SUMMARY
During protein synthesis, nascent polypeptide chains within the ribosomal tunnel can act in cis to induce ribosome stalling and regulate expression of downstream genes. The Staphylococcus aureus ErmCL leader peptide induces stalling in the presence of clinically important macrolide antibiotics, such as erythromycin, leading to the induction of the downstream macrolide resistance methyltransferase ErmC. Here, we present a cryo-electron microscopy (EM) structure of the erythromycin-dependent ErmCL-stalled ribosome at 3.9 Å resolution. The structure reveals how the ErmCL nascent chain directly senses the presence of the tunnel-bound drug and thereby induces allosteric conformational rearrangements at the peptidyltransferase center (PTC) of the ribosome. ErmCL-induced perturbations of the PTC prevent stable binding and accommodation of the aminoacyl-tRNA at the A-site leading to inhibition of peptide bond formation and translation arrest.
INTRODUCTION
Nascent polypeptide-mediated translation regulation can be an intrinsic property of the nascent chain or require an additional ligand, such as an amino acid or antibiotic (Ramu et al., 2009; Vázquez-Laslop et al., 2011). Similar to other inducible macrolide resistance genes, the Staphylococcus aureus ermC gene is controlled by programmed arrest during translation of the upstream ermCL leader peptide (Horinouchi and Weisblum, 1980; Iordanescu, 1976; Shivakumar et al., 1980) (Figure 1A): In the absence of erythromycin, ErmC expression is repressed because the ribosome-binding site (RBS) and AUG start codon of the ermC mRNA are sequestered in a stem-loop structure (Figure 1A). However, in the presence of sub-inhibitory concentrations of erythromycin, ribosomes translating the ErmCL leader peptide become stalled, leading to an alternative stem-loop structure in the mRNA that exposes the RBS and start codon of the ermC gene and thus allows ribosome binding and induction of ErmC expression (Figure 1A).
Figure 1. Cryo-EM structure of the ErmCL-SRC.
(A) Schematic for ermCL-dependent regulation of ermC translation in the presence of erythromycin (ERY). (B) Transverse section of the ErmCL-SRC, with 30S (yellow), 50S (grey), P-tRNA (green), ErmCL (teal) and ERY (red). (C) Zoom showing electron density (grey) and model for the ErmCL nascent chain (teal) attached to CCA-end of the P-tRNA (green). (D) as (B) but coloured according to local resolution. (E–H) Examples of electron density in the ErmCL-SRC map including (E) β-strands and (F) α-helix in ribosomal proteins, (G) rRNA helix and (H) coordinated Mg2+ ions. See also Figure S1.
Previous studies demonstrated that polymerization of the ErmCL nascent chain halts because the ribosome is unable to catalyze peptide bond formation between the 9 amino acid long ErmCL-tRNAIle (codon 9) in the ribosomal P-site and Ser-tRNASer (codon 10) in the A-site (Johansson et al., 2014; Vazquez-Laslop et al., 2008) (Figure 1A). Mutations of specific ErmCL amino acid residues located in the ribosomal exit tunnel of the ErmCL-SRC reduce or abolish stalling, as do mutations of certain ribosomal RNA (rRNA) nucleotides that comprise the tunnel wall (Johansson et al., 2014; Mayford and Weisblum, 1989; Vazquez-Laslop et al., 2011; Vazquez-Laslop et al., 2010; Vazquez-Laslop et al., 2008). Additionally, the chemical structure of the macrolide antibiotic can influence ribosome stalling (Vazquez-Laslop et al., 2011; Vazquez-Laslop et al., 2008). Collectively, these findings indicate that ribosome stalling results from interactions between the ErmCL leader peptide, the macrolide antibiotic and components of the ribosomal tunnel (Vázquez-Laslop et al., 2011), however, a structural basis for this complex interplay is lacking.
To elucidate the nature of these interactions and ascertain how they lead to inactivation of the PTC of the ribosome, we generated ErmCL-SRC for structural analysis by cryo-electron microscopy (EM). The structure reveals that the path of the ErmCL nascent polypeptide chain and its interactions with specific 23S rRNA nucleotides U2506, U2586 and A2062 within the ribosomal tunnel. Moreover, ErmCL is observed to directly interact with the cladinose sugar of erythromycin, thus revealing how the nascent chain monitors the presence of the tunnel-bound drug. Collectively, these interactions appear to stabilize a unique conformation of the ErmCL nascent chain that induces global rearrangements at the peptidyltransferase center (PTC) of the ribosome and prevent stable binding and accommodation of the A-tRNA, and thus induces translational arrest.
RESULTS
Cryo-EM structure of ErmCL-SRC
The ErmCL-SRC was generated by translation of a dicistronic 2XErmCL mRNA in the presence of 10 μM erythromycin using an E. coli lysate-based in vitro translation system. The ErmCL-SRC disomes were isolated by sucrose gradient purification, converted to monosomes and directly applied to cryogrids (Figure S1), as performed previously for ErmBL-SRC (Arenz et al., 2014). Data collection was performed on a Titan Krios TEM fitted with the Falcon II direct electron detector (FEI, Netherlands) and images were processed with SPIDER (Frank et al., 1996) (see Experimental Procedures). In silico sorting of cryo-EM images yielded one major homogeneous subpopulation of ribosomes bearing a P-tRNA, but lacking A-tRNA (Figure 1B,C), which contrasts with the previous cryo-EM reconstruction of the ErmBL-SRC that contained tRNAs in both the A- and P-site (Arenz et al., 2014). The ErmCL-SRC has an average resolution of 3.9 Å, while local resolution calculations indicate that the ribosomal core reaches 3.5 Å (Figure 1D and Figure S1). Rigid-body docking of crystallographic structures of the E. coli ribosome (Pulk and Cate, 2013) reveals excellent agreement with the cryo-EM map, such as strand separation in β-sheets (Figure 1E) and the pitch of α-helices (Figure 1F) within the ribosomal proteins, as well as density for the majority of the amino acid side chains (Figure 1E,F). Additionally, the rRNA backbone and nucleotides are well-resolved, as well as the position of many magnesium ions (Figure 1G,H). The distinct features of the electron density allowed the CCA-end of the P-tRNA to be accurately placed and a model for residues 3–9 of the ErmCL leader peptide to be built de novo (Figure 1C).
