Abstract
The available techniques for assessing blood cell functions are limited considering the various types of blood cells and their diverse functions. In the past decade, rapid advancement in microengineering has enabled an array of blood cell functional measurements that are difficult or impossible to achieve using conventional bulk platforms. Such miniaturized blood cell assay platforms also provide attractive capabilities of reducing chemical consumption, cost, assay time, as well as exciting opportunities of device integration, automation, and assay standardization. This review summarizes these contemporary microengineering tools and discusses their promising potential for constructing accurate in vitro models and rapid clinical diagnosis using minimal amount of whole blood samples.
Keywords: blood cell, functional analysis, phenotyping, microengineering, microfluidics
Microengineering tools for functional blood cell analysis
Human blood circulating in the body reaches and exchanges information with every tissue through the vascular network, and is therefore an important indicator of the functional status of the human body. Many life-threatening diseases either are directly caused by abnormality of the blood or blood flow (e.g. ischemic heart disease, stroke, and diabetes), or can be detected through careful examination of molecular and cellular biomarkers circulating in the blood (e.g. cancer, HIV/AIDS, and tuberculosis) [1–4]. Because of their ready availability, blood cell analysis and phenotyping is arguably the most common and important test used in the clinic to provide physiological or pathological information for disease diagnosis and staging, treatment selection, safety and efficacy monitoring, as well as drug dose adjustment.
Complementary to complete blood count and morphologic analysis, functional blood cell analysis is sometimes necessary as they provide direct information regarding the functional status of the human body. Red blood cell (RBC) fragility and deformability [5], white blood cell (WBC) immune response [6], and platelet aggregation [7] are among the most common functional tests of blood cells. However, available techniques for assessing blood cell functions are limited, especially when considering the various types of blood cells and their diverse functions involved in different physiological and pathological contexts. Moreover, conventional tools for analyzing blood cells are bulky and costly, rely on complex manual operations and sample preparation, and are designed exclusively for research or clinical settings [8, 9]. Due to these common technical limitations, traditional blood cell analysis and phenotyping tools are still difficult for standardization and do not meet the needs of modern clinical and healthcare applications, including accurate and rapid testing on diverse functions of blood cells, point-of-care diagnostics, and construction of highly reliable in vitro models [10].
Recent advances in microengineering have offered researchers and clinicians an exciting new set of tools for accurate, fast, and affordable analysis of the cellular components of the blood (Box 1) [11, 12]. Their ability to precise control and manipulate single cells in a defined environment has enabled an array of functional measurements that are difficult or impossible to achieve on conventional bulk platforms. Such miniaturized assays also provide attractive capabilities of reducing chemical consumption, cost and assay time, as well as exciting opportunities of integrating blood cell analysis with upstream blood sample preparation on a monolithic platform [13]. The aim of this review is thus to introduce the recent achievements of microengineering tools for functional analysis and phenotyping of blood cells. Examples of how microengineering tools are adapted for analysis of RBCs, WBCs, and platelets are discussed. Finally, we offer speculations on the research directions and potential opportunities for microengineered blood cell analysis tools to meet current and future challenges of clinical and laboratory diagnosis.
Box 1. The microengineering toolbox.
Laminar flow
Fluid flow in most microfluidic devices is laminar due to the small geometrical sizes of microfluidic devices. The stable and predictable flow field for laminar flow makes it easy to maintain a pre-defined shear rate, the magnitude of which can be tuned by adjusting flow rate or microchannel geometry. Laminar flow can also be manipulated to create complex flow patterns such as flow focusing [28, 87] and hydrodynamic stretching [25, 58] (Figure IA).
Constriction channel
Microfluidic constriction channels are microchannels whose width is smaller than that of cells passing through the channels (Figure IB). They have been extensively used as mechanical means to deform blood cells to assess their deformability. Due to the ease of fabrication, almost all constriction microchannels have a rectangular cross section, which is different from the circular blood vessel shape. Despite this difference, constriction microchannels have been successful in retaining in vivo blood cell functionalities [88].
Microwell array
Microwell array is used for isolation and analysis of single blood cells (Figure IC) [89]. To ensure single cell trapping, blood cell suspension with a proper cell density is placed onto the microwell array and allowed to sediment into the microwells. One microwell array thus contains up to thousands of single cells, with each single cell trapped in individual microwells. Each of the microwells creates a confined cellular environment that can effectively concentrate analytes and amplify detection sensitivity.
Microcontact printing
Microcontact printing (μCP) is a simple yet highly versatile method to pattern proteins on various kinds of substrates [90, 91]. Briefly, a monolayer of protein is coated on a micropatterned elastomeric stamp. The stamp is then brought into direct contact with the target substrate to which proteins can preferentially bind. Only proteins in direct contact with the substrate are transferred onto the substrate (Figure ID). For blood cell functional analysis, μCP is mainly used for patterning adhesion proteins for cell adhesion and aggregation.
Micropost force sensor
The micropost force sensor was originally developed to measure cell traction force (Figure IE). It contains a regular array of vertical elastomeric posts with a post diameter down to 1 – 2 μm. The tips of microposts are functionalized with adhesive proteins, with post sidewalls passivated with non-adhesive molecules to ensure cells adherent to the post tops. When a cell exerts lateral contractile forces on the underlying posts, the posts will bend and the magnitude of cell contractile forces can be inferred by displacements of post tips.
Functional analysis of RBCs
RBCs are the most abundant cells in human blood with a normal concentration of around 5 × 109 cells mL−1. With a biconcave discoid shape and a diameter of 6 – 8 μm, they are highly differentiated cells that lack nucleus and most of the organelles [5]. Two properties of RBCs, deformability and adenosine triphosphate (ATP) release, are commonly measured for clinical and laboratory diagnosis for diseases such as malaria, sickle-cell disease, and pulmonary hypertension [14–16].
Deformability of RBCs
Healthy RBCs are smooth and extremely deformable so that they can easily pass through the spleen and microcapillaries. Conversely, abnormal stiffness of RBCs is usually an indicator of diseases. For example, in Plasmodium falciparum infected malaria patients, RBCs gradually lose their deformability with the progression of infection, and late stage infected RBCs can become stiffer by a factor of 50 [14]. There is also a loss of RBC deformability due to abnormal polymerization of hemoglobin in patients with sickle-cell disease [15]. In both diseases, hardened RBCs can impair blood circulation and may eventually lead to occlusion.
