Abstract
Impressive improvements in docking performance can be achieved by applying energy bonuses to poses in which glycan hydroxyl groups occupy positions otherwise preferred by bound waters. In addition, inclusion of glycosidic conformational energies allows unlikely glycan conformations to be appropriately penalized. A method for predicting the binding specificity of glycan-binding proteins has been developed, which is based on grafting glycan branches onto a minimal binding determinant in the binding site. Grafting can be used either to screen virtual libraries of glycans, such as the known glycome, or to identify docked poses of minimal binding determinants that are consistent with specificity data. The reviewed advances allow accurate modelling of carbohydrate-protein 3D co-complexes, but challenges remain in ranking the affinity of congeners.
Introduction
The structural similarity between monosaccharides, combined with their ability to form branched oligomers, makes distinguishing between variations in linkage site or glycan composition challenging for experimental methods. These challenges are amplified by the size and diversity of the cellular glycome. And yet, it is the ability to discriminate between these subtle structural variations that defines the specificity of a given glycan-binding protein (GBP) [1].
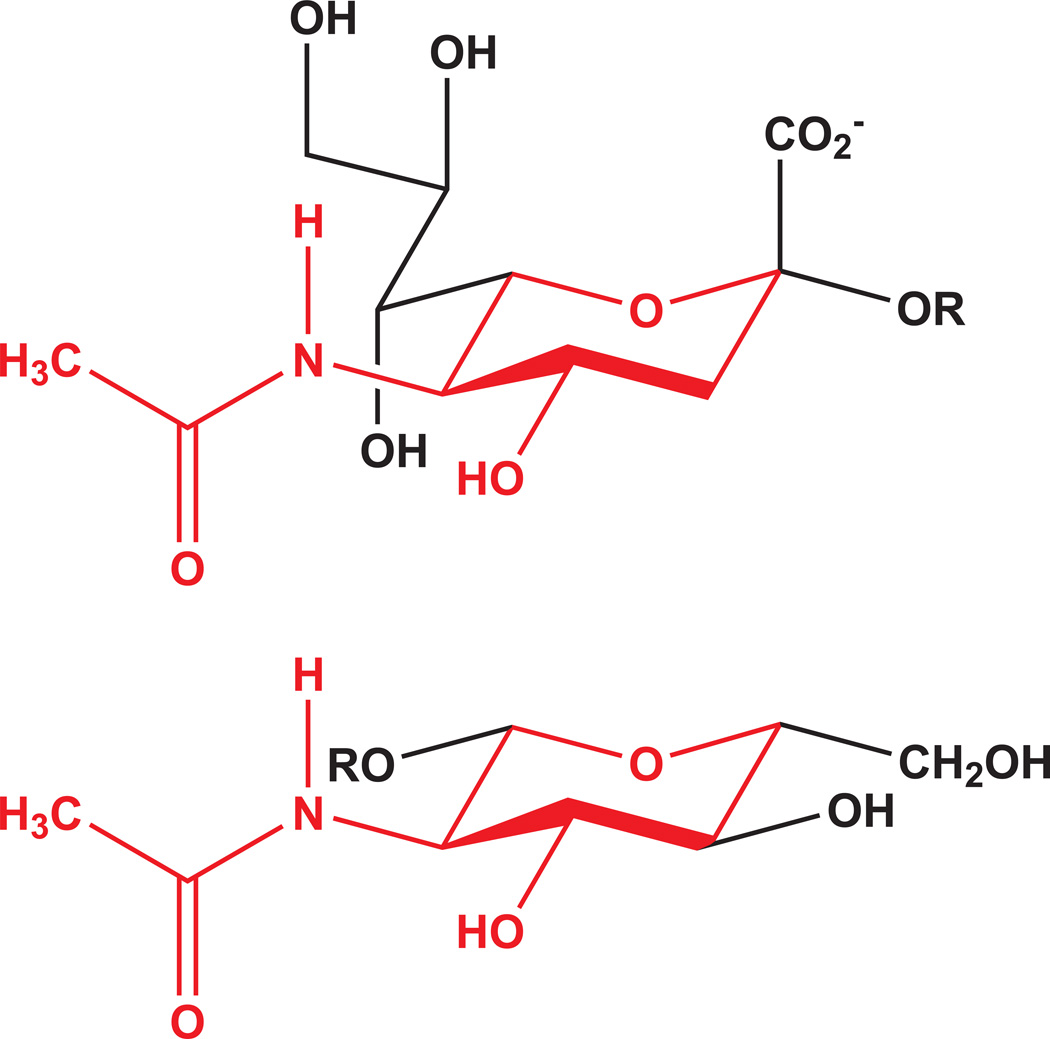
Identifying the glycan receptors preferred by a given GBP is an area of intense study [2–6], with applications ranging from biomass polysaccharide recognition [7–9], through fundamental biology [4,5,10,11] and medicine [3,5,12–15]. The introduction of glycan array screening has caused a paradigm shift in GBP specificity determination. Although subject to limitations [2,6,11,16–19], such screening produces a rich data set that allows researchers to determine the minimal binding determinant (MBD) for a given GBP, and the various glycans in which the MBD is recognized [17]. Complexities arise in identifying MBDs in glycan array data because of the potential for structural mimicry among monosaccharides. A classic example of this is the subset of structural features shared by GlcNAc and Neu5Ac which can result in cross-reactivity (Figure 1), as in the case of binding to wheat germ agglutinin isolectin II [20].
Figure 1.
Partial structural mimicry among monosaccharides (Neu5Ac, top; GlcNAc, bottom;) can confuse MBD assignments, and is challenging to detect, in part because of nomenclature limitations and established structure representation preferences.
Overall, the data from glycan array screening shows that binding specificity is determined by both the affinity of the MBD, and the context in which the MBD is presented. Context encompasses such properties as the overall structure of the glycan, the nature of the underlying aglycone (protein, lipid, etc), glycan clustering (avidity), the presentation of the glycan relative to its environment [21], the specificity of modules attached to nonspecific catalytic domains [22,23] and the composition of the cellular membrane [24].