Critical interactions of ErmCL with components of the ribosomal tunnel
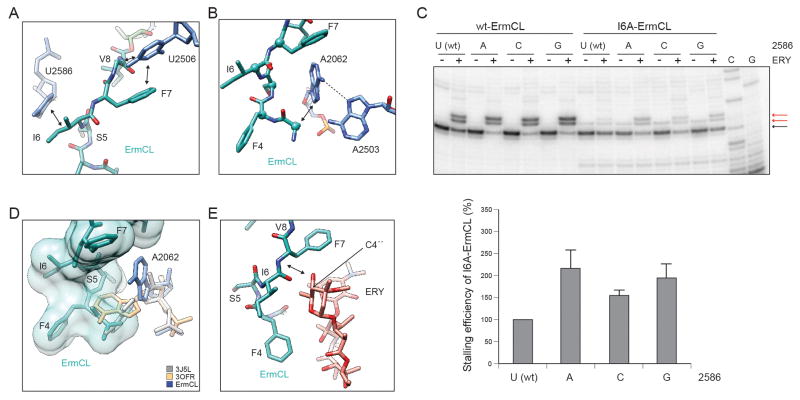
The overall path of the ErmCL nascent chain within the ribosomal tunnel is shifted towards erythromycin when compared with the path of ErmBL (Arenz et al., 2014) (Figure S2). Four sites of contact are observed between the ErmCL nascent chain and components of the ribosomal tunnel (Figure 2A,B and Figure S3): At the C-terminus of ErmCL, U2506 interacts with V8 and stacks upon the aromatic side chain of F7, whereas I6 interacts with U2586 (Figure 2A). These contacts are likely to be important since mutations to alanine in the conserved “IFVI” motif (I6–I9) of ErmCL severely reduce ribosome stalling (Johansson et al., 2014; Vazquez-Laslop et al., 2008). Although mutation of U2586 to A, C or G did not affect wild-type ErmCL stalling (Figure 2C), the stalling efficiency of the I6A-ErmCL mutant could be partially rescued by the U2586 mutations, especially U2586G (Figure 2C). This functional interplay between U2586 and I6 is consistent with the direct interaction observed in the structure (Figure 2A). To investigate this contact further, we generated all possible amino acid substitutions at position 6 of ErmCL and monitored the efficiency of stalling by toe-printing (Figure S4). The results indicated that the hydrophobic amino acids M, V and L (and to a lesser extent the charged amino acid E) at position 6 maintained efficient ribosome stalling, whereas all other substitutions further reduced the stalling efficiency.
Figure 2. Interaction of the ErmCL nascent chain within the ribosomal tunnel.
(A,B) ErmCL (teal) contacts (arrowed) with 23S rRNA nucleotides (A) U2506, U2586 and (B) A2062. (C) Toe-printing of wild-type (wt) and mutant (I6A) ErmCL translation on E. coli ribosomes with wt (U2586) or U2586A/C/G mutants, in the presence (+) or absence (−) of ERY. All reactions contained the Thr-tRNA synthetase inhibitor borrelidin which halts translation at the Thr (T11) codon in the absence of ERY-induced arrest at the Ile (I9) codon. Error bars represent one standard deviation of the mean. (D) Spacefill representation of ErmCL (teal) illustrating the steric clash with A2062 positions from PDB3OFR (Dunkle et al., 2010) (yellow) and ErmBL-SRC (PDB ID 3J5L, grey) (Arenz et al., 2014). For comparison, the A2062 position in ErmCL-SRC is shown (blue). (E) Sensing of the C3 cladinose sugar of ERY (Dunkle et al., 2010) (red) by the ErmCL nascent chain (teal). See also Figure S2–S4.
An interaction is also observed from the N-terminus of ErmCL with A2062 (Figure 2D), however, due to the poor density (indicating flexibility) of the N-terminus (Figure S3), we can only tentatively assign this interaction with A2062 to I3, and could not model the very N-terminal residues M1–G2. While alanine scanning mutagenesis indicates that the nature of the N-terminal residues G2–S5 is not critical for ribosome stalling (Johansson et al., 2014; Vazquez-Laslop et al., 2008), truncation of more than two N-terminal residues of ErmCL notably reduced stalling (Vazquez-Laslop et al., 2008), suggesting that the I3 position is at least sterically important for the stalling mechanism. In this regard, we note that in the ErmCL-SRC, A2062 is stabilized in a conformation that lies flat against the tunnel wall (Figure 2B and Figure S3), whereas the A2062 conformation that protrudes into the tunnel lumen observed in the majority of ribosome structures (Vazquez-Laslop et al., 2010), including the ErmBL-SRC (Arenz et al., 2014), would sterically clash with the ErmCL nascent chain (Figure 2D). In the flat conformation, the N7 of A2062 is within hydrogen bonding distance of the exocyclic amino group of A2503 (Figure 2B), suggesting that the steric role of the N-terminus of ErmCL may be to induce the previously noted interaction between A2062 and A2503 (Vazquez-Laslop et al., 2010). Consistently, mutations of A2062U/C or A2503G dramatically reduce ErmCL stalling (Vazquez-Laslop et al., 2010; Vazquez-Laslop et al., 2008). In the absence of conformational changes with previously proposed relays from A2062-A2503 back to the PTC (Vazquez-Laslop et al., 2010), we favor a model whereby A2062-A2503 exert an effect on the PTC via the ErmCL nascent chain.