Conventional RBC deformability assays can be classified into bulk assays that measure the average effect of a RBC population and single cell assays that assess the deformability of single RBCs [5]. In a commonly used filtration assay, anti-coagulated blood is flowed through a filtration membrane with a pore diameter of 5 μm and the pressure drop across the membrane which correlates with RBC deformability is measured [17]. Because bulk RBC deformability assays deal with a RBC population, they cannot identify subsets of RBCs or rare RBCs that have direct pathological implications [18]. In contrast, single cell deformability measurements such as micropipette aspiration [19] and ektacytometry [20] apply hydrostatic or shear force on individual RBCs while simultaneously monitoring their morphological changes using microscopy. However, due to the complexity of experimental setup and cumbersome assay procedures, single cell assays for RBC deformability measurements tend to have a low throughput (1 – 10 cells hr−1).
The recent trend of using high-throughput and multiplex microfluidic technologies to redesign traditional bulk and single cell deformability assays has significantly improved their performance. For instance, a microchannel network was constructed with a constant channel depth of 6 μm and varying channel widths from 6 – 70 μm (Figure 1A) [21]. RBC suspension with a hematocrit of 40% was perfused through the network under a constant hydraulic pressure, and the average RBC transit velocity was measured to indicate cell deformability. Compared with the conventional filtration assay that works only under 1% hematocrit, this microfluidic perfusion assay was shown to be more sensitive to small changes of RBC deformability.
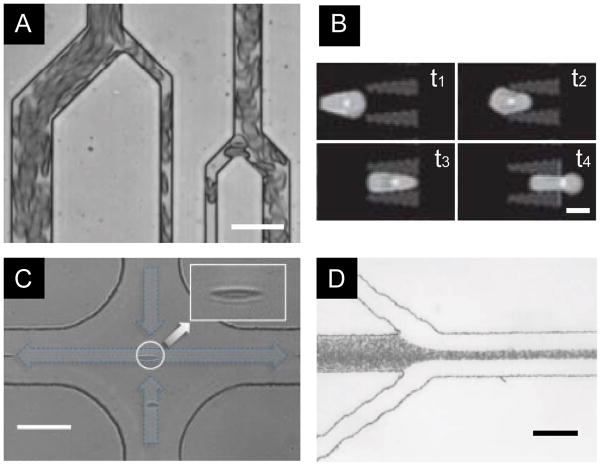
Figure 1.
Microfluidic cell deformability assays for RBCs. (A) A microfluidic vasculature network to perfuse RBC suspension, where RBCs were deformed by constriction channels [21]. (Scale bar, 30 μm.) Reproduced with permission from [21]. (B) Time-lapse images showing deformation of a single RBC moving through a constriction [22]. (Scale bar, 5 μm.) Reproduced with permission from [22]. (C) In a hydrodynamic stretching device using viscoelastic fluid, single RBCs were spontaneously focused to the center of the channel and stretched by shear force as they approached the intersection [25]. (Scale bar, 50 μm.) Reproduced with permission from [25]. (D) Flow focusing was used to deform RBCs and trigger their ATP release [28]. (Scale bar, 100 μm.) Reproduced with permission from [28].
Microfluidic constriction channels that resemble micropipettes have also been developed for measuring the deformability of single RBCs in a high throughput manner (Figure. 1B) [22]. A two-dimensional microfluidics constriction array, for example, is capable of measuring the deformability of 104 cells simultaneously [22, 23]. Such high throughput single cell measurement made it possible to identify a small subset of P. falciparum infected RBCs from a large background population of normal RBCs. Another strategy for increasing single cell assay throughput has been developed by continuously pushing single RBCs through a constriction, while measuring the cell transition time in real time using integrated electrodes based on the Coulter principle [24]. A throughput of 10 cells sec−1 was reported using such continuous flow operation.
The aforementioned microfluidic devices all utilize defined microscale constrictions to mechanically deform RBCs while the cells are passing through the constrictions under the influence of external forces. However, such confined microfluidic environment unavoidably leads to clogging. A newly developed hydrodynamic stretching device can effectively avoid clogging by using inertia focusing and fluid shear force to stretch single RBCs [25]. Shape of the deformed RBCs was recorded using high-speed camera, and a cell shape elongation index was used to describe RBC deformability (Figure 1C).
Label-free cellular biomarkers such as mechanical deformability are attractive targets for point-of-care testing, as measuring such markers may eliminate the use of chemicals which are often costly and difficult for transportation and storage. With the ability of rapid, high-throughput measurements of mechanical properties of RBCs, microfluidic deformability assays may provide promising solutions for rapid diagnostic testing of diseases such as malaria. It should be noted that however, such microfluidic deformability assay platforms need more careful characterization and validation using clinical blood samples to fully assess their sensitivity, accuracy, and clinical utility.
ATP release of RBCs
RBC-derived ATP stimulates nitric oxide (NO) synthesis by endothelial cells, which in turn induces relaxation of vascular smooth muscle cells in the blood vessel wall to facilitate passage of RBCs in narrow vascular regions [26, 27]. Impaired ATP release has been shown to correlate with pulmonary hypertension [16]. ATP concentration is commonly measured using luciferase assays which can be conveniently combined with a microfluidic constriction channel or flow focusing that deforms RBCs and triggers their ATP release (Figure 1D) [28]. An important advantage of such microfluidic ATP releasing assays is the real time detection capability that is useful for providing biological insights of ATP biosynthesis. This feature was exploited in an elegant study investigating the time delay between cell deformation and ATP release in RBCs [29]. Microchannels used in this study contained a segment of narrow constriction to locally deform and stimulate single RBCs. Deformed RBCs were allowed to recover downstream of the narrow constriction. Using this device, it was shown that the response time between RBC deformation and ATP release was tens of milliseconds, and this delay decreased with increasing shear stress and was independent of RBC stiffness.