The three-dimensional (3D) structures of both the GBP and the glycan are required in order to generate a structural rationalization of the origin of specificity. The experimental generation of these co-complexes is a hard-fought battle for crystallography, which must overcome the intrinsic resistance of glycans to crystallization, among other issues. Theoretical glycan structure prediction tools have become particularly useful in augmenting experimental data, and in surmounting some of the limitations. Online tools such as GLYCAM-Web (www.glycam.org) have evolved to provide rapid access to 3D structures of glycans and glycoproteins that can be used to assist in interpreting sparse experimental data, or to aid in developing structure-based models of binding specificity.
In contrast to the general case of proteins or peptides, the unique structural properties of oligosaccharides enable their conformations to be accurately modelled given only the primary structure (composition and linkage information) [25,26]. A key advantage for glycan structure prediction, relative to the case of proteins, is that glycan 3D structure and dynamics are driven by well-defined conformational preferences of the glycosidic angles, and by interactions with solvent, rather than by strong inter-residue interactions. These features enable approximate low energy conformations of a glycan to be generated solely by consideration of the properties of the individual linkages. Exceptions to this include highly flexible glycans, such as polysialic acids and glycosaminoglycans (GAGs), which require more sophisticated approaches, including molecular dynamics (MD) simulation, to determine their conformational preferences.
Until recently, the focus of many computational studies of glycan binding has been on predicting the 3D structure and binding free energy of particular GBP-MBD complexes, using techniques such as computational docking or analysis of MD simulations [27–36]. Such studies have been essential for developing a general understanding of the molecular features responsible for the affinity of a given MBD, but have done relatively little to address the origin of glycan specificity in a more general context. However, with the creation of virtual libraries of 3D glycan structures, and the introduction of automated workflows, such as Computational Carbohydrate Grafting (CCG, [37]) it is now possible predict whether a given MBD in the context of a larger glycan structure, or ensemble of structures, would be recognized by a particular GBP.
This review is intended to provide a summary of the latest advances in carbohydrate docking and in the computational tools used to predict glycan specificity.
Recent Advances in Computational Carbohydrate Docking
Accuracy in computational docking requires a search algorithm capable of finding the correct solution and a scoring function that is able to correctly rank this pose above the other putative solutions. When docking carbohydrates it is generally found that current algorithms are able to produce a pose that is close to known crystal structure coordinates, but that the scoring functions are unable to correctly rank this pose [27–31,35,36]. Carbohydrate ligands are particularly challenging to rank in this manner because they have low chemical diversity, and often display weak affinities. Compounding these difficulties is the fact that certain inter-glycosidic linkages are highly flexible, resulting in glycans being far from ideal ligands for molecular docking. These challenges have spurred the innovations in carbohydrate docking discussed in the following sections.
There have been several studies [27–31,34–36] assessing the performance of software for docking carbohydrates, however, quantitative or wide-ranging conclusions remain elusive for a number of reasons:
The reported performance levels are often favourably biased by docking to “positive-controls”, in which both ligand and receptor coordinates are taken from a co-crystal structure, with one or both partners retained in the bound conformation.
The study may lack “negative-controls”, such as congeners known not to bind, without which the discriminatory ability of the method is uncertain.
The study may consider too few systems to be generalizable, or is focused on a narrow class of glycan.
Successful reproduction of the pose geometry in a positive-control experiment should be considered a minimal requirement to demonstrate the capability of docking under optimal conditions. However, such an experiment does not indicate that the protocol is able to distinguish between binders and non-binders, nor does it mimic the general case of docking ligands from virtual libraries, or docking to unliganded crystal structures.
Docking Glycosaminoglycans (GAGs)
Docking GAGs is particularly challenging due to their highly charged structure, polymeric nature, heterogeneity in sulfation patterns or monosaccharide composition, varying ring conformations (particularly of iduronate residues), as well as the difficulties associated with obtaining experimental constraints to guide or validate the results. Nevertheless, the relative lack of experimental structural data for these systems, combined with a growing awareness of their biological significance, has led to a number of innovative approaches to GAG docking. Recent examples include advances in the prediction of GAG binding sites [38,39], improvement in pose prediction via inclusion of specific bound waters [40], co-complex generation via threading and superimposition [41], mimetic discovery [42], as well as combining spectroscopic data with GAG modelling [43,44]. Very recently, an online utility for the automatic generation of 3D structures for GAGs has appeared (www.glycam.org/gag, [25]) that should find application in GAG modelling.
Of the examples above, a particularly interesting approach docked sulfated moieties of increasing size, starting from methyl sulfates and progressing up to hexasaccharide GAG fragments to elucidate the GAG-binding site in αvβ3 integrin [38]. The results from docking the smaller fragments were used to focus the search regions used when docking the larger structures. Finally, multiple different conformations from a MD simulation of a hexasaccharide were docked to the subset of potential sites.
An alternative to docking is illustrated in a study of heparanase specificity, in which a homology model of heparanase was generated, and various co-crystallized substrates from similar binding sites were threaded into the active site. GAG substrates and inhibitory oligosaccharides were superimposed onto these models, and the subsequent structures were consistent with data from surface plasmon resonance and NMR spectroscopy [41].
Including Conformational Energies in Carbohydrate Docking
In docking oligosaccharides, if the glycosidic linkages are permitted to be flexible, conformations are invariably generated that are inconsistent with known preferences, based on solution NMR data or protein crystallography. Conversely, rigid docking does not account for the innate flexibility of glycosidic linkages or permit induced fit in the glycan upon binding.
