Abstract
Feline immunodeficiency virus (FIV) is a retrovirus that infects domestic cats, and is an excellent animal model for human immunodeficiency virus type 1 (HIV-1) pathogenesis. The nucleocapsid (NC) protein is critical for replication in both retroviruses. FIV NC has several structural features that differ from HIV-1 NC. While both NC proteins have a single conserved aromatic residue in each of the two zinc fingers, the aromatic residue on the second finger of FIV NC is located on the opposite C-terminal side relative to its location in HIV-1 NC. In addition, whereas HIV-1 NC has a highly charged cationic N-terminal tail and a relatively short C-terminal extension, the opposite is true for FIV NC. To probe the impact of these differences on the nucleic acid (NA) binding and chaperone properties of FIV NC, we carried out ensemble and single-molecule assays with wild-type (WT) and mutant proteins. The ensemble studies show that FIV NC binding to DNA is strongly electrostatic, with a higher effective charge than that observed for HIV-1 NC. The C-terminal basic domain contributes significantly to the NA binding capability of FIV NC. In addition, the non-electrostatic component of DNA binding is much weaker for FIV NC than for HIV-1 NC. Mutation of both aromatic residues in the zinc fingers to Ala (F12A/W44A) further increases the effective charge of FIV NC and reduces its non-electrostatic binding affinity. Interestingly, switching the location of the C-terminal aromatic residue to mimic the HIV-1 NC sequence (N31W/W44A) reduces the effective charge of FIV NC and increases its non-electrostatic binding affinity to values similar to HIV-1 NC. Consistent with the results of these ensemble studies, single-molecule DNA stretching studies show that while WT FIV NC has reduced stacking capability relative to HIV-1 NC, the aromatic switch mutant recovers the ability to intercalate between the DNA bases. Our results demonstrate that altering the position of a single aromatic residue switches the binding mode of FIV NC from primarily electrostatic binding to more non-electrostatic binding, conferring upon it NA interaction properties comparable to that of HIV-1 NC.
1. Introduction
Feline immunodeficiency virus (FIV) is a retrovirus that infects domestic cats, and is the causative agent of an AIDS-like syndrome (Pedersen et al., 1987). Its primary modes of transmission are bloodborne through deep bite wounds and scratches. FIV can be transmitted among various feline species. However, there is no evidence of FIV transmission to human beings, despite efficient infection of CD4+ T-cells through interactions with CD134 and CXCR4 (Willett and Hosie, 2013); this is possibly due to poor recognition of the FIV promoter (5′-LTR) in human cells (Mustafa et al., 2005). As the only non-primate lentivirus to cause an AIDS- like syndrome, FIV is an excellent animal model for human immunodeficiency virus type 1 (HIV-1) pathogenesis (Luttge and Freed, 2010) and an attractive system to develop anti-retroviral vaccines, drugs and non-pathogenic gene therapy vectors. However, in comparison to HIV-1, little is known about the molecular determinants of FIV replication (Kemler et al., 2012; Moscardini et al., 2002).
The HIV-1 nucleocapsid protein is a nucleic acid (NA) chaperone that is essential for several stages of HIV-1 replication, such as genomic RNA (gRNA) dimerization (Darlix et al., 1990; Feng et al., 1996; Kafaie et al., 2008; Laughrea et al., 2001), reverse transcription (Levin et al., 2005; Moscardini et al., 2002) and recombination (Anderson et al., 1998; Bampi et al., 2004; Mark-Danieli et al., 2005; Negroni and Buc, 1999; Negroni and Buc, 2001). The three major steps of reverse transcription, including tRNA primer annealing, minus strand transfer, and plus strand transfer require significant rearrangement of NA secondary structure (Auxilien et al., 1999; Guo et al., 2000; Hargittai et al., 2004; Johnson et al., 2000; Peliska et al., 1994; Rodriguez-Rodriguez et al., 1995; Wu et al., 1999; You and McHenry, 1994). HIV-1 NC facilitates these processes mainly through its chaperone activity, which includes NA aggregation, NA destabilization and rapid protein-NA interaction kinetics (Cruceanu et al., 2006a; Cruceanu et al., 2006b; Levin et al., 2005; Vo et al., 2006; Williams et al., 2002; Williams and Rouzina, 2002; Williams et al., 2001). Mutations in HIV-1 NC that alter its NA chaperone activity correlate directly with in vivo HIV-1 replication (Wu et al., 2013; Wu et al., 2014). NC has been proposed to be a potential drug target for anti-HIV-1 therapy and several strategies have been developed (Darlix et al., 2000; de Rocquigny et al., 2008; Mori et al., 2011; Musah, 2004).
While the overall structure of FIV NC is similar to that of HIV-1 NC, there are several key differences. Whereas HIV-1 NC has a highly basic N-terminal tail and a relatively short C-terminal extension, the opposite is true in the case of FIV NC, which has a long C-terminal tail with a high positive charge density. Both proteins possess two zinc finger domains, each with a conserved aromatic residue. However, the aromatic residue in the C-terminal zinc finger of FIV NC is located on the opposite side of the finger compared to HIV-1 NC. Previous studies showed that FIV NC promotes dimerization of 5′-leader RNA, as well as tRNA annealing to the primer binding site (PBS) (Moscardini et al., 2002). It also facilitates initiation of minus-strand DNA synthesis by FIV reverse transcriptase (RT) and minus-strand transfer (Moscardini et al., 2002). Deletion of the two zinc fingers caused attenuation of FIV NC’s NA chaperone activity. In particular, the zinc fingers were found to be required to facilitate FIV RNA dimerization and inhibition of non-specific initiation (i.e. self-priming) of reverse transcription by FIV RT (Moscardini et al., 2002). In another study, it was reported that the first Cys residue in the N-terminal zinc finger (C11), as well as the basic residues in the linker region between the two zinc fingers (K29/R29) play an important role in NA binding and viral particle production (Affranchino and González, 2010; Manrique et al., 2004). In contrast, the first Cys residue in the C-terminal zinc finger appears to be less important (Affranchino and González, 2010; Manrique et al., 2004). However, a detailed mechanistic study of FIV NC chaperone activity has not been reported; in particular, the role of the basic C-terminal extension domain and the impact of the unique location of the C-terminal aromatic residue on chaperone function are unknown.
Here we use ensemble NA binding and annealing assays, as well as single-molecule force spectroscopy to probe the NA chaperone activity of FIV NC. We find that the basic C-terminal domain of FIV NC plays a similar role as the N-terminal extension of HIV-1 NC. In addition, the specific location of the C-terminal zinc finger aromatic residue of FIV NC results in altered NA interaction and chaperone properties relative to those of HIV-1 NC, particularly at low protein concentrations.
2. Materials and Methods
2.1 Preparation of proteins and NAs
Cloning, expression and purification of FIV NC
Briefly, proviral DNA was isolated by the lab of Dr. Lawrence Mathes (Ohio State University) from infected cat lymphocyte cultures, and PCR was used to amplify the Gag gene (accession number AF361320.1). The FIV NC region was then amplified using primers that have restriction sites for cloning into a pET32a expression vector. The protein was expressed with an N-terminal thioredoxin tag and a TEV cleavage site was inserted between the tag and NC. FIV mutants used in this study were generated using the QuikChange Site Directed Mutagenesis kit (Agilent, Santa Clara, CA). FIV NC purification was performed as described for HIV-1 NC (Stewart-Maynard et al., 2008; Wu et al., 2014).
Preparation of nucleic acids
A single-stranded (ss) DNA 20-mer (ssDNA20) was purchased from TriLink Biotechnologies (San Diego, CA) with 6-carboxyfluorescein (FAM) at the 5′ end: 5′-FAM-CTT CTT TGG GAG TGA ATT AG-3′. Two RNAs were designed based on predicted stem-loop elements present in the 5′ region of FIV gRNA (Kenyon et al., 2008; Rizvi et al., 2010) with the following sequences: micro R: 5′-UUU GAG GAC GUU UGC GUU CUC-3′; micro SL2: 5′-UUU GCU GAC CUA AAU AGG GAA GC-3′. The predicted secondary structures of these two RNAs are shown in Fig. 1D. These RNAs were purchased with 5′-FAM labels from Dharmacon (now Thermo Scientific, Waltham, MA). HIV-1 TAR RNA was prepared as described (Vo et al., 2009b) and TAR DNA was purchased from Integrated DNA Technologies (Coralville, IA). TAR RNA was 5′-radiolabeled using γ-[32P]-GTP (Perkin Elmer) and T4 polynucleotide kinase (New England Biolabs) using standard protocols, followed by purification by denaturing PAGE. Concentrations of all NAs were determined by measuring UV absorption at 260 nm and using the following extinction coefficients: ssDNA20: 192,900 M−1cm−1, 377,800 M−1cm−1, micro R: 222,400 M−1cm−1, micro SL2: 253,700 M−1cm−1, TAR RNA: 533,700 M−1cm−1, TAR DNA: 564,800 M−1cm−1.
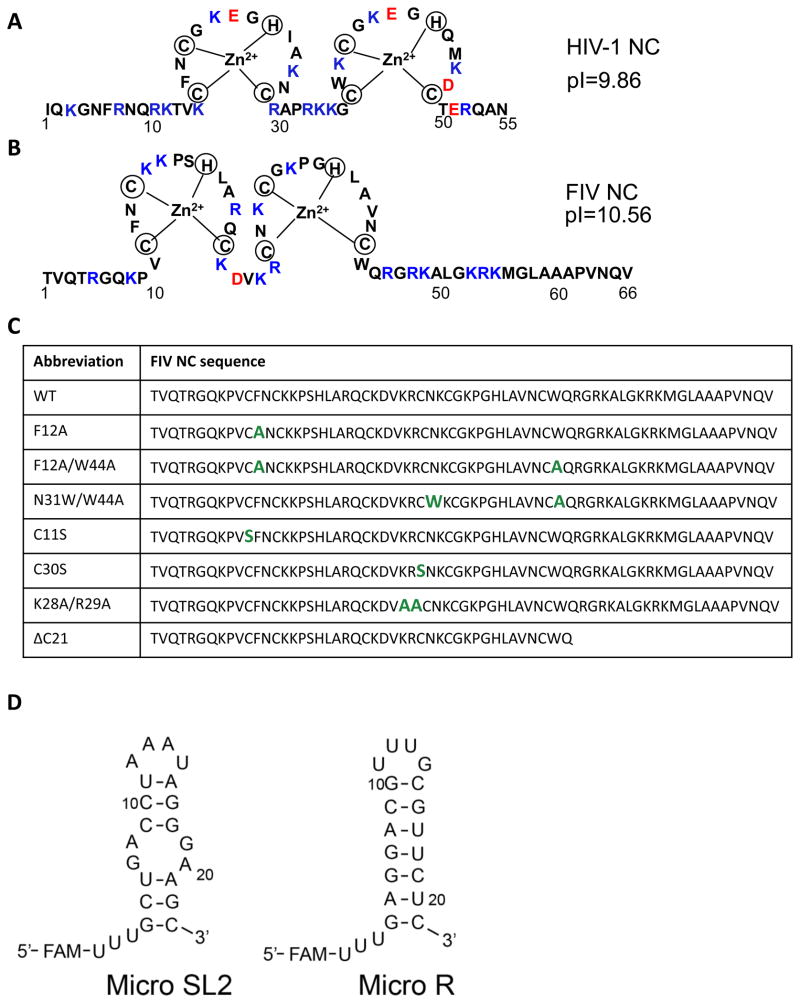
Fig. 1.
