Abstract
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DNA double-strand breaks (DSBs) can arise from internal or external sources of damage, and the rapid detection, processing, and repair of this damage is important for cell viability. Failure to repair DNA damage can result in genomic instability, ultimately increasing the frequency of lymphoid disorders, neurodegeneration, and cancer. The Mre11-Rad50-Nbs1 (Xrs2) complex plays a central and critical role in detection and repair of DSBs and is conserved in all kingdoms of life, as Mre11/Rad50 (MR) in prokaryotes and as MRN/X in eukaryotes (Lamarche et al., 2010; Stracker and Petrini, 2011). The importance of this complex is emphasized by the fact that deletion of any of the three components results in embryonic lethality in mice and loss of proliferative activity in embryonic stem cells (Buis et al., 2008; Luo et al., 1999; Xiao and Weaver, 1997; Zhu et al., 2001) which is likely related to the role of MRN/X in homologous recombination. Repair of DSBs by homologous recombination involves replication of the broken region using an undamaged template, usually a sister chromatid. Deletions of other genes important for homologous forms of repair also exhibit early embryonic lethality, including Rad51, BRCA1, BRCA2, and CtBP-interacting protein (CtIP)(Chen et al., 2005b; Gowen et al., 1996; Lim and Hasty, 1996; Sharan et al., 1997). Hypomorphic mutations in MRN components result into developmental and neurodegenerative disorders in humans, including Ataxia-Telangiectasia-Like Disorder (ATLD), Nijmegen Breakage Syndrome (NBS), and NBS-like syndrome (Matsumoto et al., 2011; Stewart et al., 1999; Varon et al., 1998; Waltes et al., 2009), which are related, at least in part, to the role of MRN/X in the activation of cell-cycle checkpoints through the Ataxia-Telangiectasia-Mutated (ATM) protein kinase (Lee and Paull, 2007; Shiloh and Ziv, 2013). The roles of MRN/X also extend to the processing of DSBs during meiosis, for which it is essential, and to telomere maintenance (Borde, 2007; Lamarche et al., 2010).
Repair of DSBs is achieved through two broadly-defined groups of pathways: nonhomologous end joining (NHEJ) and homologous recombination (HR) (Krogh and Symington, 2004). The choice between these pathways primarily depends on the cell-cycle phase and the complexity of the damage generated at the break site (Chapman et al., 2012; Schipler and Iliakis, 2013). In the classical NHEJ pathway, ends are bound by the Ku70–Ku80 heterodimer/DNA-dependent protein kinase catalytic subunit (DNA-PKcs) complex which recruits additional factors involved in end modifications and gap filling. DNA ends are ultimately ligated by the NHEJ-specific DNA ligase IV complex (Deriano and Roth, 2013). In mammalian cells, the C-NHEJ pathway is not dependent on the MRN complex, although in budding yeast MRX contributes to NHEJ pathway through interactions with Ku70-Ku80 and DNA Lig4 complexes (Lewis and Resnick, 2000). The MRN complex, in conjunction with CtIP/Sae2, also regulates the alternative NHEJ (A-NHEJ or MMEJ), which utilizes short microhomologies and can result in large deletions (Lee and Lee, 2007; Yun and Hiom, 2009). In mammalian cells MRN was also shown to interact with DNA ligaseIIIα/Xrcc1, the ligase complex implicated in alternative NHEJ, stimulating intermolecular ligation (Della-Maria et al., 2011).
In contrast to NHEJ, HR requires the 5′–3′ resection of dsDNA to generate single-stranded DNA tails, a process that is initiated by the MRN complex and CtIP (You and Bailis, 2010). Extensive resection is perfomed by exonuclease 1 (Exo1), and Dna2 (Symington and Gautier, 2011), whose activities are also promoted by MRN (Cejka et al., 2010; Nicolette et al., 2010; Niu et al., 2010; Yang et al., 2013; Zhou et al., 2014; Zhou and Paull, 2013). 3′ ssDNA tails thus generated are bound by replication protein A (RPA), which activates ATM- and Rad3-Related (ATR), promoting replication checkpoint arrest and stabilization of replication forks (Zeman and Cimprich, 2014). RPA on these 3′ ssDNA tails is then exchanged for Rad51 to create Rad51 filaments that catalyze homology search and strand invasion, ultimately priming DNA synthesis and resolution of repair intermediates.
The MRN complex plays important and diverse roles in DNA double-strand break repair and signaling. Here we review recent evidence elucidating the structures and regulation of the Mre11/Rad50 complex, focusing primarily on the enzymatic activities of MRN and the role of ATP-driven conformational changes in Rad50.
