Abstract
Introduction
Alterations inside Polycytosine tract (C-tract) of mitochondrial DNA (mtDNA) have been described in many different tumor types. The Poly-Cytosine region is located within the mtDNA D-loop region which acts as point of mitochondrial replication origin. A suggested pathogenesis is that it interferes with the replication process of mtDNA which in turn affects the mitochondrial functioning and generates disease.
Methodology
100 premalignant cases (50 leukoplakia & 50 oral submucous fibrosis) were selected and the mitochondrial DNA were isolated from the lesion tissues and from the blood samples. Polycytosine tract in mtDNA was sequenced by direct capillary sequencing.
Results
40 (25 leukoplakia & 15 oral submucous fibrosis) patients harbored lesions that displayed one additional cytosine after nucleotide thymidine (7CT6C) at nt position 316 in C-tract of mtDNA which were absent in corresponding mtDNA derived from blood samples.
Conclusion
Our results show an additional cytosine in the mtDNA at polycytosine site in oral precancer cases. It is postulated that any increase/decrease in the number of cytosine residues in the Poly-Cytosine region may affect the rate of mtDNA replication by impairing the binding of polymerase and other transacting factors. By promoting mitochondrial genomic instability, it may have a central role in the dysregulation of mtDNA functioning, for example alterations in energy metabolism that may promote tumor development. We, therefore, report and propose that this alteration may represent the early development of oral cancer. Further studies with large number of samples are needed in to confirm the role of such mutation in carcinogenesis.
Keywords: C-tract, D-loop, Mitochondria, Oral precancer
1. Introduction
Oral cancer is the 6th most common cancer globally.1 Genetic progression model predicted for many cancers point to sequential accumulation of genetic changes. The same is true for oral cancer. Though progression from normal to cancer through precancer stage in oral cavity has been worked but that is at cytogenetic level.1 Consensus about genetic changes at molecular level is lacking. Several studies have tried to find molecular signatures for oral cancer carcinogenesis2,3 but they still are deficient, first, in the non-homogeneity of the samples, and, second, in detailing molecular signatures that are inherently transient and lack temporal reproducibility. Recently, various studies have revisited the role of mitochondria in health and disease, in general.4–8
Mitochondria has an important role to play in energy metabolism, the generation of ROS, aging, and the initiation of apoptosis. Due to their role on control of cell death, mitochondria have been implicated in tumorigenesis.4,6 Since mitochondrial DNA mutations in OSCC were first described,9 today we know that changes in Mitochondrial DNA (mtDNA) include addition and small & large scale deletions in oral tumors and surrounding tissues.10–13 Most of the mutations reported have been in the D-loop region, which is a hypervariable non-coding region.14,15 Inside D-loop polycytosine tract is a stretch of mononucleotide repeats which has been reported to be associated with oral cancer progression.16 But such an association is incomplete without including precancer cases. Therefore functional significance of these mutations in tumorigenesis is still a mystery. The present study was therefore initiated to investigate the association of C-tract alterations of mitochondrial DNA with various stages of oral precancer.
2. Materials and methods
A hospital based study was conducted at King George's Medical University, Lucknow, India. The study comprised 100 oral precancer cases (50 leukoplakia, 50 oral submucous fibrosis) visiting our out-patient clinic in Department of Oral & Maxillofacial Surgery, for management of oral precancer. The diagnosis of leukoplakia of the oral cavity, was confirmed by established clinical guidelines and histopathological examinations. OSMF patients, classified on the basis of following criteria were recruited: decreased mouth opening (<20 mm interincisal distance), palpable fibrous bands in buccal mucosa, blanched oral mucosa (soft palate, buccal, labial, retromolar), reduced elasticity of mucosa, burning sensation of the oral mucosa, and restricted tongue movements. Tissue and blood samples were obtained during the routine clinical management of the cases after obtaining their informed consent. The blood was taken to serve as self matched control. Mitochondrial DNA (mtDNA) was extracted from all tissue and blood samples cases. Samples were anonymized before processing and approval for the study was obtained from the local research ethics committee.