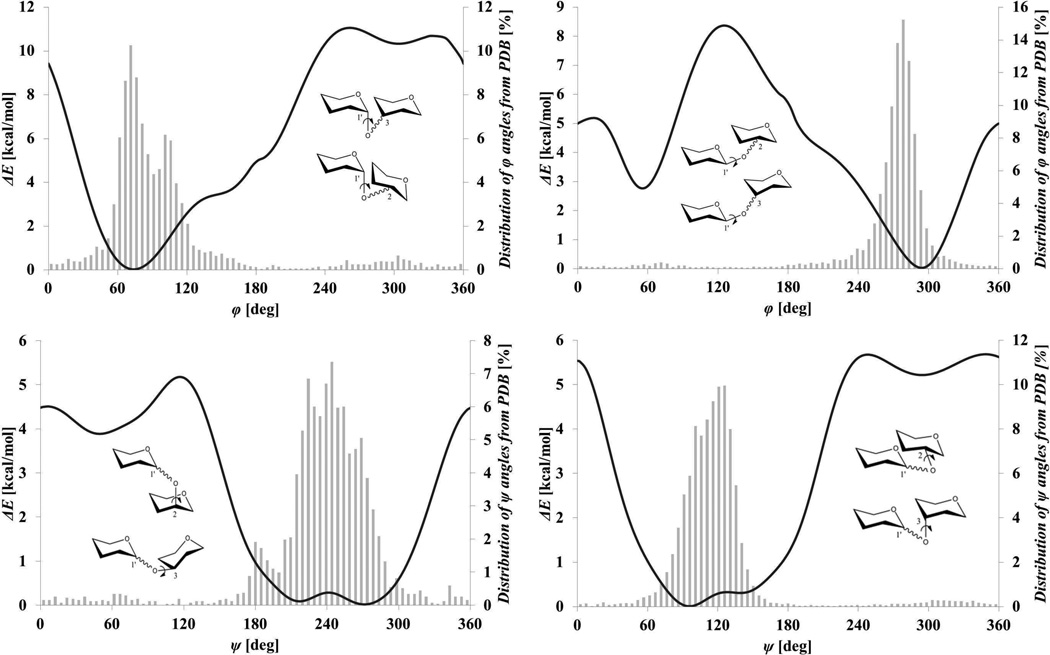
Two recent examples demonstrate how this problem can be circumvented by employing a set of post-docking filtering functions, either enabling poses to be rescored based on inclusion of their conformational energy [30], or to be rejected when their glycosidic torsion angles fall outside of expected ranges [45]. Thus in the case of Nivedha et al. [30], poses which scored well during flexible docking, but whose glycosidic linkages were distorted, received an additional penalty in their scores. An advantage of post-filtering approaches is their independence from the docking software used. The agreement between the energy profiles for the glycosidic torsions and the observed distributions of these angles in ligands in the protein data bank (PDB) (Figure 2), led Nivedha et al. [30] to conclude that the majority of proteins that recognize oligosaccharides select low-energy (solution-like) conformations. This observation suggests that only a restricted range of torsion angles may need to be considered when performing carbohydrate docking, which would significantly enhance its efficiency.
Figure 2.
Calculated glycosidic torsion rotational energies (solid lines) versus distributions of related torsion angles from glycans in the PDB [30].
It should be noted that this conclusion was based solely on the observed conformational preferences of glycosidic linkages, and does not take into account the energy required for enzymes, such as hydrolases, to distort monosaccharide rings into enzyme-active conformations [46–48]. If we assume that a hydrolase binds the most populated low-energy conformation of a glycan, any conformational change required for enzyme activity must be induced in the substrate after or during binding. This presents a significant challenge for docking since the current computational protocols do not facilitate changes in ring conformation. One approach to overcoming this limitation is to perform full-relaxed MD simulations starting with the substrate in the docked conformation. During the minimization steps or the MD simulation the substrate may spontaneously distort into a conformation similar to that required for enzyme activity [49,50]. Additionally, the ability of quantum mechanical (QM) and hybrid QM/MM techniques to study bond breaking/forming reactions can be extremely useful for detailed analysis of these systems [51,52].
Docking using MD Generated Water Sites
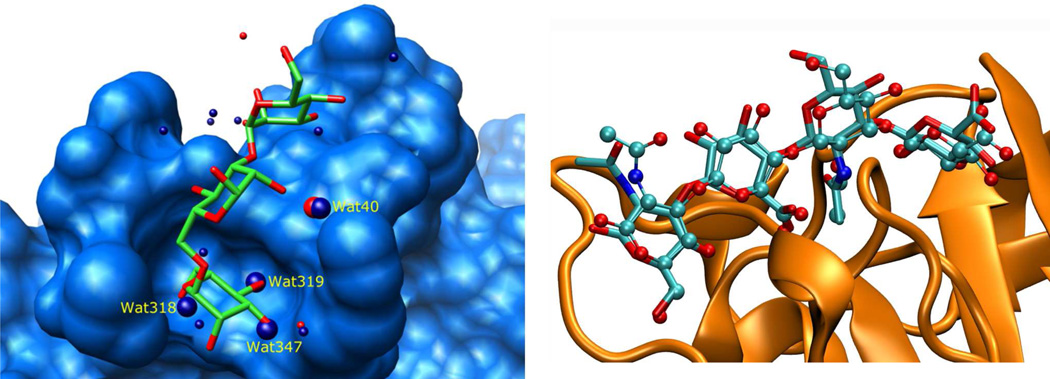
It has been shown that displaced water plays an important role in ligand binding [53–55] and that the positions of crystallographic waters are often replaced by glycan hydroxyl groups upon complex formation (Figure 3) [56,57]. These “hydration or water sites” can also be readily identified using MD simulations of the unliganded GBP [58–62]. Promising results for carbohydrate docking have been reported [63] when the score included a bonus for poses that placed ligand hydroxyl groups in water sites. The reported improvement in the accuracy of the docking results was impressive for the six reported systems.
Figure 3.
Left: solvent accessible surface of the lentil lectin Concanavalin A (light blue) in complex with the natural trisaccharide ligand (PDB: 1CVN). Waters of crystallization are shown as red spheres. Superimposed are the waters of crystallization (dark blue) from the ligand-free structure (PDB: 1GKB). Key waters, which appear to be displaced by the ligand, as well as a conserved water molecule (Wat40), are shown as larger spheres [57]. Right: results from docking a hyaluronan tetrasaccharide to CD44 (orange, ribbon model) using the water-site biased docking method. The top ranked pose (ball and stick model) has an RMSD of 0.7 Å from the reference ligand position (stick model) [63].
Additional Tips for Accurate Computational Carbohydrate Docking
Thoroughly vet crystal structures for inconsistencies between the electron density and the reported model, using for example the Coot program [64]. Note that even with reasonable resolution structures, many structural details may be ambiguous. Check for distorted monosaccharide ring conformations, unlikely glycosidic angles [30], as well as flipped or distorted amide groups in Neu5Ac, GlcNAc, etc. The pdb-care webtool (www.glycosciences.de/tools/pdb-care) can be helpful in this regard [65]. Incorrect side chain rotamers in asparagine, glutamine, and histidine are also common, and may be inferred from careful analysis of potential hydrogen bond networks. The NQ-Flipper webtool (https://flipper.services.came.sbg.ac.at) can be useful in such an analysis [66].