Drug-sensing by the ErmCL nascent chain within the ribosomal tunnel
In the ErmCL-SRC, erythromycin is bound in the canonical position (Dunkle et al., 2010; Tu et al., 2005), as observed previously in the ErmBL-SRC (Arenz et al., 2014). However, unlike the ErmBL-SRC (Arenz et al., 2014), direct contact is observed between ErmCL and the drug (Figure 2E and Figure S3), possibly between the backbone N of F7 and the C4″-OH of the cladinose sugar of erythromycin, however, higher resolution will be required to verify this. Biochemical experiments support ErmCL monitoring for the presence and the structure of the drug; specifically, ErmCL-mediated stalling was observed in the presence of other cladinose-containing macrolides, but not in the presence of ketolide antibiotics, such as telithromycin, which lack the C3-cladinose (Vazquez-Laslop et al., 2011; Vazquez-Laslop et al., 2008) (Figure S4). Moreover, macrolides bearing modifications of the C3-cladinose are also impaired for ErmCL-mediated ribosome stalling (Vazquez-Laslop et al., 2011).
Inactivation of the peptidyltransferase center of the ribosome by ErmCL
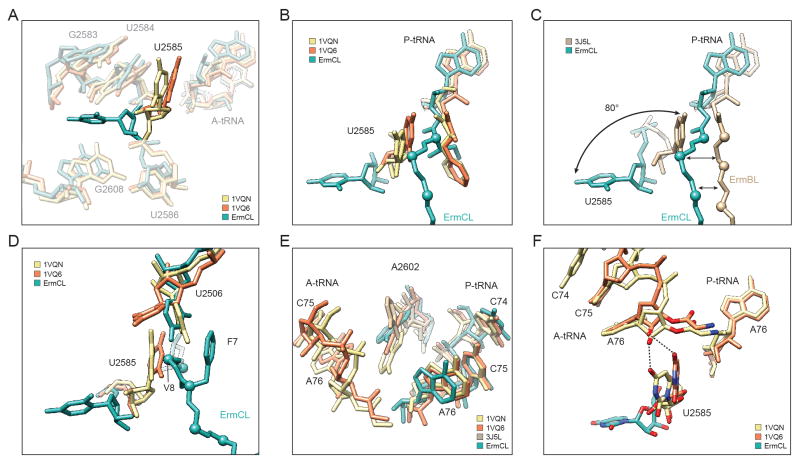
To understand how the interaction of ErmCL with erythromycin and rRNA components of the exit tunnel prevents stable binding of the A-tRNA and therefore leads to inhibition of peptidyl-transfer, we compared the PTC of the ErmCL-SRC with crystal structures of the ribosome in different states of peptide bond formation (Schmeing et al., 2005a; Schmeing et al., 2005b; Schmeing et al., 2002; Voorhees et al., 2009) (Figure 3A). While the overall conformation of the PTC is similar between ErmCL and the crystal structures, a few clear differences are evident. In the fully functional ribosome, U2585 rotates by 19° upon A-tRNA accommodation, i.e. when moving from the unaccommodated (uninduced) to the accommodated (induced) state (Schmeing et al., 2005a; Schmeing et al., 2005b; Schmeing et al., 2002) (Figure 3A). Surprisingly, U2585 adopts a dramatically different conformation in ErmCL-SRC, such that it is rotated by 80° compared to the unaccommodated state, and is flipped into a pocket formed by U2584/G2583 and G2608 (Figure 3A,B). This is reminiscent of the large rotation (180°) of U2585 observed upon streptogramin binding to the Deinococcus radiodurans 50S subunit (Harms et al., 2004), however, the final position is distinct (Figure S3). The positions of U2585 in the fully accommodated state would sterically clash with the ErmCL nascent chain, which may be the cause of the flipped state of U2585 in the ErmCL-SRC (Figure 3B). This contrasts with the ErmBL-SRC, where the path of the ErmBL nascent chain is compatible with (and even proposed to stabilize) the position of U2585 in the unaccommodated state (Arenz et al., 2014) (Figure 3C).
Figure 3. Conformational changes at the PTC of the ErmCL-SRC.
(A,B) Flipped conformation of 23S rRNA nucleotide U2585 in ErmCL-SRC (teal) compared to canonical position of U2585 in the uninduced (PDB1VQ6, orange) and induced (PDB1VQN, yellow) states of the PTC (Schmeing et al., 2005a; Schmeing et al., 2005b). (C) Comparison of ErmCL (teal) and ErmBL (PDB3J5L, tan) (Arenz et al., 2014) nascent chains and respective U2585 positions. (D,E) Relative positions of (D) U2585 and U2506, and (E) A2602 in ErmCL-SRC (teal), uninduced (PDB1VQ6, orange) and induced (PDB1VQN, yellow) states of the PTC (Schmeing et al., 2005a; Schmeing et al., 2005b). In (E), A2602 position of ErmBL-SRC (PDB3J5L, tan) (Arenz et al., 2014) is included for reference. (F) The flipped U2585 position in ErmCL-SRC (teal) prevents stabilization of the A-tRNA as observed in the uninduced (PDB1VQ6, orange) and induced (PDB1VQN, yellow) state of the PTC (Schmeing et al., 2005a; Schmeing et al., 2005b). See also Figure S3.