Functional analysis of WBCs
WBCs are functional units of the immune system that protects human body from foreign invaders. The numbers, compositions, and functional responses of WBCs change drastically in the presence of infections, malignancies, and autoimmune disorders. In particular, WBCs can secrete soluble proteins, termed “cytokines”, to regulate the growth, maturation, and responsiveness of immune cells. The production of interferon gamma (IFN-γ) by T-cells, for example, correlates with the body’s ability to mount a vigorous immune response against tuberculosis infection [30]. Thus, detection of cytokine secretion from WBCs is of great importance for both fundamental understanding of human immunity and for immune monitoring in healthy and diseased humans with allergy, asthma, autoimmunity, acquired and primary immunodeficiency, transplantation, and infection. In inflammatory response, WBCs also need to emigrate from the blood vessel to enter targeting tissues, a process dependent on WBC deformability, adhesion, and shear rate of the blood flow [31]. In this section, we focus on two types of WBC functional analyses – functional immunophenotyping and deformability measurements, which have recently been achieved using innovative microengineering tools.
Functional immunophenotyping of WBCs
Functional immunophenotyping refers to the identification of cytokine secretion levels by immune cells, and can be conducted at the whole cell population, subpopulation, or single cell level [32]. Recently, different novel microengineering tools have been developed to improve traditional functional immunophenotyping assays for WBCs.
Miniaturized ELISA/ELISpot/AlphaLISA
Enzyme-linked immunosorbent assay/spot (ELISA/ELISpot) is the current gold standard in quantifying cytokine secretion from WBCs. However, conventional ELISA/ELISpot requires multiple washing and incubation, and is notorious for tedious manual operation and prolonged assay time. Miniaturized ELISA/ELISpot was developed with the aim of reducing sample consumption and assay time while maintaining high sensitivity [33–35]. Notably, miniaturized immunoassays are convenient for integration with upstream blood cell separation, and such integrated cell isolation and biodetection microfluidic devices are ideal for assessing functional status of subsets of immune cells or even single immune cells. For example, microscale anti-CD4 and anti-CD8 antibodies were printed on poly-(ethyleneglycol) (PEG) hydrogel-coated glass slides to capture CD4+ and CD8+ T-cells directly from RBC-depleted blood samples (Figure 2A&B) [36]. On the same slide next to anti-CD4 and anti-CD8 antibody spots, interleukin-2 (IL-2) and IFN-γ specific antibodies were printed to detect IL-2 and IFN-γ secreted from captured T-cells, respectively [36]. This antibody array device was later successfully combined with a holographic imaging system for point-of-care applications [37].
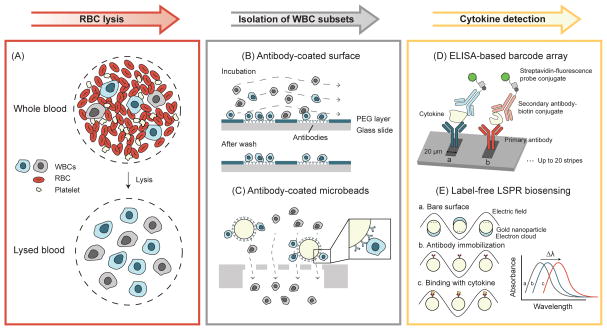
Figure 2.
Paradigm of functional immunophenotyping of WBCs using microengineering tools. Background RBCs are first removed from whole blood samples by lysis. Target WBC subsets are then isolated for cytokine secretion measurements. (A) Schematic of RBC lysis. (B) Antibody-coated surface for capturing subsets of WBCs. Target WBCs are bound to the antibodies coated on the glass slide during incubation, while unbounded cells are washed away [36]. (C) Isolation of subsets of WBCs using antibody-coated microbeads and microfilters. Target WBCs are retained on the filter as the microbead is larger than the filter pore size, while all other undesired WBCs pass through the filter freely [48]. (D) High-density antibody barcode using sandwich ELISA for multiplexed detection of secreted proteins [44, 45]. Adapted with permission from [44]. (E) Principle of LSPR biosensing on gold nanoparticle surface. The absorbance spectrum peak shift is used to quantify the amount of cytokine secreted by WBCs [51]. Adapted with permission from [51].
Cellular heterogeneity is common in an isogenic cell population. Thus, quantitative immunophenotyping of single immune cells is desirable for precise assessment of patient immune status [38, 39]. By reducing the size of antibody spots on PEG gels, single immune cells were captured, and cytokine secretions from the captured single cells were measured [40]. Another elegant technique for single cell immunophenotyping is the microwell array designed for trapping individual WBCs in a confined microwell environment [41–43]. Cytokines secreted by single WBCs were confined in the microwell, resulting in significantly increased cytokine concentrations that were readily detected by antibodies coated on the coverslip sealing the microwell. This microwell immunophenotyping assay was successfully utilized for simultaneous quantification of three different cytokines secreted from single WBCs. More recently, the microwell array was combined with a high-density antibody barcode array for simultaneous detection of 14 cytokines secreted from single WBCs (Figure 2D) [44, 45].
AlphaLISA is a newly developed homogeneous no-wash bead-based chemiluminescence immunodetection assay [46]. Using AlphaLISA, a microfluidic immunophenotyping assay (MIPA) device capable of on-chip cell trapping, endotoxin stimulation, and in situ cytokine measurement was recently developed [47]. The MIPA device incorporated a polydimethylsiloxane (PDMS) microfiltration membrane (PMM) for isolation and enrichment of WBCs. The PMM further allowed diffusion of cytokines secreted from WBCs trapped on the PMM to an underlying immunoassay chamber, where biodetection using AlphaLISA was conducted. The MIPA device required 20-fold fewer cells than traditional immunoassays, and the total assay time was 7 times shortened than conventional ELISA. More recently, the MIPA device was combined with antibody-coated microbeads for isolation and immunophenotyping of different subpopulations of WBCs from whole blood specimens (Figure 2C) [48].
Although all aforementioned methods used on-chip microwell or microfiltration structures to capture target immune cells, they still required some human intervention to carry out immunosensing. To address this limitation, a microfluidic system incorporating integrated microvalves was recently developed for single cell isolation and automation of ELISA [49]. This highly integrated and automated system minimized human intervention and could potentially be utilized for improving standardization of ELISA-based immunoassays.
