Abstract
CYP51 (sterol 14α-demethylase) is an efficient target for clinical and agricultural antifungals and an emerging target for treatment of Chagas disease, the infection that is caused by multiple strains of a protozoan pathogen Trypanosoma cruzi. Here, we analyze CYP51A from the Y strain T. cruzi. In this protein, proline 355, a residue highly conserved across the CYP51 family, is replaced with serine. The purified enzyme retains its catalytic activity, yet has been found less susceptible to inhibition. These biochemical data are consistent with cellular experiments, both in insect and human stages of the pathogen. Comparative structural analysis of CYP51 complexes with VNI and two derivatives suggests that broad-spectrum CYP51 inhibitors are likely to be preferable as antichagasic drug candidates.
Keywords: Sterol 14α-demethylase, CYP51 sequence variations, Trypanosoma cruzi, drug resistance, structure-based drug design
1. Introduction
Trypanosoma cruzi is a protozoan parasite that uses blood-sucking triatomine insects (kissing bugs) as vectors and a variety of mammals as hosts. In mammals, the pathogen multiplies intracellularly, populating different organs and tissues, though damaging predominantly the heart or/and gastrointestinal tract (1). T. cruzi was first reported as the causative agent of human infections by Carlos Chagas in 1909 (2), but since then both the disease and the pathogen have remained remarkably neglected. Current therapeutic options are limited mainly to two nitroderivatives, benznidazole and nifurtimox. Both drugs are highly toxic, cause severe side effects, and their efficiency in the chronic phase is still debated (1). Chagas disease remains endemic in Latin America (3) and is now becoming an emerging global health problem, mainly due to human/vector migration. For example, it has been reported that only in the USA there is up to one million infected (4), of them more than 260,000 patients living in Texas alone (5). Spreading the disease all over the world eventually attracted attention, and two antifungal drugs, inhibitors of fungal sterol 14α-demethylase (CYP51), posaconazole and ravuconazole, that demonstrated promising results in animal models of Chagas disease (6–8) have been advanced into clinical trials (9). The results, however, appear to be controversial (80% treatment failure (10)), thus calling for better, safer, and cost-efficient rationally designed T. cruzi CYP51 inhibitors.
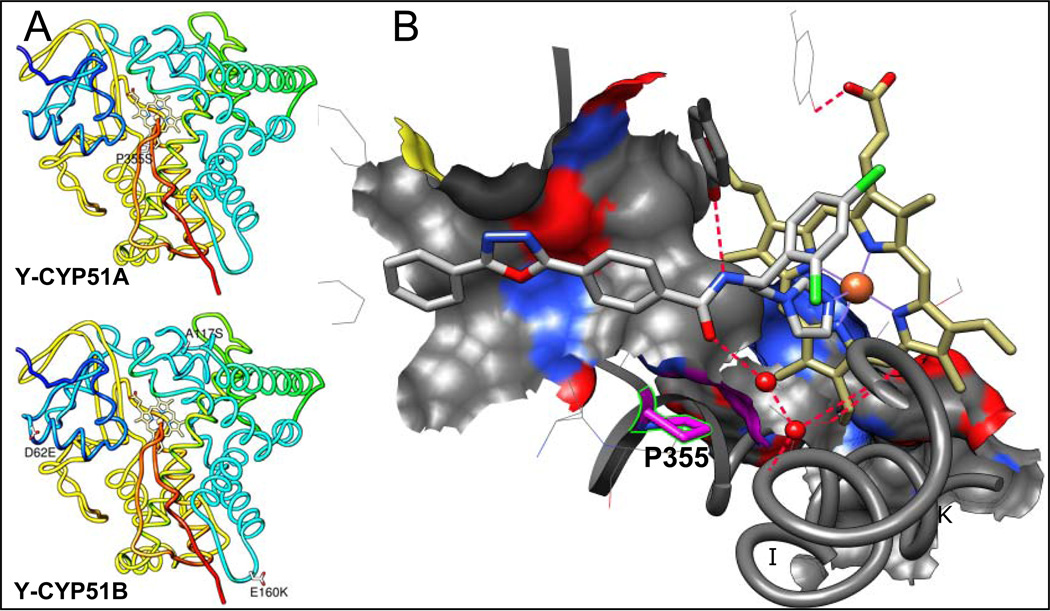
We have recently shown that VNI, the novel, nontoxic and highly potent experimental inhibitor of T. cruzi CYP51, can efficiency cure both the acute and chronic models of Chagas disease in mice infected with the Tulahuen strain of T. cruzi (11). However, T. cruzi is a highly heterogeneous population, known to represent a pool of >70 different strains (http://www.dbbm.fiocruz.br/TcruziDB/strain.html). The strains vary significantly in the disease progression (the time of parasitemia onset/peak), the severity of the acute stage, chronic symptoms (cardiac versus gastrointestinal) and particularly in drug sensitivity (12, 13). The results of VNI testing in the stringent short-term treatment protocols of mice infection with the Y and Colombiana strains of T. cruzi (medium and high resistance to benznidazole, respectively) have been inconclusive. Although VNI suppressed parasitemia and prevented from mortality, no complete parasitological cure was achieved under these conditions based on the RT-PCR analysis after immunosupression (14). Amplification of CYP51 from Colombiana revealed the presence of two genes, encoding eight (gene A) and seven (gene B) amino acid differences from Tulahuen CYP51 (A-like) (Table 1), though none of these residues is located within the enzyme substrate binding cavity (14). In this work, we analyzed CYP51 in the Y strain of T. cruzi (Table 1, Figure 1A). Again, two CYP51 genes were identified. CYP51A was of particular interest because it carries a sequence variation that results in the substitution of a highly conserved across the whole CYP51 family proline residue (P355 in the T. cruzi CYP51 sequence numbering). In the CYP51-VNI co-structure, this proline forms the surface of interaction with the VNI carboxamide fragment (Figure 1B). Replacement of this proline with serine (the variation also found in the CYP51A paralogues from some intrinsically drug resistant filamentous fungi, such as multiple species of Aspergillus (Figure 2)), was likely to increase flexibility of this portion of the CYP51 binding cavity therefore suggesting that its sensitivity to inhibition may be altered. The findings of this work support this idea, imply that it might be more preferable for CYP51 inhibitors aimed at serving as antichagasic drug candidates to have a broad-spectrum activity rather than a high target-selectivity, and outline a promising direction for the CYP51 structure-based VNI scaffold development.
