Abstract
Virus-encoded molecular signatures, such as cytosolic double-stranded or otherwise biochemically distinct RNA species, trigger cellular antiviral signaling. Cytoplasmic proteins recognize these non-self RNAs and activate signal transduction pathways that drive the expression of virus-induced genes, including the primary antiviral cytokine, IFNβ, and diverse direct and indirect antiviral effectors [1–4]. One important group of cytosolic RNA sensors known as the RIG-I like receptors (RLRs) is comprised of three proteins that are similar in structure and function. The RLR proteins, RIG-I, MDA5, and LGP2, share the ability to recognize nucleic acid signatures produced by virus infections and activate antiviral signaling. Emerging evidence indicates that RNA detection by RLRs culminates in the assembly of dynamic multimeric ribonucleoprotein (RNP) complexes. These RNPs can act as signaling platforms that are capable of propagating and amplifying antiviral signaling responses. Despite their common domain structures and similar abilities to induce antiviral responses, the RLRs differ in their enzymatic properties, their intrinsic abilities to recognize RNA, and their ability to assemble into filamentous complexes. This molecular specialization has enabled the RLRs to recognize and respond to diverse virus infections, and to mediate both unique and overlapping functions in immune regulation [5, 6].
Overview of RLR Structure and Function
RLR proteins are characterized by the fusion of RNA binding, ATP hydrolysis, and signal transduction domains into a single antiviral sentry (Figure 1). All three RLR proteins share a prominent DECH-box helicase domain that is required for dsRNA binding and ATP hydrolysis. Like other superfamily 2 helicase proteins, this domain’s catalytic core is composed of two RecA-like domains (Hel1 and Hel2), and these two fundamental helicase domains are interrupted by an intervening insertion, Hel2i, within the N-terminus of Hel2. Prominent, highly conserved helicase domain sequence motifs cluster within the two RecA subdomains and function to coordinate dsRNA binding and ATP hydrolysis [7]. These motifs are identifiable in the three RLRs, but shared variations clearly distinguish these motifs as related versions of the consensus sequences [8]. Although the term “helicase” is used to indicate unwinding of double stranded DNA and RNA, DECH-box proteins are known to have many diverse functions, including translocation along single-stranded and double-stranded nucleic acids, unwinding of double-stranded nucleic acids, annealing of complementary strands, and displacement of proteins from ribonucleoprotein complexes [9]. There is little evidence for duplex unwinding activity by the RLR proteins, but ATP hydrolysis is essential for proper RLR signal transduction. Each RLR uses ATP to power specific actions related to their RNA recognition and oligomerization properties, required for activation of antiviral signal transduction.
Figure 1. RLR family protein domain structures.
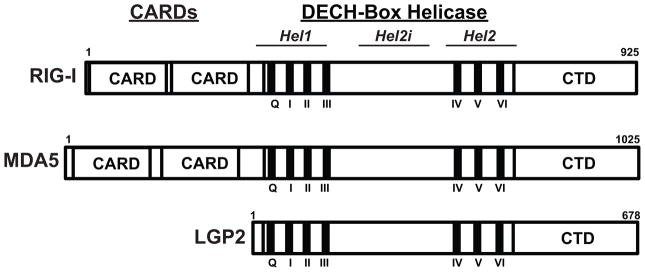
Diagram illustrating domain structure and features of RIG-I, MDA5, and LGP2. The three RLRs are composed of a central DECH-box Helicase domain that encompasses conserved helicase subdomains, Hel1 (surrounding helicase motifs Q, I, II, and III) and Hel2 (surrounding helicase motifs IV, V, and VI). Between them lies the helicase insert domain, Hel2i. A C-terminal domain (CTD) is required for auto-regulation and RNA terminus recognition by RIG-I and shares some similarity with MDA5 and LGP2. RIG-I and MDA5 contain tandem caspase activation and recruitment domain (CARD) regions at their N-termini, which are essential for interactions with MAVS to promote downstream signaling.