In the Haloarcula marismortui 50S crystal structures, U2506 is also observed to undergo a shift upon A-tRNA accommodation, such that the base interacts with U2585 (Schmeing et al., 2005a; Schmeing et al., 2005b; Schmeing et al., 2002) (Figure 3D). In the ErmCL-SRC, U2506 adopts a position similar to (but distinct from) U2506 in the accommodated state, which establishes interactions with residues V8 and F7 of ErmCL and is thereby presumably stabilized by the shifted conformation of the nascent chain (Figure 3D). Furthermore, A2602 is observed to undergo a slight shift upon A-tRNA accommodation (Figure 3E). In the ErmCL-SRC, A2602 appears to adopt a defined position (Figure S3), similar to the unaccommodated state but slightly shifted towards the A-tRNA (Figure 3E), which may also influence A-tRNA stability. During binding and accommodation of the A-tRNA, the ribose 2′OH of A76 maintains hydrogen bonding distance to the C4 oxygen of U2585 (Schmeing et al., 2005a; Schmeing et al., 2005b; Schmeing et al., 2002), which is also observed for the unaccommodated A-tRNA in the ErmBL-SRC (Arenz et al., 2014). In contrast, this contact is not possible with the flipped position of U2585 in the ErmCL-SRC (Figure 3F).
DISCUSSION
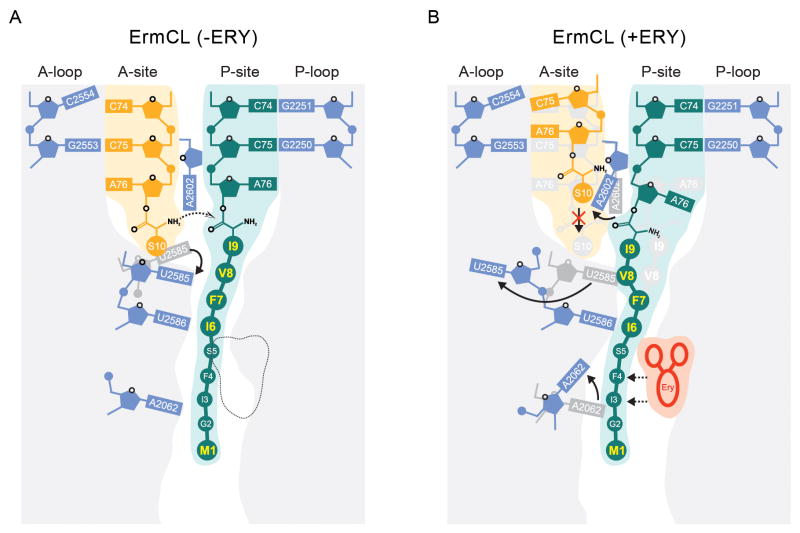
Collectively, our biochemical and structural findings lead us to propose a model for ErmCL-mediated drug-dependent ribosome stalling (Figure 4): While the exact conformation of the ErmCL peptide within the ribosomal tunnel in the absence of erythromycin has yet to be determined, biochemical studies indicate that peptide bond formation between Ser-tRNA in the A-site and the ErmCL-tRNA in the P-site can occur (Johansson et al., 2014; Vazquez-Laslop et al., 2008) (Figure 4A).
Figure 4. Schematic model of ErmCL-mediated translation arrest.
(A) In the absence of erythromycin (−ERY), Ser-tRNA accommodates at the A-site enabling peptide bond formation with the ErmCL-tRNA in the P-site. (B) In contrast, the presence of erythromycin, the ErmCL nascent chain adopts a distinct conformation that promotes conformational rearrangements of 23S rRNA nucleotides A2062, A2602 and most dramatically, flipping of U2585. Collectively, this global rearrangement of the peptidyltransferase center prevents stable binding and accommodation of Ser-tRNA at the ribosomal A-site and thus results in translational arrest.
However, in the presence of erythromycin, the ErmCL nascent chain interacts with the drug and adopts a unique conformation that is stabilized via interactions with specific rRNA components of the tunnel such as U2586, U2506 and A2062 (Figure 4B). In this stalled state, the ErmCL nascent chain precludes the canonical position of U2585 and promotes an alternative flipped conformation, which together with shifted P-tRNA and A2602 positions remodels the PTC making it unfavorable for the A-tRNA to fully accommodate and therefore leads to dissociation and translation arrest (Figure 4B). The absence of Ser-tRNA in A-site of the ErmCL-SRC (Figure 1B) is consistent with previous single molecule fluorescence experiments demonstrating that the A-site of ErmCL-SRC has a reduced capability to stably bind A-tRNA (Johansson et al., 2014). This contrasts with the structures of ErmBL- and SecM-stalled ribosomes, where stable tRNA binding was observed at the ribosomal A-site (Arenz et al., 2014; Bhushan et al., 2011).
In conclusion, our study illustrates how the ribosome can employ the ErmCL nascent chain to monitor the presence of erythromycin and induce allosteric conformational rearrangements within the PTC active site, leading to translational stalling and regulation of expression of downstream genes. The distinct mechanisms of drug-sensing and active site perturbation utilized by ErmCL and ErmBL (Arenz et al., 2014) raises the question as to whether similar mechanisms are utilized by other drug-dependent stalling systems.