Mre11 nuclease activity
The Mre11 protein is related to a family of phosphoesterases that includes lambda phosphatase, protein phosphatase-2B, PP2A, PP1, calcineurin, and purple acid phosphatases (Koonin, 1994)(Fig. 1). This family of enzymes binds two metal ions in the active site and cleaves either phosphomonoester or phosphodiester bonds. Mre11 is conserved in all species and exhibits manganese-dependent 3′ to 5′ exonuclease and endonuclease activities on double-stranded DNA in vitro (Connelly et al., 1999; Hopkins and Paull, 2008; Paull and Gellert, 1998; Trujillo and Sung, 2001; Trujillo et al., 1998). The roles of these activities in cells have been widely debated and it is still not entirely clear what the biologically relevant activity is, but it is likely that this depends on the structure and context of the DNA ends. Experiments in budding yeast have shown that the nuclease activity of Mre11 is dispensable for the resection of enzymatically-generated DSBs but is absolutely required for meiosis when DSBs are covalently bound by the Spo11 protein, as well as for the processing of cruciform structures in vegetatively growing cells (Lobachev et al., 2002; Moreau et al., 1999; Rattray et al., 2001). Mre11 nuclease activity also contributes to (but is not essential for) the survival of radiation damage and topoisomerase conjugates in budding yeast (Moreau et al., 1999), although at least some of its activities are redundant with Dna2 (Budd and Campbell, 2009). In vitro experiments show that a nuclease-deficient MRN complex can promote Exo1-mediated resection in the presence of Ku and DNA-PKcs equivalently to that of a wild-type complex (Yang et al., 2013; Zhou and Paull, 2013), further suggesting that Mre11 nuclease activity is not essential for resection of enzymatically generated DSBs. Similarly, the nuclease activity of MRN not necessary for compatible end ligation by DNA ligaseIIIα/Xrcc1, but is critical for the ligation of incompatible DNA ends that require processing (Della-Maria et al., 2011).
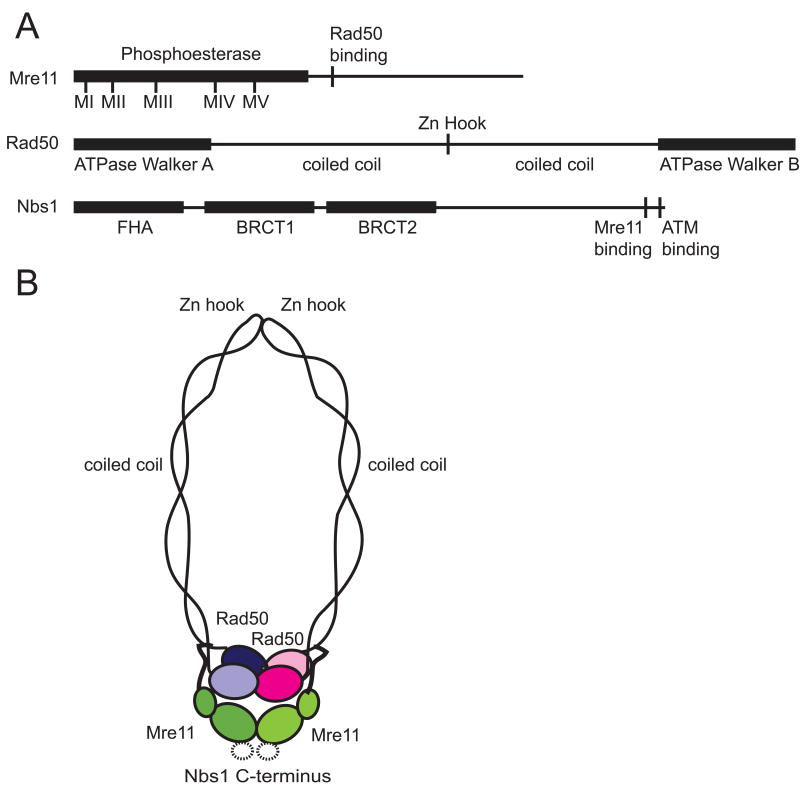
Figure 1.
Components of the MRN complex. (A) Linear maps of Mre11, Rad50 and Nbs1 showing functional domains, including the conserved phosphoesterase motifs in Mre11, Walker A and B ATP-binding motifs in Rad50 and the FHA and BRCT domains in Nbs1. (B) Interactions among the components of the MRN dimer. The globular domain is comprised of Mre11 phosphoesterase/nuclease domains, Rad50 ATPase domains, and Nbs1. The two Rad50 molecules also interact with a Zn hook at the tip of coiled coils. Binding site for Nbs1 is based on crystallographic data (Park et al., 2011; Schiller et al., 2012).
