Abstract
Cyclic di-AMP (c-di-AMP) is a signaling molecule that has been shown to play important roles in bacterial physiology and infections. Currently, c-di-AMP detection and quantification relies mostly on the use of high-performance liquid chromatography (HPLC) or liquid chromatography-mass spectrometry (LC-MS). In this study, a competitive enzyme-linked immunosorbent assay (ELISA) for the quantification of c-di-AMP was developed, which utilizes a novel pneumococcal c-di-AMP binding protein (CabP) and a newly commercialized c-di-AMP derivative. With this new method, c-di-AMP concentrations in biological samples can be quickly and accurately quantified. Furthermore, this assay is much more efficient than current methods as it requires less overall cost and training while processing many samples at once. Therefore, this assay can be extensively used in research into c-di-AMP signaling.
Keywords: c-di-AMP, ELISA, c-di-AMP binding protein, Detection
1. Introduction
Cyclic nucleotides have been well studied over the past few decades and are integral to signal transduction in both eukaryotic and prokaryotic organisms (Kalia et al., 2012, Romling et al., 2013). However, cyclic di-AMP (c-di-AMP) was only recently discovered and the roles of cdi-AMP in various organisms have just started to be recognized (Corrigan & Grundling, 2013, Kalia et al., 2012, Witte et al., 2008). Currently, c-di-AMP is linked to bacterial infections, biofilm formation, potassium uptake, cell wall homeostasis, and resistance to antibiotics (Bai et al., 2013, Bai et al., 2014, Barker et al., 2013, Corrigan et al., 2011, Corrigan et al., 2013, Griffiths & O'Neill, 2012, Luo & Helmann, 2012, Mehne et al., 2013, Pozzi et al., 2012, Schwartz et al., 2012, Yamamoto et al., 2012, Yang et al., 2014, Ye et al., 2014). Additionally, c-di-AMP produced by Listeria monocytogenes has been shown to activate a host type I interferon response (Woodward et al., 2010) mediated by host effector proteins STING and DDX41 (Bowie, 2012, Burdette et al., 2011, Jin et al., 2011, Parvatiyar et al., 2012, Sauer et al., 2011). This widespread influence has made the study of c-di-AMP a quickly developing area of research for the foreseeable future.
Enzymatically, c-di-AMP is synthesized from two molecules of ATP, which is catalyzed by diadenylate cyclase (DAC). This enzyme was first discovered through a structural study of DisA in Bacillus subtilis and Thermotoga maritima (Witte et al., 2008). DAC selectively converts either ATP or ADP to c-di-AMP, and does not utilize GTP, ITP, CTP, or UTP as its substrate (Bai et al., 2012, Witte et al., 2008). In B. subtilis, DisA is named after DNA integrity scanning protein A and it has three interdependent domains: an N-terminal DAC domain (known as DUF147), a linker domain, and an RuvA-like C-terminal DNA-binding helix-hairpin-helix (HhH) domain. Furthermore, two additional proteins with DAC domains, CdaA (also referred to as YbbP) and CdaS (also referred to as YojJ), were also found to produce c-di-AMP in B. subtilis (Mehne et al., 2013, Romling, 2008). It has been noticed that the DAC domain is widespread in bacteria and archaea (Corrigan & Grundling, 2013, Romling, 2008). In addition to B. subtilis and T. maritima, the presence of functional DAC has also been reported in L. monocytogenes (Woodward et al., 2010), Staphylococcus aureus (Corrigan et al., 2011), Streptococcus pyogenes (Kamegaya et al., 2011), Mycobacterium tuberculosis (Bai et al., 2012), Mycobacterium smegmatis (Zhang & He, 2013), Chlamydia trachomatis (Barker et al., 2013), and Streptococcus pneumoniae (Bai et al., 2013). However, little is known about the signaling pathways mediated by c-di-AMP.