2.1. DNA extraction
For tissue sample, DNA was extracted from preselected regions of precancer lesion using 20 mg of formalin fixed tissue. Briefly, tissue sections were washed 6 times with 500 μl 1× GTE (100 mM glycine; 10 mM Tris–HCL pH 8.0; 1 mM EDTA) every 12 h and subsequently put with 300 μl Cell Lysis Solution (10 mM Tris–HCl pH 8.0, 0.1 mM EDTA pH 8.0; 0.5% SDS) for 1 h at 55 °C. To the solution 10 μl of proteinase K (10 mg/ml) were added every 12 h and kept at 55 °C upto 48 h. DNA was subsequently isolated by phenol/chloroform method and re-suspended in 200 ul TE buffer. The whole genomic DNA was extracted from blood using standard phenol chloroform method.
2.2. Mitochondrial DNA C-tract amplification & mitochondrial DNA C-tract sequencing
The Mitochondrial DNA C-tract amplification was done using set of primer (Table 1) yielding a 109-bp fragment.11 Bidirectional direct capillary was done using BigDye® Direct Cycle Sequencing, Applied Biosystems, USA using manufacturer's recommended protocol. Both Forward and Reverse primers were used to analyze and confirm the sequences in region of interest.
Table 1.
Primer sequence details along with PCR conditions followed.
| Primer | Sequence | Amplification conditions |
|---|---|---|
| Forward Reverse |
5′ ACAATTGAATGTCTGCACAGCCACTT 3′ 5′ GGCAGAGATGTGTTTAAGTGCTG 3′ |
94 °C for 5 min, 35 cycles of 94 °C for 45 s, 58 °C for 45 s and 72 °C for 45 s, followed by 72 °C for 10 min. |
2.3. Statistical methods
The main statistical objective in this study was the association of mitochondrial DNA mutations with histopathological diagnosis of premalignant oral precancer lesions. Patients were categorized into three groups according to lesion progression: (a) mild dysplasia; (b) moderate dysplasia; and (c) severe dysplasia. The association of mitochondrial alteration with increasing severity of dysplasia was based on a cross-tabulation and a logistic regression model. All statistical computations were performed using the SAS system version 16, and all Ps reported are two sided.
3. Results
DNA from 100 premalignant lesions and corresponding blood samples with no history of prior or concurrent malignancy was analyzed for mitochondrial C-tract alterations. There were 50 cases of oral leukoplakia and 50 cases of oral submucous fibrosis. After histopathological review, there were 31 mild dysplasia, 33 moderate dysplasias, and 36 severe dysplasias. To avoid overestimation the mutation rate in low grade lesions the most histologically severe sample was analyzed. Overall, 40 of 100 (40%) patients harbored premalignant lesions that exhibited an additional cytosine at nt position 316 that falls inside polycytosine tract of mitochondrial DNA when compared with uninvolved DNA from corresponding blood sample. Fig. 1 demonstrates sequencing chart with additional C-tract region in mtDNA from lesion while Fig. 2 demonstrates normal number of cytosines in corresponding blood sample. Of the 40 samples that had additional cytosine, there were 25 sample from oral leukoplakia and 15 from oral submucous fibrosis (Table 2). The probability of having additional cytosine mitochondrial increased with increasing severity of dysplasia in both cases when group is subdivided into leukoplakia & oral submucous fibrosis and when as a whole (Tables 3 and 4). When taken as whole, with reference to the mild dysplasia group, there was modest, though statistically not significant increased risk for having additional cytosine in mtDNA C-tract, ORs 1.22 (95% CI: 0.42–3.53). The risk for having additional cytosine in mtDNA C-tract increased to 3.05 (95% CI: 01.11–8.44) in the severe dysplasia group which was statistically significant, P < 0.05.
Fig. 1.
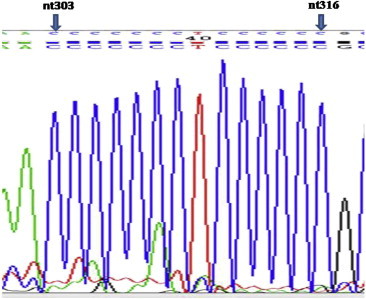
Tissue sample mitochondrial C-tract sequence of showing 6th (additional cytosine) at nt316.
Fig. 2.
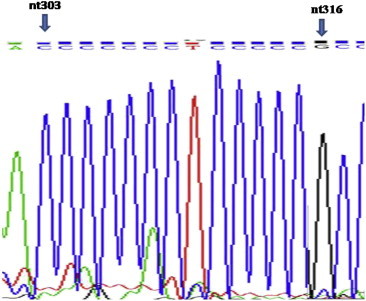
Blood sample mitochondrial C-tract sequence of same case showing normal cytosine number.