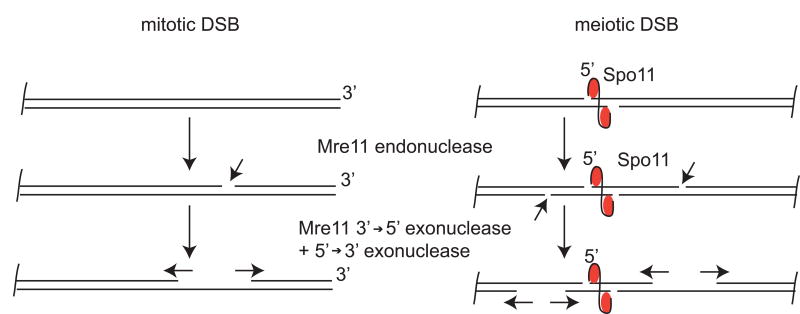
The role of Mre11 3′ to 5′ exonuclease activity has been unclear, in part because it requires millimolar levels of manganese in vitro (higher than would ever be encountered in vivo) and because the polarity of the exonuclease is opposite to the 5′ to 3′ resection required for creation of long 3′ ssDNA tails that are ultimately bound by Rad51. Recent support for a physiological role of the exonuclease activity came from a study of meiotic DSB repair in which breaks are created by and covalently linked to Spo11 on the 5′ strands (Garcia et al., 2011). Although Mre11 nuclease activity was known to be required for removal of Spo11, Neale and colleagues showed in this work that the 3′ to 5′ exonucleolytic activity of Mre11 is specifically required to resect the Spo11-linked strand after an endonucleolytic break is first made, approximately 300 nt from the Spo11 cut; 5′ to 3′ exonucleolytic degradation from the nick was found to be dependent on Exo1 (Fig. 2).
Figure 2.
Model for the role of Mre11 catalytic activities during repair of DSBs in mitotic homology directed repair (left) and meiotic recombination (right) based on studies in budding yeast and mammalian cells (Garcia et al., 2011; Shibata et al., 2014). An Mre11-dependent endonucleolytic cut is made at a distance from DSB. Mre11 exonuclease activity is proposed to process the single-strand break in the 3′ to 5′ direction towards the DSB and other nucleases (for instance Exo1) would continue resection in the 5′-3′ direction away from the DSB.
A similar model was proposed for HR in mammalian cells, where the roles of Mre11 exo-and endonuclease activites were assessed through the use of small molecule inhibitors that specifically affect each of these activities (Shibata et al., 2014). It was suggested that Mre11 endonucleolytic activity initiates resection, followed by Mre11-dependent 3′ to 5′ exonuclease and Exo1/BLM-dependent bidirectional resection from the site of the nick. The catalytic functions of Mre11 appear to be much more important in mammals than they are in yeast, as a transgenic mouse model expressing nuclease-deficient Mre11 exhibited early embryonic lethality (Buis et al., 2008). In further support of a physiological role of Mre11 exonucleolytic activity in mammalian cells, Mre11 was found to be responsible for the degradation of nascent strands at stalled replication forks in the absence of BRCA2 (Schlacher et al., 2011). Mre11 endonucleolytic activity in manganese has been observed in vitro with all Mre11 orthologs studied (Connelly et al., 1998; Herdendorf et al., 2011; Paull and Gellert, 1999; Trujillo and Sung, 2001). A weak but detectable endonuclease activity was also observed on 5′ strands of DSBs in the presence of magnesium using P. furiosus MR (PfMR)(Hopkins and Paull, 2008). The oligonucleotide products of this activity were found to be 10 to 50 nt long and were eliminated by mutations that inactivate the exonuclease activity, confirming that the activity resides in the same active site. pfMR was shown in this work to process DSB ends cooperatively with the helicase nuclease complex HerA/NurA that is expressed from the same operon in P. furiosus and other archaea species (Constantinesco et al., 2004). Magnesium-dependent nuclease activity has also been observed with gp46/47 (phage T4 Mre11/Rad50) in vitro (Herdendorf et al., 2011). Interestingly, with T4 MR, other proteins were found to promote the endonucleolytic activity of Mre11 in magnesium, including the recombination mediator protein UvsY and the ssDNA binding protein gp32 (Herdendorf et al., 2011).