EXPERIMENTAL PROCEDURES
Generation and purification of ErmCL-SRC
ErmCL-SRC were generated following the same procedure as previously described (Arenz et al., 2014). The 2XermCL construct was synthesized (Eurofins, Martinsried, Germany) such that it contained a T7 promoter followed by a strong ribosome binding site (RBS) spaced by 7 nucleotides (nts) to the ATG start codon of the first ermCL cistron. A linker of 22 nts separated the stop codon of the first ermCL cistron and the start codon of the second ermCL cistron. The linker also comprised the strong RBS 7 nts upstream of the ATG start codon of the second ermCL cistron, enabling initiation of translation independent from the first ermCL cistron. Each ermCL cistron encoded amino acids 1–19 corresponding to ErmCL leader peptide (Genbank accession number V01278) present on macrolide resistance plasmid pE194 (Iordanescu, 1976; Narayanan and Dubnau, 1985). The complete sequence of 2XermCL construct is: 5′-TAATACGACTCACTATAGGGAGTTTTATAAGGAGGAAAAAATATGGGCATTTTTAGTATTTTTGTAATCAGCACAGTTCATTATCAACCAAACAAAAAATAAAGTTTTATAAGGAGGAAAAAATATGGGCATTTTTAGTATTTTTGTAATCAGCAC AGTTCATTATCAACCAAACAAAAAATAA-3′ (T7 Promoter, italics; RBS, bold; ErmCL ORF, shaded grey with ATC codon in P-site of stalled ribosome shown in bold; Annealing site for complementary DNA oligonucleotide, underlined). In vitro translation of the 2xermCL construct was performed using the Rapid Translation System RTS 100 E. coli HY Kit (5PRIME; Cat. No. 2401110).
Translation reactions were analyzed on sucrose density gradients (10%–55% sucrose in a buffer A, containing 50 mM HEPES-KOH, pH 7.4, 100 mM KOAc, 25 mM Mg(OAc)2, 6 mM β-mercaptoethanol, 10 μM erythromycin and 1× Complete EDTA-free Protease Inhibitor cocktail (Roche)) by centrifugation at 154,693 x g (SW-40 Ti, Beckman Coulter) for 3 h at 4 °C. For ErmCL-SRC purification, disome fractions were collected using a Gradient Station (Biocomp) with an Econo UV Monitor (Biorad) and a FC203B Fraction Collector (Gilson). Purified ErmCL-SRC disomes were concentrated by centrifugation through Amicon Ultra-0.5 mL Centrifugal Filters (Merck-Millipore) according to the manufacturer’s protocol. To obtain monosomes of the ErmCL-SRC, a short DNA oligonucleotide (5′-ttcctccttataaaact-3′, Metabion) was annealed to the linker between the ermCL cistrons of the disomes, generating a DNA-RNA hybrid that could be cleaved by RNase H (NEB) treatment in buffer A at 25°C for 1h. After cleavage of the disomes, ErmCL-SRC monosomes were again purified and concentrated by centrifugation through Amicon Ultra-0.5 mL Centrifugal Filters (Merck-Millipore) according to the manufacturer’s protocol.
Negative-stain electron microscopy
Ribosomal particles were diluted in buffer A to final concentrations of 0.5 A260/ml up to 5 A260/ml in order to determine the optimal ribosome density for cryo-EM. One drop of each sample was deposited on a carbon-coated grid. After 30 seconds, grids were washed with distilled water and then stained with 3 drops of 2% aqueous uranyl acetate for 15 seconds. The remaining liquid was removed by touching the grid with filter paper. Micrographs were taken using a Morgagni transmission electron microscope (FEI), 80 kV, wide angle 1K CCD at direct magnifications of 72K.
Cryo-electron microscopy and single particle reconstruction
4 A260/ml monosomes of the ErmCL-SRC were applied to 2 nm pre-coated Quantifoil R3/3 holey carbon supported grids and vitrified using a Vitrobot Mark IV (FEI Company). Data collection was performed at NeCEN (Leiden, Netherlands) on a Titan Krios transmission electron microscope (TEM) (FEI, Eindhoven, Netherlands) equipped with a Falcon II direct electron detector at 300 kV with a magnification of 125,085x, a pixel size of 1.108 Å and a defocus range of 0.7–1.2 μm. The data are provided as a series of seven frames (dose per frame of 4 e−/Å2) from which we summed frames 2–5 (accumulated dose of 28 e−/Å2) after alignment using Motion Correction software (Li et al., 2013). Images were processed using a frequency-limited refinement protocol that helps prevent over-fitting (Scheres and Chen, 2012), specifically by truncation of high frequencies (in this case at 8 Å). As reported and expected (Scheres and Chen, 2012), we find that using this processing regime the 0.143 FSC value provides a good indicator for the true average resolution of the map. Additionally, the local resolution of the map was calculated using ResMap (Kucukelbir et al., 2014). Power-spectra and defocus values were determined using the SPIDER TF ED command and recorded images were manually inspected for good areas and power-spectra quality. Data were processed further using the SPIDER software package (Frank et al., 1996), in combination with an automated workflow as described previously (Becker et al., 2012). After initial, automated particle selection based on the program SIGNATURE (Chen and Grigorieff, 2007), initial alignment was performed with 419,113 particles, using E. coli 70S ribosome as a reference structure (Arenz et al., 2014). After removal of noisy particles (47,340 particles; 11%), the dataset could be sorted into two main subpopulations using an incremental K-means-like method of unsupervised 3D sorting (Loerke et al., 2010): The major subpopulation (285,841 particles; 68%) was defined by the presence of stoichiometric densities for P-tRNA and could be refined to an average resolution of 3.9 Å (0.143 FSC) and a local resolution extending to 3.5 Å for the core of the 30S and 50S subunit as computed using ResMap (Kucukelbir et al., 2014) (Figure S1). The final map was subjected to the program EM-BFACTOR (Fernandez et al., 2008) in order to apply an automatically determined negative B-factor for sharpening of the map.