Table 1.
Sequence variations in CYP51s from different T.cruzi strains*
|
T. cruzi strain |
CYP51 (GenBank Protein ID) |
Amino acids | DNA | ||
|---|---|---|---|---|---|
| % identity | # of differences (amino acid substitutions) | % identity | # of differences | ||
| CL-Brener | A (XP_820210) | 100 | 0 (none) | >99 | 2 (2 silent) |
| B (XP_821219) | 99 | 4 (G9A, D62E, A117S, E160K) | 97 | 35 (31 silent) | |
| Y | A (AFW98339) | 99 | 1 (P355S) | >99 | 3 (2 silent) |
| B (AFW98340) | 99 | 4 (G9A, D62E, A117S, E160K) | 97 | 33 ( 29 silent ) | |
| Colombiana | A (AGF25233) | 98 | 8 (A117S, E160K, H196R, K203Q, V245I, K314E, D405E, P480S) | 98 | 19 (11 silent) |
| B (AGF25234) | 98 | 7 (G9A; L13P, P32S, D62E, A117S, E160K A288S) | 97 | 39 (32silent) | |
| Sylvio | Single, A-like (EKG07251) | 98 | 6 (G9C, A117S, E160K, H196R, V245I, D405E) | ||
| Marinkellei | Single, B-like (EKF39305) | 95 | 24 | ||
compared with Tulahuen CYP51
Figure 1. Amino acid differences between CYP51s from Tulahuen and Y strains of T. cruzi.
A. Mapping Y-CYP51A amino acid variations on the Tulahuen CYP51 structure [3k1o]. Distal view. The protein backbone is rainbow colored, from the N terminus (blue) to the C-terminus (red). The residues that are different in the Y strain CYP51s are shown as white carbon sticks, the variations are labeled. B. Enlarged view of the substrate binding cavity in the in the CYP51 co-structure with VNI [3gw9]. P355 (K/β1–4 turn) and its surface of interaction with VNI are colored in pink. The C atoms of VNI are grey. The C atoms of the heme are yellow. The water molecules are displayed as red spheres. The hydrogen bonds are indicated as red dashes. The I and K helices are outlined and marked.
Figure 2. A fragment of multiple sequence alignment of CYP51 from different biological kingdoms in the region of P355 (T. cruzi numbering).
P355 is marked with black arrow. P/S variation is also found in CYP51A in several filamentous ascomycetes, whose constitutively expressed CYP51B paralogues always have P in this position. More extended alignment can be seen as Supplemental Figure 1.
2. Materials and Methods
T. cruzi and mammalian cell cultures
Epithelial cells (Vero cell line) and cardiomyoblasts (H9C2 line) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics at 37°C in an atmosphere of 5% CO2. Y strain T. cruzi epimastigotes expressing GFP (Y-GFP) (kindly provided by Dr. S. Schenkman, Universidade Federal de Sao Paulo (Sao Paulo, Brazil)) were maintained in Diamond medium (0.1 M NaCl, 0.05 M K2HPO4 pH 7.2, 0.625% tryptose, 0.625% tryptone, 0.625% yeast extract, 12.5 µg/mL Hemin) supplemented with 10% inactivated fetal bovine serum (Gibco), at 28 °C. Y-GFP trypomastigotes were obtained by in vitro metacyclogenesis of epimastigotes as described (15) and maintained in culture of Vero cells in DMEM supplemented with 3% fetal bovine serum (FBS) and antibiotics at 37°C in an atmosphere of 5% CO2.
T. cruzi cellular infection assays. Human stage (amastigotes)
The assay was perform using the conditions previously described for testing antiparasitic activity of VNI in the Tulahuen strain of T. cruzi (11). Briefly, Y-GFP trypomastigotes were used to infect Vero cells or cardiomyoblasts (10 parasites per cell) for 24 h. Unbound trypomastigotes were removed by washing with DMEM. Infected mammalian cells were incubated with VNI or VFV dissolved in DMSO/DMEM in triplicate and cocultured in DMEM + 3% FBS for 48 h to observe parasite multiplication. 72 h post-infection, the cells were washed with phosphate-buffered saline, fixed with 10% paraformaldehyde and stained with Hoechst to visualize DNA and with TRITC phalloidin (Invitrogen) to visualize cardiomyocyte or Vero actin myofibrils. The number of parasites in each cell was quantified by confocal microscopy using a FV1000 Confocal Olympus microscope. Insect stage (epimastigotes). 1×106 epimastigotes of Y-GFP were cultured in Diamond medium in the presence of 1 µM VNI; VFV, or 1% DMSO (Control). Each 48 h the medium was replaced by fresh medium at the same conditions. Aliquots were collected every 24 h, and were mixed with 2% p-formaldehyde in PBS (dilution 1:10). The parasites were counted in Neubauer hemocytometer. The number of dead parasites was determined by the dye exclusion method (0.1% of eosin in PBS).
CYP51 gene sequencing
Total DNA was isolated from Y strain T. cruzi epimastigotes as described (16). The CYP51 gene was then PCR amplified using a FailSafe PCR Premix Selection Kit (Epicentre). The upstream primer 5’-CGCCATATGTTCATTGAAGCCATTGTATTGG –3’ contained a unique Nde I cloning site (underlined) and was complimentary to the Tulahuen T. cruzi CYP51 cDNA (GenBank accession number AY856083 (17) from 1 to 25 bp. The downstream primer 5’-CGCAAGCTTCAGTGATGGTGATGCGAGGGCAATTTCTTCTTGCG - 3’ included a unique Hind III cloning site (underlined) followed by a stop codon (bold) and C-terminal 4-histidine tag (italics), and was complementary to the Tulahuen T. cruzi CYP51 sequence from 1443 to 1423 bp. Amplification was carried out as described previously (17). The PCR products were subcloned into pGEM-T Easy vector (Promega) and sequenced. The Y-CYP51A and Y-CYP51B cDNA and protein sequences were deposited into the NCBI GenBank (http://www.ncbi.nih.gov/Genbank), nucleotide accession numbers JQ434483 and JQ434484, respectively.