In order to extensively investigate c-di-AMP mediated signaling pathways and the biological roles of c-di-AMP, it is essential to establish a simple and sensitive assay to determine the levels of this dinucleotide. Recently, a c-di-AMP binding protein (CabP) in S. pneumoniae was identified using c-di-AMP affinity chromatography with 2’-AHC-c-di-AMP-agarose resin (BioLog) (Bai et al., 2014). We have demonstrated that CabP directly interacts with a potassium transporter and plays a role in pneumococcal potassium uptake. The physical interaction between the two proteins can be impaired by elevated bacterial c-di-AMP. Additionally, CabP displays a high affinity for c-di-AMP, and the c-di-AMP binding site in this protein is extremely specific. The interaction between CabP and c-di-AMP could not be interrupted by an excessive amount (1,000-fold) of c-di-GMP, cAMP, ATP, ADP, AMP, pApA, NAD+, NADH, NADP+, and NADPH (Bai et al., 2014). This protein was therefore utilized as the initial coating protein to establish a competitive enzyme-linked immunosorbent assay (ELISA) for the detection of c-di-AMP. Additionally, biotin-labeled c-di-AMP (B-c-di-AMP) has recently become commercially available, such as “2’-[biotin]-AHC-c-di-AMP” (BioLog). The advent of these developments has allowed for the creation of a novel assay utilizing a competitive-ELISA technique to quantify cdi-AMP levels in biological samples, as we have reported (Bai et al., 2013, Bai et al., 2014, Yang et al., 2014). The principle of this assay is using B-c-di-AMP to compete with c-di-AMP in a sample or standard for binding with CabP. Therefore, higher c-di-AMP concentration presents in a sample will be reflected by less B-c-di-AMP that can be detected. Here we provide a detailed protocol for evaluating the concentrations of c-di-AMP by measuring c-di-AMP bound to pneumococcal CabP in a competitive ELISA assay (Fig. 1). This assay has been optimized and is shown to have an extreme specificity (Fig. 2) and high sensitivity (Fig. 3) for c-di-AMP, making it ideal to fill the need in the field of c-di-AMP research.
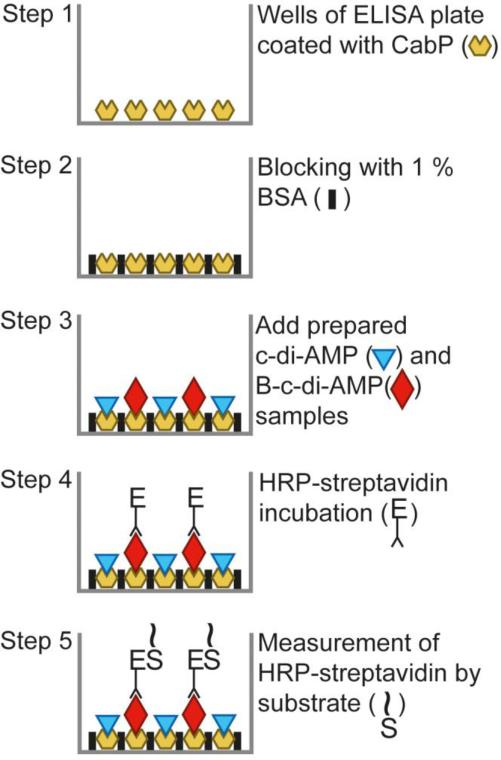
Fig. 1.
Diagrammatic representation of the competitive ELISA for detection of c-di-AMP. Symbols are defined where appropriate. The first part of the assay relies on the coating of wells with CabP protein followed by a standard blocking step with BSA. A mixture of biotin-labeled and unlabeled c-di-AMP in standards or samples is then added to create the competitive condition in each well. Finally, the bound B-c-di-AMP is detected by incubation with HRP-conjugated streptavidin and followed by reaction with an OPD substrate.
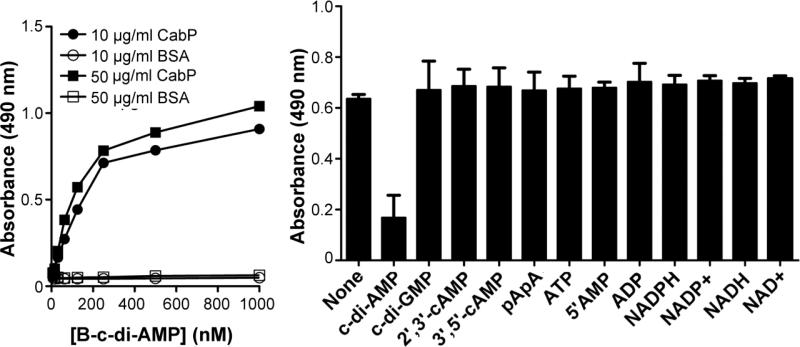
Fig. 2.
