/s10620-009-1005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dellon ES, Aderoju A, Woosley JT, Sandler RS, Shaheen NJ. Variability in diagnostic criteria for eosinophilic esophagitis: A systematic review. Am J Gastroenterol. 2007;102:2300–13. doi: 10.1111/j.1572-0241.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 13.Dellon ES, Bower JJ, Keku TO, Chen X, Miller CR, Woosley JT, Orlando RC, Shaheen NJ. Markers of tyrosine kinase activity in eosinophilic esophagitis: a pilot study of the FIP1L1-PDGFRalpha fusion gene, pERK 1/2, and pSTAT5. Dis Esophagus. 2012;25:166–74. doi: 10.1111/j.1442-2050.2011.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharya B, Carlsten J, Sabo E, Kethu S, Meitner P, Tavares R, Jakate S, Mangray S, Aswad B, Resnick MB. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol. 2007;38:1744–1753. doi: 10.1016/j.humpath.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, Alexander ES, Butz BK, Jameson SC, Kaul A, Franciosi JP, Kushner JP, Putnam PE, Abonia JP, Rothenberg ME. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–17. 217 e1–7. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, Jameson SC, Kirby C, Konikoff MR, Collins MH, Cohen MB, Akers R, Hogan SP, Assa’ad AH, Putnam PE, Aronow BJ, Rothenberg ME. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta SK, Fitzgerald JF, Kondratyuk T, HogenEsch H. Cytokine expression in normal and inflamed esophageal mucosa: a study into the pathogenesis of allergic eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2006;42:22–6. doi: 10.1097/01.mpg.0000188740.38757.d2. [DOI] [PubMed] [Google Scholar]
- 18.Justinich CJ, Ricci A, Jr, Kalafus DA, Treem WR, Hyams JS, Kreutzer DL. Activated eosinophils in esophagitis in children: a transmission electron microscopic study. J Pediatr Gastroenterol Nutr. 1997;25:194–8. doi: 10.1097/00005176-199708000-00011. [DOI] [PubMed] [Google Scholar]
- 19.Lucendo AJ, Navarro M, Comas C, Pascual JM, Burgos E, Santamaria L, Larrauri J. Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am J Surg Pathol. 2007;31:598–606. doi: 10.1097/01.pas.0000213392.49698.8c. [DOI] [PubMed] [Google Scholar]
- 20.Mueller S, Aigner T, Neureiter D, Stolte M. Eosinophil infiltration and degranulation in oesophageal mucosa from adult patients with eosinophilic oesophagitis: a retrospective and comparative study on pathological biopsy. J Clin Pathol. 2006;59:1175–80. doi: 10.1136/jcp.2005.031922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller S, Neureiter D, Aigner T, Stolte M. Comparison of histological parameters for the diagnosis of eosinophilic oesophagitis versus gastro-oesophageal reflux disease on oesophageal biopsy material. Histopathology. 2008;53:676–84. doi: 10.1111/j.1365-2559.2008.03187.x. [DOI] [PubMed] [Google Scholar]
- 22.Colombo JM, Neilan NA, Schurman JV, Friesen CA. Validation of methods to assess potential biomarkers in pediatric patients with esophageal eosinophilia. World J Gastrointest Pharmacol Ther. 2013;4:113–9. doi: 10.4292/wjgpt.v4.i4.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abonia JP, Blanchard C, Butz BB, Rainey HF, Collins MH, Stringer K, Putnam PE, Rothenberg ME. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:140–9. doi: 10.1016/j.jaci.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108:954–61. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 25.Kirsch R, Bokhary R, Marcon MA, Cutz E. Activated mucosal mast cells differentiate eosinophilic (allergic) esophagitis from gastroesophageal reflux disease. J Pediatr Gastroenterol Nutr. 2007;44:20–6. doi: 10.1097/MPG.0b013e31802c0d06. [DOI] [PubMed] [Google Scholar]
- 26.Molina-Infante J, Rivas MD, Rodriguez GV, Alonso MH, Duenas-Sadornil C, Rodriguez JMM, Gallardo BP, Banares R, Zamorano J. Remission in proton pump inhibitors-responsive esophageal eosinophilia correlates with downregulation of eotaxin-3 and TH2 cytokines, similarly to eosinophilic esophagitis after steroids. Gastroenterology. 2013;144 (Suppl 1):S484. (Su1828) [Google Scholar]
- 27.Veerappan GR, Perry JL, Duncan TJ, Baker TP, Maydonovitch C, Lake JM, Wong RK, Osgard EM. Prevalence of Eosinophilic Esophagitis in an Adult Population Undergoing Upper Endoscopy: A Prospective Study. Clin Gastroenterol Hepatol. 2009;7:420–426. doi: 10.1016/j.cgh.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Moawad FJ, Veerappan GR, Dias JA, Baker TP, Maydonovitch CL, Wong RK. Randomized controlled trial comparing aerosolized swallowed fluticasone to esomeprazole for esophageal eosinophilia. Am J Gastroenterol. 2013;108:366–72. doi: 10.1038/ajg.2012.443. [DOI] [PubMed] [Google Scholar]