It is still an open question what metal ions exist in Mre11 proteins in vivo, and thus what nuclease activities the proteins possess under physiological conditions. We recently performed partial proteolysis experiments with human Mre11 in vitro and found that oxidative cleavage of the protein occurred in the presence of ascorbic acid and hydrogen peroxide but without any added metal (Makharashvili et al., 2014). We furthermore mapped the sites of cleavage within Mre11 to amino acids 131 and 217, both residues directly contacting metal ion 2 in the structure of human Mre11 (Park et al., 2011). This evidence strongly suggests that there is a stably-bound transition metal ion bound to Mre11 in site 2, and is similar to previous experiments showing a high affinity manganese binding site in lambda phosphatase at metal position 2 (White et al., 2001). It will be informative to identify what metals are bound to sites 1 and 2 and to determine the functional consequences of these different states.
Rad50 ATP binding
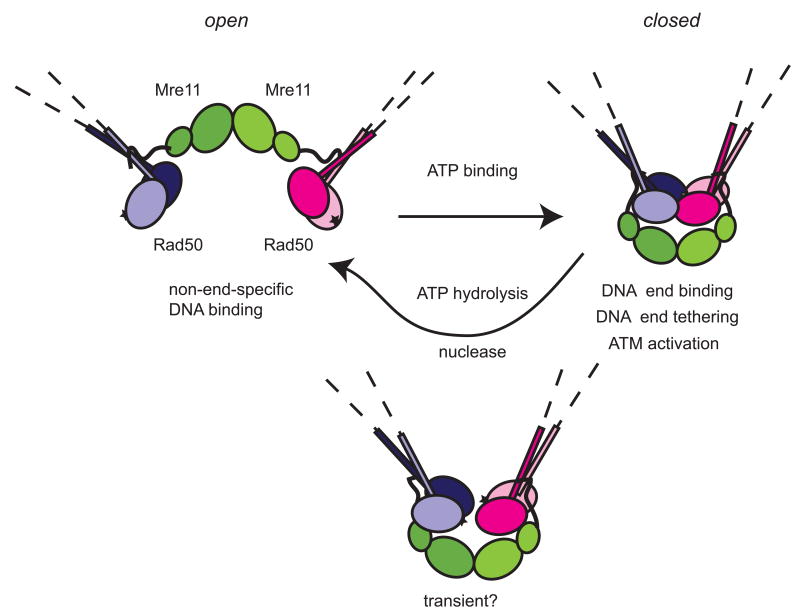
Rad50 contains Walker A and Walker B ATP-binding motifs and is similar in domain configuration to the Structural Maintenance of Chromosomes family of ATPases that regulate the activities and topology of chromosomal DNA (Williams et al., 2007)(Fig. 1). Two Rad50 catalytic domains come together to form the ATP-bound structure, with the coiled-coils of each protein extending away from the globular domains (Fig. 3). The ATP-related activities of the Rad50 protein are essential for all of its roles in DNA repair and signaling, as mutations in the Walker A or signature motifs exhibit phenotypes equivalent to a rad50 deletion in vivo (Alani et al., 1990; Bhaskara et al., 2007; Chen et al., 2005a; Moncalian et al., 2004). Even though Rad50 ATPase activity has long been known to be essential for MRN function, the exact role of this activity has not been very clear. In recent years, significant advances were made in understanding the structure of the Rad50 catalytic domain in the absence and presence of ATP, which showed that the protein undergoes a dramatic conformational change between these states (Lammens et al., 2011; Lim et al., 2011; Mockel et al., 2011; Williams et al., 2011). The catalytic head of Mre11-Rad50 complexes is composed of Mre11 nuclease domains and Rad50 ATPase domains. A crystal structure of the catalytic domains of bacterial MR in the absence of ATP shows that Mre11 holds the Rad50 ATPase domains near the base of the coiled coils (Lammens et al., 2011)(Fig. 3). In this configuration, the ATPase domains are separated and facing away from each other, forming an “open” structure. In contrast, the ATP-bound form of MR shows the ATPase domains together, where two ATP molecules are bound within the Rad50 dimer, occluding the Mre11 nuclease active sites and forming a “closed” conformation (Lim et al., 2011; Mockel et al., 2011). A large cavity within the Rad50 domain in the unbound, “open” configuration is important for the large conformational change to the “closed” state and mutations made in this cavity have effects on the relative stability of each state (Deshpande et al., 2014).
Figure 3.
ATP induced conformational changes in the catalytic head of MRN complex. The globular domain comprised of Rad50 ATPase and Mre11 nuclease domains is depicted as revealed from crystal structures (Lammens et al., 2011; Lim et al., 2011; Mockel et al., 2011). Mre11 binds Rad50 at the base of coiled coils. In the ATP unbound form, the structure is “open”, with Mre11 nuclease active sites accessible. ATP binding sites are shown as stars. In this state, the complex can engage DNA in a non-end specific manner (Deshpande et al., 2014). Binding of ATP brings the ATPase domains together forming a “closed” state. This form promotes end specific DNA binding and DNA tethering by MR/MRN complex and ATM checkpoint activation (Lee et al., 2013). Although this form blocks the nuclease site, ATP hydrolysis followed by separation of the ATPase domains is required for nuclease activity of Mre11, likely through a transient intermediate, although the structure of this theoretical conformation is unknown.