Protein expression, purification and spectral characterization
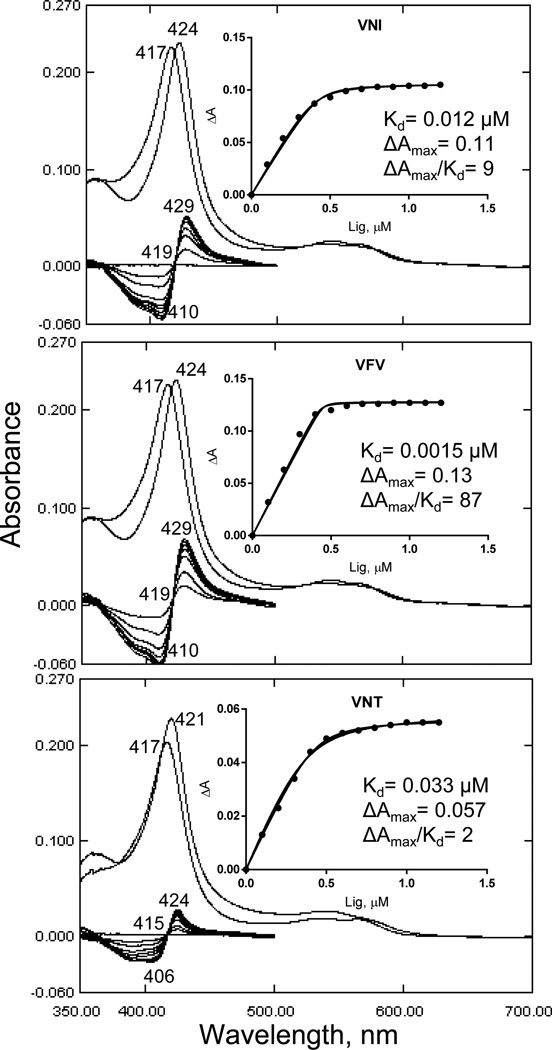
To obtain the expression construct, the Y-CYP51A gene insert was subcloned into pCW as described (17). The protein was coexpressed with GroEL/ES in E. coli DH5α and purified using metal affinity chromatography on Ni-NTA Agarose followed by anion-exchange chromatography (CM-Sepharose). The absorption spectra were recorded on a dual-beam Shimadzu UV-240IPC spectrophotometer. P450 concentration was determined from the absolute absorbance (ϵ417 = 117 mm−1 cm−1) and reduced CO difference spectra (Δϵ450–490 = 91 mm−1 cm−1). Substrate binding parameters were calculated from the difference type I spectral response (low-to-high spin transition of the P450 heme iron) at the conditions described for Tulahuen T.cruzi CYP51 (17). Binding of VNI and its derivatives VNT and VFV was monitored as type 2 spectral response reflecting coordination of the P450 heme iron to the azole nitrogen. The apparent Kds were calculated in Prism (GraphPad Software) using a quadratic function for tight binding ligands (18).
CYP51 activity assay
Enzymatic activities of Tulahuen CYP51 and Y-CYP51A were reconstituted as described previously (17). The reaction products were analyzed using a reverse-phase HPLC system (Waters) equipped with β-RAM detector (INUS Systems, Inc.). Potencies of the compounds to inhibit CYP51 activity were compared as inhibition of the substrate conversion in a one hour enzymatic reaction (18, 19).
X-ray crystallography
Crystallographic analysis was performed using Tulahuen T. cruzi CYP51 structure (PDB ID 3k10) (20). CYP51 co-structures with VNI (21), VNT and VFV were determined using the orthologous enzyme from T. brucei (PDB codes 3gw9, 4g3j, and 4g7g, respectively), because these complexes diffract at atomic resolution (<2.0 Å).
3. Results
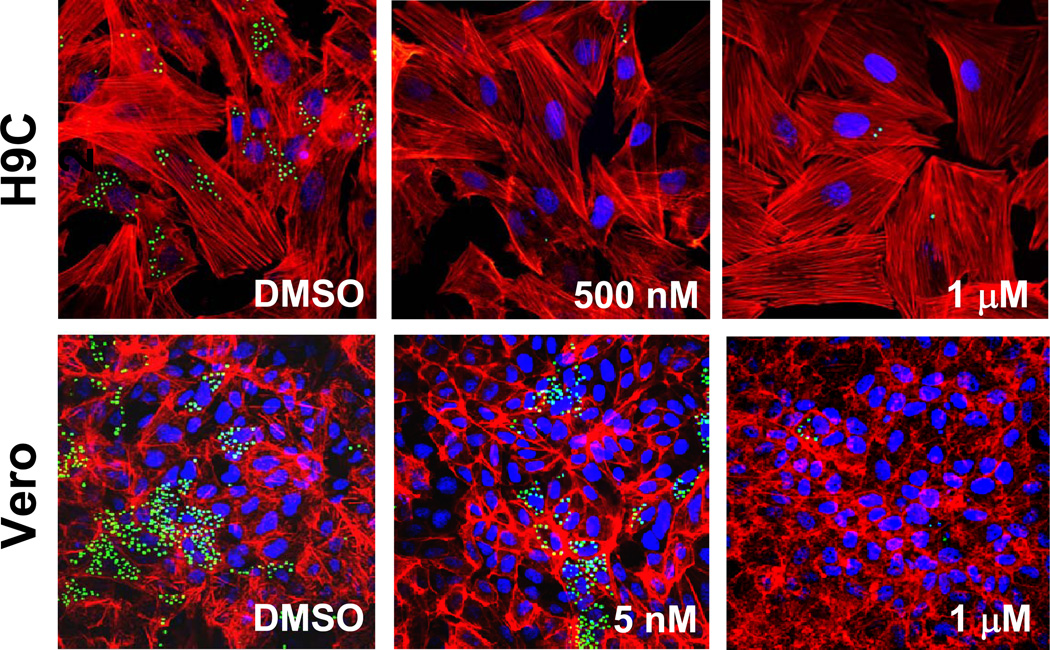
While in the cells infected by Tulahuen T. cruzi VNI completely eradicates the parasite at 8 nM concentration (EC50 =1.3 nM) (11), in the Y strain infection its EC50 was found to be >5 nM, some trace of amastigotes being observed within cardiomyoblasts and Vero cells even after the treatment with up to 1 µM VNI (Figure 3). Amplification of CYP51 from the Y strain DNA has shown that the effect of VNI may possibly be weakened by the presence of two CYP51 genes (higher CYP51 protein abundance, although it remains to be studied whether Y T. cruzi expresses both CYP51 paralogues constitutively, or in both replicative stages of the life cycle). The former can potentially be tested in the CL strain of T. cruzi, which also carries two CYP51 genes (Table 1), CL-CYP51B being 100% identical to Y-CYP51B, and CL-CYP51A (100% identical to Tulahuen-CYP51) having one amino acid difference: P355 vs. S355 in Y-CYP51A.