Reduce the size of the docked ligand so that it contains only the MBD; the structures for larger glycans can be obtained via grafting after a successful MBD pose has been generated.
Enabling glycosidic linkages to be flexible is preferred, but unlikely rotamers should be expected and filtered post-docking [30,45] as outlined in the Including Conformational Energies in Carbohydrate Docking section.
When docking glycans that are normally linked via their reducing terminus, disregard poses where the reducing terminal is occluded upon binding to the receptor.
Defining the Context for MBD Recognition with Computational Carbohydrate Grafting
The presence of an MBD in a glycan is not sufficient for recognition by the cognate GBP; the MBD must also be accessible for binding to the GBP. That is, the context in which the MBD occurs is critical for recognition.
Context has been examined by explicitly modelling the larger glycan into the binding site via filtered docking [45], or by manual superimposition [32,67,68]. The CCG approach builds on these concepts by completely automating the grafting process, allowing 3D structural libraries as large as the known glycome to be screened [37]. The predictions from this high-throughput virtual screening can be validated by comparison with experimental data, particularly from glycan array screening [37]. Predictions from CCG can be used for a great many purposes, from identifying putative cross-reactivities for anti-carbohydrate antibodies, to rational design of GBPs with tailored specificity, to guiding the design of experimental glycan arrays. In a recent extension of the CCG approach, explicit modelling of glycans conjugated to hypothetical array surfaces provided a structural rationale for false-negative binding resulting from surface and linker effects [19].
Assessing Putative Docking Poses with Glycan Array Data
Consistency with data from glycan array screening can also be used, in conjunction with CCG, to identify poses of the MBD generated by docking that are unable to explain the observed specificity. This approach was demonstrated in the case of the binding of the Thomsen Friedenreich (TF) disaccharide (Galβ1-3GalNAcα) to a monoclonal antibody (JAA-F11) [37]. Four putative poses were generated by docking the disaccharide, however, grafting to each pose revealed that only one was consistent with data from glycan array screening (Table 1).
Table 1.
Using CCG to identify poses consistent with observed binding specificity for the TF-disaccharide in complex with the mAb JAA-F11
| Predicted Overlaps Consistent with Experimenta |
||||
|---|---|---|---|---|
| Experimental Bindersb | Pose 1 | Pose 2 | Pose 3 | Pose 4 |
| Galβ1-3GalNAcα | Y | Y | Y | Y |
| Neu5Acβ2-6(Galβ1-3)GalNAcα | Y | Y | -c | Y |
| Neu5Acα2-6(Galβ1-3)GalNAcα | Y | - | - | Y |
| Galβ1-4GlcNAcβ1-6(Galβ1-3)GalNAcα | Y | Y | - | Y |
| GlcNAcβ1-6(Galβ1-3)GalNAcα | Y | Y | - | Y |
| Experimental Non-Bindersb | ||||
| Fucα1-2Galβ1-3GalNAcα | Y | Y | - | Y |
| GlcNAcβ1-3Galβ1-3GalNAcα | Y | Y | - | Y |
| GlcNAcβ1-2Galβ1-3GalNAcα | Y | Y | - | Y |
| (3S)Galβ1-3GalNAcα | Y | - | - | Y |
| Neu5Acα2-6(Neu5Acα2-3Galβ1-3)GalNAcα | Y | Y | Y | Y |
| 6S(Neu5Acα2-3Galβ1-3)GalNAcα | Y | Y | - | Y |
| Neu5Acα2-3Galβ1-3GalNAcα | Y | Y | - | Y |
| GalNAcβ1-3GalNAcα | Y | - | - | Y |
| Galβ1-3GalNAcβ | Y | Y | Y | - |
| Galβ1-3GalNAcβ1-4Galβ1-4Glcβ | Y | Y | Y | - |
A relative buried surface area of greater than 1 (relative to the surface are of a carbon atom (~36 Å2)) was used to categorise a predicted glycan as being a non-binder.
Experimental binding was based on a mean relative fluorescence signal greater than 10% of the maximum signal at each protein concentration.
A dash denotes that the predicted overlaps were inconsistent with the experimental array data.
The ability to discriminate between acceptable and unacceptable poses relied equally on both the presence of binding and non-binding glycans that contain the MBD. Thus this approach could fail to identify a unique pose if there were too few glycans in the experimental array. Notably, for use as specificity constraints, the glycans need not be naturally occurring, and data from synthetic deoxy-analogs might be particularly informative [69]. In this case, comparing the docked poses to data from Saturation Transfer Difference NMR (STD-NMR) was used to further confirm the validity of pose 1 [37].
Molecular Dynamics as a Tool to Refine Ligand Placement
The use of MD to probe the specificity of GBPs is well established and widely used, and has been recently reviewed [70,71]. Force fields for carbohydrates in many of the popular software packages have reached a generally reliable level of predictive accuracy, and continue to be refined [72]. Advances in high-performance computing, and particularly in the development of algorithms for use on graphics processing units (GPUs), are allowing MD simulations to be performed for significantly longer periods, and on larger systems, than was previously feasible with CPU-based implementations. Particular strengths of MD simulation in regard to carbohydrate specificity assessment are its ability to remove inadvertent bias from ligand docking, and to permit induced fit in both binding partners. Recent examples of ligand refinement by MD simulation include an analysis of the binding specificity of Galectin-8 [32], O-GlcNAc hydrolase [33], and Galectin-3 [73]. A further particularly useful feature of post-docking MD simulation is that unlikely ligand poses, or unlikely ligands, may be detected based on their instability during the simulation [74–77].
Future perspectives
Molecular modelling is transitioning from anecdotal studies of MBD – GBP complexes into a high-throughput methodology for determining the specificity of GBPs. Virtual screening employing methods such as CCG offer an opportunity to advance the understanding of glycan context on specificity. Potential applications of virtual screening include defining and predicting antibody antigenicity, host or pathogen receptor preferences, and enzyme substrate preferences, with therapeutic design implications.
The innovations in docking methodology and CCG have yet to be widely evaluated, but initial results in terms of improved structure and large-scale specificity prediction are promising. However, improving scoring functions for carbohydrate docking remains an underdeveloped area that would benefit from the creation of a reference suite of high resolution 3D structures for co-complexes with associated binding energies.