(A) Sequence of WT HIV-1 NC (NL4-3 isolate). (B) Sequence of WT FIV NC (MD isolate). Positively and negatively charged residues are highlighted in blue and red, respectively, in both (A) and (B). (C) Sequence of WT and mutant FIV NC, with altered residues highlighted in green. (D) Predicted secondary structures of micro SL2 and micro R RNAs derived from the FIV genome. Three U’s that are not encoded in the genome and a 5′-FAM label are also indicated.
2.2 Fluorescence Anisotropy (FA) binding experiments and salt titration assays
FA binding experiments were performed as described (Stewart-Maynard et al., 2008). RNAs (micro R and micro SL2) were first refolded in a buffer containing 25 mM HEPES, pH 7.5 and 100 mM NaCl by heating to 80 °C for 2 min, cooling to 60 °C for 2 min and adding MgCl2 to a final concentration of 10 mM before placement on ice. ssDNA20 was used without refolding. For binding measurements, fluorescently-labeled NAs (20 nM) were incubated with increasing concentrations of NC (0–2000 nM) in the presence of 20 mM HEPES, pH 7.5, 50 mM NaCl, 100 μM tris(2-carboxyethyl)phosphine (TCEP), 5 mM β-mercaptoethanol (BME) and 1 μM Zn(OAc)2 at room temperature for 30 min in the dark before FA measurements were recorded on a MD SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA).
Salt-titration binding assays were carried out as described (Webb et al., 2013). Briefly, 400 nM of FIV NC was incubated with 20 nM ssDNA20 in the presence of increasing NaCl concentrations (0 mM to 1 M) in 20 mM HEPES, pH 7.5, 100 μM TCEP, 5 mM BME, and 1 μM Zn(OAc)2. The reactions were incubated at room temperature in the dark for 30 min prior to FA measurements. The FA change at high salt concentration was corrected by subtracting the background (i.e., change in FA observed with increasing salt in the absence of NC). The data were analyzed as described (Webb et al., 2013) to obtain Zeff, the number of positive charges involved directly in the NC-NA interaction, and Kd,1M, which is the dissociation constant in the presence of 1 M NaCl. These parameters describe the non-specific (i.e. electrostatic) and specific (i.e. nonelectrostatic) binding component of protein-NA interaction, respectively, with
| (Eq. 1) |
2.3 Gel-shift annealing assays
Gel-shift annealing assays were performed as described (Vo et al., 2009b). Prior to each experiment, 32P-labeled TAR RNA and TAR DNA were first refolded as described above. For most assays, 32P-labeled TAR RNA (15 nM) and 45 nM TAR DNA were mixed with 0.88 μM FIV NC (corresponding to 4:1 nt: protein) in a buffer containing 50 mM HEPES, pH 7.5, 50 mM NaCl, 1 mM MgCl2 and 5 mM DTT on ice. At desired time points, aliquots (30 μl) were removed from the reaction and quenched with 1% SDS. Following two phenol-chloroform extraction steps, the reactions were mixed with dyes and run on a 12% denaturing gel. Quantification was performed on a Typhoon Trio phosphorimager (GE Healthcare, Pataskala, OH).
To determine the reaction rates for annealing, we use two-exponential fits to the measured data and analyze the resulting rates in terms of a two-step TAR RNA-TAR DNA reaction (Vo et al., 2006; Vo et al., 2009a; Vo et al., 2009b), as described by the following reaction scheme:
| (Eq. 2) |
Here the first step is a fast bi-molecular reaction of complex nucleation, which involves partial annealing of the TAR RNA and DNA via the hairpin loops or the stem ends. As a bi-molecular process, this step of annealing is greatly facilitated by NA aggregation activity of NC, which increases the local NA concentration. The second step involves a much slower strand exchange between the TAR RNA and DNA hairpin stems, and is mostly sensitive to the duplex-destabilizing activity of NC.
In our gel-shift annealing assays, the annealed complex band is a combination of both partially and completely annealed TAR RNA/DNA molecules, such that the kinetics of the appearance of the annealed fraction contains both fast and slow components, as described by two-exponential kinetics:
| (Eq. 3) |
The fitted parameters of the annealing kinetics, kf, ks and f are the fast and the slow rates, and the fraction of the fast component, respectively. These fitted parameters are related to the basic reaction rates introduced in Eq. (3) in the following way:
| (Eq. 4) |
| (Eq. 5) |
| (Eq. 6) |
These basic reaction rates can be found from the values of the fitted parameters as follows:
| (Eq. 7) |
| (Eq. 8) |
| (Eq. 9) |
2.4 Single-molecule DNA stretching experiments
Two laser beams are overlapped to form an optical trap, trapping one bead while another bead is attached to a glass micropipette. Bacteriophage λ DNA, labeled with biotin on both 5′ ends, is caught between the two beads due to strong non-covalent bonds between biotin and streptavidin (Cruceanu et al., 2006a; Morellet et al., 1998), as previously described (McCauley and Williams, 2007). By manipulating streptavidin-coated polystyrene beads (Bangs Laboratories, Inc., Fishers, IN) with optical tweezers and the micropipette, we can stretch λ DNA up to 300 pN. During the experiment, we capture a single DNA molecule in standard buffer without protein. We then rinse out the DNA solution with clean buffer, leaving a single DNA molecule captured between the beads. Next, a specific concentration of FIV NC is flowed into the experimental chamber to interact with the single DNA molecule. All stretching experiments were performed at a pulling rate of 100 nm/s in 10 mM HEPES, 50 mM Na+, pH 7.5 buffer.
3. Results
3.1 Design of FIV NC variants
Fig. 1A and B show the sequences of HIV-1 (NL4-3 isolate) and FIV NC (MD isolate), respectively. Several FIV NC mutants were designed to test the contribution of specific sequence elements to NA binding and chaperone function, and these are shown in a table in Fig. 1C. In the case of HIV-1 NC, it is well established that the highly conserved aromatic residues in each zinc finger form a hydrophobic platform that participates in stacking interactions with unpaired guanosines of ss NAs (Amarasinghe et al., 2000; Amarasinghe et al., 2001; De Guzman et al., 1998; Lee et al., 1998; Mely et al., 1993; Morellet et al., 1994; Morellet et al., 1998; Morellet et al., 1992; Summers et al., 1990; Tisne et al., 2001); removing one or both of the aromatic residues greatly weakens NC’s chaperone activity (Cruceanu et al., 2006a; Fisher et al., 1998; Paoletti et al., 2002; Vuilleumier et al., 1999; Wu et al., 2013). Moreover, these stacking interactions contribute to the ability of NC to recognize and package its cognate gRNA (Amarasinghe et al., 2000; De Guzman et al., 1998). To determine whether a similar role is played by FIV NC’s aromatic residues, we prepared an F12A mutant and a F12A/W44A double mutant. Whereas HIV-1 NC’s two aromatic residues are located between the first two Cys residues of each finger and FIV NC’s N-terminal aromatic residue, F12, has a position identical to that of HIV-1 NC, the C-terminal aromatic residue, W44, is located on the opposite side of the second zinc finger at the start of the long C-terminal tail (Fig. 1C). To investigate the extent to which the location of this aromatic residue affects NC’s chaperone activity, we constructed an FIV NC aromatic switch variant, N31W/W44A, which changes the location of the Trp residue to mimic the location found in HIV-1 NC (Fig. 1C).
Even subtle changes to the Cys and His residues that coordinate the zinc ions have been shown to affect HIV-1 NC’s chaperone function (Guo et al., 2000; Williams et al., 2002). Because Ser cannot form a bond with the zinc ion, replacing Cys with Ser is expected to alter the zinc finger structure and slightly influence the position of the critical aromatic residues. To establish if FIV NC is sensitive to these subtle changes, we constructed two Cys-to-Ser mutants, C11S and C30S. In the primary sequence, C11 is adjacent to aromatic residue F12, but C30 is far removed from both of the aromatics. Therefore, replacing C11 is expected to have a more pronounced effect on aromatic residue positioning.
There are 16 basic residues distributed throughout the FIV NC protein. These positively charged residues are expected to facilitate NA binding via electrostatic interactions. In HIV-1 NC it is known that the N-terminal cationic tail and the basic residues in the linker domain contribute significantly to binding (Heath et al., 2003; Narayanan et al., 2006; Vuilleumier et al., 1999; Wu et al., 2014). In order to determine if the linker residues and C-terminal tail of FIV NC play a similar role, we constructed two additional variants: K28A/R29A and ΔC21 (Fig. 1C).