At the C-terminus of the RLRs is a regulatory domain that has been implicated in recognition of non-self RNA termini and both cis and trans regulation of antiviral signaling [10–12]. Analysis of the RLR CTDs demonstrated that the analogous regions of LGP2 and MDA5 exhibit some affinity for the termini of dsRNA, but only the RIG-I CTD specifically recognizes RNA 5′ end tri-phosphorylation [10, 13–17]. The RLR proteins differ in their intrinsic abilities to interact with RNA substrates, and recent structural and biochemical experiments have revealed that RIG-I and MDA5 are able to assemble into filamentous structures in association with dsRNA templates. This common ordered-aggregation mechanism facilitates the oligomerization of their N-terminal CARD domains, protein interaction modules that can associate with and nucleate the oligomerization of the essential mitochondria-localized antiviral signaling protein, MAVS. Organization of the MAVS CARD by RIG-I or MDA5 can initiate the formation of detergent-resistant, high-molecular-weight MAVS polymers. These activated MAVS fibers can seed further activation of MAVS molecules, perpetuating and amplifying the antiviral signal [18, 19]. Current information suggests that the initiation of MAVS fibril assembly is a central outcome of RNA recognition by the RLR proteins, and is critical for the efficient activation of downstream antiviral signaling cascades. Here, fundamental aspects of RLR-mediated RNA recognition, assembly, and signal transduction are reviewed, with particular emphasis on the enzymatic and functional distinctions between RIG-I, MDA5, and LGP2.
RIG-I
RIG-I is the first recognized RLR, and serves as a prototype [20]. RIG-I is auto-inhibited at steady state, ensuring low activity in the absence of high-affinity ligand RNAs. Crystal structures have revealed that in the absence of suitable RNA ligands, hydrophobic interactions and salt bridges between the second CARD and the Hel2i domain form a repressed structure, leaving the CTD free to scan the cytoplasm for suitable RNA ligands. Engagement of an appropriate RNA ligand by the CTD conformationally activates RIG-I to assume signaling competence by releasing the auto-repressed CARDs and exposing a functional ATP-binding helicase domain [12, 21, 22]. Thus, RNA interaction induces RIG-I’s ATP hydrolysis activity, which is required for signaling. RIG-I is activated by interactions with short dsRNA containing a 5′ tri- or di- phosphate and base-paired ends, a non-self signature common to many virus genomes, replication intermediates, and defective-interfering particles, including poly (I:C), viral hairpins and DI genomes, and HCV poly U-rich UTR RNA [23]. Cells derived from RIG-I knockout mice fail to initiate antiviral signaling programs, resulting in reduced production of IFNβ, failure of antiviral immunity, and broad susceptibility to virus infections [24]. These diverse activating ligands reflect the broad range of RNA viruses that have been demonstrated to be susceptible to detection by RIG-I.
After RNA recognition, the exposed CARDs are able to interact with the MAVS CARD, seeding its oligomerization and initiating the assembly of antiviral signaling complexes. ATP hydrolysis is essential for RIG-I signal transduction, and ATPase-deficient mutants have a dominant-negative phenotype able to suppress signaling by wild-type RIG-I [8]. The precise roles played by ATP hydrolysis in RIG-I signaling remain to be fully accounted for, but it is likely that enzymatic activity enables the protein to more effectively achieve its activated conformational state for downstream signaling. Intriguingly, the ATP hydrolysis requirement for RIG-I signaling can be bypassed by mutations to conserved helicase motifs that inactivate enzymatic activity but render the protein constitutively active. These hyperactive alleles likely encode RIG-I proteins that constitutively assume de-repressed conformations that expose the CARDs, allowing their oligomerization in the absence of virus infection or ligand stimulation [8].
Single molecule analysis of RIG-I-RNA interactions has demonstrated that it can use the energy from ATP hydrolysis to translocate along the dsRNA duplex [25]. Though the biological impact of RIG-I translocation is yet to be clarified, this movement was suggested to enable RIG-I to discriminate templates based on the length of time spent in translocation, or to displace viral RNA-binding proteins that obscure substrate recognition. More recent results indicate that ATP-dependent translocation may also increase the frequency or efficiency of RIG-I assembly into high molecular weight aggregates on RNA templates. RIG-I interacts with short target dsRNAs as a monomer, with the CTD mediating end recognition, and there is little evidence for cooperative binding under these conditions [12, 21, 22], though it must be noted that many contributing studies analyzed RIG-I fragments that may not accurately represent native interactions, or were carried out in conditions that did not include ATP hydrolysis [26]. Monomeric RIG-I binds to a minimal 9–10 base pair dsRNA with a 5′ triphosphate [27], but oligomerization on longer base-paired stretches of RNA was observed under ATP hydrolysis conditions. Oligomerization was further correlated with enhanced signaling ability, suggesting that a higher order structure represents the most active conformation [28].