Label-free methods
Conventional immunoassays require labeling analytes with fluorescent molecules for detection, rendering such immunoassays as endpoint detection methods. To investigate dynamic immune responses, semiconductor nanowires were fabricated to enable label-free detection of low concentrations of antibodies and were further applied for real-time monitoring of immune responses of WBCs [50]. More recently, a label-free immunoassay based on localized surface plasmon resonance (LSPR) was developed to detect tumor necrosis factor (TNF)-α secreted by CD45+ cells isolated directly from human blood (Figure 2E) [51]. The assay time of these label-free immunosensing methods is much shorter than conventional ELISA; however, their clinical utility is still limited due to necessary manual sample preparation before immunosensing [52]. Further optimization and integration of such label-free immunosensing methods with on-chip sample preparation components would be required for these label-free methods to fulfill their potential for clinical applications [53].
Deformability of WBCs
Abnormality in WBC deformability is an important indicator of diseases such as sepsis [54] and diabetes [55]. However, the low-throughput associated with traditional deformability assays hamper their use in clinical applications. An on-chip micropipette aspiration system was developed to improve the throughput of tradition micropipette aspiration, where single WBCs were trapped and deformed at a constriction by a carefully designed microfluidic circuit [56]. The threshold pressure allowing the cell to pass through the microscale constriction was recorded. Using this device the authors were able to conduct single cell deformability assays at a throughput of 30 cells hr−1. An astonishingly high throughput of 2,000 cell sec−1was achieved by the deformability cytometer based on inertial focusing and fluidic shear force for cell deformation [57, 58]. Interestingly, scatter plots of cell size and deformability (here defined as the ratio of the long axis length of the cell body to its short axis length) for a population of WBCs resembled a cytometry plot, suggesting that cell size and deformability were completely decoupled traits for WBCs.
Functional analysis of platelets
Platelets are the smallest and second most abundant corpuscle in the blood. Platelets are derived from fragments of the cytoplasm of megakaryocytes and thus do not contain nucleus. Normal platelets are discoid-shaped cells with a diameter of 2 – 4 μm and a thickness of about 0.5 μm. Upon activation, platelets change into a more round shape with long dendritic extensions to facilitate adhesion [59]. Primary functions of platelets are hemostasis and thrombosis, both of which are related to coagulation, a complex and highly regulated process [60]. Platelet adhesion and contraction are two important functional phenotypes involved in coagulation that have been assessed using newly developed microengineered tools.
Platelet adhesion
Platelet adhesion is the first step in clot formation and is mediated by various plasma and extracellular matrix (ECM) proteins (e.g. von Willebrand factor, fibrinogen, and collagen) and mechanical cues such as shear force of the blood flow. Failure of platelet adhesion is considered as a marker of various coagulation disorders, including von Willebrand disease (vWD), hemophilia A and B, and afibrinogenemia [60]. To reconstruct the process of platelet adhesion during hemostasis, collagen stripes and dots were deposited in microfluidic channels using microfluidic patterning and microcontact printing to mimic a wound formed in vivo (Figure 3A) [61–63]. The latest version of the device incorporated a parallel channel design to generate a broad range of shear rates (50 – 920 s−1) on the same chip to investigate the effect of shear stress on platelet adhesion and clot formation, with results suggesting a threshold ECM pattern size of 20 μm required for platelet adhesion and faster platelet clotting under higher shear stress conditions [63]. This threshold ECM pattern size for clot formation was also observed in another study using tissue factor (TF) patches of predefined sizes coated on lipid bilayers [64, 65]. Another important factor regulating hemostasis is the transthrombus pressure gradient that drives bleeding. This parameter was specially examined in a microfluidic device that could independently control shear rate and transthrombus pressure by incorporating multiple fluid/pressure inlets and outlets on one single microfluidic chip (Figure 3B) [66].
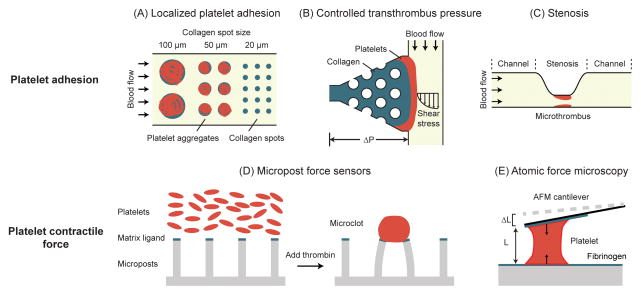
Figure 3.
Microengineered devices for measurements of platelet adhesion and contraction. (A) Platelet aggregates on collagen spots of different sizes after perfusion with whole blood. A threshold collagen pattern size of 20 μm was necessary for platelet adhesion [63]. Adapted with permission from [63]. (B) Microfluidic thrombosis device to independently control transthrombus pressure and wall shear stress. Localized collagen plug surrounding posts defines platelet adhesion site. In addition, its permeability allows for control of transthrombus pressure [66]. Adapted with permission from [66]. (C) A microfluidic device with high shear stenosis region for inducing thrombus formation in whole blood [67]. Adapted with permission from [67]. (D) Micropost array for measurement of platelet contractile force (PCF). Platelets form microclots and exert contractile forces on the micropost force sensors after adding thrombin [80]. Adapted with permission from [80]. (E) AFM for measuring the PCF of single platelets. As a single platelet contracts, its length shortens and cause the AFM cantilever to deflect [83]. Adapted with permission from [83].
In another study targeting thrombosis, a stenosis was included in a microfluidic channel to assess the time needed for a clot to form and completely block the stenosis (i.e. occlusion time) (Figure 3C) [67]. Real time blood clot formation was monitored by measuring light transmission through thrombus - as thrombus grew, light transmission would increase. By integrating multiple channels on the same device, the authors varied flow rates from physiological to pathological conditions (500 – 13,000 s−1) and observed that occlusion would occur only with shear rate above 4,000 s−1 [67]. Similarly, a microfluidic device containing microscale constrictions was constructed to mimic stenosis formed in arteriole [68, 69]. Combining this in vitro microfluidic system with intravital microscopy, it was demonstrated that hydrodynamics could play a pivotal role in the formation of thrombus at the stenosis.
In addition to shear rate, pressure, and ECM pattern size, microfluidic channels were also utilized to study the effect of agonist/antagonist on platelet adhesion. Microchannels made of NO releasing polymers were fabricated to independently tune NO concentration and shear rate [70]. These assays revealed that platelet adhesion and aggregation would diminish when NO flux exceeded a certain threshold and would completely disappear if NO flux became too high.