3.2 Binding affinity of WT and mutant FIV NC
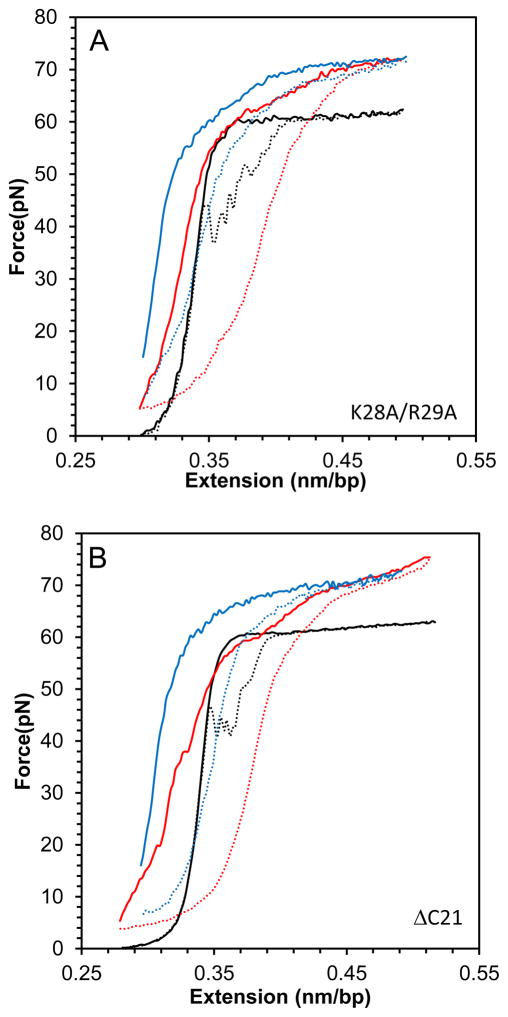
The binding affinities of FIV NC variants to ssDNA20, micro R and micro SL2 (see Fig. 1D), were measured using FA. At 50 mM NaCl, WT FIV NC binds all three oligonucleotides with dissociation constants (Kd) ≤ 30 nM (Table 1). This value is close to the lower limit that can be measured given the 20 nM NA concentration used in these studies. Most NC variants displayed similar high-affinity binding. Two FIV NC mutants, the double aromatic mutant F12A/W44A and the aromatic position switch mutant N31W/W44A, had dissociation constants in the range of 30–70 nM; ΔC21 FIV NC binds ssDNA20 with a Kd of ~545 nM, while the Kds measured for the two RNAs are even higher (~1 μM). Thus, the basic C-terminal domain is critical for high-affinity FIV NC-NA interaction, similar to the N-terminal domain of HIV-1 NC.
Table 1.
Apparent dissociation constants of WT and mutant FIV NC binding to DNA and RNA.
| FIV NC variant | ssDNA20 (nM)a | micro R (nM)a | micro SL2 (nM)a | ssDNA20 (nM)b |
|---|---|---|---|---|
| WT | 30 ± 2 | 23 ± 5 | 31 ±10 | 1095±137 |
| C11S | 8 ± 2 | 13 ± 1 | 21 ± 2 | 1076±159 |
| C30S | 14 ± 2 | 21 ± 1 | 13 ± 0.9 | 1119±142 |
| F12A | 2.2 ± 1 | ND | 19 ± 7 | ND |
| F12A/W44A | 13 ± 2 | 76 ± 9 | 57 ± 18 | >2000 |
| N31W/W44A | 72 ± 14 | 42 ± 14 | 50 ± 8 | 99±13 |
| K28A/R29A | 24 ± 3 | 31 ± 3 | 49 ± 4 | >2000 |
| ΔC21 | 545 ± 41 | 1395 ± 1110 | 938 ±488 | ND |
Kd values were determined by FA performed in the presence of 50 mM NaCl.
Kd values were determined by FA performed in the presence of 150 mM NaCl.
All values are averages of at least three experiments with standard deviations indicated. ND is ‘not determined’.
In an attempt to differentiate the relative binding affinities of the NC mutants that bound too strongly for us to measure accurately at low salt, binding of some variants to ssDNA20 was carried out in the presence of higher salt concentration (150 mM NaCl). WT, C11S, and C30S FIV NC bind ssDNA20 with a similar affinity under these conditions (Kd ~1 μM; Table 1). Strikingly, one FIV NC variant, N31W/W44A, binds ssDNA20 significantly tighter than WT FIV NC (Kd ~100 nM). At high NaCl concentration, F12A/W44A and K28A/R29A FIV NC bind at least 2-fold more weakly than the rest of FIV NC variants (Kd > 2 μM), suggesting a significant role for these residues in NA binding.
Under the two salt conditions tested in the direct binding assay, the affinity appeared to be either too strong or too weak to measure; thus, the complete salt dependence of ssDNA20 binding was determined for several of the most interesting FIV NC variants, namely WT, F12A/W44A and N31W/W44A. In these experiments, the NC proteins were first incubated with fluorescently-labeled ssDNA20 to form a complex. Various concentrations of NaCl were then added to compete with NA for NC binding and the data were analyzed as described in the Materials and Methods section to yield two parameters, Kd,1M and Zeff. Kd,1M is the Kd at 1 M NaCl, when all electrostatic interactions are screened by salt, and is a measure of salt-independent, specific binding interactions. Zeff is the effective positive charge of the protein, which reports on the number of direct protein-NA contacts established during binding that lead to the release of Na+ cations.
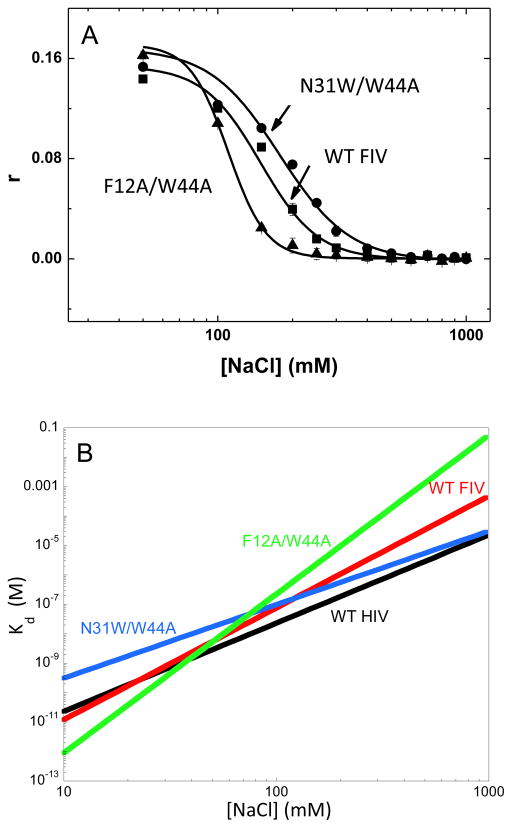
The salt titration results are presented in Fig. 2A and the fitted binding parameters are reported in Table 2. Fig. 2B shows the salt dependence of the Kd values calculated according to Eq. 1. These results suggest that WT FIV NC binds ssDNA20 in a more electrostatic manner than HIV-1 NC. Compared to HIV-1 NC (Zeff=3.0±0.3), FIV NC has slightly higher effective charge (Zeff=3.8±0.3) and an ~10-fold weaker specific binding component (Kd,1M =(4.8±2.6) X 10−4 for FIV NC vs. (2.3±0.7) X 10−5 for HIV-1 NC). Switching the location of the second aromatic residue in FIV NC (N31W/W44A) leads to an increase in the non-electrostatic component of binding (Kd,1M = (3.1±1.1) X 10−5) to a level very similar to that of HIV-1 NC. A concomitant decrease in the effective protein charge to Zeff=2.5±0.2 is also observed. These results support a key role for the specific aromatic residue position within the zinc finger structure in the mode of the NC-NA interaction. Consistent with this result is the observation that mutation of both aromatic residues to generate F12A/W44A FIV NC leads to a significant loss of specific NA interaction, reflected by an ~100-fold increase in Kd,1M and an increase in the protein’s effective charge to 5.4.
Fig. 2.
(A) FA as a function of salt concentration for WT, N31W/W44A, and F12A/W44A FIV NC binding to ssDNA20. (B) Salt dependence of the dissociation constant, Kd([Na]), of several NC variants from ssDNA20, for WT FIV NC (red line), N31W/W44A FIV NC (blue line), F12A/W44A FIV NC (green line), and HIV-1 NCp7 (black line, (Wang et al., 2014). The Kd([Na]) were calculated according to Eq. (1) using parameters Kd,(1M) and Zeff (Table 2) fit from the salt titration curves (data in panel (A)).
Table 2.
Binding parameters for WT and mutant FIV NC based on salt-titration assays with ssDNA20.
| FIV NC | Zeffa | Kd,1Mb |
|---|---|---|
| WT | 3.8±0.3 | (4.8±2.6)·10−4 |
| N31W/W44A | 2.5±0.2 | (3.1±1.1)·10−5 |
| F12A/W44A | 5.4±0.4 | (5.6±4.2)·10−2 |
| HIV-1 NCp7c | 3.0±0.3 | (2.3±0.7)·10−5 |
Zeff is the effective charge of the protein during its binding interaction with the DNA and reflects the number of Na+ ions that are displaced during binding.
Kd, 1M is the affinity of the specific, non-electrostatic component of binding.
Data from (Wang et al., 2014). For these assays a 40-nt unstructured ssDNA was used.
All values represent the average of three or more trials with the associated standard deviation.
3.3 Annealing activity of WT and mutant FIV NC
The NA annealing activity of these FIV NC variants was next compared using ensemble TAR RNA/DNA annealing assays (Vo et al., 2009b). At standard annealing reaction temperatures of 37 °C or 22 °C, the FIV NC-facilitated annealing reactions were too fast to be resolved with our gel-shift assay. This is in contrast to the analogous reaction facilitated by saturating concentrations of HIV-1 NC (Vo et al., 2009a) for which annealing rates of ~1 min−1 (37 °C) and ~0.2 min−1 (22°C) were determined. The efficient TAR RNA/DNA annealing activity of FIV NC may be attributed to its strong nucleic acid aggregating activity, consistent with the higher effective charge of this NC protein relative to HIV-1 NC (Table 2). In order to distinguish between the annealing activities of the WT and mutant FIV NC proteins we performed the TAR RNA/DNA annealing experiments at 0 °C.