The importance of the ATP-bound “closed state”
We have known for many years that the ATPase activity of Rad50 is quite slow. Estimates of the ATPase activity of Rad50 from bacteria, yeast, and humans have yielded rates of 0.03 to 14 moles ATP hydrolyzed per mole of protein per minute, with the eukaryotic complexes generally showing much slower rates of hydrolysis (0.03 to 0.1) (Bhaskara et al., 2007; de Jager et al., 2002) compared to the phage, bacterial, and archaea complexes for which most measurements yield estimates of 1 to 4 moles per minute (Deshpande et al., 2014; Herdendorf et al., 2011; Lammens et al., 2011; Majka et al., 2012). These observations suggest that the complex may occupy the closed state for relatively long periods of time in vivo. Recent evidence shows that many important functions of Rad50 take place with the complex in the ATP-bound but unhydrolyzed state, however, perhaps explaining this long occupancy. We showed several years ago that stable binding of DNA fragments by human MRN is supported by non-hydrolyzable analogs of ATP (Lee et al 2003), and recent evidence suggests that ATP binding but not hydrolysis also promotes DNA end tethering by the pfMR complex (Deshpande et al, 2014). Using single-molecule FRET, ATP-bound human MRN was shown to bind to DSBs in a manner that stably unwinds 15-20 base pairs at the end of duplex, holding the branched structure open for minutes at a time at room temperature (Cannon et al., 2013). This unwound form of DNA present in the closed conformation of Rad50 was predicted from earlier ensemble experiments (Lee and Paull, 2005; Paull and Gellert, 1999) and is required for ATM activation as well as DSB resection in human cells (Cannon et al., 2013; Lee et al., 2013). It is still unclear what stimulates ATP hydrolysis by Rad50, although DNA as well as other DNA-binding proteins were reported to have a marked effect on the ATPase activity of T4 Rad50 (Herdendorf et al., 2011).
Release from the closed state with ATP hydrolysis
An important aspect of the Rad50 closed state is the fact that the Mre11 nuclease active sites are completely occluded by the Rad50 catalytic domains in this conformation (Lim et al., 2011; Mockel et al., 2011). Consistent with the structural predictions, we have found that stabilization of the closed state (either by mutation or by crosslinking) results in loss of Mre11 nuclease activity (Deshpande et al., 2014). These observations are also consistent with early data from analysis of E. coli SbcCD (E. coli MR) and pfMR showing that ATP hydrolysis is essential for the exonuclease activity of Mre11 when bound to Rad50 (Connelly et al., 1997; Hopfner et al., 2000). More recent studies with T4 MR have provided further detail for this model, showing that ATP hydrolysis is required for repetitive nucleotide removal but not required for removal of first nucleotide by nuclease activity, suggesting that ATP hydrolysis is likely involved in translocation of the complex (Herdendorf et al., 2011). Human Mre11 can act independently from Rad50 as a 3′ to 5′ exonuclease (Paull and Gellert, 1998), unlike the prokaryotic enzymes, again confirming that Rad50 ATP hydrolysis is not essential for Mre11 nuclease activity per se but serves to restrain and regulate the nuclease functions of Mre11.
Although a structure of PfMre11 bound to two DNA molecules has been published in which the Mre11 dimer interface adopts at least two different states during the exonuclease reaction (Williams et al., 2008), it is not yet clear how the closed state of MR transitions to a conformation in which the ends of DNA are accessible to the Mre11 active site. We envision that an intermediate state occurs after ATP hydrolysis that positions the Mre11 active sites such that they form productive complexes with DNA (Fig. 3). Understanding these details will likely require structural analysis of MR intermediates with DNA ends.