Molecular modeling and map-docking procedures
The molecular model for the ribosomal proteins and rRNA of the ErmCL-SRC is based on the molecular model for the 50S subunit from the recent crystal structure of the E. coli 70S ribosome (PDB ID 4KIX) (Pulk and Cate, 2013) and obtained by performing a rigid body fit into the cryo-EM density map of the ErmCL-SRC using UCSF Chimera (Pettersen et al., 2004) (fit in map function). Similarly, the position of erythromycin was identical to that observed previously (Dunkle et al., 2010; Tu et al., 2005) and obtained by rigid body fit of PDB3OFR (Dunkle et al., 2010) into the cryo-EM density map of the ErmCL-SRC. The overall fit of the crystal structures were in very good agreement with the electron density of ErmCL-SRC and since exclusively the conformations of important nucleotides of the PTC and the exit tunnel are crucial to interpret the molecular mechanism leading to stalling on ErmCL, only these 23S rRNA nucleotides (A2602, G2583, U2584, U2585, U2586, U2506, A2062), were manually shifted/rotated where necessary into their respective densities using Coot (Emsley and Cowtan, 2004), whereas the rest of the rigid-body-fitted crystal structure remained unchanged. The conformation of A2602 observed in ErmCL-SRC is distinct from the “up” conformation observed in ErmBL-SRC (PDB3J5L) (Arenz et al., 2014) and from the “down” conformation in (PDB1VQN) (Schmeing et al., 2005a; Schmeing et al., 2005b) as such its conformation resembles an intermediate state between “up” and “down”. The conformation of U2506 is similar to its conformation in (PDB1VQN) (Schmeing et al., 2005a; Schmeing et al., 2005b), however slight manual rotation of the base was required to get an optimal fit. Different conformations of A2062, e.g. the “down” (PDB2WRJ) (Gao et al., 2009) and “tunnel-in” (PDB3I8F) (Jenner et al., 2010) conformations sterically clash with the ErmCL nascent chain, whereas the “up” conformation present in the Haloarcula marismortui 50S subunit in complex with model peptide bond substrates (e.g. PDB1VQN) (Schmeing et al., 2005a; Schmeing et al., 2005b) was similar to the conformation of A2062 observed in the ErmCL-SRC. Most notably, U2585 adopts a unique and novel “flipped out” position, which is very different to previously observed positions for U2585, i.e. U2585 in the uninduced (PDB1VQ6) or (PDB1VQN) induced state of the PTC (Schmeing et al., 2005a; Schmeing et al., 2005b). In the novel, “flipped-out” conformation U2585 is rotated approximately 80° when compared to its canonical position. In order to regularize the rRNA backbone surrounding U2585, neighboring nucleotides G2583, U2584 and U2586 were slightly adjusted in the model for ErmCL-SRC. Besides 23S rRNA nucleotides, the CCA-end of the P-tRNA, in particular A76, was manually adjusted in order to fit the density, since previously reported conformations observed in crystal structures of the Haloarcula marismortui 50S subunit in complex with model peptide bond substrates did not fit the density when fitted as a rigid body together with the 50S subunit. The molecular model for the ErmCL nascent polypeptide chain was modeled and refined into the density using Coot (Emsley and Cowtan, 2004).
Figure preparation
Figures showing electron densities and atomic models were generated using UCSF Chimera (Pettersen et al., 2004).
Toe-printing assay
The DNA templates containing T7 promoter, ribosome binding site and the ErmCL coding ORF (wild type or the mutants), were generated by crossover PCR. The toeprinting analysis of drug-dependent ribosome stalling was carried out as described (Vazquez-Laslop et al., 2008). Briefly, the DNA templates (0.1 pmol) were used in a total volume of 5 μl of PURExpress (NEB) cell-free transcription-translation reactions. Samples were incubated for 15 min at 37°C, followed by addition of the [32P]-labeled NV1 toe-printing primer designed to anneal ~100 nucleotides downstream from the anticipated ribosome stalling site. The primer was extended by reverse transcriptase and the reaction products were analyzed in sequencing gels. In experiments with mutant ribosomes, the “Δ ribosome” version of the PUREXpress kit was supplemented with ribosomes (10 pmol per reaction) isolated from the SQ171 E. coli strain carrying the plasmid expressing E. coli rrnB operon with the engineered mutations in the 23S rRNA gene. Mutant ribosomes were purified as described (Ohashi et al., 2007).
Supplementary Material
Figure S1 Average and local resolution determination of the ErmCL-SRC, related to Figure 1. (A) Schematic for ermCL-dependent regulation of ermC translation in the presence of erythromycin (ERY). (B,C) The bicistronic 2XermCL mRNA was translated in vitro in the presence of 10 μM ERY in order to generate (B) ErmCL-SRC disomes, which were (C) converted to monosomes by annealing of complementary DNA oligo and RNase H cleavage, as shown by sucrose density centrifugation and negative stain EM. (D) Local resolution (Kucukelbir et al., 2014) displayed on the ErmCL-SRC with a transverse section through the 50S subunit (as depicted in the insert), 30S (yellow), P-tRNA (green) and erythromycin (red). (E) Average resolution of the ErmCL-SRC was 4.6 Å using the Fourier shell correlation (FSC) cut-off value of 0.5. Since the microscopy images were processed in the absence of spatial frequencies higher than 8 Å, the FSC cutoff value of 0.143 was used for average resolution determination of 3.9 Å (Scheres and Chen, 2012). The overall fit of the 50S model to the ErmCL-SRC map is indicated by the FSC calculation between the model and map (red curve). (F) Histogram generated by ResMap (Kucukelbir et al., 2014) showing that the local resolution of the final map predominantly ranges from 3.5–4.0 Å.