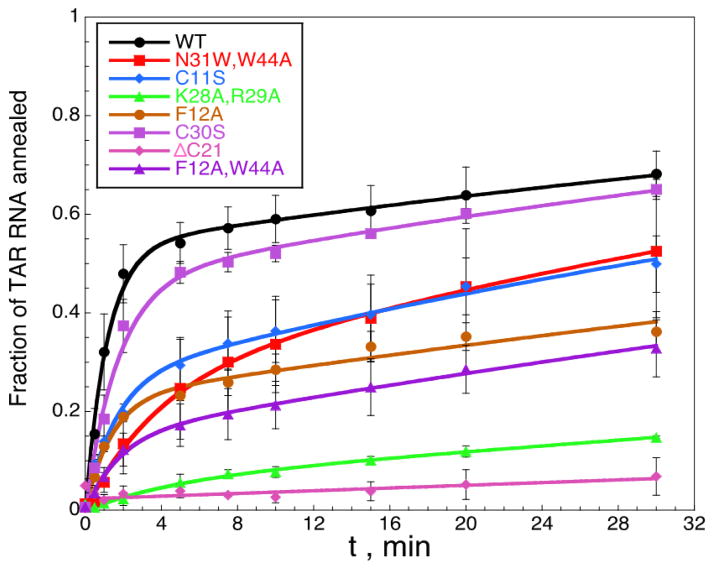
Presented in Fig. 3 are the time courses for the annealing of 15 nM of TAR RNA to 45 nM TAR DNA in the presence of 880 nM WT or mutant FIV NC in a buffer containing 50 mM HEPES, pH 7.5, 50 mM NaCl, 1 mM MgCl2 and 5 mM DTT on ice. We use two-exponential fits to describe the time dependence of the observed reaction, obtaining fast and slow rates kf and ks (Eq. 3), and these fits are shown in Fig. 3. These observed rates from these fits are then used to determine the fundamental reaction rates k1[DNA], k−1, and k2, as described in Materials and Methods. These parameters describe the bimolecular on, k1[DNA], and off, k−1, rates for initial complex nucleation and the slower rate of strand exchange, k2, between TAR RNA and TAR DNA hairpins leading to formation of the final annealed structure, which requires NA destabilization.
Fig. 3.
TAR RNA/DNA annealing in the presence of WT and mutant FIV NC. 32P-labeled TAR RNA (15 nM) and TAR DNA (45 nM) were annealed by 0.88 μM NC in a buffer containing 50 mM HEPES, pH 7.5, 50 mM NaCl, 1 mM MgCl2 and 5 mM DTT on ice. Lines correspond to fits to Eq. 3.
The results of the fits of the annealing data to the two-step reaction are presented in Table 3. The least defective FIV NC mutants are the C11S, C30S and N31W/W44A. All of these variants have an ~1.2 to 2-fold reduced fast fraction, f, suggesting slightly less efficient complex nucleation, and the strand exchange rate, k2, is about the same or a few-fold higher than for WT FIV NC. The highest value of k2 is observed in the presence of N31W/W44A (~3-fold greater than WT), consistent with its increased duplex destabilizing ability relative to WT FIV NC.
Table 3.
Parameters describing WT and mutant FIV NC annealing of TAR RNA/DNAa.
| FIV NC | f | kf | ks | k1·[DNA] | k−1 | k2 |
|---|---|---|---|---|---|---|
| WT | 0.53±0.05 | 0.87±0.3 | 0.013±0.003 | 0.46±0.2 | 0.40±0.1 | 0.023±0.01 |
| C11S | 0.27±0.03 | 0.63±0.2 | 0.013±0.003 | 0.17±0.1 | 0.46±0.1 | 0.050±0.02 |
| C30S | 0.46±0.05 | 0.58±0.2 | 0.014±0.003 | 0.27±0.1 | 0.31±0.1 | 0.030±0.02 |
| N31W,W44A | 0.25±0.02 | 0.27±0.1 | 0.015±0.003 | 0.067±0.1 | 0.20±0.1 | 0.060±0.01 |
| F12A | 0.23±0.02 | 0.73±0.3 | 0.007±0.004 | 0.17±0.1 | 0.56±0.1 | 0.033±0.02 |
| F12A,W44A | 0.15±0.01 | 0.56±0.2 | 0.008±0.004 | 0.084±0.1 | 0.48±0.1 | 0.053±0.02 |
| K28A,R29A | 0.06±0.03 | - | 0.003±0.004 | - | - | - |
| ΔC21 | 0.02±0.02 | - | 0.001±0.002 | - | - | - |
Values were obtained from a two-exponential fit of the annealing data as described in the Methods. Fundamental reaction rates are in units of min−1, and a dash indicates that values could not be obtained from fits due to low amplitudes. Uncertainties are determined from the fits to the data.
The two aromatic mutations, F12A and F12A/W44A, result in an even greater reduction in the fast fraction, while maintaining approximately the same or 2-fold greater strand-exchange rate as WT FIV NC. However, our ability to determine the strand exchange rate is more limited in this case, as the overall annealing is slower and we only measured the very beginning of the annealing process. Finally, the two cationic mutants, K28A/R29A and ΔC21, are so defective in annealing that they almost lack the fast annealing component, and only their slow rate can be characterized, making a determination of the basic reaction rates unreliable. The pronounced decrease in the fraction of the fast component relative to WT FIV NC reflects the low NA binding affinity of these proteins, leading, in turn, to their inability to aggregate NAs.
3.4 Single molecule DNA stretching in the presence of WT and mutant FIV NC
3.4.1 Effects of protein binding on DNA stretch and release curves
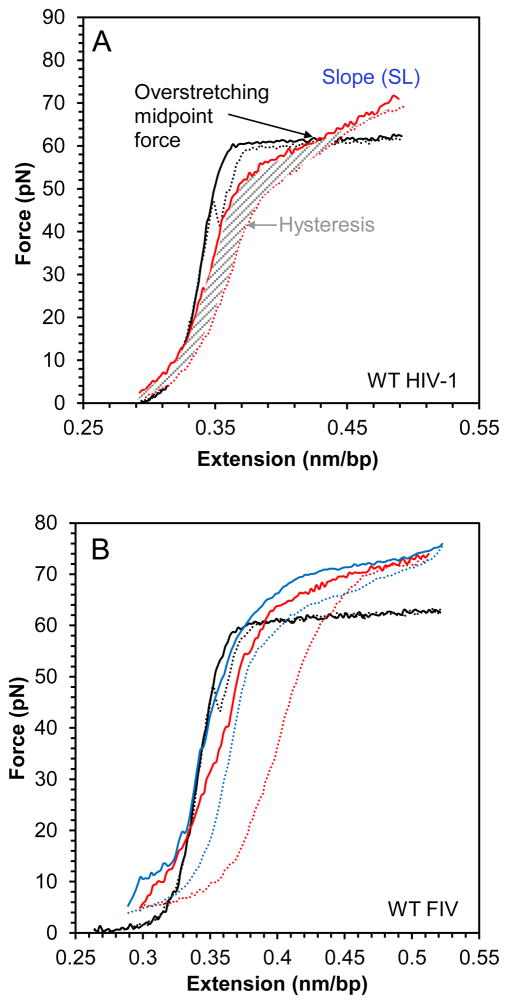
The NA interaction and chaperone properties of WT and mutant FIV NC were also compared using single-molecule DNA stretching by optical tweezers. This method is very sensitive to the mode of protein-DNA interaction. Fig. 4A shows stretching and release cycles for effectively random sequence polymeric DNA (from bacteriophage λ) in the absence of protein and in the presence of WT HIV-1 NC, for comparison with our results on FIV NC. In the absence of protein, a small force is applied to stretch dsDNA to its B-form contour length (0.34 nm/bp, presented as the total DNA end-to-end extension divided by the number of bp in the molecule). As the contour length is approached, the force increases dramatically, reflecting the elasticity of the double helix. The subsequent plateau that occurs at a constant force of about 62 pN, referred to as the DNA overstretching transition, represents a force-induced DNA strand separation transition from dsDNA to ssDNA under the solution conditions used here (Chaurasiya et al., 2010; King et al., 2013; Rouzina and Bloomfield, 2001; Shokri et al., 2008; van Mameren et al., 2009; Williams et al., 2001). This force plateau is very cooperative, as it represents a phase transition in DNA, with an almost doubling of the DNA length occurring over an interval of ~4 pN force increase, and is characterized by a small slope (SL) of the force-extension (F-x) curve of SL~20 pN/nm (Fig. 4A). Without any binding protein, as the DNA is released back to lower extensions at the moderate salt concentration used here (50 mM Na+), the ssDNA strands immediately re-anneal back to form dsDNA, as evidenced by the very small difference between the DNA stretch and release curves, or small hysteresis of the F-x cycle.
Fig. 4.
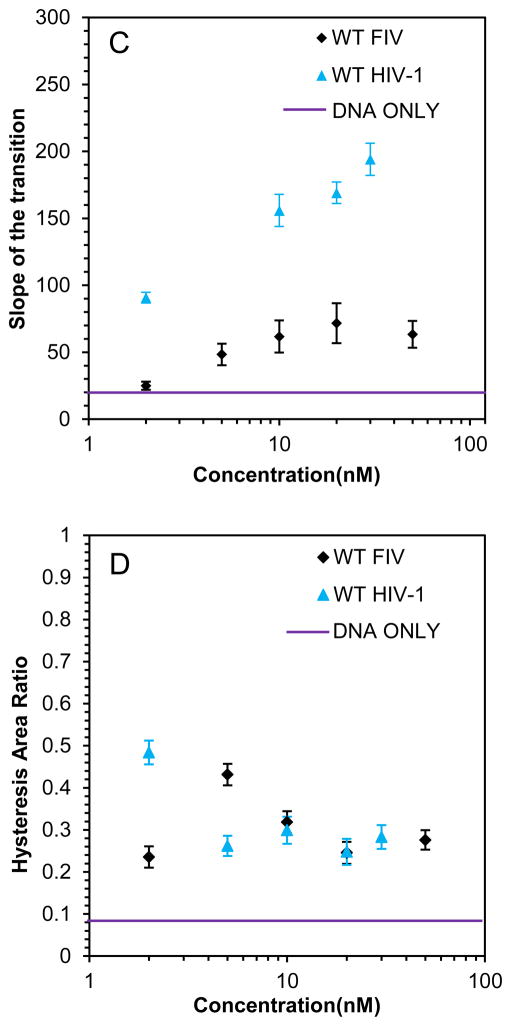
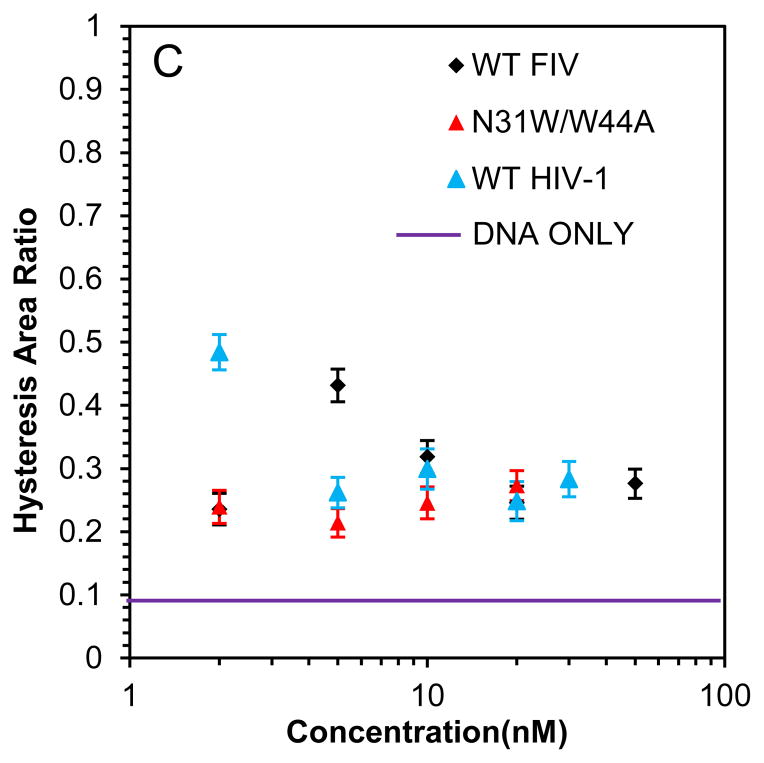
(A) Stretch (solid line) and release (dotted line) curves for dsDNA in the absence (black) and presence (red) of 10 nM WT HIV-1 NC. A blue dashed line illustrates how the slope (SL) is calculated form the stretching curve, while the area covered by thin gray lines is the hysteresis area. The overstretching midpoint force, which shows how NC alters DNA stability, is also shown. (B) DNA stretch (solid line) and release (dotted line) curves in the absence of protein (black) and in the presence of 5 nM (red) and 20 nM (blue) WT FIV NC. (C) Slope of the transition of WT FIV NC (black) and WT HIV-1 NC (blue). (D) Hysteresis versus concentration of WT FIV NC (black) and WT HIV-1 NC (blue).