Figure 3. Antiparasitic effect of VNI in cardiomyoblasts and Vero cells infected with the Y strain of T. cruzi.
Fluorescence microscopic observation of GFP-expressing amastigotes multiplying within the host cells 72 h after treatment with different concentrations of VNI versus control (DMSO). T. cruzi amastigotes are green, H9C2/Vero cells nuclei are blue, and actin myofibrils are red.
In the CYP51 molecule P355 forms the surface of the active site cavity (cytochrome P450 SRS5 (22)). Its substitution with serine in Y-CYP51A was likely to increase the local flexibility in the region. Because it is our belief that high rigidity of the binding cavity provides the structural basis for CYP51 druggability and catalytic conservation (23), a more detailed analysis of Y-CYP51A was chosen as the subject for this study.
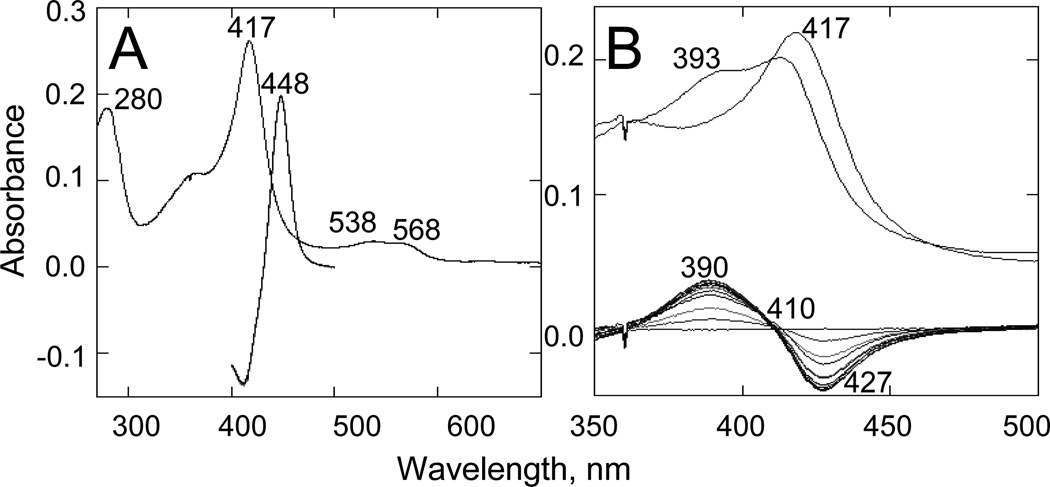
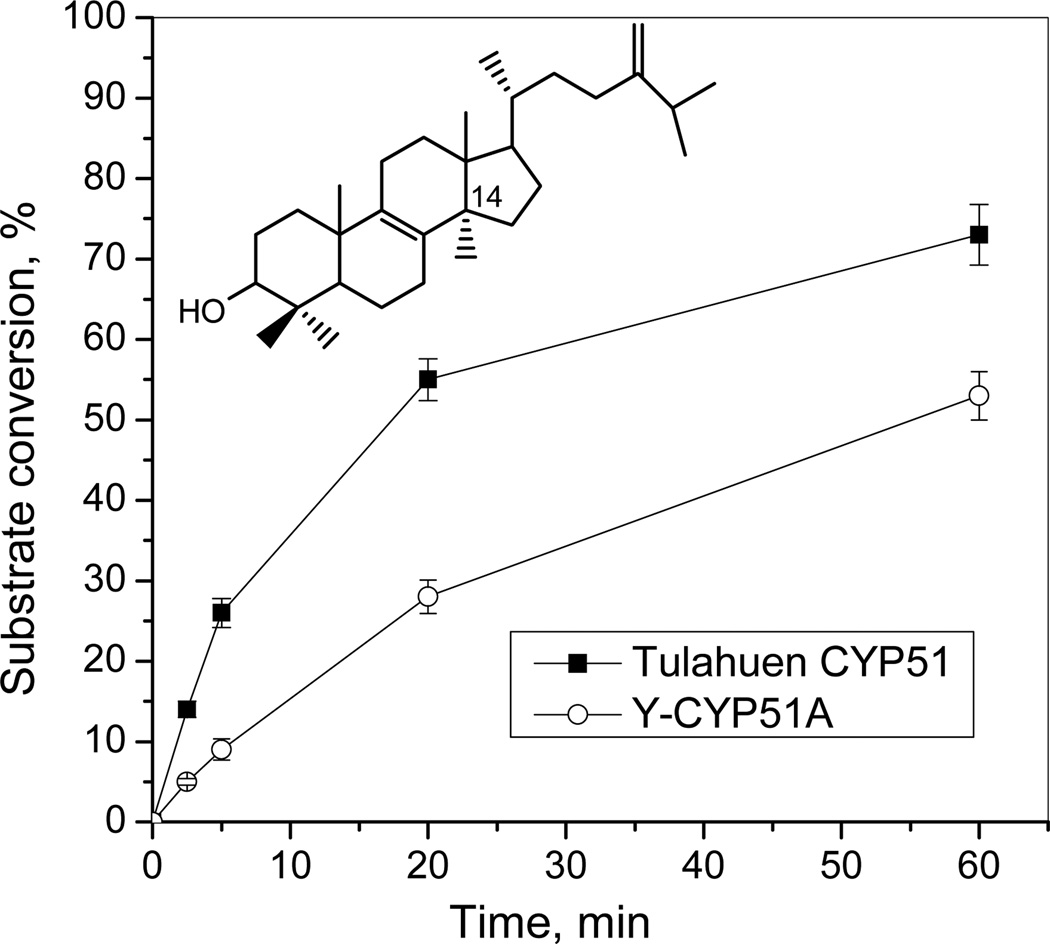
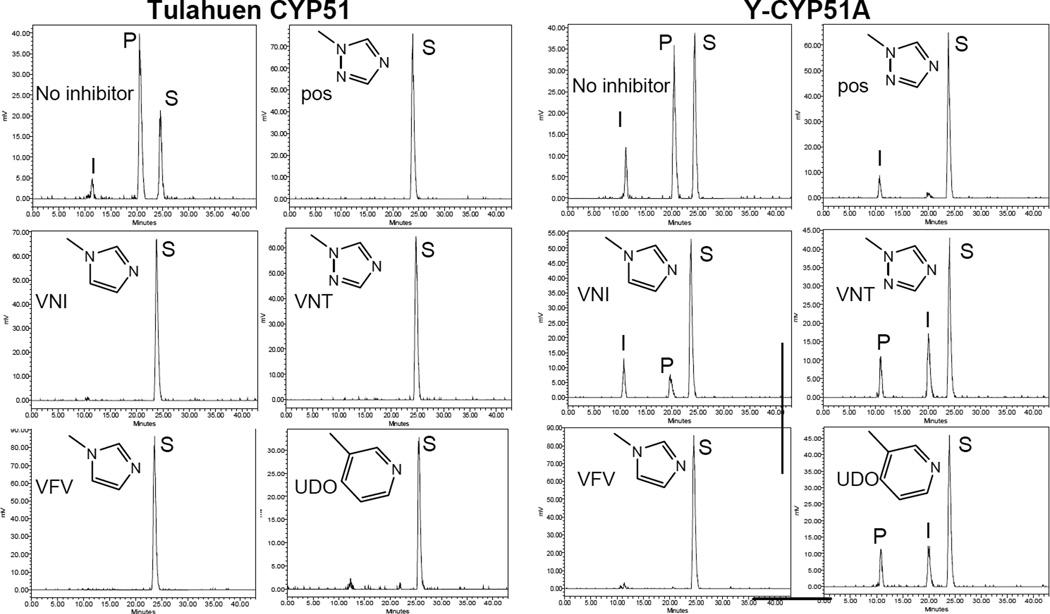
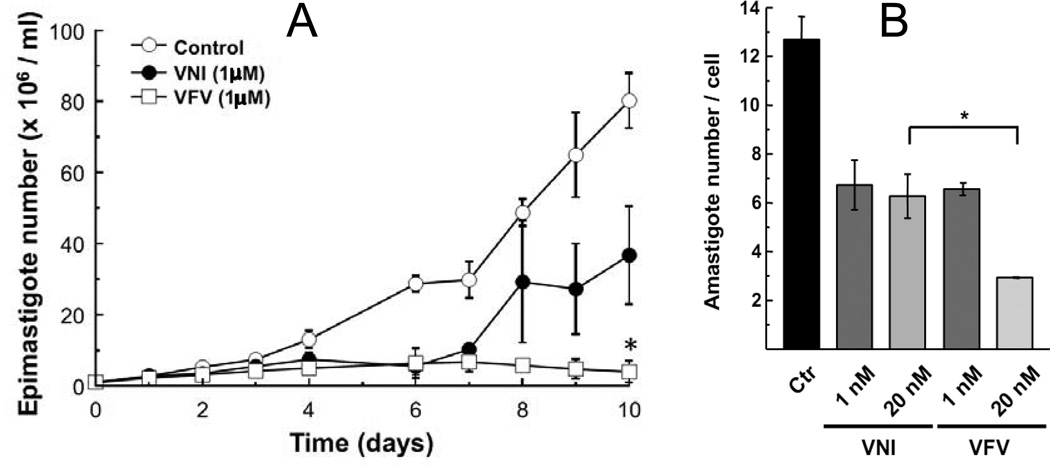
The purified protein did not display any differences from Tulahuen-CYP51 in its absolute absorbance or CO-binding spectra (Figure 4A). Titration with the substrate produced a characteristic type 1 spectral response (Figure 4B); though both the Kd and the amplitude of the low-to-high spin transition in the heme iron were slightly higher (Table 2). Their ratio (ΔA/Kd) indicated lower apparent binding efficiency, meaning that the Y-CYP51A ability to hold the substrate molecule tightly during the three catalytic steps of its 14α-demethylation (24) might be mildly affected. Lower binding efficiency correlated well with the about 3-fold slower