Biochemical and electron microscopic analysis of the oligomeric state of the full length RIG-I revealed that it is capable of assembling into a filamentous structure on dsRNA in an ATP-dependent process [29]. Current evidence suggests a recognition and activation process (Figure 2A) in which monomers of RIG-I initially interact with dsRNA ends (5′-PPP) that are recognized by the CTD. This substrate recognition simultaneously exposes the CARDs, while the helicase domain engages the dsRNA template and uses ATP hydrolysis to translocate away from the end and toward the dsRNA interior. Thus, the monomeric recognition of RNA is ATP-independent and produces end-capping structures prior to translocation. ATP-mediated translocation results in tighter stacking of RIG-I oligomers into a more filamentous architecture. RIG-I translocation is not very processive under the conditions tested, and these RIG-I RNP structures have limited ability to spread, resulting in short, end-directed filaments on dsRNA. Direct contact between monomers is required for appropriate, signaling-competent RIG-I filament packing [29].
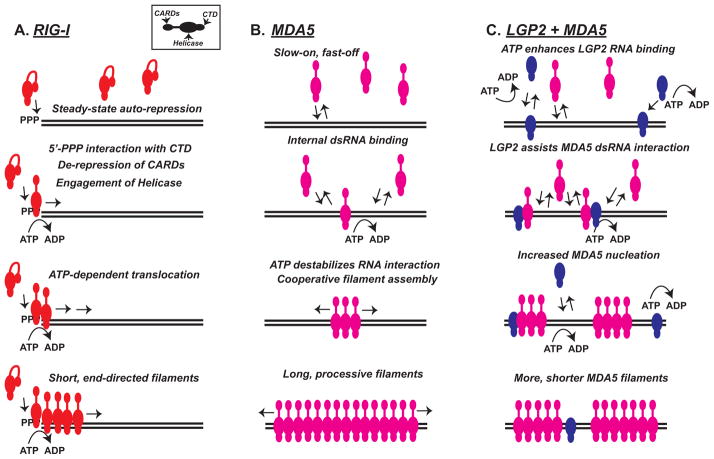
Figure 2. Comparison of RNA filament assembly by RLR proteins.
Panels illustrate features of RLR assembly onto RNA templates. Each RLR protein is illustrated as a colored module, and inset box demonstrates positions of CARD, Helicase and CTD regions. Black lines indicate dsRNA.
A) RIG-I (red) is auto-repressed at steady state by CARD-Hel2i interactions. Recognition of a 5′-triphosphorylated dsRNA terminus (5′-PPP) by the CTD results in CARD de-repression. The helicase domain mediates ATP-dependent translocation away from the RNA terminus with limited processivity, and leads to packaging of RIG-I into short, end-directed filaments.
B) MDA5 (magenta) has a low monomeric RNA binding affinity that is insensitive to terminal structures and characterized by a slow on-rate. ATP-mediated destabilization prevents MDA5 accumulation on dsRNA, but cooperative interactions between monomers can enhance processive filament assembly. Long MDA5 filaments are observed in vitro.
C) LGP2 (blue) has a high affinity for dsRNA and uses ATP hydrolysis to enable high-efficiency binding to diverse RNA species. LGP2 is able to facilitate and stabilize MDA5-dsRNA interactions leading to increased filament nucleation, producing a greater number of shorter MDA5 filaments.