We have previously stretched single DNA molecules in the presence of WT and mutant HIV-1 NC (Wu et al., 2013; Wu et al., 2014). These studies showed that WT HIV-1 NC interacts with dsDNA by intercalating in between base pair stacks, thereby elongating the DNA duplex far beyond its B-form contour length. Therefore, the effects due to HIV-1 NC intercalation at sufficiently high protein concentrations completely replace the force-induced DNA strand separation, leading to a very high slope of the DNA F-x curves approaching SL~300 pN/nm (Fig. 4A). Also, strong DNA intercalation by the protein is typically associated with small hysteresis of the stretch-release cycle, as the two DNA strands never separate, and the same DNA/protein complex is stretched and released during the cycle. The two major features of strong protein-DNA intercalative binding are therefore: (i) the F-x overstretching transition slope increases with increasing protein concentration beyond its protein-free value of ~20 pN/nm; and (ii) the hysteresis of the F-x stretch-release cycle remains small, but will typically first increase with protein concentration relative to its value in the absence of protein because proteins bound to ssDNA slow strand reannealing. However, further increase in protein concentration leads to extensive intercalation that prevents strand separation, leading to decreasing hysteresis in the F-x cycle. The observation of these features in the WT HIV-1 NC-DNA stretch-release cycles (Fig. 4A, (Wu et al., 2013; Wu et al., 2014)), demonstrates the intercalative nature of this interaction. According to previous studies, the capability of HIV-1 NC to intercalate requires stacking between the aromatic residues and unpaired NA bases. While HIV-1 NC seems incapable of intercalating into stable DNA duplexes (as demonstrated by the lack of DNA elongation upon NC binding at forces less than 10 pN), HIV-1 NC intercalates extensively at higher forces. Similarly, in the absence of force, HIV-1 NC is capable of forming intercalative or stacking interactions with NA duplexes locally destabilized by structural defects, such as mismatches, loops or bulges (Bazzi et al., 2012; Cosa et al., 2004; Godet et al., 2013; Godet et al., 2011; Wang et al., 2013).
When a protein is capable of binding to either ss or ds DNA or both, but is unable to intercalate, the DNA stretching curve is characterized by a small slope of the overstretching transition, similar to the behavior observed in the absence of protein. However, the transition force can be either decreased or increased relative to the protein-free force, depending on whether the protein is a duplex stabilizer or destabilizer, respectively. In addition, the F-x stretch-release cycle of such a protein-DNA complex typically shows a much larger hysteresis relative to that observed in the absence of protein, as it takes some time for the protein to dissociate from ssDNA before the strands can reanneal upon DNA release. If the non-intercalating protein is a highly charged cation (with Zeff >~ +3), then at saturating concentrations of protein, non-specific aggregation of both ss and dsDNA may occur (Bloomfield, 1997; Nguyen et al., 2000). Such protein-induced aggregation, in turn, can promote faster DNA strand re-annealing (Vo et al., 2006; Vo et al., 2009a), leading to an apparent decrease in the F-x hysteresis at higher protein concentrations. In summary, the main features of the F-x stretch-release cycle of a non-intercalative protein-DNA complex are: (i) a small transition slope ~20 pN/nm at all protein concentrations; (ii) a relatively large F-x cycle hysteresis, which may or may not decrease at saturating protein concentrations, depending on the protein’s DNA strand aggregating ability and its dissociation kinetics.
3.4.2 Effects of WT and mutant FIV NC on DNA stretching and release curves
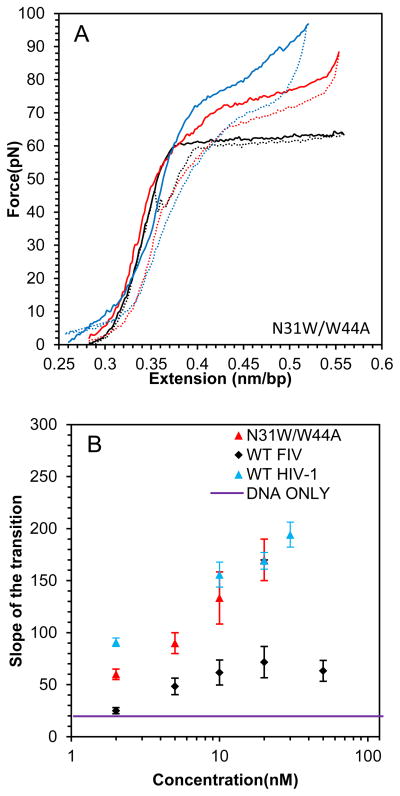
Presented in Figs. 4A and 4B are the F-x stretch-release curves for DNA in the presence of WT HIV-1 and FIV NC, respectively. Also shown in Fig. 4 are the overstretching transition slopes of the F-x curves (Fig. 4C) and the hysteresis of the stretch-release cycles of the DNA-protein complexes as a function of protein concentration (Fig. 4D). Interestingly, the maximum slope of the DNA-FIV NC extension curves is SL~70 pN/nm, which is much higher than the overstretching slope for DNA in the absence of the protein (SL~20 pN/nm), but much smaller than the saturated transition slope in the presence of HIV-1 NC (SL~300 pN/nm) (Fig. 4C). This result shows the ability of FIV NC to intercalate into dsDNA, but to an extent much smaller than observed for HIV-1 NC. The hysteresis of the F-x cycles for the DNA complex saturated with either FIV or HIV-1 NC is similar to the hysteresis observed in the absence of NC, suggesting that the two DNA strands do not separate significantly during the stretching of either complex. However, in the presence of low FIV NC concentrations (5 nM), hysteresis is observed. Thus, higher concentrations of FIV NC are required to strongly intercalate into DNA and to prevent strand separation (Fig. 4D). The weaker intercalative capability of WT FIV NC compared to HIV-1 NC is consistent with the ~10-fold higher Kd,1M for FIV NC binding to ssDNA20 relative to HIV-1 NC (see Table 2). This demonstrates the weaker ability of FIV NC to interact with unpaired DNA bases in high salt.