The coiled-coil domains of the Rad50 protein
Rad50 has structural features similar to SMC proteins and harbors a long coiled-coil that coordinates a Zn atom with its tip (Fig. 1). There is some variation in the length of the coiled-coil region across complexes from different species (de Jager et al., 2004). As observed with AFM, the average contour length of the complex varies from 39 nm observed with the E. coli SbcC/D MR complex (660 amino acids) to 47 nm (960 amino acids) in human MR. By primary sequence comparison the coiled-coils of T4 Rad50 are among the shortest, consisting of only 330 amino acids. AFM and EM imaging of the complexes shows that the arms appear in various conformations from completely splayed to closed and the coils can be either straight, bent, or kinked (de Jager et al., 2004; Hopfner et al., 2002). Time resolved analysis of single molecules of MR showed joining and separation of the arms at the tip, as well as changes in curvature indicating these arms to be highly flexible (Moreno-Herrero et al., 2005). The coiled-coil domains clearly respond to ATP binding and hydrolysis through large changes in their conformation, as visualized by crystal structures and AFM. A large swivel of Rad50 coiled coil domains accompanies ATP binding, changing the angle between the coiled-coils domains from 120 to 90 degrees (Figure 3)(Deshpande et al., 2014; Lammens et al., 2011; Lim et al., 2011; Williams et al., 2011).
The mechanistic function of the coils has been elusive, although it is clear that they are essential for Rad50 function in vivo. Ablation of Zn chelation in yeast Rad50 by mutation of the Cys residues in the Zn hook region leads to an increase in IR sensitivity similar to that of a null strain (Hopfner et al., 2002). Complete deletion of the Zn hook abolishes telomere maintenance and meiotic DSB formation, and severely impairs HR as well as NHEJ (Hohl et al., 2011). Similar results were seen for coiled coil truncation mutants, stressing the importance of the length of Rad50 coiled coils, although shortening the coiled coil region by 243 amino acids (retaining the zinc hook) was substantially tolerated for homologous recombination. Using live cell imaging, the Rad50 zinc hook was shown to be important for human MRN localization to the sites of DSBs and for the DNA damage response including homologous recombination and both ATM and ATR activation pathways (He et al., 2012). In addition, cohesin enrichment at replication forks depends on Rad50, and the zinc hook and coiled coils are important for this loading (Tittel-Elmer et al., 2009).
Experiments in vitro have also established the critical role of the coiled-coil domains. Removal of the coils impairs the DNA binding ability of MRN as well as DNA end tethering and ATPase activity (Deshpande et al., 2014; Lee et al., 2013). In addition, loss of the Zn hook-mediated connection between Rad50 molecules results in loss of Nbs1 binding and ATM activation (Lee et al., 2013). Why is the coiled-coil region so important for activities that take place in the globular domains of Rad50 and Mre11? From our in vitro experiments it has become clear that one of the essential roles of the coils and hook structure is to physically link the Rad50 catalytic heads together. In the absence of the coils/hook, Rad50 catalytic heads do not stably form a complex together with ATP (Deshpande et al., 2014). This makes intuitive sense because the Rad50 catalytic heads need to have a low affinity for each other in order to release from the closed state with ATP hydrolysis. The presence of the zinc hook connection increases the local concentration of the Rad50 catalytic heads such that the association between the heads is strongly favored and can even occur in the absence of ATP (Deshpande et al., 2014).
More difficult to answer is the question: why are the Rad50 coiled-coils so long? Results from coil deletion experiments in which shortened coils have been connected together either by the zinc hook itself or a heterologous linkage show that the proper length of the coils is essential for normal function (Hohl et al., 2011). One possibility that is often depicted in diagrams of MRN is that the coils physically connect two Rad50 molecules that are each bound to different DNA ends; however, it is difficult to determine this from the resolution of AFM images. MR proteins have been visualized tethering multiple linear DNA molecules into large, protein-bound complex, perhaps with the coiled-coil domains mediating these interactions (de Jager et al., 2001). These observations are consistent with the known DNA tethering activity of the complex and would allow for flexible, multipartite interactions between DNA ends. Another theoretical possibility is that the Rad50 coils actually encircle multiple DNA molecules, similar to cohesins, although there is currently no data supporting such a role.
Roles of Nbs1 in Regulation of MR
The Nbs1/Xrs2 component of MRN in eukaryotic cells does not possess enzymatic activity, but regulates the activities of Mre11 and Rad50 (Paull and Gellert, 1999) and is responsible for localizing MR to the nucleus in mammalian cells (Desai-Mehta et al., 2001). Recent structures of Mre11 from humans and fission yeast revealed parts of the Nbs1 interaction region on Mre11 (Park et al., 2011; Schiller et al., 2012), showing that two Nbs1 subunits stretch around the outside of nuclease domains of Mre11. We do not have structural information about the entire MRN catalytic head domain complex, but it is clear from in vitro biochemistry that Nbs1 stabilizes the ATP-bound form of MR and appears to be required for the ATP-dependent functions of the complex (Lee et al., 2003; Lee et al., 2013; Paull and Gellert, 1999). Nbs1 also contains an ATM-binding region at its C-terminus that is critical for ATM activation via MRN and DNA DSBs (Falck et al., 2005). It is not clear what the mechanistic role of this peptide is in activating ATM but in vitro studies with recombinant ATM also show this requirement for the Nbs1 C-terminus (Lee et al., 2010). Interestingly, we also found in this study that the mediator protein 53BP1 can compensate for the loss of the Nbs1 C-terminus in vitro in promoting ATM activity, most likely due to its ability to bind to both ATM and MRN. MR can also bind to ATM independently of Nbs1 (Lee and Paull, 2004) through a binding site in the Rad50 protein (JH Lee and T.Paull, unpublished observations).