Several studies have indicated that ubiquitin has a fundamental function in RIG-I activation [30, 31]. It has been demonstrated that K63-linked ubiquitin can be conjugated to the RIG-I CARD by the TRIM25 protein, and that this modification facilitates downstream antiviral signaling [32]. In addition, it has been demonstrated that unattached K63-linked ubiquitin chains can promote RIG-I activation through a non-covalent mechanism in vitro and in vivo [33]. Structural studies have revealed the non-covalent K63 ubiquitin chains can organize RIG-I CARD-CARD interactions to form a tetrameric structure that is bridged by three K63-ubiquitin chains. These tetrameric CARDs are sufficient to interact with and organize the assembly of the MAVS CARD, promoting the initiation of MAVS filament formation and its subsequent prion-like aggregation [34]. The efficacy of RIG-I CARDs to simulate MAVS filament assembly was enhanced by covalent K63 ubiquitin conjugation. As such, three mechanisms can non-exclusively contribute to RIG-I signaling by organizing the CARDs into higher-order oligomers: (i) non-covalent interactions with ubiquitin chains, (ii) covalent modification by ubiquitin, and (iii) ATP-mediated filament formation. Together, these mechanisms ensure efficient and specific RIG-I activation by diverse pathogens that can present appropriate non-self RNA species.
MDA5
MDA5 shares the overall domain structure of RIG-I with tandem CARDs fused to homologous helicase and CTD regions (Figure 1), and is thought to signal through a similar CARD-mediated, MAVS-dependent system to activate antiviral gene expression. Unlike the auto-inhibited RIG-I, expression of MDA5 alone is sufficient to activate the IFNβ gene in the absence of specific RNA recognition [8, 35], though its activity is regulated in vivo by CARD ubiquitination, phosphorylation, and direct or indirect association with other antiviral mediators [3, 36–39]. MDA5 deficient mice fail to respond to treatment with the synthetic dsRNA analog, poly(I:C), and exhibit greater susceptibility to certain positive-sense single-stranded RNA viruses, including the picornaviruses, poliovirus and encephalomyocarditis virus (EMCV), and murine norovirus [24, 40, 41], all of which are poorly recognized by RIG-I.
There is a relative lack of detailed information regarding MDA5 RNA recognition substrates, in part due to the apparently poor RNA binding activity of MDA5. However, a few studies have elucidated potential RNA features, modifications, or specific viral RNA regions that are discriminated by MDA5. MDA5 was found to be activated by enzymatically digested or sheared populations of RNA longer than 2kbp [42], and high molecular weight RNAs extracted from virus-infected cells were shown to preferentially activate MDA5-mediated signaling [43]. It was proposed that structural features, such as RNA branches found in RNAs with both single-stranded and double-stranded regions might be required for recognition by MDA5. This is consistent with the observation that poly (I:C) is able to activate MDA5 in vitro and in vivo [24, 40]. MDA5 may also be able to discriminate some features specific to virus-derived mRNAs, including 2′-O-methylation or secondary structures [44, 45]. EMCV is a virus that effectively escapes RIG-I detection by masking its RNA 5′ ends and replicates more efficiently in the absence of either MDA5 or LGP2. A region of the transcribed negative-strand RNA acts as a physiological agonist of MDA5 -dependent signaling that is necessary and sufficient for MDA5-mediated antiviral responses to EMCV infection [46]. However, this EMCV-encoded MDA5 agonist RNA was identified based on its association with LGP2, and binds poorly to MDA5. Therefore, while RNA features specific for MDA5 recognition were not revealed, this study added to the increasing evidence that LGP2 acts as a collaborator for MDA5 RNA recognition [47]. Further attempts to isolate MDA5-specific RNA ligands have exploited photo-activated RNA crosslinking of measles virus-infected cells to preserve low-affinity interactions during subsequent immunoprecipitation. The captured RNA sequences were evaluated by statistical approaches to determine if MDA5 binds particular viral motifs. Although RNA features specific for MDA5 recognition were not clearly identified, the informatic analysis revealed a correlation between RNA A/U content and the activation of both RIG-I and MDA5 signal transduction. Curiously, A/U-rich RNAs that stimulate MDA5 signaling were found to be less effective at stimulating MDA5 ATP hydrolysis. ATP hydrolysis is essential for MDA5 signal transduction [8], and mutations that disrupt MDA5 ATP hydrolysis are inactive but, unlike RIG-I, do not interfere with wild-type MDA5 in trans. The reasons for this difference are unclear but may be related to the unique ways that MDA5 and RIG-I utilize ATP hydrolysis in generating signaling-competent oligomers or filamentous assemblies.