Given the importance of glycobiology to issues of human health, and the difficulties associated with experimental structural glycobiology, there is a clear and growing demand for effective modelling tools. In the field of computational glycoscience there is a growing trend toward web-based implementations, with user-friendly interfaces, such as GlycomeDB [78], UniCarbKB [79] or GLYCAM-Web [25]. Online resources offer incomparable user access but place a correspondingly high level of responsibility on the software developers to ensure that the tools are robust and accurate. Nevertheless, fully leveraging the benefits of the emerging computational methods for 3D structure generation will require that they are made accessible and understandable to nonspecialists.
Highlights.
Recent innovations in carbohydrate docking enhance 3D structure prediction accuracy
Glycan array screening data can be used to filter and validate docking results
Computational Carbohydrate Grafting can predict specificity within the human glycome
Acknowledgements
The authors are grateful to the National Institutes for Health (GM094919 (EUREKA)) and the Science Foundation of Ireland (08/IN.1/B2070) for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Citations
- 1.Drickamer K, Taylor ME. Convergent and divergent mechanisms of sugar recognition across kingdoms. Current Opinion in Structural Biology. 2014 doi: 10.1016/j.sbi.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Cummings RD, Smith DF, Huflejt M, Campbell CT, Gildersleeve JC, Gerlach JQ, Kilcoyne M, Joshi L, Serna S, et al. Cross-Platform Comparison of Glycan Microarray Formats. Glycobiology. 2014;24:507–517. doi: 10.1093/glycob/cwu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geissner A, Anish C, Seeberger PH. Glycan arrays as tools for infectious disease research. Current Opinion in Chemical Biology. 2014;18:38–45. doi: 10.1016/j.cbpa.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Wei J, Zheng L, Lv X, Bi Y, Chen W, Zhang W, Shi Y, Zhao L, Sun X, Wang F, et al. Analysis of Influenza Virus Receptor Specificity Using Glycan-Functionalized Gold Nanoparticles. ACS Nano. 2014 doi: 10.1021/nn5002485. [DOI] [PubMed] [Google Scholar]
- 5.Stowell SR, Arthur CM, McBride R, Berger O, Razi N, Heimburg-Molinaro J, Rodrigues LC, Gourdine JP, Noll AJ, von Gunten S, et al. Microbial glycan microarrays define key features of host-microbial interactions. Nature Chemical Biology. 2014;10:470–476. doi: 10.1038/nchembio.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song X, Heimburg-Molinaro J, Cummings RD, Smith DF. Chemistry of natural glycan microarrays. Current Opinion in Chemical Biology. 2014;18:70–77. doi: 10.1016/j.cbpa.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen HL, Fangel JU, McCleary B, Ruzanski C, Rydahl MG, Ralet MC, Farkas V, von Schantz L, Marcus SE, Andersen MC, et al. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. Journal of Biological Chemistry. 2012;287:39429–39438. doi: 10.1074/jbc.M112.396598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cid M, Pedersen HL, Kaneko S, Coutinho PM, Henrissat B, Willats WG, Boraston AB. Recognition of the helical structure of beta-1,4-galactan by a new family of carbohydrate-binding modules. Journal of Biological Chemistry. 2010;285:35999–36009. doi: 10.1074/jbc.M110.166330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert HJ, Knox JP, Boraston AB. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Current Opinion in Structural Biology. 2013;23:669–677. doi: 10.1016/j.sbi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Gabius HJ. Cell surface glycans: the why and how of their functionality as biochemical signals in lectin-mediated information transfer. Critical Reviews in Immunology. 2006;26:43–79. doi: 10.1615/critrevimmunol.v26.i1.30. [DOI] [PubMed] [Google Scholar]
- 11.Rillahan CD, Paulson JC. Glycan microarrays for decoding the glycome. Annual Review of Biochemistry. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525–546. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 13.Heimburg-Molinaro J, Lum M, Vijay G, Jain M, Almogren A, Rittenhouse-Olson K. Cancer vaccines and carbohydrate epitopes. Vaccine. 2011;29:8802–8826. doi: 10.1016/j.vaccine.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connaris H, Govorkova EA, Ligertwood Y, Dutia BM, Yang L, Tauber S, Taylor MA, Alias N, Hagan R, Nash AA, et al. Prevention of influenza by targeting host receptors using engineered proteins. Proceedings of the National Academy of Sciences. 2014;111:6401–6406. doi: 10.1073/pnas.1404205111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. Emerging principles for the therapeutic exploitation of glycosylation. Science. 2014;343:1235681. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- 16.Taylor ME, Drickamer K. Structural insights into what glycan arrays tell us about how glycan-binding proteins interact with their ligands. Glycobiology. 2009;19:1155–1162. doi: 10.1093/glycob/cwp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith DF, Song X, Cummings RD. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods in Enzymology. 2010;480:417–444. doi: 10.1016/S0076-6879(10)80033-3. [DOI] [PubMed] [Google Scholar]
- 18.Oyelaran O, Gildersleeve JC. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grant OC, Smith HMK, Firsova D, Fadda E, Woods RJ. Presentation, Presentation, Presentation! Molecular Level Insight into Linker Effects on Glycan Array Screening Data. Glycobiology. 2014;24:17–25. doi: 10.1093/glycob/cwt083. A molecular modelling approach is adopted to explore the impact of linker choice on glycan array screening data. The study provides a structure-based rationale for detecting false negatives arising from linker effects on binding of a monoclonal antibody to glycans containing the TF-disaccharide on an array surface. This work illustrates the importance of glycan presentation on recognition.
- 20.Wright CS. Structural comparison of the two distinct sugar binding sites in wheat germ agglutinin isolectin II. Journal of Molecular Biology. 1984;178:91–104. doi: 10.1016/0022-2836(84)90232-8. [DOI] [PubMed] [Google Scholar]
- 21.Gabius HJ, Andre S, Jimenez-Barbero J, Romero A, Solis D. From lectin structure to functional glycomics: principles of the sugar code. Trends in Biochemical Sciences. 2011;36:298–313. doi: 10.1016/j.tibs.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 22. Cuskin F, Flint JE, Gloster TM, Morland C, Basle A, Henrissat B, Coutinho PM, Strazzulli A, Solovyova AS, Davies GJ, et al. How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20889–20894. doi: 10.1073/pnas.1212034109. The authors identify a carbohydrate binding function for a module within SacC from Bacillus subtilis, an exo-acting levanase. They demonstrate that the combination of this module with the nonspecific catalytic domain of SacC confers specificity towards highly branched fructans. The study has broad implications for how glycan binding specificity is acheived and also for strategies directed at manipulating specificity.