Specificity of the ELISA assay. (A) CabP binds c-di-AMP specifically. Using coating concentrations of 10 and 50 μg/ml, CabP and BSA were coated into each well similarly. Subsequently, a titration of B-c-di-AMP ranging from 0 nM to 1,000 nM was performed in order to optimize efficiency and lower overall use of B-c-di-AMP. Our results from this titration indicate that a concentration of >1.5 nM was required for proper detected. (B) c-di-AMP binding site in CabP is specific for c-di-AMP. Using a 10 μg/ml coat of CabP and a standard 25 nM B-cdi-AMP tracer, 200 nM of various closely related nucleotides were used to challenge the performance of this assay. The only nucleotide that significantly reduced the signal was c-di-AMP, indicating that this assay is specific for c-di-AMP and not the other nucleotides tested. Data shown are the means of two independent experiments. The error bars denote the standard deviation (SD). Data set labeled “Control” represents repeats without any nucleotide challenge.
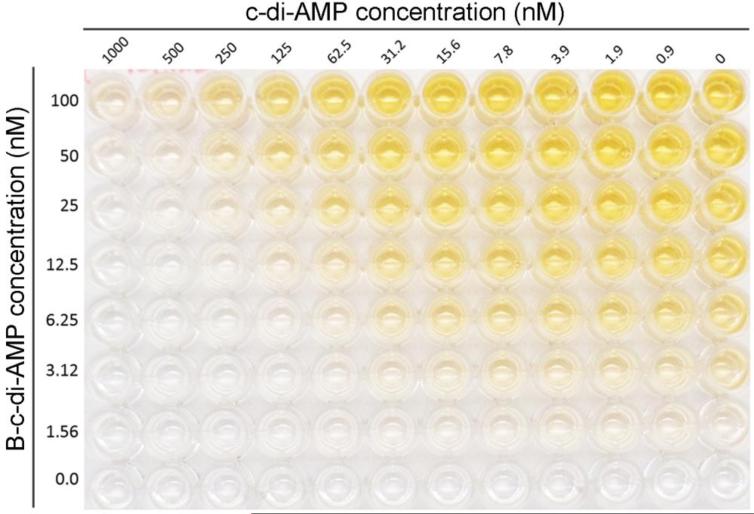
Fig. 3.
Sensitivity of CabP with titration of B-c-di-AMP and c-di-AMP standards. Using standard coating conditions, B-c-di-AMP was 2-fold serially diluted from 100 nM and c-di-AMP was titrated 2-fold up to 1,000 nM. This representation provides photographic evidence for the optimization of B-c-di-AMP in the assay.
2. Materials and Methods
2.1. Reagents and materials
Both c-di-AMP and B-c-di-AMP were purchased from BioLog and were resuspended as 5 mM stocks. Aliquots were stored at −20 °C until use.
The following buffers and solutions were used in sample preparation and the assay. The coating buffer contains 50 mM Na2CO3 and 50 mM NaHCO3 with a final pH of 9.6. Phosphate buffered saline (PBS, pH 7.4) contains 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl. PBST wash buffer consists of 0.05 % (v/v) Tween 20 in PBS (pH 7.4). Citric acid buffer contains 0.1 M citric acid titrated with 0.2 M Na2HPO4 to a final pH of 5.0. Substrate buffer is prepared by dissolving a tablet of o-phenylenediamine dihydrochloride (OPD, 10 mg/tablet; Sigma) and 20 μl H2O2 in 20 ml citric acid buffer, which is mixed thoroughly at room temperature and used immediately. All buffers and solutions were stored at 4°C until use unless otherwise specified.
2.2. Preparation of c-di-AMP samples from bacteria
To create bacterial samples for evaluating this assay, Escherichia coli was grown in 10 ml of Luria-Bertani (LB) media to the expected growth phase. Testing conditions vary based on c-di-AMP levels present in the bacterium being studied and various testing scenarios. The cell suspension to be collected is processed by centrifuge for 2 min at 13,000 rpm. The resulting bacterial pellet is then washed three times with 1 ml of PBS (pH 7.4). Once washed, the pellet was then resuspended in 0.5 ml of 50 mM Tris-HCl (pH8.0) and further disrupted using a sonic dismembrator (Model 100, Fischer Scientific) for periods of 10 sec with a 10 sec interval on ice. Samples were then heated at 95 °C for 10 min and subsequently centrifuged for 10 min at 13,000 rpm. Supernatants were collected and used immediately or stored at −20 °C until use. The sample preparation methods for S. pneumoniae and M. tuberculosis has been reported earlier (Bai et al., 2013, Yang et al., 2014). In all these studies, no more than 10 ml of bacterial culture was needed for three repeats in the assay of bacterial c-di-AMP.