Despite its relatively low solution RNA binding affinity compared to RIG-I or LGP2, electron microscopy has revealed that MDA5 is able to assemble cooperatively into filaments on dsRNA, with ring-like asymmetric units that form helical twists [48–53]. The unit structures of MDA5 and RIG-I are similar, with the helicase domains surrounding the dsRNA core. However, while RIG-I uses its CTD to bind tightly to the 5′-triphosphate ends, MDA5 does not preferentially recognize dsRNA ends and instead the MDA5 CTD associates with its helicase domain to form a more complete ring around the dsRNA duplex. Data indicate that MDA5 initially binds slowly, with low affinity, to dsRNA. Once engaged, MDA5 oligomerization is mediated by cooperative binding, and long RNP filaments can be assembled in vitro (Figure 2B). In the presence of the ATP transition state analogue, ADP-AlF4, these structures can assemble processively into long head-to-tail filaments, but are destabilized by MDA5 ATP hydrolysis [51, 53–55]. Physiological ATP levels promote MDA5 dissociation from RNA, and these long RNP filaments have yet to be detected inside living cells. The observed long MDA5 filaments may represent a captured transition state formed during MDA5 signaling rather than a stable intracellular structure. Moreover, it remains likely that additional proteins or post-translational modifications regulate the extent of MDA5 filament assembly in vivo during virus infections.
Recent investigation of mouse and human genetic abnormalities that contribute to autoimmune syndromes has revealed a novel clinical connection to MDA5. Early identification of MDA5 characterized it as a melanoma- differentiation associated gene [56, 57], and it was also implicated in the regulation of nuclear remodeling occurring during apoptosis [58]. These biological regulatory properties implicate MDA5 in biological actions beyond innate antiviral signaling. An early intersection of MDA5 with autoimmune disease was established by a genome-wide association linking the MDA5 locus as a risk factor for type 1 diabetes [59]. MDA5 connections to autoimmune disease were extended to lupus susceptibility [60], and other diseases that are characterized by chronic IFN stimulated gene expression signatures [61].
Molecular insights into the ability of MDA5 to participate in autoimmune signaling were acquired recently from a study of mutagenized mice that acquired a lupus-like phenotype, including chronic multi-organ inflammation, widespread lymphocyte infiltration, and unregulated cytokine expression. These mice featured widespread IFN and IFN-stimulated gene expression, and this was dependent on MDA5 signaling through MAVS [62]. A single point mutation in MDA5 (G821S) was found to produce an MDA5 protein defective in ATP hydrolysis activity, but constitutively active for signal transduction. As a result, the mouse cells homozygous for the mutant MDA5 express high constitutive levels of IFNβ but cannot properly recognize and respond to the challenge of infection with EMCV. Although these findings may seem to present a paradox for RLR signaling, prior studies clearly established a precedent for interpreting the mutant phenotype. Engineered mutations targeting the human MDA5 helicase domain can produce MDA5 proteins with constitutive and hyperactive interferon signal transduction capacity in the absence of RNA binding and ATP hydrolysis [8]. Paradoxically, while MDA5 does require ATP hydrolysis for its antiviral activity, some catalytically-inactive MDA5 mutants are capable of constitutive signaling. The discovery of a signaling-competent, ATPase-deficient MDA5 mutant as the basis mammalian autoimmunity [62] suggests that a greater appreciation of the mechanisms underlying MDA5 signaling is needed to reconcile its roles in regulating both autoimmunity and antiviral signaling.
The mouse mutant demonstrated the potential for MDA5 to mediate autoimmunity, and the discovery of MDA5 variants in human patients underscores its clinical importance to autoimmunity. Aicardi-Goutieres Syndrome (AGS) is a hereditary autoimmune disease that is characterized by early onset of neurological degeneration and deterioration of myelinated nerve fibers [63]. Most relevantly, AGS is also characterized by constitutive expression of IFN response gene signatures. Applying whole-exome sequencing determined the molecular basis underpinning genetically uncharacterized cases of AGS to be gain-of-function mutations in MDA5. Analysis of these mutant MDA5 proteins determined that they have normal or increased RNA binding activity and are competent for ATP hydrolysis. It will be interesting to analyze the connections and distinctions among these altered MDA5 alleles to determine the factors that lead to increased IFN signaling and heightened antiviral immunity versus the autoimmune manifestations of SLE, AGS, or autoimmune diabetes. These new findings bring to mind a comment from the late Dr. Jurg Tschopp, that “undoubtedly, MDA5 has not yet disclosed all its secrets” [64].