Figure S2 Comparison of the path of the ErmCL and ErmBL nascent chains through the ribosomal tunnel, related to Figure 2. (A–D) Comparison of cryo-EM structures of (A,B) ErmCL-SRC and (C,D) ErmBL-SRC (Arenz et al., 2014) with 30S (yellow) and transverse section of the 50S subunit (grey), illustrating the path of the respective nascent polypeptide chains trough the ribosomal tunnel. Enlargements with density (grey mesh) and molecular models for the ErmCL (teal) and ErmBL (green) nascent chains, erythromycin (ERY, red) and 23S rRNA nucleotides (blue).
Figure S3 Electron density for selected regions of the PTC and tunnel, related to Figure 2 and 3. (A–C) Views of the ErmCL nascent chain (teal), 23S rRNA nucleotides (blue), and erythromycin (Dunkle et al., 2010) (red) in the ribosomal tunnel of the ErmCL-SRC. Views identical to those shown in Fig. 2A,B,E, respectively, but including electron density (grey mesh). (D) Electron density (grey mesh) for the defined position of A2602 in the ErmCL-SRC. (E) Unique conformation of 23S rRNA nucleotide U2585 in ErmCL-SRC (teal) compared to its positions observed in the uninduced (PDB1VQ6, orange) and induced (PDB1VQN, yellow) state of the PTC (Schmeing et al., 2005a; Schmeing et al., 2005b). Insert show electron density for nucleotides G2583-U2586 coloured according to local resolution (Kucukelbir et al., 2014). (F,G) Electron density for G2583-U2586 in (G) ErmBL-SRC (Arenz et al., 2014) and (G) TnaC-SRC indicating the absence of electron density for the flipped position of U2585 and presence of electron density for canonical position of U2585. (H) Comparison of U2585 conformations in the uninduced (PDB1VQ6, orange) and induced (PDB1VQN, yellow) state of the PTC (Schmeing et al., 2005a; Schmeing et al., 2005b), as well as in ErmCL-SRC (teal) and streptogramin-bound ribosomes (PDB1SM1) (Harms et al., 2004).
Figure S4 Superimposition of telithromycin antibiotic relative to ErmCL, related to Figure 2. (A) Interaction between I6 and U2586 in the ErmCL-SRC. (B) Quantification of the stalling efficiency of mutations at position I6 of ErmCL based on toe-printing assays. (C,D) Chemical structures of (C) erythromycin (ERY) and (D) telithromycin (TEL) showing, various alterations of the lactone ring and the absence of the C3 cladinose sugar in telithromycin. (E–H) Relative position of (E,F) erythromycin and (G,H) telithromycin to ErmCL. Spacefill representations of ErmCL and (F) ERY (Dunkle et al., 2010) and (H) TEL (Dunkle et al., 2010) illustrate the proximity of the C3 cladinose sugar of erythromycin to ErmCL, which is absent in the superimposition of telithromycin due to the lack of the C3 cladinose.
Acknowledgments
We thank Rishi Matadeen and Sacha DeCarlo for data collection at the NeCEN facility (Leiden, Netherlands) and Charlotte Ungewickell and Dorota Klepacki for expert technical assistance. This research was supported by grants from the NIH (GM104370 to N.V-L. and R01GM095737 to D.N.W.) and the Deutsche Forschungsgemeinschaft (FOR1805, WI3285/3-1 and GRK1721 to D.N.W.).
Footnotes
Supplemental Information includes Figures S1–S4 and Supplemental References can be found with this article online at http://...