- 23.Abbott DW, Bueren ALV. Using Structure to Inform Carbohydrate Binding Module Function. Current Opinion in Structural Biology. 2014 doi: 10.1016/j.sbi.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Lingwood D, Binnington B, Rog T, Vattulainen I, Grzybek M, Coskun U, Lingwood CA, Simons K. Cholesterol modulates glycolipid conformation and receptor activity. Nature Chemical Biology. 2011;7:260–262. doi: 10.1038/nchembio.551. [DOI] [PubMed] [Google Scholar]
- 25.GLYCAM Web, editor. Woods Group. Athens, GA: Complex Carbohydrate Research Center, University of Georgia; 2005–2014. ( http://www.glycam.com) [Google Scholar]
- 26.Engelsen SB, Cros S, Mackie W, Perez S. A molecular builder for carbohydrates: application to polysaccharides and complex carbohydrates. Biopolymers. 1996;39:417–433. doi: 10.1002/(SICI)1097-0282(199609)39:3%3C417::AID-BIP13%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Mishra SK, Adam J, Wimmerova M, Koca J. In silico mutagenesis and docking study of Ralstonia solanacearum RSL lectin: performance of docking software to predict saccharide binding. Journal of Chemical Information and Modeling. 2012;52:1250–1261. doi: 10.1021/ci200529n. [DOI] [PubMed] [Google Scholar]
- 28.Agostino M, Jene C, Boyle T, Ramsland PA, Yuriev E. Molecular docking of carbohydrate ligands to antibodies: structural validation against crystal structures. Journal of Chemical Information and Modeling. 2009;49:2749–2760. doi: 10.1021/ci900388a. [DOI] [PubMed] [Google Scholar]
- 29.Agostino M, Yuriev E, Ramsland PA. Antibody recognition of cancer-related gangliosides and their mimics investigated using in silico site mapping. PLoS ONE. 2012;7:e35457. doi: 10.1371/journal.pone.0035457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nivedha AK, Makeneni S, Foley BL, Tessier MB, Woods RJ. The Importance of Ligand Conformational Energies in Carbohydrate Docking: Sorting the Wheat from the Chaff. Journal of Computational Chemistry. 2013;35:526–539. doi: 10.1002/jcc.23517. A set of functions are developed to post-filter carbohydrate docking results by penalizing the score of poses according to the conformational energies of the glycosidic linkages. The resulting poses have improved shapes relative to crystal structures from the protein data bank (PDB).
- 31.Nurisso A, Kozmon S, Imberty A. Comparison of docking methods for carbohydrate binding in calcium-dependent lectins and prediction of the carbohydrate binding mode to sea cucumber lectin CEL-III. Molecular Simulation. 2008;34:469–479. [Google Scholar]
- 32. Kumar S, Frank M, Schwartz-Albiez R. Understanding the specificity of human Galectin-8C domain interactions with its glycan ligands based on molecular dynamics simulations. PLoS ONE. 2013;8:e59761. doi: 10.1371/journal.pone.0059761. The study demonstrates how superimposition of protein crystal structures and ligand swapping, followed by refinement with MD simulations, can provide a structure-based rationalization of experimentally observed GBP specificity. The differing specificities of Galectin 8 for blood groups determinants A and B and lactosamines are explained.
- 33. Martin JC, Fadda E, Ito K, Woods RJ. Defining the structural origin of the substrate sequence independence of O-GlcNAcase using a combination of molecular docking and dynamics simulation. Glycobiology. 2014;24:85–96. doi: 10.1093/glycob/cwt094. Ligand (O-GlcNAc) superimposition followed by MD simulation was employed to demonstrate that O-GlcNAcase (OGA) interacts primarily with the peptide backbone atoms, thus explaining the ability of OGA to hydrolyse O-GlcNAc independent of the peptide sequence proximal to the glycosylation site.
- 34.Bolia A, Woodrum BW, Cereda A, Ruben MA, Wang X, Ozkan SB, Ghirlanda G. A flexible docking scheme efficiently captures the energetics of glycan-cyanovirin binding. Biophysical Journal. 2014;106:1142–1151. doi: 10.1016/j.bpj.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerzmann A, Fuhrmann J, Kohlbacher O, Neumann D. BALLDock/SLICK: a new method for protein-carbohydrate docking. Journal of Chemical Information and Modeling. 2008;48:1616–1625. doi: 10.1021/ci800103u. [DOI] [PubMed] [Google Scholar]
- 36.Adam J, Kriz Z, Prokop MP, Wimmerova M, Koca J. In silico mutagenesis and docking studies of Pseudomonas aeruginosa PA-IIL lectin predicting binding modes and energies. Journal of Chemical Information and Modeling. 2008;48:2234–2242. doi: 10.1021/ci8002107. [DOI] [PubMed] [Google Scholar]
- 37. Tessier MB, Grant OC, Heimburg-Molinaro J, Smith D, Jadey S, Gulick AM, Glushka J, Deutscher SL, Rittenhouse-Olson K, Woods RJ. Computational screening of the human TF-glycome provides a structural definition for the specificity of antitumor antibody JAA-F11. PLoS One. 2013;8:e54874. doi: 10.1371/journal.pone.0054874. This study introduces a program for automated carbohydrate grafting and demonstrates its utility by screening a mAb against each TF-containing glycan in the known glycome. It also illustrates the use of glycan array data to validate theoretical specificity predictions.
- 38. Ballut L, Sapay N, Chautard E, Imberty A, Ricard-Blum S. Mapping of heparin/heparan sulfate binding sites on alphavbeta3 integrin by molecular docking. Journal of Molecular Recognition. 2013;26:76–85. doi: 10.1002/jmr.2250. Potential GAG-binding sites are identified on the αvβ3 integrin surface by docking a series of sulfated moieties of increasing size. The results from docking smaller fragments are used to narrow the search region when docking hexasaccharides. A sequence conservation analysis shows that basic amino acids found within the putative binding sites are semi-conserved across the human integrin family.