The MRN complex associates with DSB sites through at least two different mechanisms. Binding to the chromatin domain containing the break requires phosphorylation of the histone variant H2AX by ATM and DNA-PKcs, which is subsequently bound by the mediator protein Mdc1 (Stucki et al., 2005). Nbs1 contains N-terminal FHA and BRCT domains that bind to Mdc1 at constitutively phosphorylated CK2 sites, localizing MRN to sites of DNA damage (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008). MRN also is thought to localize directly to break sites in a much smaller region of chromatin visible by immunofluorescence, independent of Mdc1 (Lukas et al., 2004); this localization may be dependent at least in part on the replication checkpoint protein Rad17 (Wang et al., 2014). The N-terminal FHA domain of Nbs1 in fission yeast also associates with the DSB repair factor Ctp1, the ortholog of CtIP, which is required for the efficient repair of breaks in S. pombe (Dodson et al., 2010; Williams et al., 2009). Direct binding between human MRN and CtIP has been well-documented (Chen et al., 2008; Sartori et al., 2007; Yuan and Chen, 2009).
Overall, the MRN complex is a multifunctional enzyme assembly with several distinct activities in the DNA damage response. Mre11 nuclease activity is tightly regulated by the binding and ATP hydrolysis of the Rad50 protein, and these two enzymatic components are regulated in turn by Nbs1. The association of MR and Nbs1 with ATM and the regulation of ATM by MRN serve to link this central DNA end recognition complex with the primary signaling kinase in eukaryotic cells to coordinate DSB repair with checkpoint activation. There are many aspects of this relationship that we still do not understand but the recent and ongoing structural and biochemical insights into the complex bode well for this goal being achieved.
Acknowledgments
We are grateful for helpful comments from the Paull laboratory and support from NIH R01 CA094008 and P01 CA092584.
References
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Bhaskara V, Dupre A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian VA, Paull TT. Rad50 Adenylate Kinase Activity Regulates DNA Tethering by Mre11/Rad50 complexes. Molecular Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PloS one. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Kuhnlein J, Yang SH, Cheng A, Schindler D, Stark JM, Russell R, Paull TT. Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18868–18873. doi: 10.1073/pnas.1309816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO reports. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem. 2005a;280:2620–2627. doi: 10.1074/jbc.M410192200. [DOI] [PubMed] [Google Scholar]
- Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EY, Lee WH. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005b;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DRF. DNA cleavage and degradation by the SbcCD protein complex from escherichia coli. Nucleic acids research. 1999;27:1039–1046. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Okely EA, Leach DR. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinesco F, Forterre P, Koonin EV, Aravind L, Elie C. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic acids research. 2004;32:1439–1447. doi: 10.1093/nar/gkh283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, Trujillo KM, Sung P, Hopfner KP, Carney JP, Tainer JA, Connelly JC, Leach DR, Kanaar R, Wyman C. Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. J Mol Biol. 2004;339:937–949. doi: 10.1016/j.jmb.2004.04.014. [DOI] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- de Jager M, Wyman C, van Gent DC, Kanaar R. DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Res. 2002;30:4425–4431. doi: 10.1093/nar/gkf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Maria J, Zhou Y, Tsai MS, Kuhnlein J, Carney J, Paull T, Tomkinson A. hMre11/hRad50/Nbs1 and DNA ligase III{alpha}/XRCC1 act together in an alternative non-homologous end joining pathway. J Biol Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annual review of genetics. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol. 2001;21:2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee JH, Classen S, Guenther G, Russell P, Tainer JA, et al. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson GE, Limbo O, Nieto D, Russell P. Phosphorylation-regulated binding of Ctp1 to Nbs1 is critical for repair of DNA double-strand breaks. Cell cycle. 2010;9:1516–1522. doi: 10.4161/cc.9.8.11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- He J, Shi LZ, Truong LN, Lu CS, Razavian N, Li Y, Negrete A, Shiloach J, Berns MW, Wu X. Rad50 zinc hook is important for the Mre11 complex to bind chromosomal DNA double-stranded breaks and initiate various DNA damage responses. J Biol Chem. 2012;287:31747–31756. doi: 10.1074/jbc.M112.384750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdendorf TJ, Albrecht DW, Benkovic SJ, Nelson SW. Biochemical characterization of bacteriophage T4 Mre11-Rad50 complex. J Biol Chem. 2011;286:2382–2392. doi: 10.1074/jbc.M110.178871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Kwon Y, Galvan SM, Xue X, Tous C, Aguilera A, Sung P, Petrini JH. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat Struct Mol Biol. 2011;18:1124–1131. doi: 10.1038/nsmb.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP. Mre11 and rad50 from pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionary conserved multiprotein machine. J Bacteriol. 