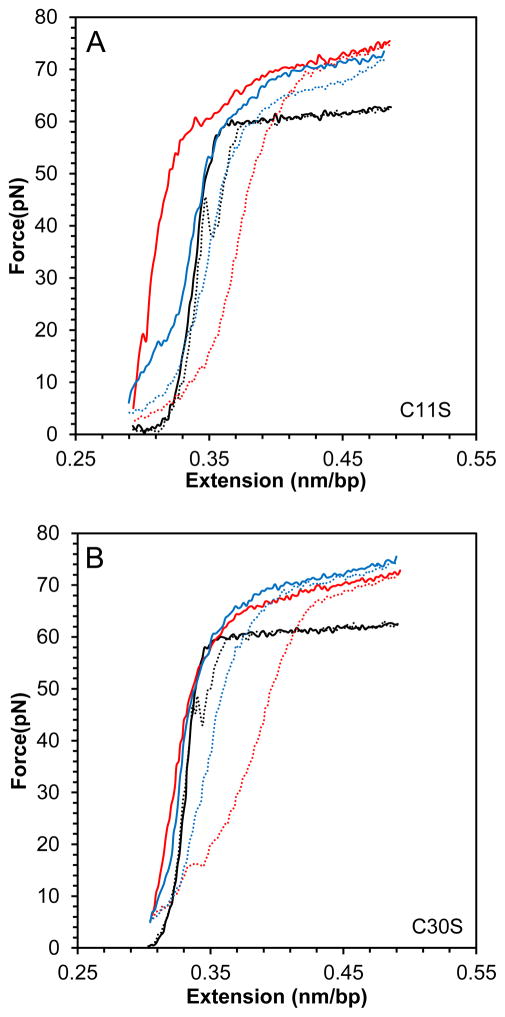
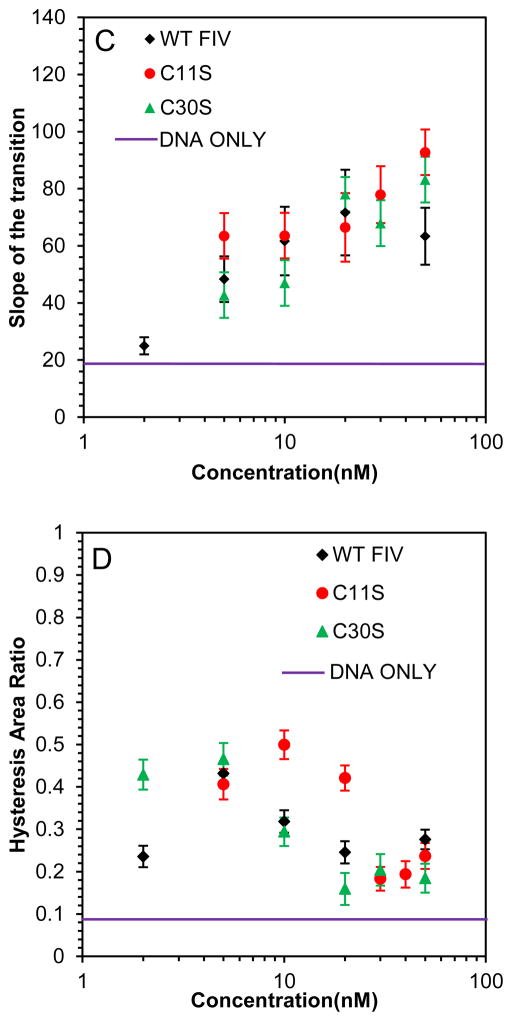
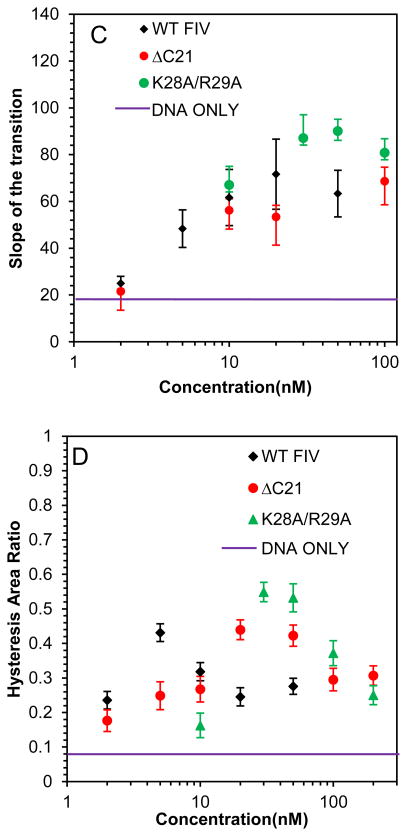
Presented in Fig. 5 are the DNA stretching and release curves for DNA in the presence of two single point mutants of FIV NC: C11S and C30S, which are expected to alter the zinc coordination in the first and second zinc finger, respectively. It appears that the effect of this mutation on the NC-DNA stretching curves compared to WT FIV NC is minimal, with just slightly weaker binding, as demonstrated by the hysteresis vs NC concentration dependence (Fig. 5D), and a slightly higher saturated slope (Fig. 5C), showing marginally stronger intercalation by these FIV NC mutants.
Fig. 5.
(A) DNA stretch (solid line) and release (dotted line) curves in the absence of protein (black) and in the presence of 10 nM C11S FIV NC (red) and 50 nM C11S FIV NC (blue). (B) DNA stretch (solid line) and release (dotted line) curves in the absence of protein (black) and in the presence of 5 nM C30S FIV NC (red) and 50 nM C11S FIV NC (blue). (C) Slope of the transition in the presence of WT FIV NC (black), C11S FIV NC (red) and C30S FIV NC (green). (D) Hysteresis versus concentration in the presence of WT FIV NC (black), C11S FIV NC (red) and C30S FIV NC (green). Error bars are propagated from the uncertainty in the force-extension measurements.
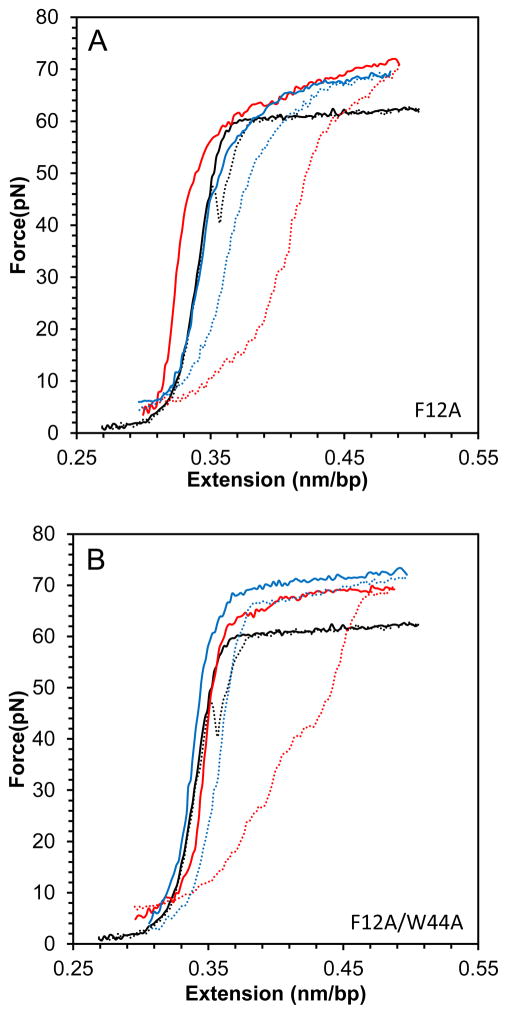
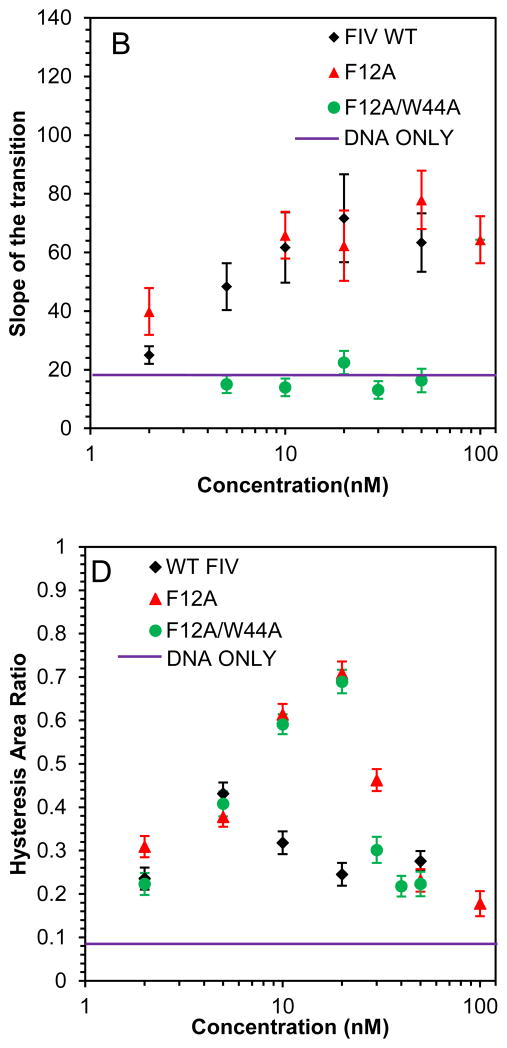
In contrast, the DNA stretching and release curves in the presence of the double aromatic mutant F12A/W44A FIV NC presented in Fig. 6, show a complete loss of duplex intercalation by this protein, as demonstrated by the small slope of DNA overstretching in the presence of this protein (Fig.6C). The single aromatic substitution mutant, F12A, can still intercalate to an extent typical of WT FIV NC. Although the F12A/W44A and F12A FIV NC mutants have very different intercalation capabilities, they both lead to small hysteresis at high concentrations due to the strong NA aggregating capability of these FIV NC variants (Fig. 6D).
Fig. 6.
(A) DNA stretch (solid line) and release (dotted line) curves in the absence of protein (black) and in the presence of 20n M FIV NC F12A (red) and 50 nM FIV NC F12A (blue). (B) DNA stretch (solid line) and release (dotted line) curves in the absence of protein (black) and in the presence of 20 nM FIV NC F12A/W44A (red) and 50 nM FIV NC F12A/W44A (blue). (C) Slope of the transition of FIV NC WT (black), F12A (red) and F12A/W44A (green). (D) Hysteresis versus concentration of FIV NC WT (black), F12A (red) and F12A/W44A (green). Error bars are propagated from the uncertainty in the force-extension measurements.
The two cationic FIV NC mutants, ΔC21 and K28A/R29A, both lead to an overstretching slope increase similar to that observed for WT FIV NC, but they require higher protein concentrations to reach saturation (Fig. 7A–C). Similarly, small hysteresis is observed with these FIV NC mutants, showing their ability to prevent strand separation, but only at higher concentrations than are required for WT FIV NC (Fig. 7D). We conclude that the loss of cationic residues either in the C-terminal domain of FIV NC or in the linker between its two zinc fingers, both lead to weaker NC binding, but the proteins retain the ability to intercalate DNA to the same extent as WT FIV NC at saturated concentration.
Fig. 7.
(A) DNA stretch (solid line) and release (dotted line) curves in the absence of protein (black) and in the presence of 50 nM FIV NC K28A/R29A (red) and 200 nM FIV NC K28A/R29A (blue). (B) DNA stretch (solid line) and release (dotted line) curves in the absence of protein (black) and in the presence of 50 nM FIV NC ΔC21 (red) and 200 nM FIV NC ΔC21 (blue). (C) Slope of the transition of FIV NC WT (black), ΔC21 (red) and K28A/R29A (green). (D) Hysteresis versus concentration of FIV NC WT (black), ΔC21 (red) and K28A/R29A (green). Error bars are propagated from the uncertainty in the force-extension measurements.
Finally, presented in Fig. 8 A–C are the analogous results for the DNA stretching with N31W/W44A FIV NC. With this aromatic residue switch variant, there is a striking increase in the slope of the DNA overstretching transition to ~180 pN/nm, which far exceeds the saturated slope of ~70 pN in the presence of WT FIV NC, and approaches the slope observed in the presence of HIV-1 NC (Fig. 8B). In addition, the hysteresis of the DNA stretch-release cycle with N31W/W44A FIV NC is always small, demonstrating its strong intercalative ability, such that even low protein concentrations are capable of preventing DNA strand separation (Fig. 8C). We thus conclude that the DNA intercalation capability of N31W/W44A FIV NC is much stronger than that of WT FIV NC, and comparable to that of HIV-1 NC. This conclusion is in full agreement with the stronger specific ssDNA binding observed for this mutant in high salt (Fig. 2 and Table 2).
Fig. 8.