catalytic turnover (Figure 5, Table 2). Regardless of its lower enzymatic activity, in the presence of equimolar amounts of VNI or its triazole derivative VNT Y-CYP51A retained the ability to metabolize eburicol (26 and 40% conversion per hour, respectively), while Tulahuen CYP51 was completely inhibited at these conditions (Figure 6, Table 2). In fact, amongst all tested inhibitors that are known to be highly potent against Tulahuen CYP51 (18, 20, 25), only VFV maintained essentially the same strength against both CYP51 orthologs. The apparent binding efficiencies for VNI, VFV and VNT, calculated from the Y-CYP51A titration curves (9, 87 and 2, respectively, Figure 7), are in good agreement with their inhibitory effects on the enzymatic activity. Cellular experiments in insect and human stages of Y T. cruzi also reflected higher antiparasitic potency of VFV (Figure 8). Collaborative studies on comparative testing of VNI and VFV in animal models of Chagas infection with the Y strain T. cruzi are currently underway and strongly support this observation (the results will be reported elsewhere).
Figure 4. Spectral characteristics of Y-CYP51A.
A. Absolute absorbance spectrum (270–700 nm; spectrophotometric index A417/A280=1.42; ΔA(390–460)/ΔA(417–460)=0.41) and difference reduced carbon monoxide binding spectrum (400500 nm). [P450] = 2.19 µM. B. Type 1 spectral response to the binding of eburicol. [P450] = 1.6 µM. Upper: absolute absorbance spectra, the Soret band maximum shifts to the left. Lower: difference spectra upon titration; titration step 0.5 µM; titration range 0.5–5.0 µM. Optical path length 1 cm.
Table 2.