Platelet contraction
In the final step of blood coagulation, the platelet-fibrin clot retracts, resulting in decreased fluid drag [71] and stiffening of blood clots [72, 73]. Blood clot retraction is mediated by actomyosin-based contraction of actin microfilaments in platelets, giving rise to platelet contractile force (PCF) [74]. PCF plays an essential role in coagulation, and can serve as a functional marker for platelet dysfunction [75, 76]. Various conventional methods (e.g. platelet clot strip [77] and clot retractometry [78]) have been developed to measure PCF. However, these techniques are designed for bulk studies and therefore cannot recapitulate microscale platelet-fibrin interactions and the delicate dynamics of blood clot formation.
Since the concept of PCF is similar to that of cell traction force exerted by adherent mammalian cells on the ECM, well-established methods for cell traction force measurements have been adapted for assessing PCF. For example, traction force microscopy was employed to study dynamic evolution of PCF during platelet activation, where microbeads were incorporated into the surface of a compliant polymer substrate, and platelets were allowed to adhere and exert PCF to the substrate [79]. When platelets started contracting, displacements of beads were observed and recorded to deduce the force field. Another popular cell traction force measurement tool termed micropost force sensors was also utilized to measure PCF [80]. Platelets were seeded on top of PDMS microposts pre-coated with fibrinogen or fibronectin to facilitate platelet adhesion. Once clotting was triggered by addition of thrombin, deflections of PDMS microposts were observed and recorded to calculate PCF based on the Euler-Bernoulli beam theory (Figure 3D). Variations of this method had been developed to study the effects of glycoprotein Ib, von Willebrand factor VWF [81], as well as shear force [82] on platelet contraction. In another recent study, a custom-built side-view atomic force microscopy (AFM) was used to measure PCF generated by single platelets bridging the AFM cantilever and a substrate (Figure 3E) [83].
The microengineering tools discussed above for PCF measurements have significantly improved the measurement accuracy and sensitivity to an unprecedented level. However, these tools were designed primarily for mechanistic studies of PCF and thus are complicated and suboptimal for direct clinical applications. Automated microengineering tools that are convenient to use for measurements of PCF is yet to be developed for identifying disorders in hemostasis and thrombosis.
Concluding remarks and future perspectives
It is clear that microengineering tools have already made a positive impact on transforming the functional analysis and phenotyping of blood cells. First, microengineering tools consumes much less blood samples and assay chemicals, making them highly desirable for applications where such resources are limited or costly. Prominent examples include the microfluidic immunoassays for pediatric and neonatal patients [47] and platelet functional assays for high-throughput screening (HTS) of antiplatelet drugs [84]. Second, microengineering tools allow for precise control and manipulation of blood cells and their microenvironment at a scale comparable to single cells and microcapillaries. This enables accurate and sensitive functional analysis of blood cells, such as RBC deformation and ATP release, WBC deformability, platelet adhesion and contraction, to an unprecedented level. Finally, microengineering tools brings about exciting opportunities for assay automation by integrated device designs, paving the way for assay standardization and point-of-care applications.
With an increasing number of studies utilizing microengineering tools to reveal fundamental blood cell physiology and mechanisms of blood-related diseases, the majority of current microdevices are designed for a single type of blood cells only. In reality each of the blood cells resides in a complex environment where multiple types of cells coexist and influence each other. To construct more reliable in vitro models, a promising approach is the organ-on-a-chip devices that can be adapted to construct blood vessel mimics and incorporate multiple blood cell types [85]. Such organ-on-a-chip devices are promising platforms to study endothelium-blood cell and RBC-WBC-platelet interactions. As for practical applications such as rapid disease diagnosis of malaria and HIV/AIDS, microengineered blood cell analysis tools still need to be further simplified and automated [86]. Particularly, integration with on-chip blood sample preparation will be required for a much higher degree of device automation.
The field of microengineering is quickly evolving to address the challenges ahead. We believe that future microengineering tools will continue to offer new functionalities for revealing novel insights of blood cell physiology and exciting opportunities for advanced clinical diagnosis using minimal blood samples.
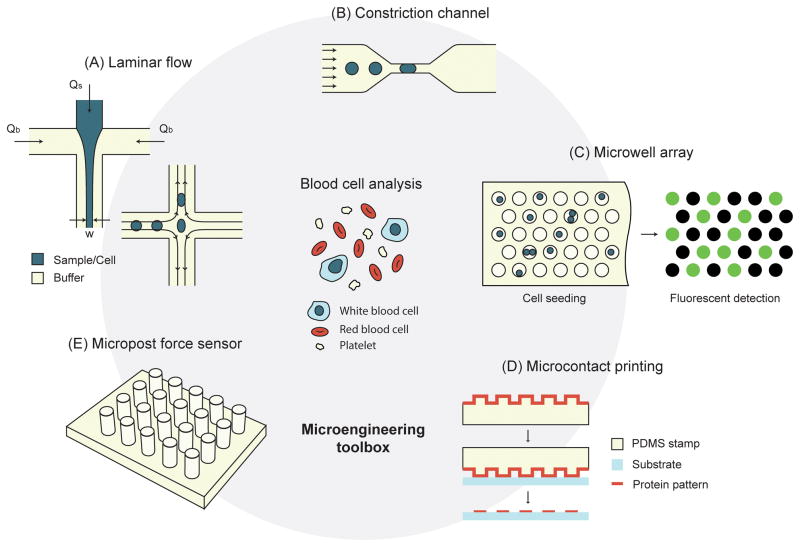
Box 1, Figure I. The microengineering toolbox.
(A) Microfluidic control of laminar flow. In one example, sample flow is focused by buffer flows to a width of w, which is determined by the ratio of buffer flow rate (Qb) to sample flow rate (Qs). In another example, extensional shear force was generated at the intersection of four perpendicular flows. (B) Microfluidic constriction channel for cell deformation assays. (C) Microwell array for simultaneous capture and analysis of thousands of single cells. Fluorescence-based biodetection (e.g. ELISA) can be used to measure the amount of proteins secreted by each single cell trapped in the microwell. (D) Microcontact printing for selective surface functionalization. The protein is absorbed on the micropatterned surface of a PDMS stamp before transferred onto a substrate by direct contact of the two surfaces. (E) Micropost force sensor for measuring contractile force of platelets and blood clots.