(A) DNA stretch (solid line) and release (dotted line) curves in the absence of protein (black) and in the presence of 2 nM FIV NC N31W/W44A (red) and 20 nM FIV NC N31W/W44A (blue). (B) Slope of the transition of FIV NC WT (black) and N31W/W44A (red). (C) Hysteresis versus concentration of FIV NC WT (black) and N31W/W44A (red). Error bars are propagated from the uncertainty in the force-extension measurements.
4. Discussion
In this work, we used bulk and single molecule measurements to characterize the NA interactions and chaperone activity of WT and mutant FIV NC. WT FIV NC exhibits more electrostatic and less specific NA binding characteristics than HIV-1 NC. This is illustrated by the results of salt titration binding studies, which show a larger effective charge for WT FIV NC binding to ssDNA (Zeff ~4) relative to HIV-1 NC (Zeff ~3). Interestingly, mutation of the aromatic residues modulates FIV NC’s effective charge. A mutant that lacks both aromatic residues (F12A/W44A) has a stronger electrostatic binding character (Zeff=5.4), whereas the aromatic residue switch mutant (N31W/W44A) has a reduced electrostatic binding component (Zeff=2.5) more similar to that of HIV-1 NC. The cationic residues contributing to FIV NC’s effective charge are distributed throughout the protein, as demonstrated by the significant increase in Kd observed for the C-terminal truncation mutant, ΔC21 FIV NC, and the linker mutant, K28A/R29A FIV NC (Table 1).
Although the electrostatic component of FIV NC binding is stronger than that of HIV-1 NC, the specific non-electrostatic component of FIV NC binding to NAs, as measured by Kd,1M, is ~10-fold weaker than that of HIV-1 NC (Table 2). As for the Zeff, this difference in specific binding is due to the unusual position of the Trp residue in the second zinc finger (Fig. 1). Switching the position of this residue to mimic the location found in HIV-1 NC (N31W/W44A FIV NC) leads to an enhancement of the non-electrostatic component of FIV NC-DNA interaction to a level approaching that of HIV-1 NC. N31W/W44A FIV NC also shows a 3-fold enhanced ability to destabilize NA and facilitate the strand exchange step of TAR RNA and DNA annealing relative to WT FIV NC (k2, Table 3), demonstrating improved chaperone activity with this single aromatic residue position change. In contrast, F12A/W44A FIV NC displays reduced binding affinity at high salt (Table 1) and an ~100-fold increased Kd,1M relative to WT FIV NC (Table 2). Thus, as for HIV-1 NC, specific FIV NC-NA interactions appear to be largely due to stacking between the aromatic residues in the zinc fingers and NA bases.
A previous study using northwestern blotting showed that C11S is deficient in binding to RNA (Manrique et al., 2004), whereas in the current study, C11S, C30S and WT FIV NC proteins bind model NAs with comparable affinity. This apparent discrepancy could be attributed to several factors. First, FA experiments measure the binding of proteins to NA in solution at equilibrium. Northwestern blotting measures NA binding on a membrane and is not a true equilibrium method. Moreover, a long RNA was used in the northwestern blotting experiment (732-nt) but short DNA or RNA oligonucleotides were studied here. Finally, the previous study was conducted under relative high salt conditions (400 mM), while the present study was carried out in 50 mM NaCl. All of these factors could contribute to the differences in reported binding results.
In our previous DNA stretching studies, we showed that the ability of HIV-1 NC to intercalate stretched dsDNA is due to the stacking interactions of its aromatic residues with DNA bases (Wu et al., 2013). In this work, we show that WT FIV NC has a much weaker propensity to intercalate the DNA duplex compared to HIV-1 NC. However, switching the position of the second aromatic residue in FIV NC to the analogous position in HIV-1 NC leads to a pronounced enhancement of this protein’s intercalative ability (see Fig. 8B). In contrast, the F12A/W44A double mutant lacks intercalation capability (Fig. 6C). In addition, our DNA stretching assay also demonstrates the strong capability of FIV NC to aggregate dsDNA, as shown by the additional stretching force needed to unwind dsDNA (at low extensions) from its FIV NC-aggregated state (Fig. 4B, (Wu et al., 2010)). Also, in contrast to HIV-1 NC, which destabilizes the DNA duplex by lowering the overstretching transition midpoint, FIV NC appears to stabilize the duplex, as evidenced by the increased overstretching transition midpoint force relative to that observed in the absence of protein (Fig. 4B).
Interestingly, the DNA stretching curves in the presence of FIV NC strongly resemble those observed in the presence of RSV NC (Stewart-Maynard et al., 2008). The first aromatic residue in both proteins is located in the same position as in HIV-1 NC. However, in contrast to HIV-1 NC, the second aromatic residue of RSV NC is also located in the N-terminal zinc finger. Thus, it is possible that both FIV and RSV NC exhibit significantly weaker specific interactions with NA as compared to HIV-1 NC, due to the non-optimal positioning of their second aromatic residue. These specific NA interactions are also responsible for the ability of retroviral NC proteins to recognize their cognate gRNA in the context of Gag. Interestingly, both RSV (Lochmann et al., 2013; Parent, 2011) and FIV (Kemler et al., 2012) have been shown to shuttle their Gag proteins through the nucleus. This suggests the possibility that gRNA selection by FIV and RSV Gag may occur in the nucleus, prior to gRNA export into the cytoplasm. This is in contrast to HIV-1 Gag, which has been shown to select its gRNA in the cytoplasm prior to membrane binding (Fogarty et al., 2014; Jouvenet et al., 2011). Thus, it is possible that the different cellular environment for gRNA selection by HIV-1 Gag (in the cytoplasm) vs. FIV or RSV Gag (in the nucleus) may require different NC-NA binding interactions. Such different gRNA binding specificities for FIV and HIV-1 Gag would be fully consistent with the quantitative differences in the FIV NC and HIV-1 NC DNA binding modes observed here.
FIV NC-nucleic acid interactions are probed using ensemble and single molecule approaches
Structure of FIV and HIV-1 NC differ primarily by location of highly basic tail and one aromatic residue
Non-electrostatic FIV NC interactions with nucleic acids are much weaker than HIV-1 NC
Switching the aromatic residue location in FIV NC to mimic HIV-1 NC restores non-electrostatic binding
Acknowledgments
This work was supported by the National Institutes of Health [GM065056 to K.M.-F. and GM072462 to M.C.W.] and the National Science Foundation [MCB-1243883 to M.C.W]. This project was also supported by the National Cancer Institute, National Institutes of Health [contract no. HHSN261200800001E with Leidos Biomedical Research, Inc. to R.J.G.]. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We thank Drs. Lawrence Mathes and Kathleen Hays (Ohio State University) for the DNA plasmid encoding FIV Gag. We also thank Donald G. Johnson and Catherine V. Hixson of the AIDS and Cancer Virus Program for their assistance in cloning, and producing and purifying several of the recombinant proteins used in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affranchino J, González S. In vitro assembly of the feline immunodeficiency virus Gag polyprotein. Virus Res. 2010;150:153–157. doi: 10.1016/j.virusres.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- Amarasinghe GK, Zhou J, Miskimon M, Chancellor KJ, McDonald JA, Matthews AG, Miller RR, Rouse MD, Summers MF. Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein. structural studies and implications for genome recognition. J Mol Biol. 2001;314:961–970. doi: 10.1006/jmbi.2000.5182. [DOI] [PubMed] [Google Scholar]
- Anderson JA, Teufel RJ, 2nd, Yin PD, Hu WS. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J Virol. 1998;72:1186–1194. doi: 10.1128/jvi.72.2.1186-1194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auxilien S, Keith G, Le Grice SF, Darlix JL. Role of post-transcriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J Biol Chem. 1999;274:4412–4420. doi: 10.1074/jbc.274.7.4412. [DOI] [PubMed] [Google Scholar]
- Bampi C, Jacquenet S, Lener D, Decimo D, Darlix JL. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Curr HIV Res. 2004;2:79–92. doi: 10.2174/1570162043485022. [DOI] [PubMed] [Google Scholar]
- Bazzi A, Zargarian L, Chaminade F, De Rocquigny H, Rene B, Mely Y, Fosse P, Mauffret O. Intrinsic nucleic acid dynamics modulates HIV-1 nucleocapsid protein binding to its targets. PLoS One. 2012;7:e38905. doi: 10.1371/journal.pone.0038905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Chaurasiya KR, Paramanathan T, McCauley MJ, Williams MC. Biophysical characterization of DNA binding from single molecule force measurements. Phys Life Rev. 2010;7:299–341. doi: 10.1016/j.plrev.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosa G, Harbron EJ, Zeng Y, Liu HW, O’Connor DB, Eta-Hosokawa C, Musier-Forsyth K, Barbara PF. Secondary structure and secondary structure dynamics of DNA hairpins complexed with HIV-1 NC protein. Biophys J. 2004;87:2759–2767. doi: 10.1529/biophysj.104.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein’s nucleic acid chaperone function. J Mol Biol. 2006a;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Stephen AG, Beuning PJ, Gorelick RJ, Fisher RJ, Williams MC. Single DNA molecule stretching measures the activity of chemicals that target the HIV-1 nucleocapsid protein. Anal Biochem. 2006b;358:159–170. doi: 10.1016/j.ab.2006.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix JL, Cristofari G, Rau M, Péchoux C, Berthoux L, Roques B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Advances in Pharmacology (San Diego, Calif) 2000;48:345–372. doi: 10.1016/s1054-3589(00)48011-7. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Gabus C, Nugeyre M, Clavel F, Barré-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. Journal of Molecular Biology. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- de Rocquigny H, Shvadchak V, Avilov S, Dong C, Dietrich U, Darlix J-L, Mély Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini-Rev Med Chem. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- Feng Y, Copeland T, Henderson L, Gorelick RJ, Bosche W, Levin JG, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty KH, Berk S, Grigsby IF, Chen Y, Mansky LM, Mueller JD. Interrelationship between Cytoplasmic Retroviral Gag Concentration and Gag-Membrane Association. J Mol Biol. 2014;426:1611–1624. doi: 10.1016/j.jmb.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet J, Kenfack C, Przybilla F, Richert L, Duportail G, Mely Y. Site-selective probing of cTAR destabilization highlights the necessary plasticity of the HIV-1 nucleocapsid protein to chaperone the first strand transfer. Nucleic Acids Res. 2013;41:5036–5048. doi: 10.1093/nar/gkt164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet J, Ramalanjaona N, Sharma KK, Richert L, de Rocquigny H, Darlix JL, Duportail G, Mely Y. Specific implications of the HIV-1 nucleocapsid zinc fingers in the annealing of the primer binding site complementary sequences during the obligatory plus strand transfer. Nucleic Acids Res. 2011;39:6633–6645. doi: 10.1093/nar/gkr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- Heath MJ, Derebail SS, Gorelick RJ, DeStefano JJ. Differing roles of the N- and C-terminal zinc fingers in human immunodeficiency virus nucleocapsid protein-enhanced nucleic acid annealing. J Biol Chem. 2003;278:30755–30763. doi: 10.1074/jbc.M303819200. [DOI] [PubMed] [Google Scholar]
- Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Simon SM, Bieniasz PD. Visualizing HIV-1 assembly. J Mol Biol. 2011;410:501–511. doi: 10.1016/j.jmb.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafaie J, Song R, Abrahamyan L, Mouland A, Laughrea M. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology. 2008;375:592–610. doi: 10.1016/j.virol.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Kemler I, Saenz D, Poeschla E. Feline immunodeficiency virus Gag is a nuclear shuttling protein. J Virol. 2012;86:8402–8411. doi: 10.1128/JVI.00692-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon JC, Ghazawi A, Cheung WK, Phillip PS, Rizvi TA, Lever AM. The secondary structure of the 5′ end of the FIV genome reveals a long-range interaction between R/U5 and gag sequences, and a large, stable stem-loop. RNA. 2008;14:2597–2608. doi: 10.1261/rna.1284908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GA, Gross P, Bockelmann U, Modesti M, Wuite GJ, Peterman EJ. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy. Proc Natl Acad Sci USA. 2013;110:3859–3864. doi: 10.1073/pnas.1213676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughrea M, Shen N, Jetté L, Darlix JL, Kleiman L, Wainberg M. Role of distal zinc finger of nucleocapsid protein in genomic RNA dimerization of human immunodeficiency virus type 1; no role for the palindrome crowning the R-U5 hairpin. Virology. 2001;281:109–116. doi: 10.1006/viro.2000.0778. [DOI] [PubMed] [Google Scholar]
- Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J Mol Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Lochmann TL, Bann DV, Ryan EP, Beyer AR, Mao A, Cochrane A, Parent LJ. NC-mediated nucleolar localization of retroviral gag proteins. Virus Res. 2013;171:304–318. doi: 10.1016/j.virusres.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttge BG, Freed EO. FIV Gag: virus assembly and host-cell interactions. Vet Immunol Immunopathol. 2010;134:3–13. doi: 10.1016/j.vetimm.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique M, Rauddi M, González S, Affranchino J. Functional domains in the feline immunodeficiency virus nucleocapsid protein. Virology. 2004;327:83–92. doi: 10.1016/j.virol.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Mark-Danieli M, Laham N, Kenan-Eichler M, Castiel A, Melamed D, Landau M, Bouvier NM, Evans MJ, Bacharach E. Single point mutations in the zinc finger motifs of the human immunodeficiency virus type 1 nucleocapsid alter RNA binding specificities of the gag protein and enhance packaging and infectivity. J Virol. 2005;79:7756–7767. doi: 10.1128/JVI.79.12.7756-7767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley MJ, Williams MC. Mechanisms of DNA binding determined in optical tweezers experiments. Biopolymers. 2007;85:154–168. doi: 10.1002/bip.20622. [DOI] [PubMed] [Google Scholar]
- Mely Y, Piemont E, Sorinas-Jimeno M, De Rocquigny H, Jullian N, Morellet N, Roques BP, Gerard D. Structural and dynamic characterization of the aromatic amino acids of the human immunodeficiency virus type I nucleocapsid protein zinc fingers and their involvement in heterologous tRNA(Phe) binding: a steady-state and time-resolved fluorescence study. Biophys J. 1993;65:1513–1522. doi: 10.1016/S0006-3495(93)81222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N, de Rocquigny H, Mely Y, Jullian N, Demene H, Ottmann M, Gerard D, Darlix JL, Fournie-Zaluski MC, Roques BP. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J Mol Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- Morellet N, Demene H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournie-Zaluski MC, Roques BP. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J Mol Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO Journal. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Manetti F, Botta M. Targeting protein-protein and protein-nucleic acid interactions for anti-HIV therapy. Curr Pharm Des. 2011;17:3713–3728. doi: 10.2174/138161211798220972. [DOI] [PubMed] [Google Scholar]
- Moscardini M, Pistello M, Bendinelli M, Ficheux D, Miller JT, Gabus C, Le Grice SF, Surewicz WK, Darlix JL. Functional interactions of nucleocapsid protein of feline immunodeficiency virus and cellular prion protein with the viral RNA. J Mol Biol. 2002;318:149–159. doi: 10.1016/S0022-2836(02)00092-X. [DOI] [PubMed] [Google Scholar]
- Musah R. The HIV-1 nucleocapsid zinc finger protein as a target of antiretroviral therapy. Curr Top Med Chem. 2004;4:1605–1622. doi: 10.2174/1568026043387331. [DOI] [PubMed] [Google Scholar]
- Mustafa F, Jayanth P, Phillip PS, Ghazawi A, Schmidt RD, Lew KA, Rizvi TA. Relative activity of the feline immunodeficiency virus promoter in feline and primate cell lines. Microbes Infect. 2005;7:233–239. doi: 10.1016/j.micinf.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Narayanan N, Gorelick RJ, DeStefano JJ. Structure/function mapping of amino acids in the N-terminal zinc finger of the human immunodeficiency virus type 1 nucleocapsid protein: residues responsible for nucleic acid helix destabilizing activity. Biochemistry. 2006;45:12617–12628. doi: 10.1021/bi060925c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negroni M, Buc H. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J Mol Biol. 1999;286:15–31. doi: 10.1006/jmbi.1998.2460. [DOI] [PubMed] [Google Scholar]
- Negroni M, Buc H. Mechanisms of retroviral recombination. Annu Rev Genet. 2001;35:275–302. doi: 10.1146/annurev.genet.35.102401.090551. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Rouzina I, Shklovskii BI. Reentrant condensation of DNA induced by multivalent counterions. J Chem Phys. 2000;112:2562–2568. [Google Scholar]
- Paoletti AC, Shubsda MF, Hudson BS, Borer PN. Affinities of the nucleocapsid protein for variants of SL3 RNA in HIV-1. Biochemistry. 2002;41:15423–15428. doi: 10.1021/bi026307n. [DOI] [PubMed] [Google Scholar]
- Parent LJ. New insights into the nuclear localization of retroviral Gag proteins. Nucleus. 2011;2:92–97. doi: 10.4161/nucl.2.2.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NC, Ho EW, Brown ML, Yamamoto JK. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- Peliska JA, Balasubramanian S, Giedroc DP, Benkovic SJ. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- Rizvi T, Kenyon J, Ali J, Aktar S, Phillip P, Ghazawi A, Mustafa F, Lever A. Optimal packaging of FIV genomic RNA depends upon a conserved long-range interaction and a palindromic sequence within gag. J Mol Biol. 2010;403:103–119. doi: 10.1016/j.jmb.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes GM, Bambara RA, Fay PJ. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix 1. Thermodynamic analysis. Biophys J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokri L, McCauley MJ, Rouzina I, Williams MC. DNA overstretching in the presence of glyoxal: structural evidence of force-induced DNA melting. Biophys J. 2008;95:1248–1255. doi: 10.1529/biophysj.108.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers MF, South TL, Kim B, Hare DR. High resolution structure of an HIV zinc finger via a new NMR-based distance geometry aproach. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- Tisne C, Roques B, Dardel F. Heteronuclear NMR studies of the interaction of tRNA(Lys)3 with HIV-1 nucleocapsid protein. J Mol Biol. 2001;306:443–454. doi: 10.1006/jmbi.2000.4391. [DOI] [PubMed] [Google Scholar]
- van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJ, Peterman EJ. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc Natl Acad Sci USA. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J Mol Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Effect of Mg(2+) and Na(+) on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J Mol Biol. 2009a;386:773–788. doi: 10.1016/j.jmb.2008.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper”. J Mol Biol. 2009b;386:789–801. doi: 10.1016/j.jmb.2008.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier C, Bombarda E, Morellet N, Gerard D, Roques BP, Mely Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- Wang H, Musier-Forsyth K, Falk C, Barbara PF. Single-molecule spectroscopic study of dynamic nanoscale DNA bending behavior of HIV-1 nucleocapsid protein. J Phys Chem B. 2013;117:4183–4196. doi: 10.1021/jp3018259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Naiyer N, Mitra M, Li J, Williams MC, Rouzina I, Gorelick RJ, Wu Z, Musier-Forsyth K. Distinct nucleic acid interaction properties of HIV-1 nucleocapsid protein precursor NCp15 explain reduced viral infectivity. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku335. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JA, Jones CP, Parent LJ, Rouzina I, Musier-Forsyth K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA. 2013;19:1078–1088. doi: 10.1261/rna.038869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett BJ, Hosie MJ. The virus-receptor interaction in the replication of feline immunodeficiency virus (FIV) Curr Opin Virol. 2013;3:670–675. doi: 10.1016/j.coviro.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MC, Gorelick RJ, Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein’s nucleic acid chaperone function. Proc Natl Acad Sci USA. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MC, Rouzina I. Force spectroscopy of single DNA and RNA molecules. Curr Opin Struct Biol. 2002;12:330–336. doi: 10.1016/s0959-440x(02)00340-8. [DOI] [PubMed] [Google Scholar]
- Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein’s nucleic acid chaperone activity and retroviral replication. Virus Res. 2013;171:263–277. doi: 10.1016/j.virusres.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, Naufer MN, McCauley MJ, Gorelick RJ, Rouzina I, Musier-Forsyth K, Williams MC. Differential contribution of basic residues to HIV-1 nucleocapsid protein’s nucleic acid chaperone function and retroviral replication. Nucleic Acids Res. 2014;42:2525–2537. doi: 10.1093/nar/gkt1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Rouzina I, Williams MC. Single-molecule stretching studies of RNA chaperones. RNA Biol. 2010;7:712–723. doi: 10.4161/rna.7.6.13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Guo J, Bess J, Henderson LE, Levin JG. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J Virol. 1999;73:4794–4805. doi: 10.1128/jvi.73.6.4794-4805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JC, McHenry CS. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J Biol Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]