Substrate binding, catalytic activity and inhibition of Y-CYP51A comparatively to Tulahuen CYP51.
| CYP51 ortholog |
Substrate binding parameters | Turnover number (nmol substrate/nmol P450 /min) |
Inhibition at I/E/S molar ratio 1:1:100, one hour reaction (%) |
||||
|---|---|---|---|---|---|---|---|
| Kd, (µM) |
ΔA (% low-to- high spin transition) |
Binding efficiency (ΔA/Kd) |
VNI | VNT | VFV | ||
| Tulahuen | 0.8 | 36 | 45 | 5.6±0.3 | 100 | 100 | 100 |
| Y-CYP51A | 1.54 | 48 | 31 | 2.0±0.2 | 51±2 | 35±3 | 99±0.8 |
Figure 5. Catalytic activity of Y-CYP51A in comparison with Tulahuen CYP51, time-course.
[P450]= 0.5 µM; [eburicol]=50 µM.
Figure 6. HPLC profiles of eburicol 14α-demethylation by Tulahuen CYP51 and Y-CYP51A.
1 h reaction; molar ratio enzyme: inhibitor substrate = 1 : 1 : 100. Triazole posaconazole (pos) and a pyridine derivative UDO [3zg2] were used as controls. S-substrate; I, 14α-carboxyaldehyde intermediate; P-14α-demethylated product.
Figure 7. Spectral responses of Y-CYP51A to the binding of VNI, VFV and VNT.
[P450] = 0.4 µM. Optical path length 5 cm. Upper: absolute absorbance spectra, the Soret band maximum shifts to the right. Lower: difference spectra upon titration with the inhibitors; titration step 0.2 µM, titration range 0.1–1.3 µM. Compounds were added to the sample cuvette from 0.2 mM stock solutions in DMSO. At each step, the corresponding volume of DMSO was added to the reference cuvette. Insets: titration curves.
Figure 8. Comparative effects of VNI and VFV in Y strain epimastigotes (A) and amastigotes (B).
A. 1×106 epimastigotes were cultured in the presence of 1 µM VNI, VFV or 1% DMSO (Ctr). Each 48 h medium was replaced by fresh medium at the same conditions. Parasites were counted each day in a Neubauer chamber. B. H9C2 cells were infected with trypomastigotes (MOI=10). After 24 h the medium was replaced by fresh medium containing VNI or VFV. After 48 h of incubation with the compounds the cells were fixed and prepared for microscopic analysis.
4. Discussion
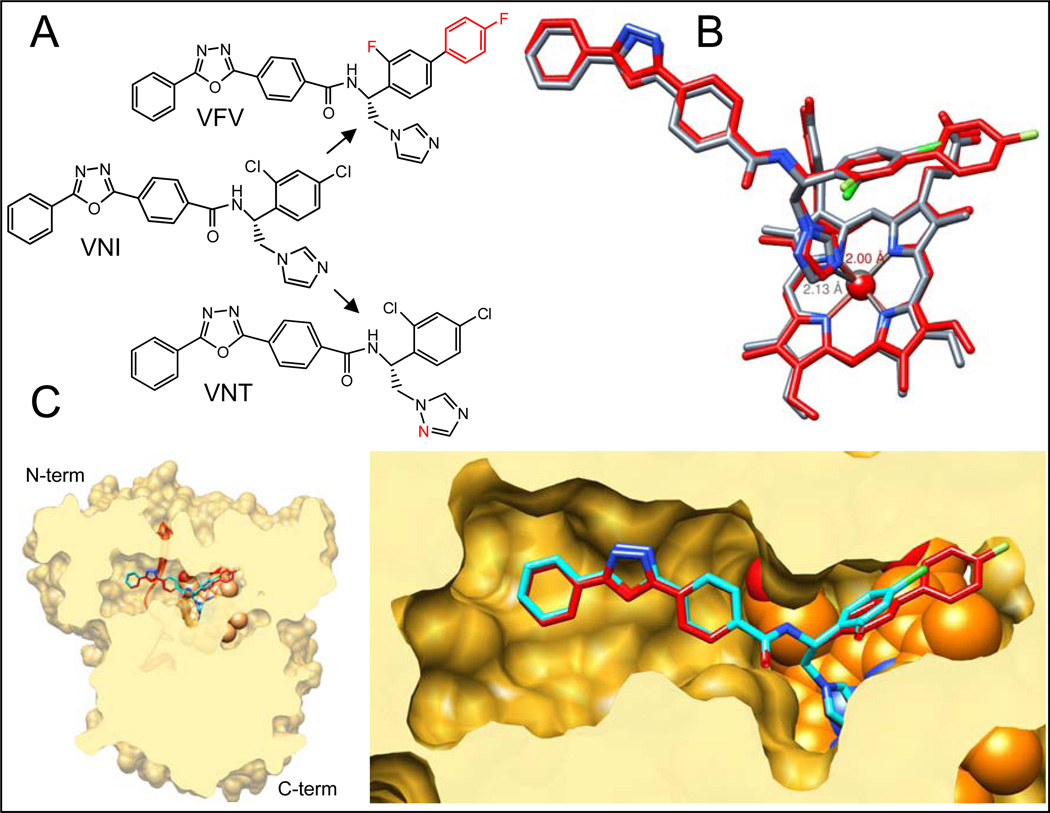
VNT and VFV (Figure 9A) were designed with different purposes. Lower basicity of the VNT triazole nitrogen weakens the Fe-N coordination bond (Figure 9B) therefore increasing the role of van der Waals contacts and topological fit between the inhibitor and apoprotein (18) and leading to higher target selectivity. VNT was proven to be as potent as VNI for Tulahuen T. cruzi CYP51, yet has weaker inhibitory effect on the enzymes from T. brucei and L. infantum (25). The second aromatic ring of the VFV biphenyl fragment was added to the molecule in order to fill the deepest segment of the CYP51 binding cavity, which we believed should broaden its spectrum of activity. The inhibitor was indeed confirmed as equally potent for all three tested protozoan CYP51s (25). P355S substitution in Y-CYP51A"softening” the surface of interaction around the inhibitor carboxamide fragment and its hydrogen bond network with the enzyme I helix (Figure 1), appears to somewhat increase the impact of the Fe-N coordination bond in the inhibitor-enzyme interaction. As a result VNI shows higher potency than VNT, yet additional contacts with the protein provided by the distal ring of the biphenyl moiety of VFV (Figure 9C) are required to completely prevent the substrate from being able to replace the inhibitor in the reconstituted enzymatic reaction.
Figure 9. CYP51 structure based VNI scaffold development.