Highlights.
Microengineering tools are capable of precise manipulation of single cells to achieve rapid, high-resolution, multi-parameter functional analysis and phenotyping of blood cells.
Microfluidics enables high-throughput deformability measurement on single RBCs and WBCs, as well as easy assessment of ATP release by RBCs.
Microengineered functional immunoassays enhance the performance of traditional immunoassays and allow for label-free detection of cytokines.
Microengineering tools enable easy and accurate evaluation on a variety of platelet functions from platelet adhesion to platelet contraction.
Acknowledgments
We acknowledge financial support from the National Science Foundation (ECCS 1231826 and CBET 1263889), the National Institutes of Health (1R01HL119542), the American Heart Association (13PRE16510018), the UM-SJTU Collaboration on Biomedical Technologies, the UM Comprehensive Cancer Center Prostate SPORE Pilot Project, the Michigan Institute for Clinical & Health Research (MICHR) Pilot Program (UL1RR024986), the Michigan Center for Integrative Research in Critical Care (M-CIRCC), and the Department of Mechanical Engineering at the University of Michigan, Ann Arbor. Finally, we extend our apologies to all colleagues in the field whose work we are unable to discuss formally because of space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Willmot M, et al. High blood pressure in acute stroke and subsequent outcome: a systematic review. Hypertension. 2004;43:18–24. doi: 10.1161/01.HYP.0000105052.65787.35. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara G, et al. Use in routine clinical practice of two commercial blood tests for diagnosis of infection with Mycobacterium tuberculosis: a prospective study. Lancet. 2006;367:1328–1334. doi: 10.1016/S0140-6736(06)68579-6. [DOI] [PubMed] [Google Scholar]
- 3.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. The New England Journal of Medicine. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 4.Piot P, Quinn TC. Response to the AIDS pandemic--a global health model. The New England Journal of Medicine. 2013;368:2210–2218. doi: 10.1056/NEJMra1201533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien S. Red cell deformability and its relevance to blood flow. Annu Rev Physiol. 1987;49:177–192. doi: 10.1146/annurev.ph.49.030187.001141. [DOI] [PubMed] [Google Scholar]
- 6.Bhorade SM, et al. Cylex ImmuKnow assay levels are lower in lung transplant recipients with infection. The Journal of Heart and Lung Transplantation. 2008;27:990–994. doi: 10.1016/j.healun.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Favaloro EJ. Seminars in Thrombosis and Hemostasis. Thieme Medical Publishers; 2008. Clinical utility of the PFA-100; pp. 709–733. ©. [DOI] [PubMed] [Google Scholar]
- 8.Yager P, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 9.Chin CD, et al. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab Chip. 2007;7:41–57. doi: 10.1039/b611455e. [DOI] [PubMed] [Google Scholar]
- 10.Sackmann EK, et al. The present and future role of microfluidics in biomedical research. Nature. 2014;507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 11.Folch A, Toner M. MICROENGINEERING OF CELLULAR INTERACTIONS. Annual Review of Biomedical Engineering. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 12.Chen CS, et al. Microengineering the Environment of Mammalian Cells in Culture. MRS Bulletin. 2005;30:194–201. [Google Scholar]
- 13.El-Ali J, et al. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 14.Cranston HA, et al. Plasmodium falciparum maturation abolishes physiologic red cell deformability. Science. 1984;223:400–403. doi: 10.1126/science.6362007. [DOI] [PubMed] [Google Scholar]
- 15.Barabino GA, et al. Sickle cell biomechanics. Annu Rev Biomed Eng. 2010;12:345–367. doi: 10.1146/annurev-bioeng-070909-105339. [DOI] [PubMed] [Google Scholar]
- 16.Sprague RS, et al. Impaired release of ATP from red blood cells of humans with primary pulmonary hypertension. Experimental Biology and Medicine. 2001;226:434–439. doi: 10.1177/153537020122600507. [DOI] [PubMed] [Google Scholar]
- 17.Reid H, et al. A simple method for measuring erythrocyte deformability. Journal of Clinical Pathology. 1976;29:855–858. doi: 10.1136/jcp.29.9.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlo DD, Lee LP. Dynamic single-cell analysis for quantitative biology. Analytical chemistry. 2006;78:7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 19.Hochmuth RM. Micropipette aspiration of living cells. J Biomech. 2000;33:15–22. doi: 10.1016/s0021-9290(99)00175-x. [DOI] [PubMed] [Google Scholar]
- 20.Groner W, et al. New optical technique for measuring erythrocyte deformability with the ektacytometer. Clinical Chemistry. 1980;26:1435–1442. [PubMed] [Google Scholar]
- 21.Shevkoplyas SS, et al. Direct measurement of the impact of impaired erythrocyte deformability on microvascular network perfusion in a microfluidic device. Lab Chip. 2006;6:914–920. doi: 10.1039/b601554a. [DOI] [PubMed] [Google Scholar]
- 22.Bow H, et al. A microfabricated deformability-based flow cytometer with application to malaria. Lab Chip. 2011;11:1065–1073. doi: 10.1039/c0lc00472c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S, et al. Dynamic deformability of Plasmodium falciparum-infected erythrocytes exposed to artesunate in vitro. Integr Biol (Camb) 2013;5:414–422. doi: 10.1039/c2ib20161e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y, et al. Electrical measurement of red blood cell deformability on a microfluidic device. Lab Chip. 2013;13:3275–3283. doi: 10.1039/c3lc50427a. [DOI] [PubMed] [Google Scholar]
- 25.Cha S, et al. Cell stretching measurement utilizing viscoelastic particle focusing. Anal Chem. 2012;84:10471–10477. doi: 10.1021/ac302763n. [DOI] [PubMed] [Google Scholar]
- 26.Sprague RS, et al. ATP: the red blood cell link to NO and local control of the pulmonary circulation. American Journal of Physiology-Heart and Circulatory Physiology. 1996;271:H2717–H2722. doi: 10.1152/ajpheart.1996.271.6.H2717. [DOI] [PubMed] [Google Scholar]
- 27.Sprague RS, et al. Deformation-induced ATP release from red blood cells requires CFTR activity. American Journal of Physiology-Heart and Circulatory Physiology. 1998;275:H1726–H1732. doi: 10.1152/ajpheart.1998.275.5.H1726. [DOI] [PubMed] [Google Scholar]