Table 2.
Cross-tabulation according to precancer type having additional mt C-tract cytosine.
| Precancer type | Additional cytosine in tissue samples present or absent |
Total | |
|---|---|---|---|
| Present | Absent | ||
| Leukoplakia | 25 | 25 | 50 |
| Oral submucous fibrosis | 15 | 35 | 50 |
| Total | 40 | 60 | 100 |
Table 3.
Distribution of additional mt C-tract cytosine histopathological gradewise in each precancer type.
| Precancer type | Histopathological grade | Number of cases having additional cytosine | Percentage | ORa (95% CI) |
|---|---|---|---|---|
| Leukoplakia | Mild | 5 | 33.3% | 1.00 |
| Moderate | 8 | 47% | 1.78 (0.42–7.46) | |
| Severe | 12 | 66.7% | 4 (0.93–17.11) | |
| Oral submucous fibrosis | Mild | 4 | 25% | 1.00 |
| Moderate | 3 | 18.8% | 0.69 (0.12–3.75) | |
| Severe | 8 | 44.4% | 2.4 (0.55–10.38) |
ORs determined by comparing each histologic category with mild dysplasia.
Table 4.
Distribution of additional mt C-tract cytosine histopathological gradewise in total cases.
| Histopathological grading | Total cases | Additional cytosine in tissue samples |
ORa (95% CI) | |
|---|---|---|---|---|
| Number | Overall frequency | |||
| Mild | 31 | 9 | 9/31 (29%) | 1.00 |
| Moderate | 33 | 11 | 11/33 (33%) | 1.22 (0.42–3.53) |
| Severe | 36 | 20 | 20/36 (55%) | 3.05b (1.06–8.44) |
| Total | 40 | 40/100 (40%) | ||
ORs determined by comparing each histologic category with mild dysplasia.
Statistically significant: P < 0.05.
4. Discussion
OSCC is often preceded by a premalignant lesion which is a morphologically altered tissue in which cancer is more likely to occur than in its apparently normal counterpart. Histologically, oral precancer shows features of epithelial dysplasia. Moderate and severe epithelial dysplasia carries the highest risk for malignant transformation. Malignant transformation occurs in around 4%–8% of oral leukoplakias and the same of OSMF ranges from 3% to 19%.17 The clinical criteria by which a white patch may be judged as premalignant are far from clear, and it is only after a study of the histological appearance of the lesion that any attempt at prognosis can be made. However, once cancer develops and progresses, prognosis gets poorer. Prognosis depends on the early detection of the lesion before the involvement of cervical nodes. Five-year survival rate is effectively doubled if the lesion is detected early. But to date, there are no reliable clinical or histological markers that can be used to predict whether the lesion will regress spontaneously, remain the same, or progress to cancer.
Oral premalignant lesions provide an excellent model of cancer progression because of the well described molecular progression pathways and defined histopathological appearance. There have been numerous reports delineating genetic changes in oral cancer progression including that of mitochondrial genome10–15,18; but only a few of them focus on premalignant phase.16 The data in this paper points to the fact that genetic changes in mitochondrial DNA are early events in the progression of oral premalignant lesions. Overall, 40%of premalignant lesions harbored a C-tract alteration. The alteration was in the form of an additional cytosine at nucleotide position 316. The prevalence of this alteration increased from mild hyperplasia to severe dysplasia meaning that the mitochondrial C-tract mutation can be a malignancy progression marker, although additional studies need to be performed.
Mitochondrial DNA represents less than 1% of total cellular DNA, but its gene products are essential for normal cell function. Mitochondria are semi-autonomous organelles performing essential functions of cellular metabolism, free radical generation, apoptosis, and regulation of cell death. Mitochondrial DNA mutations/deletions occur at a frequency of less than 1% and increase with advancing age.19 Defects in mitochondrial function have long been suspected to contribute to the development and progression of cancer. Most cancer cells rely on aerobic glycolysis, a phenomenon termed “the Warburg effect.” The Warburg effect may simply be a consequence of damage to the mitochondria in cancer, or an adaptation to low-oxygen environments within tumors, or a result of cancer genes shutting down the mitochondria because they are involved in the cell's apoptosis program which would otherwise kill cancerous cells. Considering mtDNA lacks introns, most mutations occur in the coding sequences and are thus, likely to be of biological consequence.20 mtDNA mutations can be either deleterious (pathogenic), neutral or beneficial (adaptive). Positive selection of an adaptive mutation in the mtDNA leads to its enrichment in the population.16 Moreover, computer models and in vitro studies have shown that mitochondrial homoplasmicity can occur simply by chance due to clonal selection of a particular cell.16,21,22 Probably, this can be an explanation for the homoplasmic nature of mitochondrial DNA alterations in the form an additional cytosine in C-tract observed in our samples.