A. Structural formulas of VNI and its derivatives VNT and VFV. B. Superimposition of CYP51 complexes with VNT (grey) and VFV (red). The length of the Fe-N coordination bond in each complex is marked respectively. C. Slice through the binding cavity of CYP51 complexes with VNI (cyan) and VFV (red); surface representation. Left: overall structure, distal view. The SRS5 area (the C-terminal portion of helix K – β strand 1–4) is depicted as red ribbon. Right: enlarged view of the active site. The heme is shown as orange spheres.
CYP51 is a highly conserved housekeeping gene found in all biological kingdoms. It is constitutively expressed in all organisms that produce endogenous sterols (26). Animals have one CYP51 gene. Some filamentous ascomycetes (such as Aspegrillus, Figure 2) have two or even three CYP51 genes (Fusarium (27)). While gene B is expressed constitutively, gene A was reported to be inducible, over-expressed at the conditions when a faster sterol flow is required (27, 28). Multiple CYP51 genes are found in some plants (e.g. rice, potato), though the reasons for that remain to be understood. While across the kingdoms CYP51s have low sequence similarity (26), the identity in closely related species is very high: e.g., CYP51s from human and chimpanzee have only three amino acid differences, CYP51s from human and dog differ in 9 residues (Supplemental Figure 2). Leishmania and T. brucei species have only one CYP51 gene. CYP51s from L. infantum and L. donovani (both pathogens cause visceral leishmaniasis) are identical. CYP51s from T. brucei brucei (causes nagana in cattle) and T. brucei gambiense (causes sleeping sickness in humans) have one amino acid difference (29).
Surprisingly high variability observed in CYP51s of T. cruzi (Table 1, 16 amino acid differences between Y-CYP51A and Marinkellei CYP51 (Supplemental Figure 3)) suggests that these organisms are likely to represent different species rather than strains (which in turn would logically explain the profound differences in the disease progression), and CYP51 can potentially serve as one of genetic markers. Sequencing of CYP51 genes from multiple T. cruzi organisms should be helpful in estimating the evolutionary distances between them and might be used for their more meaningful classification.
Heterogeneity of T. cruzi population may explain both the controversy of the outcomes from posaconazole clinical trials and the striking differences in the potencies of pyridine derivatives UDO and UDD (highly selective for Tulahuen T. cruzi CYP51 (18)), recently observed upon their testing in various T. cruzi strains (30). Although more studies are needed for establishing to what extent the genetic variability of T. cruzi correlates with response to drugs, it appears that the design of CYP51 inhibitors aimed at serving as antichagasic chemotherapies should be more successful if directed toward their broader spectrum of action rather than in pursuit of single-target selectivity. Ideally, new drugs should be active against all circulating variety of the parasite.
Supplementary Material
Highlights.
VNI that cures Tulahuen T. cruzi infection was found less potent against strain Y.
Amplification of CYP51 from the Y strain revealed two genes, A and B.
Y-CYP51A has a P355S substitution, which decreases its sensitivity to inhibition.
Weaker drug sensitivity of Y-CYP51A may be due to its elevated flexibility.
CYP51 structure based VNI modification produces a derivative of higher efficiency.
Acknowledgements
This work was supported by National Institutes of Health, grant R01 GM067871 (to G.I.L.)
Abbreviations
- CYP
cytochrome P450, gene or protein
- T. cruzi, Trypanosoma cruzi, EC50
drug concentration that gives half-maximal response in cellular growth reduction
- GFP
green fluorescent protein
- SRS
substrate recognition site
- VNI
((R)-N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide)
- VNT
((R)-N-(1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide
- VFV
((R)-N-(1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lepesheva GI. Design or screening of drugs for the treatment of Chagas disease: what shows the most promise? Expert Opin Drug Discov. 2013;8:1479–1489. doi: 10.1517/17460441.2013.845554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chagas C. Nova trypanozomiaze humana: estudos sobre a morfolojia e o ciclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbid do homem. Mem. Inst. Oswaldo Cruz. 1909;1:159–218. [Google Scholar]
- 3.WHO. 2014 ( http://www.who.int/mediacentre/factsheets/fs340/en/) Chagas disease (American trypanosomiasis); Fact sheet 340: Updated May.
- 4.Hotez PJ. Neglected infections of poverty in the United States of America. PLoS Negl Trop Dis. 2008;2:e256. doi: 10.1371/journal.pntd.0000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanford EJ, Zhan FB, Lu Y, Giordano A. Chagas disease in Texas: Recognizing the significance and implications of evidence in the literature. Soc Sci Med. 2007;65:60–79. doi: 10.1016/j.socscimed.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 6.Urbina JA, Payares G, Sanoja C, Lira R, Romanha AJ. In vitro and in vivo activities of ravuconazole on Trypanosoma cruzi, the causative agent of Chagas disease. Int J Antimicrob Agents. 2003;21:27–38. doi: 10.1016/s0924-8579(02)00273-x. [DOI] [PubMed] [Google Scholar]
- 7.Urbina JA. Ergosterol biosynthesis and drug development for Chagas disease. Mem Inst Oswaldo Cruz. 2009;104(Suppl 1):311–318. doi: 10.1590/s0074-02762009000900041. [DOI] [PubMed] [Google Scholar]
- 8.Urbina JA, Payares G, Contreras LM, Liendo A, Sanoja C, Molina J, Piras M, Piras R, Perez N, Wincker P, Loebenberg D. Antiproliferative effects and mechanism of action of SCH 56592 against Trypanosoma (Schizotrypanum) cruzi: in vitro and in vivo studies. Antimicrob Agents Chemother. 1998;42:1771–1777. doi: 10.1128/aac.42.7.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton J. Chagas disease: pushing through the pipeline. Nature. 2010;465:S12–S15. doi: 10.1038/nature09224. [DOI] [PubMed] [Google Scholar]