- 28.Moehlenbrock MJ, et al. Use of microchip-based hydrodynamic focusing to measure the deformation-induced release of ATP from erythrocytes. Analyst. 2006;131:930–937. doi: 10.1039/b605136g. [DOI] [PubMed] [Google Scholar]
- 29.Wan J, et al. Dynamics of shear-induced ATP release from red blood cells. Proceedings of the National Academy of Sciences. 2008;105:16432–16437. doi: 10.1073/pnas.0805779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flynn JL, et al. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. The Journal of Experimental Medicine. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong C, et al. Mechanics of leukocyte deformation and adhesion to endothelium in shear flow. Annals of Biomedical Engineering. 1999;27:298–312. doi: 10.1114/1.143. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, et al. Emerging microfluidic tools for functional cellular immunophenotyping: a new potential paradigm for immune status characterization. Frontiers in Oncology. 2013:3. doi: 10.3389/fonc.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrmann M, et al. Microfluidic ELISA on non-passivated PDMS chip using magnetic bead transfer inside dual networks of channels. Lab on a chip. 2007;7:1546–1552. doi: 10.1039/b707883h. [DOI] [PubMed] [Google Scholar]
- 34.Eteshola E, Leckband D. Development and characterization of an ELISA assay in PDMS microfluidic channels. Sensors and Actuators B: Chemical. 2001;72:129–133. [Google Scholar]
- 35.Herrmann M, et al. Quantification of low-picomolar concentrations of TNF-α in serum using the dual-network microfluidic ELISA platform. Analytical Chemistry. 2008;80:5160–5167. doi: 10.1021/ac800427z. [DOI] [PubMed] [Google Scholar]
- 36.Zhu H, et al. A microdevice for multiplexed detection of T-cell-secreted cytokines. Lab Chip. 2008;8:2197–2205. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]
- 37.Stybayeva G, et al. Lensfree holographic imaging of antibody microarrays for high-throughput detection of leukocyte numbers and function. Analytical Chemistry. 2010;82:3736–3744. doi: 10.1021/ac100142a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W, et al. Emerging Microfluidic Tools for Functional Immunophenotyping: A New Potential Paradigm for Immune Status Characterization. Frontiers in Oncology. 1 doi: 10.3389/fonc.2013.00098. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Bodovitz S. Single cell analysis: the new frontier in ‘omics’. Trends in Biotechnology. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu H, et al. Detecting cytokine release from single T-cells. Analytical Chemistry. 2009;81:8150–8156. doi: 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love JC, et al. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nature Biotechnology. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 42.Han Q, et al. Polyfunctional responses by human T cells result from sequential release of cytokines. Proceedings of the National Academy of Sciences. 2012;109:1607–1612. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varadarajan N, et al. Rapid, efficient functional characterization and recovery of HIV-specific human CD8+ T cells using microengraving. Proceedings of the National Academy of Sciences. 2012;109:3885–3890. doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan R, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nature Biotechnology. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Y, et al. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal Chem. 2013;85:2548–2556. doi: 10.1021/ac400082e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beaudet L, et al. AlphaLISA immunoassays: the no-wash alternative to ELISAs for research and drug discovery. Nature Methods. 2008;5 [Google Scholar]
- 47.Huang NT, et al. An integrated microfluidic platform for in situ cellular cytokine secretion immunophenotyping. Lab Chip. 2012;12:4093–4101. doi: 10.1039/c2lc40619e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W, et al. Surface-Micromachined Microfiltration Membranes for Efficient Isolation and Functional Immunophenotyping of Subpopulations of Immune Cells. Advanced Healthcare Materials. 2013;2:965–975. doi: 10.1002/adhm.201200378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eyer K, et al. Implementing Enzyme-Linked Immunosorbent Assays on a Microfluidic Chip To Quantify Intracellular Molecules in Single Cells. Analytical Chemistry. 2013;85:3280–3287. doi: 10.1021/ac303628j. [DOI] [PubMed] [Google Scholar]
- 50.Stern E, et al. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature. 2007;445:519–522. doi: 10.1038/nature05498. [DOI] [PubMed] [Google Scholar]
- 51.Oh BR, et al. Integrated Nanoplasmonic Sensing for Cellular Functional Immunoanalysis Using Human Blood. ACS Nano. 2014;8:2667–2676. doi: 10.1021/nn406370u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stern E, et al. Label-free biomarker detection from whole blood. Nature Nanotechnology. 2009;5:138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, et al. Continuous-flow microfluidic blood cell sorting for unprocessed whole blood using surface-micromachined microfiltration membranes. Lab Chip. 2014 doi: 10.1039/c4lc00350k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drost EM, et al. Increased rigidity and priming of polymorphonuclear leukocytes in sepsis. American Journal of Respiratory and Critical Care Medicine. 1999;159:1696–1702. doi: 10.1164/ajrccm.159.6.9803061. [DOI] [PubMed] [Google Scholar]
- 55.Pécsvarády Z, et al. Decreased polymorphonuclear leukocyte deformability in NIDDM. Diabetes Care. 1994;17:57–63. doi: 10.2337/diacare.17.1.57. [DOI] [PubMed] [Google Scholar]
- 56.Guo Q, et al. Microfluidic micropipette aspiration for measuring the deformability of single cells. Lab Chip. 2012;12:2687–2695. doi: 10.1039/c2lc40205j. [DOI] [PubMed] [Google Scholar]
- 57.Di Carlo D. Inertial microfluidics. Lab Chip. 2009;9:3038–3046. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 58.Gossett DR, et al. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proceedings of the National Academy of Sciences. 2012;109:7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.George JN. Platelets. Lancet. 2000;355:1531–1539. doi: 10.1016/S0140-6736(00)02175-9. [DOI] [PubMed] [Google Scholar]