As the mtDNA does not recombine, all of the sequence variants linked to adaptive mutations are also enriched, a phenomenon known as ‘Hitchhiking’.23 The collection of linked variants on an individual mtDNA molecule is called a mtDNA haplotype, and the descendants of an original mutant mtDNA will acquire additional variants to generate a group of related haplotypes, referred to as a haplogroup. These haplogroups frequently constitute region-specific branches of the mtDNA tree. C-tract region is a known polymorphic site and variations in population vary from seven to nine cytosine residues. Moreover, this region lies inside D-loop which is itself highly unstable part of mitochondrial DNA. In our observation, there is an additional cytosine at a unique position 316, different from usual polymorphic site i.e. 306–10 which still has not been reported widely. To the best of our knowledge, we report it for first time in Indian population. However, the potential role of this alteration in oral tumorigenesis is still unknown.
In all samples a 1 bp expansion was observed, which can be considered as wild type distribution. Therefore functional significance of this is unclear. It is postulated that alteration in the number of cytosine residues in the Poly-Cytosine region may affect the mtDNA replication rate by impairing the binding of polymerase and other transacting factors.20,24 Mitochondrial DNA is 10–100 times more vulnerable to mutations than nuclear DNA due to lack of histone protection, limited repair capacity and close proximity to the electron transport chain which constantly generates superoxide radicals. Tobacco carcinogens are known source of oxidative stress and thus may have central role in promoting mitochondrial genomic instability.10,12,13 All our subjects had history of tobacco consumption. Therefore any alteration in polycytosine site is a result of mitochondrial genomic instability, and therefore it may have a role in the dysregulation of mtDNA functioning, for example alterations in energy metabolism may promote tumor development. In this context, Mitochondrial C-tract alterations can be seen as result of alterations in the maintenance of genetic stability. Therefore, C-tract alterations can occur indicating functionality of the mitochondria.
Though it is possible that at least some of the ‘tumor-specific’ variants can be sequencing errors, but the strong association between ‘tumor-specific’ variants and population polymorphisms cannot be completely spurious. The biological relevance of the apparent association between ‘tumor-specific somatic mutations’ and population variants is supported by a review of selected individual tumor mutations that alter the amino-acid sequence of mitochondrial mRNA genes. The most compelling evidence is a population variant seen for the nt 13708 G to A missense mutation in ND5 (A458T) reported in a breast cancer.25 Such compelling evidence is still lacking in the case of oral cancer. The data here shows that there is an increased chance of having mitochondrial alteration with increased histological progression toward malignancy. Our data shows that the mitochondrial C-tract alteration is an early cellular response to carcinogens, with an increased likelihood of alteration as the histological grade increases in oral premalignant lesions. However, more studies with larger sample size are needed to detail the exact mechanism for this additional cytosine, and in general other alterations, that may play in oral precancer progression and assess their potential clinical significance.
Conflicts of interest
All authors have none to declare.
Acknowledgments
Rahul Pandey and Divya Mehrotra are thankful to the grant from Indian Council of Medical Research, New Delhi.