- 10.Molina I, Gómez i Prat J, Salvador F, Treviño B, Sulleiro E, Serre N, Pou D, Roure S, Cabezos J, Valerio L, Blanco-Grau A, Sánchez-Montalvá A, Vidal X, Pahissa A. Randomized Trial of Posaconazole and Benznidazole for Chronic Chagas' Disease. New England Journal of Medicine. 2014;370:1899–1908. doi: 10.1056/NEJMoa1313122. [DOI] [PubMed] [Google Scholar]
- 11.Villalta F, Dobish MC, Nde PN, Kleshchenko YY, Hargrove TY, Johnson CA, Waterman MR, Johnston JN, Lepesheva GI. VNI cures acute and chronic experimental Chagas disease. J Infect Dis. 2013;208:504–511. doi: 10.1093/infdis/jit042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filardi LS, Brener Z. Susceptibility and natural resistance of Trypanosoma cruzi strains to drugs used clinically in Chagas disease. TransRSoc. Trop. Med. Hyg. 1987;81:755–759. doi: 10.1016/0035-9203(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 13.Murta SMF, Gazzinelli RT, Brener Z, Romanha AJ. Molecular characterization of susceptible and naturally resistant strains of Trypanosoma cruzi to benznidazole and nifurtimox. Mol Biochem Parasitol. 1998;93:203–214. doi: 10.1016/s0166-6851(98)00037-1. [DOI] [PubMed] [Google Scholar]
- 14.Soeiro MdNC, de Souza EM, da Silva CF, Batista DdGJ, Batista MM, Pavão BP, Araújo JS, Lionel J, Britto C, Kim K, Sulikowski G, Hargrove TY, Waterman MR, Lepesheva GI. In vitro and in vivo Studies of the Antiparasitic Activity of Sterol 14α-Demethylase (CYP51) Inhibitor VNI against Drug-Resistant Strains of Trypanosoma cruzi. Antimicrob Agents Chemother. 2013;57:4151–4163. doi: 10.1128/AAC.00070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barclay JJ, Morosi LG, Vanrell MC, Trejo EC, Romano PS, Carrillo C. Trypanosoma cruzi Coexpressing Ornithine Decarboxylase and Green Fluorescence Proteins as a Tool to Study the Role of Polyamines in Chagas Disease Pathology. Enzyme Research. 2011;2011:10. doi: 10.4061/2011/657460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medina-Acosta E, Cross GA. Rapid isolation of DNA from trypanosomatid protozoa using a simple 'mini-prep' procedure. Mol Biochem Parasitol. 1993;59:327–330. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- 17.Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, Liu J, Hill GC, Waterman MR. CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B' helix defines substrate preferences of sterol 14alpha-demethylase. J Biol Chem. 2006;281:3577–3585. doi: 10.1074/jbc.M510317200. [DOI] [PubMed] [Google Scholar]
- 18.Hargrove TY, Wawrzak Z, Alexander PW, Chaplin JH, Keenan M, Charman SA, Waterman MR, Chatelain E, Lepesheva GI. Complexes of Trypanosoma cruzi Sterol 14α-Demethylase (CYP51) with Two Pyridine-based Drug Candidates for Chagas Disease: Structural basis for pathogen selectivity. J Biol Chem. 2013;288:31602–31615. doi: 10.1074/jbc.M113.497990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lepesheva GI, Ott RD, Hargrove TY, Kleshchenko YY, Schuster I, Nes WD, Hill GC, Villalta F, Waterman MR. Sterol 14 alpha-demethylase as a potential target for antitrypanosomal therapy: Enzyme inhibition and parasite cell growth. Chem Biol. 2007;14:1283–1293. doi: 10.1016/j.chembiol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lepesheva GI, Hargrove TY, Anderson S, Kleshchenko Y, Furtak V, Wawrzak Z, Villalta F, Waterman MR. Structural insights into inhibition of sterol 14 alphademethylase in the human pathogen Trypanosoma cruzi. J Biol Chem. 2010;285:25582–25590. doi: 10.1074/jbc.M110.133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lepesheva GI, Park HW, Hargrove TY, Vanhollebeke B, Wawrzak Z, Harp JM, Sundaramoorthy M, Nes WD, Pays E, Chaudhuri M, Villalta F, Waterman MR. Crystal structures of Trypanosoma brucei sterol 14 alpha-demethylase and implications for selective treatment of human infections. J Biol Chem. 2010;285:1773–1780. doi: 10.1074/jbc.M109.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 23.Lepesheva GI, Waterman MR. Sterol 14alpha-demethylase (CYP51) as a therapeutic target for human trypanosomiasis and leishmaniasis. Curr Top Med Chem. 2011;11:2060–2071. doi: 10.2174/156802611796575902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lepesheva GI, Villalta F, Waterman MR. Targeting Trypanosoma cruzi sterol 14α-demethylase (CYP51) Adv Parasitol. 2011;75:65–87. doi: 10.1016/B978-0-12-385863-4.00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hargrove TY, Kim K, de Nazaré Correia Soeiro M, da Silva CF, da Gama Jaen Batista D, Batista MM, Yazlovitskaya EM, Waterman MR, Sulikowski GA, Lepesheva GI. CYP51 structures and structure-based development of novel, pathogen-specific inhibitory scaffolds. Int J Parasitol Drugs Drug Resist. 2012;2:178–186. doi: 10.1016/j.ijpddr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepesheva GI, Waterman MR. Structural basis for conservation in the CYP51 family. Biochim Biophys Acta. 2011;1814:88–93. doi: 10.1016/j.bbapap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan J, Urban M, Parker JE, Brewer HC, Kelly SL, Hammond-Kosack KE, Fraaije BA, Liu X, Cools HJ. Characterization of the sterol 14α-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytologist. 2013;198:821–835. doi: 10.1111/nph.12193. [DOI] [PubMed] [Google Scholar]
- 28.Hawkins NJ, Cools HJ, Sierotzki H, Shaw MW, Knogge W, Kelly SL, Kelly DE, Fraaije BA. Paralog Re-Emergence: A Novel, Historically Contingent Mechanism in the Evolution of Antimicrobial Resistance. Mol Biol Evol. 2014;31:1793–1802. doi: 10.1093/molbev/msu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, Lepesheva GI. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14α-demethylase (CYP51) from Leishmania infantum. J Biol Chem. 2011;286:26838–26848. doi: 10.1074/jbc.M111.237099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moraes CB, Giardini MA, Kim H, Franco CH, Araujo-Junior AM, Schenkman S, Chatelain E, Freitas-Junior LH. Nitroheterocyclic compounds are more efficacious than CYP51 inhibitors against Trypanosoma cruzi: implications for Chagas disease drug discovery and development. Sci. Rep. 2014;4 doi: 10.1038/srep04703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.