- 60.Harrison P. Platelet function analysis. Blood Rev. 2005;19:111–123. doi: 10.1016/j.blre.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 61.Colace T, et al. Analysis of morphology of platelet aggregates formed on collagen under laminar blood flow. Ann Biomed Eng. 2011;39:922–929. doi: 10.1007/s10439-010-0182-4. [DOI] [PubMed] [Google Scholar]
- 62.Neeves KB, et al. Microfluidic focal thrombosis model for measuring murine platelet deposition and stability: PAR4 signaling enhances shear-resistance of platelet aggregates. J Thromb Haemost. 2008;6:2193–2201. doi: 10.1111/j.1538-7836.2008.03188.x. [DOI] [PubMed] [Google Scholar]
- 63.Hansen RR, et al. High content evaluation of shear dependent platelet function in a microfluidic flow assay. Ann Biomed Eng. 2013;41:250–262. doi: 10.1007/s10439-012-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kastrup CJ, et al. Characterization of the threshold response of initiation of blood clotting to stimulus patch size. Biophysical Journal. 2007;93:2969–2977. doi: 10.1529/biophysj.107.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kastrup CJ, et al. Modular chemical mechanism predicts spatiotemporal dynamics of initiation in the complex network of hemostasis. Proceedings of the National Academy of Sciences. 2006;103:15747–15752. doi: 10.1073/pnas.0605560103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muthard RW, Diamond SL. Side view thrombosis microfluidic device with controllable wall shear rate and transthrombus pressure gradient. Lab Chip. 2013;13:1883–1891. doi: 10.1039/c3lc41332b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li M, et al. Microfluidic system for simultaneous optical measurement of platelet aggregation at multiple shear rates in whole blood. Lab Chip. 2012;12:1355–1362. doi: 10.1039/c2lc21145a. [DOI] [PubMed] [Google Scholar]
- 68.Nesbitt WS, et al. A shear gradient-dependent platelet aggregation mechanism drives thrombus formation. Nature Medicine. 2009;15:665–673. doi: 10.1038/nm.1955. [DOI] [PubMed] [Google Scholar]
- 69.Tovar-Lopez FJ, et al. A microfluidics device to monitor platelet aggregation dynamics in response to strain rate micro-gradients in flowing blood. Lab Chip. 2010;10:291–302. doi: 10.1039/b916757a. [DOI] [PubMed] [Google Scholar]
- 70.Sylman JL, et al. Transport limitations of nitric oxide inhibition of platelet aggregation under flow. Ann Biomed Eng. 2013;41:2193–2205. doi: 10.1007/s10439-013-0803-9. [DOI] [PubMed] [Google Scholar]
- 71.Basmadjian D. The hemodynamic forces acting on thrombi, from incipient attachment of single cells to maturity and embolization. Journal of Biomechanics. 1984;17:287–298. doi: 10.1016/0021-9290(84)90139-8. [DOI] [PubMed] [Google Scholar]
- 72.Jen CJ, McIntire LV. The structural properties and contractile force of a clot. Cell motility. 1982;2:445–455. doi: 10.1002/cm.970020504. [DOI] [PubMed] [Google Scholar]
- 73.Storm C, et al. Nonlinear elasticity in biological gels. Nature. 2005;435:191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 74.Geiger B, et al. Transmembrane crosstalk between the extracellular matrix and the cytoskeleton. Nature Reviews Molecular Cell Biology. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 75.Collet J, et al. Altered fibrin architecture is associated with hypofibrinolysis and premature coronary atherothrombosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26:2567–2573. doi: 10.1161/01.ATV.0000241589.52950.4c. [DOI] [PubMed] [Google Scholar]
- 76.HVAS AM, et al. Tranexamic acid combined with recombinant factor VIII increases clot resistance to accelerated fibrinolysis in severe hemophilia A. Journal of Thrombosis and Haemostasis. 2007;5:2408–2414. doi: 10.1111/j.1538-7836.2007.02755.x. [DOI] [PubMed] [Google Scholar]
- 77.Cohen I, de Vries A. Platelet contractile regulation in an isometric system. Nature. 1973;246:36–37. doi: 10.1038/246036a0. [DOI] [PubMed] [Google Scholar]
- 78.Carr ME., Jr Development of platelet contractile force as a research and clinical measure of platelet function. Cell Biochem Biophys. 2003;38:55–78. doi: 10.1385/CBB:38:1:55. [DOI] [PubMed] [Google Scholar]
- 79.Henriques SS, et al. Force field evolution during human blood platelet activation. Journal of Cell Science. 2012;125:3914–3920. doi: 10.1242/jcs.108126. [DOI] [PubMed] [Google Scholar]
- 80.Liang XM, et al. Platelet retraction force measurements using flexible post force sensors. Lab Chip. 2010;10:991–998. doi: 10.1039/b918719g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feghhi S, et al. E-Beam Nanopost Arrays Reveal That Glycoprotein Ib-IV-X Complex and Von Willebrand Factor Interactions Transmit Platelet Cytoskeletal Forces. ASME 2013 Summer Bioengineering Conference; American Society of Mechanical Engineers; 2013. pp. V01BT50A004–V001BT050A004. [Google Scholar]
- 82.Ting LH, et al. Platelet Retraction Forces Induced Under High Shear Gradient Activation. ASME 2013 Summer Bioengineering Conference; American Society of Mechanical Engineers; 2013. pp. V01BT49A003–V001BT049A003. [Google Scholar]
- 83.Lam WA, et al. Mechanics and contraction dynamics of single platelets and implications for clot stiffening. Nature Materials. 2010;10:61–66. doi: 10.1038/nmat2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Colace TV, et al. Microfluidics and coagulation biology. Annu Rev Biomed Eng. 2013;15:283–303. doi: 10.1146/annurev-bioeng-071812-152406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huh D, et al. Microengineered physiological biomimicry: Organs-on-Chips. Lab Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 86.Chin CD, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nature Medicine. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 87.Dudani JS, et al. Pinched-flow hydrodynamic stretching of single-cells. Lab Chip. 2013;13:3728–3734. doi: 10.1039/c3lc50649e. [DOI] [PubMed] [Google Scholar]
- 88.Shelby JP, et al. A microfluidic model for single-cell capillary obstruction by Plasmodium falciparum-infected erythrocytes. Proceedings of the National Academy of Sciences. 2003;100:14618–14622. doi: 10.1073/pnas.2433968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Di Carlo D, et al. Dynamic single cell culture array. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 90.Bernard A, et al. Microcontact printing of proteins. Advanced Materials. 2000;12:1067–1070. [Google Scholar]
- 91.Kane RS, et al. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]