2.3. Procedure of the assay
Recombinant CabP was purified as described previously (Bai et al., 2014) and was diluted to 10 μg/ml or 50 μg/ml in coating buffer and coated at 100 μl/well in a Nunc MaxiSorp plate (Thermo Scientific). The plate was sealed with plastic wrap and incubated overnight (14-18 h) at 4 °C. The coating buffer was decanted from the wells, and the plate was washed three times with 200 μl PBST wash buffer. After washing, the wells were blocked with 100 μl of 1% BSA in PBS for 1 h at room temperature. The blocking solution was then decanted from the wells, and the plate was washed 3 times with 200 μl PBST wash buffer. Samples, controls, and calibrator dilutions were prepared in sample diluent containing 25 nM of B-c-di-AMP. An aliquot of 100 μl of this prepared sample, control, or calibrator was directly added to the 96-well plate and incubated for 2 h at room temperature. After washing the wells 3 times each with 200 μl PBST wash buffer, 100 μl of 1:10,000 HRP-conjugated streptavidin (Thermo Scientific) was added. The plate was then incubated for 1 h at room temperature. After washing with PBST buffer for 3 times, 100 μl of substrate buffer was added to each well and incubated for 30 min at room temperature. The colorimetric reaction was then stopped with 100 μl of 2 M H2SO4. The absorbance of each well was determined at 490 nm using a plate reader.
2.4. Analysis of data collected
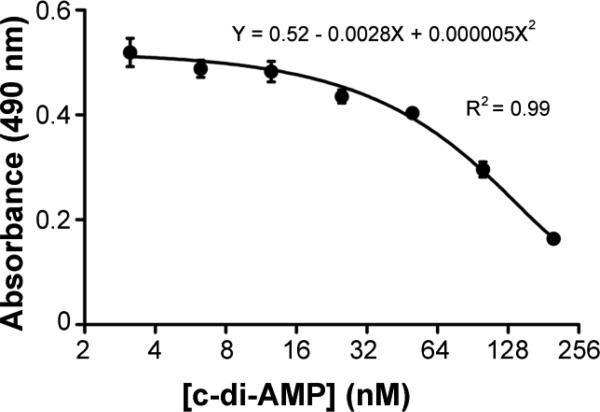
The use of a standard curve and proper calibrator wells is important to obtain reproducible results. First the average values of the blank calibrator wells were subtracted from those of the samples and the standards. At least 6 standard dilutions were used to cover low (≤ 10 nM), medium (25-75 nM), and high (≥ 200 nM) concentrations of c-di-AMP. A standard curve was generated by plotting the concentrations of the standards using a log2 scale (x axis) against the readings of optical density at 490 nm (OD490) (y axis) and then fitted with a second-order polynomial curve as shown in Fig. 4. A standard curve was repeated if the correlation coefficient was < 0.90. The c-di-AMP concentration of each sample was determined by taking the absorbance readings at OD490, which was corrected from control wells, and calculated with the equation generated from the standard curve. The amount of c-di-AMP in the samples was subsequently corrected for dilution factors and normalized against either the bacterial numbers or the OD readings of the culture.
Fig. 4.
Example of a standard curve. The curve was generated using standard conditions and performing a 1:2 dilution scheme from 200 nM down to 6.25 nM c-di-AMP in 25 nM B-c-di-AMP sample diluent media. Data shown are the means of absorbance of duplicate wells. The error bars denote SD.