- 39.Chabrol E, Nurisso A, Daina A, Vassal-Stermann E, Thepaut M, Girard E, Vives RR, Fieschi F. Glycosaminoglycans are interactants of Langerin: comparison with gp120 highlights an unexpected calcium-independent binding mode. PLoS ONE. 2012;7:e50722. doi: 10.1371/journal.pone.0050722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samsonov SA, Teyra J, Pisabarro MT. Docking glycosaminoglycans to proteins: analysis of solvent inclusion. Journal of Computer-Aided Molecular Design. 2011;25:477–489. doi: 10.1007/s10822-011-9433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gandhi NS, Freeman C, Parish CR, Mancera RL. Computational analyses of the catalytic and heparin-binding sites and their interactions with glycosaminoglycans in glycoside hydrolase family 79 endo-beta-D-glucuronidase (heparanase) Glycobiology. 2012;22:35–55. doi: 10.1093/glycob/cwr095. Structural models are generated to explain heparanase specificity. The co-complexes are generated by threading GAG ligands onto known carbohydrate substrates and inhibitor ligands in the binding site of homology-modeled heparinase.
- 42.Sidhu PS, Mosier PD, Zhou Q, Desai UR. On scaffold hopping: challenges in the discovery of sulfated small molecules as mimetics of glycosaminoglycans. Bioorganic & Medicinal Chemistry Letters. 2013;23:355–359. doi: 10.1016/j.bmcl.2012.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mobius K, Nordsieck K, Pichert A, Samsonov SA, Thomas L, Schiller J, Kalkhof S, Teresa Pisabarro M, Beck-Sickinger AG, Huster D. Investigation of lysine side chain interactions of interleukin-8 with heparin and other glycosaminoglycans studied by a methylation-NMR approach. Glycobiology. 2013;23:1260–1269. doi: 10.1093/glycob/cwt062. [DOI] [PubMed] [Google Scholar]
- 44.Nordsieck K, Pichert A, Samsonov SA, Thomas L, Berger C, Pisabarro MT, Huster D, Beck-Sickinger AG. Residue 75 of interleukin-8 is crucial for its interactions with glycosaminoglycans. Chembiochem. 2012;13:2558–2566. doi: 10.1002/cbic.201200467. [DOI] [PubMed] [Google Scholar]
- 45. Topin J, Arnaud J, Sarkar A, Audfray A, Gillon E, Perez S, Jamet H, Varrot A, Imberty A, Thomas A. Deciphering the glycan preference of bacterial lectins by glycan array and molecular docking with validation by microcalorimetry and crystallography. PLoS One. 2013;8:e71149. doi: 10.1371/journal.pone.0071149. The study demonstrates the use of crystal structures of carbohydrate co-complexes (from the BIOLOGO database) as starting structures for docking calculations. Docking poses were filtered based on the ligand RMSD relative to known coordinates for Fucose in the receptor binding site. Additionally, poses were filtered by comparision with adiabatic maps of the glycosidic linkages, removing any unreasonable glycosidic conformations.
- 46.Davies GJ, Ducros VM, Varrot A, Zechel DL. Mapping the conformational itinerary of beta-glycosidases by X-ray crystallography. Biochemical Society Transactions. 2003;31:523–527. doi: 10.1042/bst0310523. [DOI] [PubMed] [Google Scholar]
- 47.Davies GJ, Mackenzie L, Varrot A, Dauter M, Brzozowski AM, Schulein M, Withers SG. Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase. Biochemistry. 1998;37:11707–11713. doi: 10.1021/bi981315i. [DOI] [PubMed] [Google Scholar]
- 48. Speciale G, Thompson AJ, Davies GJ, Williams SJ. Dissecting conformational contributions to glycosidase catalysis and inhibition. Current Opinion in Structural Biology. 2014 doi: 10.1016/j.sbi.2014.06.003. Experimental and computational methods for studying the catalysis reaction coordinate for glycosidic hydrolases are discussed.
- 49.Mark P, Baumann MJ, Eklof JM, Gullfot F, Michel G, Kallas AM, Teeri TT, Brumer H, Czjzek M. Analysis of nasturtium TmNXG1 complexes by crystallography and molecular dynamics provides detailed insight into substrate recognition by family GH16 xyloglucan endo-transglycosylases and endo-hydrolases. Proteins. 2009;75:820–836. doi: 10.1002/prot.22291. [DOI] [PubMed] [Google Scholar]
- 50.Mark P, Zhang Q, Czjzek M, Brumer H, Ågren H. Molecular dynamics simulations of a branched tetradecasaccharide substrate in the active site of a xyloglucan endotransglycosylase. Molecular Simulation. 2011;37:1001–1013. [Google Scholar]
- 51.Davies GJ, Planas A, Rovira C. Conformational analyses of the reaction coordinate of glycosidases. Accounts of Chemical Research. 2012;45:308–316. doi: 10.1021/ar2001765. [DOI] [PubMed] [Google Scholar]
- 52.Tvaroška I, Kozmon S, Wimmerová M, Koča J. A QM/MM Investigation of the Catalytic Mechanism of Metal-Ion-Independent Core 2 β1,6-N-Acetylglucosaminyltransferase. Chemistry – A European Journal. 2013;19:8153–8162. doi: 10.1002/chem.201300383. [DOI] [PubMed] [Google Scholar]
- 53.Lemieux RU, Delbaere LT, Beierbeck H, Spohr U. Involvement of water in host-guest interactions. Ciba Foundation Symposium. 1991;158:231–245. doi: 10.1002/9780470514085.ch15. discussion 245-238. [DOI] [PubMed] [Google Scholar]
- 54.Chervenak MC, Toone EJ. A Direct Measure of the Contribution of Solvent Reorganization to the Enthalpy of Binding. Journal of the American Chemical Society. 1994;116:10533–10539. [Google Scholar]
- 55.Clarke C, Woods RJ, Gluska J, Cooper A, Nutley MA, Boons GJ. Involvement of water in carbohydrate-protein binding. Journal of the American Chemical Society. 2001;123:12238–12247. doi: 10.1021/ja004315q. [DOI] [PubMed] [Google Scholar]
- 56.Saenger W. Structure and dynamics of water surrounding biomolecules. Annual Review of Biophysics and Biophysical Chemistry. 1987;16:93–114. doi: 10.1146/annurev.bb.16.060187.000521. [DOI] [PubMed] [Google Scholar]
- 57.Kadirvelraj R, Foley BL, Dyekjaer JD, Woods RJ. Involvement of water in carbohydrate-protein binding: concanavalin A revisited. Journal of the American Chemical Society. 2008;130:16933–16942. doi: 10.1021/ja8039663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mishra NK, Kulhanek P, Snajdrova L, Petrek M, Imberty A, Koca J. Molecular dynamics study of Pseudomonas aeruginosa lectin-II complexed with monosaccharides. Proteins. 2008;72:382–392. doi: 10.1002/prot.21935. [DOI] [PubMed] [Google Scholar]