2000;182:6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins B, Paull TT. The P. furiosus Mre11/Rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Conserved sequence pattern in a wide variety of phosphoesterases. Protein science: a publication of the Protein Society. 1994;3:356–358. doi: 10.1002/pro.5560030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annual review of genetics. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS letters. 2010;584:3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ghirlando R, Bhaskara V, Hoffmeyer MR, Gu J, Paull TT. Regulation of Mre11/Rad50 by Nbs1: effects on nucleotide-dependent DNA binding and association with ATLD mutant complexes. J Biol Chem. 2003;278:45171–45181. doi: 10.1074/jbc.M308705200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Goodarzi AA, Jeggo PA, Paull TT. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J. 2010;29:574–585. doi: 10.1038/emboj.2009.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Mand MR, Deshpande RA, Kinoshita E, Yang SH, Wyman C, Paull TT. Ataxia Telangiectasia-Mutated (ATM) Kinase Activity Is Regulated by ATP-driven Conformational Changes in the Mre11/Rad50/Nbs1 (MRN) Complex. J Biol Chem. 2013;288:12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Resnick MA. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutation research. 2000;451:71–89. doi: 10.1016/s0027-5107(00)00041-5. [DOI] [PubMed] [Google Scholar]
- Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. Crystal structure of the Mre11-Rad50-ATPgammaS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Alford B, Ausio J, Finn RM, McMurray CT. ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. J Biol Chem. 2012;287:2328–2341. doi: 10.1074/jbc.M111.307041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, et al. Catalytic and Noncatalytic Roles of the CtIP Endonuclease in Double-Strand Break End Resection. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Miyamoto T, Sakamoto H, Izumi H, Nakazawa Y, Ogi T, Tahara H, Oku S, Hiramoto A, Shiiki T, et al. Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair. 2011;10:314–321. doi: 10.1016/j.dnarep.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. The Journal of cell biology. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockel C, Lammens K, Schele A, Hopfner KP. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic acids research. 2011;40:914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT. The rad50 signature motif: essential to ATP binding and biological function. J Mol Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Chae J, Kim YC, Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes & Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray AJ, McGill CB, Shafer BK, Strathern JN. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics. 2001;158:109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipler A, Iliakis G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic acids research. 2013;41:7589–7605. doi: 10.1093/nar/gkt556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nature reviews. Molecular cell biology. 2013;14:197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. The Journal of cell biology. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMre11 is mutated in individuals with an Ataxia-Telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews. Molecular cell biology. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annual review of genetics. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M, Alabert C, Pasero P, Cobb JA. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J. 2009;28:1142–1156. doi: 10.1038/emboj.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50/Mre11 complex. J Biol Chem. 2001;13:13. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. American journal of human genetics. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Alexander P, Goldstein M, Wakeman TP, Sun T, Feng J, Lou Z, Kastan MB, Wang XF. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 2014 doi: 10.1002/embj.201386064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DJ, Reiter NJ, Sikkink RA, Yu L, Rusnak F. Identification of the high affinity Mn2+ binding site of bacteriophage lambda phosphoprotein phosphatase: effects of metal ligand mutations on electron paramagnetic resonance spectra and phosphatase activities. Biochemistry. 2001;40:8918–8929. doi: 10.1021/bi010637a. [DOI] [PubMed] [Google Scholar]
- Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, SilDas S, Hammel M, Russell P, et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat Struct Mol Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Zhou R, Campbell J, Chen J, Ha T, Paull TT. The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 2013;32:126–139. doi: 10.1038/emboj.2012.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends in cell biology. 2010;20:402–409. doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen J. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J Biol Chem. 2009;284:31746–31752. doi: 10.1074/jbc.M109.023424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic acids research. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Paull TT. DNA-dependent Protein Kinase Regulates DNA End Resection in Concert with Mre11-Rad50-Nbs1 (MRN) and Ataxia Telangiectasia-mutated (ATM) J Biol Chem. 2013;288:37112–37125. doi: 10.1074/jbc.M113.514398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]