References
- 1.Zhang L., Poh C.F., Williams M. Loss of heterozygosity (LOH) profiles–validated risk predictors for progression to oral cancer. Cancer Prev Res (Phila) 2012;5(9):1081–1089. doi: 10.1158/1940-6207.CAPR-12-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan M., Myers J.N., Agrawal N. Oral cavity and oropharyngeal squamous cell carcinoma genomics. Otolaryngol Clin North Am. 2013;46(4):545–566. doi: 10.1016/j.otc.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.da Silva S.D., Ferlito A., Takes R.P. Advances and applications of oral cancer basic research. Oral Oncol. 2011;47(9):783–791. doi: 10.1016/j.oraloncology.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Gasparre G., Porcelli A.M., Lenaz G., Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol. 2013;5(2) doi: 10.1101/cshperspect.a011411. pii: a011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Hattab A.W., Scaglia F. Mitochondrial DNA depletion syndromes: review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics. 2013;10(2):186–198. doi: 10.1007/s13311-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace D.C. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grzybowska-Szatkowska L., Slaska B. Mitochondrial DNA and carcinogenesis. Mol Med Rep. 2012;6(5):923–930. doi: 10.3892/mmr.2012.1027. [DOI] [PubMed] [Google Scholar]
- 8.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fliss M.S., Usadel H., Caballero O.L. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287(5460):2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 10.Mondal R., Ghosh S.K. Accumulation of mutations over the complete mitochondrial genome in tobacco-related oral cancer from northeast India. Mitochondrial DNA. 2013;24(4):432–439. doi: 10.3109/19401736.2012.760551. [DOI] [PubMed] [Google Scholar]
- 11.Prior S.L., Griffiths A.P., Baxter J.M. Mitochondrial DNA mutations in oral squamous cell carcinoma. Carcinogenesis. 2006;27(5):945–950. doi: 10.1093/carcin/bgi326. [DOI] [PubMed] [Google Scholar]
- 12.Tan D.J., Chang J., Chen W.L. Somatic mitochondrial DNA mutations in oral cancer of betel quid chewers. Ann N Y Acad Sci. 2004;1011:310–316. doi: 10.1007/978-3-662-41088-2_30. [DOI] [PubMed] [Google Scholar]
- 13.Lee H.C., Yin P.H., Yu T.N. Accumulation of mitochondrial DNA deletions in human oral tissues – effects of betel quid chewing and oral cancer. Mutat Res. 2001;493(1–2):67–74. doi: 10.1016/s1383-5718(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 14.Sun Y., Yuan R.T., Chen W.T., Bu L.X., Jia M.Y. Screening of the gene mutation in D-loop region of mitochondrial DNA in oral squamous cell carcinoma. Zhonghua Kou Qiang Yi Xue Za Zhi. 2013;48(5):285–287. doi: 10.3760/cma.j.issn.1002-0098.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Liu S.A., Jiang R.S., Chen F.J., Wang W.Y., Lin J.C. Somatic mutations in the D-loop of mitochondrial DNA in oral squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2012;269(6):1665–1670. doi: 10.1007/s00405-011-1806-5. [DOI] [PubMed] [Google Scholar]
- 16.Ha P.K., Tong B.C., Westra W.H. Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation. Clin Cancer Res. 2002;8(7):2260–2265. [PubMed] [Google Scholar]
- 17.Murti P.R., Bhonsle R.B., Pindborg J.J., Daftary D.K., Gupta P.C., Mehta F.S. Malignant transformation rate in oral submucous fibrosis over a 17-year period. Community Dent Oral Epidemiol. 1985;13(6):340–341. doi: 10.1111/j.1600-0528.1985.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 18.Lai C.H., Huang S.F., Liao C.T., Chen I.H., Wang H.M., Hsieh L.L. Clinical significance in oral cavity squamous cell carcinoma of pathogenic somatic mitochondrial mutations. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0065578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michikawa Y., Mazzucchelli F., Bresolin N., Scarlato G., Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999 Oct;286(5440):774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee A., Dasgupta S., Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res (Phila) 2011;4(5):638–654. doi: 10.1158/1940-6207.CAPR-10-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Xing L., Mi J., Chen L., Tian Y. Mitochondrial DNA mutations may not be frequent in patients with oral cancer. Mitochondrial DNA. 2013 doi: 10.3109/19401736.2013.834427. PMID: 24047158 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22.Bandelt H.J., Salas A. Contamination and sample mix-up can best explain some patterns of mtDNA instabilities in buccal cells and oral squamous cell carcinoma. BMC Cancer. 2009;9:113. doi: 10.1186/1471-2407-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishmar D., Ruiz-Pesini E., Golik P. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100(1):171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Cespedes M., Parrella P., Nomoto S. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res. 2001;61(19):7015–7019. [PubMed] [Google Scholar]
- 25.Parrella P., Xiao Y., Fliss M. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61(20):7623–7626. [PubMed] [Google Scholar]