3. Results and Discussion
3.1. Detection of c-di-AMP in the samples
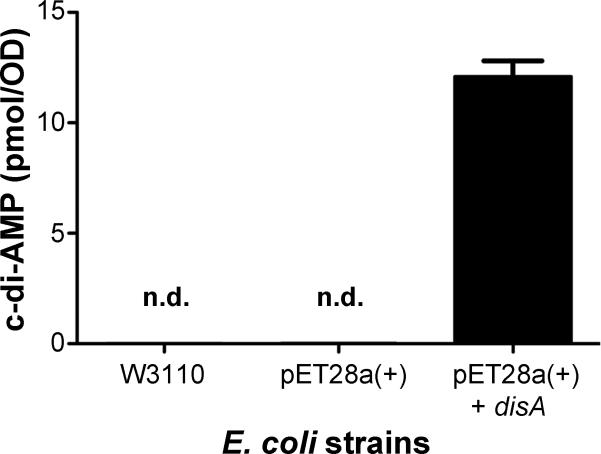
To evaluate the specificity of this assay, we prepared different E. coli samples to detect cdi-AMP. Previously, it has been reported that no c-di-AMP existed in the lysates of E. coli BL21 (DE3) harboring the empty vector pET28a+ (Novagen) or pET28b+ (Novagen) (Corrigan et al., 2011, Mehne et al., 2013). In contrast, c-di-AMP has previously been detected in the lysate of E. coli expressing S. aureus dacA, B. subtilis cdaA or cdaS using LC-MS (Corrigan et al., 2011, Mehne et al., 2013). In this study, E. coli W3110 was grown in 10 ml LB broth overnight to an OD600 of 1.0. E. coli BL21 (DE3) harboring either the empty pET28a+ vector (Novagen) or the plasmid expressing M. tuberculosis disA (also referred to as dacA) (Bai et al., 2012) were grown in 10 ml LB broth to an OD600 of 0.5 and then induced at room temperature for 3 hours with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation, and each bacterial pellet was resuspended into 0.5 ml 50 mM Tris-HCl (pH 8.0). Samples were then prepared and c-di-AMP was detected following the established protocol. Our results displayed that the E. coli W3110 and BL21 (DE3) strains did not produce c-di-AMP, which is consistent with the observations described in the other reports (Corrigan et al., 2011, Mehne et al., 2013). In contrast, the E. coli strain expressing disA exhibited high levels of c-di-AMP (Fig. 5), which is consistent with the result that was previously reported and analyzed using LC-MS (Mehne et al., 2013). This indicates that the novel assay can be used to directly quantify c-di-AMP levels from a biological sample. These findings also suggest that this assay is highly specific for c-di-AMP rather than other molecules present in the E. coli lysate.
Fig. 5.
Detection of intracellular c-di-AMP in E. coli strains. E. coli W3110, BL21 with empty vector pET28a+ or the plasmid expressing M. tuberculosis disA. The samples were processed by following the protocol described in the Experimental section. Data shown are the means of three independent experiments. The error bars denote the SD. Abbreviations: n.d. indicates not detectable.
3.2. Comparison of the assay with other current available methods
As the study of c-di-AMP evolves, simple and sensitive methods for the detection of this molecule in biological samples will be needed. Currently, very few methods for quantifying c-di-AMP have been reported and they rely primarily on high-performance liquid chromatography (HPLC) (Bai et al., 2012, Oppenheimer-Shaanan et al., 2011), liquid chromatography–mass spectrometry (LC-MS) (Barker et al., 2013, Burhenne & Kaever, 2013, Corrigan et al., 2011, Mehne et al., 2013), or complex formation between c-di-AMP and coralyne (Zhou et al., 2014). HPLC and LC-MS both have a relatively short analysis time for single samples, strong resolving power, reproducibility, and acceptable sensitivity of results. However, training and equipment costs for each technique make these methods expensive and inaccessible for many laboratories. Additionally, these current techniques either utilize large volumes of culture (Oppenheimer-Shaanan et al., 2011), or need [13C,15N]-c-di-AMP to generate internal standards (Burhenne & Kaever, 2013, Corrigan et al., 2011, Mehne et al., 2013). Because of limitations with the current methods, we developed the ELISA that is inexpensive, reproducible, and easily performed. Moreover, each 96-well plate has the capacity to measure up to 80 distinct samples, or 40 samples in duplicate, on the same plate under identical condition. This is a massive advantage over current quantification methods that rely on individually measured samples. Similar detection methods have been developed for the study of cyclic AMP (cAMP), which propelled the field forward as evident by the myriad of commercially available cAMP ELISA-based detection kits. The analogous development of the ELISA-based c-di-AMP detection assay would similarly make quantification of c-di-AMP in biological samples assessable to broader users.