- 59.Li Z, Lazaridis T. The effect of water displacement on binding thermodynamics: concanavalin A. Journal of Physical Chemistry B. 2005;109:662–670. doi: 10.1021/jp0477912. [DOI] [PubMed] [Google Scholar]
- 60.Gauto DF, Di Lella S, Estrin DA, Monaco HL, Marti MA. Structural basis for ligand recognition in a mushroom lectin: solvent structure as specificity predictor. Carbohydrate Research. 2011;346:939–948. doi: 10.1016/j.carres.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 61.Gauto DF, Di Lella S, Guardia CM, Estrin DA, Marti MA. Carbohydrate-binding proteins: Dissecting ligand structures through solvent environment occupancy. Journal of Physical Chemistry B. 2009;113:8717–8724. doi: 10.1021/jp901196n. [DOI] [PubMed] [Google Scholar]
- 62.Saraboji K, Hakansson M, Genheden S, Diehl C, Qvist J, Weininger U, Nilsson UJ, Leffler H, Ryde U, Akke M, et al. The carbohydrate-binding site in galectin-3 is preorganized to recognize a sugarlike framework of oxygens: ultra-high-resolution structures and water dynamics. Biochemistry. 2012;51:296–306. doi: 10.1021/bi201459p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gauto DF, Petruk AA, Modenutti CP, Blanco JI, Di Lella S, Marti MA. Solvent structure improves docking prediction in lectin-carbohydrate complexes. Glycobiology. 2013;23:241–258. doi: 10.1093/glycob/cws147. Sites of high solvent occupancy on the surface of unliganded GBPs were identified using MD simulations. The scoring function in AutoDock4 was modifed to include a bonus for poses in which glycan hydroxyls occupy these water sites. The subsequent docking results show an impressive improvement in docking accuracy for each of the tested structures.
- 64.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lutteke T, von der Lieth CW. pdb-care (PDB carbohydrate residue check): a program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics. 2004;5:69. doi: 10.1186/1471-2105-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weichenberger CX, Sippl MJ. NQ-Flipper: recognition and correction of erroneous asparagine and glutamine side-chain rotamers in protein structures. Nucleic Acids Research. 2007;35:W403–W406. doi: 10.1093/nar/gkm263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richichi B, Imberty A, Gillon E, Bosco R, Sutkeviciute I, Fieschi F, Nativi C. Synthesis of a selective inhibitor of a fucose binding bacterial lectin from Burkholderia ambifaria. Organic & Biomolecular Chemistry. 2013;11:4086–4094. doi: 10.1039/c3ob40520f. [DOI] [PubMed] [Google Scholar]
- 68. Satoh T, Suzuki K, Yamaguchi T, Kato K. Structural basis for disparate sugar-binding specificities in the homologous cargo receptors ERGIC-53 and VIP36. PLoS ONE. 2014;9:e87963. doi: 10.1371/journal.pone.0087963. The article presents an excellent example of combining crystallography and modelling (manual carbohydrate grafting) to define glycan binding specificity.
- 69.Costello C, Bundle DR. Synthesis of three trisaccharide congeners to investigate frame shifting of beta1,2-mannan homo-oligomers in an antibody binding site. Carbohydrate Research. 2012;357:7–15. doi: 10.1016/j.carres.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 70.Sapay N, Nurisso A, Imberty A. Simulation of carbohydrates, from molecular docking to dynamics in water. Methods Mol Biol. 2013;924:469–483. doi: 10.1007/978-1-62703-017-5_18. [DOI] [PubMed] [Google Scholar]
- 71.Fadda E, Woods RJ. Molecular simulations of carbohydrates and protein-carbohydrate interactions: motivation, issues and prospects. Drug Discovery Today. 2010;15:596–609. doi: 10.1016/j.drudis.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foley BL, Tessier MB, Woods RJ. Carbohydrate force fields. Wiley Interdisciplinary Reviews: Computational Molecular Science. 2012;2:652–697. doi: 10.1002/wcms.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yongye AB, Calle L, Arda A, Jimenez-Barbero J, Andre S, Gabius HJ, Martinez-Mayorga K, Cudic M. Molecular recognition of the Thomsen-Friedenreich antigen-threonine conjugate by adhesion/growth regulatory galectin-3: nuclear magnetic resonance studies and molecular dynamics simulations. Biochemistry. 2012;51:7278–7289. doi: 10.1021/bi300761s. [DOI] [PubMed] [Google Scholar]
- 74.Proctor EA, Yin S, Tropsha A, Dokholyan NV. Discrete molecular dynamics distinguishes nativelike binding poses from decoys in difficult targets. Biophysical Journal. 2012;102:144–151. doi: 10.1016/j.bpj.2011.11.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ford MG, Weimar T, Kohli T, Woods RJ. Molecular dynamics simulations of galectin-1-oligosaccharide complexes reveal the molecular basis for ligand diversity. Proteins. 2003;53:229–240. doi: 10.1002/prot.10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alonso H, Bliznyuk AA, Gready JE. Combining docking and molecular dynamic simulations in drug design. Medicinal Research Reviews. 2006;26:531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 77.Okimoto N, Futatsugi N, Fuji H, Suenaga A, Morimoto G, Yanai R, Ohno Y, Narumi T, Taiji M. High-performance drug discovery: computational screening by combining docking and molecular dynamics simulations. PLoS Computational Biology. 2009;5:e1000528. doi: 10.1371/journal.pcbi.1000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ranzinger R, Herget S, von der Lieth CW, Frank M. GlycomeDB--a unified database for carbohydrate structures. Nucleic Acids Research. 2011;39:D373–D376. doi: 10.1093/nar/gkq1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, Packer NH. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Research. 2014;42:D215–D221. doi: 10.1093/nar/gkt1128. [DOI] [PMC free article] [PubMed] [Google Scholar]