ACCESSION NUMBERS
The cryo-EM map and associated atomic coordinates have been deposited in the EMDB and PDB with the accession codes EMDB-6057 and PDB ID 3J7Z, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arenz S, Ramu H, Gupta P, Berninghausen O, Beckmann R, Vazquez-Laslop N, Mankin AS, Wilson DN. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun. 2014;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, Karcher A, Thomm M, Hopfner KP, Green R, Beckmann R. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, Berninghausen O, Wilson DN, Beckmann R. SecM-stalled ribosomes adopt an altered geometry at the peptidyltransferase center. PLoS Biol. 2011;19:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JZ, Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci U S A. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallographica Section D - Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernandez JJ, Luque D, Caston JR, Carrascosa JL. Sharpening high resolution information in single particle electron cryomicroscopy. J Struct Biol. 2008;164:170–175. doi: 10.1016/j.jsb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Fucini P, Bartels H, Yonath A. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2004;2:4. doi: 10.1186/1741-7007-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S, Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980;77:7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Archives roumaines de pathologie experimentales et de microbiologie. 1976;35:111–118. [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- Johansson M, Chen J, Tsai A, Kornberg G, Puglisi JD. Sequence-Dependent Elongation Dynamics on Macrolide-Bound Ribosomes. Cell reports. 2014 doi: 10.1016/j.celrep.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke J, Giesebrecht J, Spahn CM. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- Mayford M, Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J Mol Biol. 1989;206:69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- Narayanan CS, Dubnau D. Evidence for the translational attenuation model: ribosome-binding studies and structural analysis with an in vitro run-off transcript of ermC. Nucleic Acids Res. 1985;13:7307–7326. doi: 10.1093/nar/13.20.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi H, Shimizu Y, Ying BW, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochem Biophys Res Commun. 2007;352:270–276. doi: 10.1016/j.bbrc.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005a;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005b;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Seila AC, Hansen JL, Freeborn B, Soukup JK, Scaringe SA, Strobel SA, Moore PB, Steitz TA. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat Struct Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- Shivakumar AG, Hahn J, Grandi G, Kozlov Y, Dubnau D. Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc Natl Acad Sci U S A. 1980;77:3903–3907. doi: 10.1073/pnas.77.7.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu D, Blaha G, Moore P, Steitz T. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Klepacki D, Mulhearn DC, Ramu H, Krasnykh O, Franzblau S, Mankin AS. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc Natl Acad Sci U S A. 2011;108:10496–10501. doi: 10.1073/pnas.1103474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Laslop N, Ramu H, Mankin AS. Nascent peptide-mediated ribosome stalling promoted by antibiotics. In: Rodnina MV, Wintermeyer W, Green R, editors. Ribosomes Structure, function, evolution. Wien, New York: Springer-Verlag; 2011. pp. 377–392. [Google Scholar]
- Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Average and local resolution determination of the ErmCL-SRC, related to Figure 1. (A) Schematic for ermCL-dependent regulation of ermC translation in the presence of erythromycin (ERY). (B,C) The bicistronic 2XermCL mRNA was translated in vitro in the presence of 10 μM ERY in order to generate (B) ErmCL-SRC disomes, which were (C) converted to monosomes by annealing of complementary DNA oligo and RNase H cleavage, as shown by sucrose density centrifugation and negative stain EM. (D) Local resolution (Kucukelbir et al., 2014) displayed on the ErmCL-SRC with a transverse section through the 50S subunit (as depicted in the insert), 30S (yellow), P-tRNA (green) and erythromycin (red). (E) Average resolution of the ErmCL-SRC was 4.6 Å using the Fourier shell correlation (FSC) cut-off value of 0.5. Since the microscopy images were processed in the absence of spatial frequencies higher than 8 Å, the FSC cutoff value of 0.143 was used for average resolution determination of 3.9 Å (Scheres and Chen, 2012). The overall fit of the 50S model to the ErmCL-SRC map is indicated by the FSC calculation between the model and map (red curve). (F) Histogram generated by ResMap (Kucukelbir et al., 2014) showing that the local resolution of the final map predominantly ranges from 3.5–4.0 Å.
Figure S2 Comparison of the path of the ErmCL and ErmBL nascent chains through the ribosomal tunnel, related to Figure 2. (A–D) Comparison of cryo-EM structures of (A,B) ErmCL-SRC and (C,D) ErmBL-SRC (Arenz et al., 2014) with 30S (yellow) and transverse section of the 50S subunit (grey), illustrating the path of the respective nascent polypeptide chains trough the ribosomal tunnel. Enlargements with density (grey mesh) and molecular models for the ErmCL (teal) and ErmBL (green) nascent chains, erythromycin (ERY, red) and 23S rRNA nucleotides (blue).
Figure S3 Electron density for selected regions of the PTC and tunnel, related to Figure 2 and 3. (A–C) Views of the ErmCL nascent chain (teal), 23S rRNA nucleotides (blue), and erythromycin (Dunkle et al., 2010) (red) in the ribosomal tunnel of the ErmCL-SRC. Views identical to those shown in Fig. 2A,B,E, respectively, but including electron density (grey mesh). (D) Electron density (grey mesh) for the defined position of A2602 in the ErmCL-SRC. (E) Unique conformation of 23S rRNA nucleotide U2585 in ErmCL-SRC (teal) compared to its positions observed in the uninduced (PDB1VQ6, orange) and induced (PDB1VQN, yellow) state of the PTC (Schmeing et al., 2005a; Schmeing et al., 2005b). Insert show electron density for nucleotides G2583-U2586 coloured according to local resolution (Kucukelbir et al., 2014). (F,G) Electron density for G2583-U2586 in (G) ErmBL-SRC (Arenz et al., 2014) and (G) TnaC-SRC indicating the absence of electron density for the flipped position of U2585 and presence of electron density for canonical position of U2585. (H) Comparison of U2585 conformations in the uninduced (PDB1VQ6, orange) and induced (PDB1VQN, yellow) state of the PTC (Schmeing et al., 2005a; Schmeing et al., 2005b), as well as in ErmCL-SRC (teal) and streptogramin-bound ribosomes (PDB1SM1) (Harms et al., 2004).
Figure S4 Superimposition of telithromycin antibiotic relative to ErmCL, related to Figure 2. (A) Interaction between I6 and U2586 in the ErmCL-SRC. (B) Quantification of the stalling efficiency of mutations at position I6 of ErmCL based on toe-printing assays. (C,D) Chemical structures of (C) erythromycin (ERY) and (D) telithromycin (TEL) showing, various alterations of the lactone ring and the absence of the C3 cladinose sugar in telithromycin. (E–H) Relative position of (E,F) erythromycin and (G,H) telithromycin to ErmCL. Spacefill representations of ErmCL and (F) ERY (Dunkle et al., 2010) and (H) TEL (Dunkle et al., 2010) illustrate the proximity of the C3 cladinose sugar of erythromycin to ErmCL, which is absent in the superimposition of telithromycin due to the lack of the C3 cladinose.