3.3. Limitation of the assay
This method has a detection limit of approximately 10 nM. It is simple to determine c-di-AMP levels within bacteria, as detailed in the Methods and Results sections. The c-di-AMP levels vary from different bacteria and their growth stages. For S. pneumoniae and M. tuberculosis, by collecting bacteria from 1 to 10 ml of culture at their end logarithmic phase and preparing samples in 0.5 ml Tris-HCl, the c-di-AMP levels were ranging from 25 to 100 nM. However, it is more complicated in measuring c-di-AMP levels secreted by bacteria, either in culture media or in host cells. c-di-AMP levels in culture media may be undetectable directly for some bacteria; such samples might need to be concentrated prior to detection. Moreover, components in media may affect the assay, thus chemically defined media (CDM) are preferable for analysis of c-di-AMP secretion using this assay. As an example, Sauton's broth was used for determination of secreted c-di-AMP by M. tuberculosis (Yang et al., 2014).
Highlights.
□ We have established a simple assay to determine bacterial c-di-AMP levels
□ The assay is highly specific to c-di-AMP
□ The assay is easy to perform
□ Many samples can be processed in one assay
Acknowledgments
This work is partly supported by National Institutes of Health grant DC006917.
Abbreviations
- CDM
chemically defined medium
- c-di-AMP
cyclic di-AMP
- B-c-di-AMP
biotin-labeled c-di-AMP
- DAC
diadenylate cyclase
- ELISA
enzyme-linked immunosorbent assay
- HhH
helix-hairpin-helix
- HPLC
high-performance liquid chromatography
- LC-MS
liquid chromatography-mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests statement
The authors declare no competing financial interests.
References
- Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol. 2013;195:5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai G. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol. 2014;196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yang J, Zhou X, Ding X, Eisele LE, Bai G. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS One. 2012;7:e35206. doi: 10.1371/journal.pone.0035206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. STING-Dependent Recognition of Cyclic di-AMP Mediates Type I Interferon Responses during Chlamydia trachomatis Infection. MBio. 2013;4:e00018–00013. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie AG. Innate sensing of bacterial cyclic dinucleotides: more than just STING. Nat Immunol. 2012;13:1137–1139. doi: 10.1038/ni.2469. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhenne H, Kaever V. Quantification of cyclic dinucleotides by reversed-phase LC MS/MS. Methods Mol Biol. 2013;1016:27–37. doi: 10.1007/978-1-62703-441-8_3. [DOI] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP Is a New Second Messenger in Staphylococcus aureus with a Role in Controlling Cell Size and Envelope Stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Grundling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- Griffiths JM, O'Neill AJ. Loss of function of the GdpP protein leads to joint beta-lactam/glycopeptide tolerance in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:579–581. doi: 10.1128/AAC.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev. 2012;42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- Kamegaya T, Kuroda K, Hayakawa Y. Identification of a Streptococcus pyogenes SF370 gene involved in production of c-di-AMP. Nagoya J Med Sci. 2011;73:49–57. [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis sigma(M) in beta-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stulke J. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high-level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem. 2013;288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O'Gara JP. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012;8:e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci Signal. 2008;1:pe39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz KT, Carleton JD, Quillin SJ, Rollins SD, Portnoy DA, Leber JH. Hyperinduction of host beta interferon by a Listeria monocytogenes strain naturally overexpressing the multidrug efflux pump MdrT. Infect Immun. 2012;80:1537–1545. doi: 10.1128/IAI.06286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Hara H, Tsuchiya K, Sakai S, Fang R, Matsuura M, Nomura T, Sato F, Mitsuyama M, Kawamura I. Listeria monocytogenes strain-specific impairment of the TetR regulator underlies the drastic increase in cyclic di-AMP secretion and beta interferon-inducing ability. Infect Immun. 2012;80:2323–2332. doi: 10.1128/IAI.06162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bai Y, Zhang Y, Gabrielle VD, Jin L, Bai G. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol Microbiol. 2014;93:65–79. doi: 10.1111/mmi.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhang JJ, Fang X, Lawlis GB, Troxell B, Zhou Y, Gomelsky M, Lou Y, Yang XF. DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect Immun. 2014;82:1840–1849. doi: 10.1128/IAI.00030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, He ZG. Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein A, to negatively affect its cyclic-di-AMP synthesis activity in Mycobacterium smegmatis. J. Biol. Chem. 2013;288:22426–22436. doi: 10.1074/jbc.M113.464883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Sayre DA, Zheng Y, Szmacinski H, Sintim HO. Unexpected complex formation between coralyne and cyclic diadenosine monophosphate providing a simple fluorescent turn-on assay to detect this bacterial second messenger. Anal Chem. 2014;86:2412–2420. doi: 10.1021/ac403203x. [DOI] [PMC free article] [PubMed] [Google Scholar]