LGP2
The third member of the RLR family shares sequence conservation within the helicase domain and the CTD, but lacks CARD or other known signaling domains. Despite its recognition as a close relative of MDA5 and RIG-I, the functions of LGP2 in innate antiviral immunity remain to be completely understood. A number of experiments carried out in vivo and in vitro have revealed antithetic activities for LGP2 as both an activator and an inhibitor of RLR-mediated antiviral signaling [47, 65], and three independent LGP2 knockout mouse lines were reported with distinct but overlapping phenotypes [11, 66–70]. LGP2 can mediate cellular responses related to viral RNA recognition and antiviral signaling, and participates in antiviral T cell expansion [69], responses triggered by cytosolic dsDNA [71], and cancer cell resistance to ionizing radiation [72].
The LGP2 mRNA is transcribed in response to virus infections or stimulation with antiviral mediators including poly(I:C) and IFNs [11, 66, 67, 73–75]. It has been established that LGP2 expressed from plasmid vectors can act as a negative regulator of RLR signaling [11, 66, 67], suggesting that LGP2 functions as a feedback inhibitor of antiviral responses. However, while the mechanistic basis for LGP2-mediated feedback inhibition remains unclear, it has been attributed to PAMP RNA sequestration [67], inhibitory protein interactions [11], and direct interference with MAVS signaling [66]. These mechanisms may or may not be mutually exclusive and could represent temporally or spatially separated functions in RLR signaling.
Accumulating results indicate that LGP2 also participates in virus detection and the positive regulation of antiviral signaling. Mice lacking LGP2 are more susceptible to picornaviruses that had been previously linked to detection by MDA5 [24, 68]. Co-expression of LGP2 with MDA5 synergistically increases antiviral signaling activity, and it has been demonstrated that LGP2 can enhance MDA5-dependent responses [35, 68, 76, 77]. LGP2 positive regulation of antiviral signaling through MDA5 is further supported by virological implications. It is specifically noted that the EMCV-derived MDA5 agonist RNA was identified through its association with LGP2, and has little affinity for MDA5 [46]. In addition, a number of RNA viruses in the Paramyxovirus family encode antagonist proteins that interfere with ATP hydrolysis by directly binding to the Hel2 domain of both MDA5 and LGP2, but not RIG-I [78].
The connections between MDA5 and LGP2 are yet to be fully deciphered, but the positive and negative properties of LGP2 can be distinguished from one another on the basis of their requirements for RNA binding and ATP hydrolysis. The negative regulatory activity of LGP2 is independent of enzymatic activity and remains intact despite mutations that target key helicase domain residues [8, 35, 47, 79]. In contrast, helicase domain mutations disrupt LGP2’s ability to co-activate MDA5 signaling, which requires both ATP hydrolysis and RNA binding [35, 47, 79]. Unlike the RNA-induced activity of other RLRs, LGP2 has a high basal ATP hydrolysis activity that is independent of RNA binding [8, 35]. RNA interaction further stimulates LGP2 enzymatic activity, and LGP2 uses this basal ATP hydrolysis to enhance its ability to scan the cytoplasm and efficiently engage diverse dsRNA species. This ATP-powered RNA interaction is connected to the ability of LGP2 to potentiate MDA5-mediated signal transduction, as mutations that block basal ATP hydrolysis also prevent its ability to enhance MDA5.
Understanding these features of LGP2 have expanded our understanding of this innate immune sensor and enabled the generation of new models to help reconcile its dual roles in RLR regulation [8, 80, 81]. For example, we have proposed a working model in which LGP2 switches between MDA5-specific enhancement and a more general RLR negative regulation [47]. Titrating LGP2 expression into MDA5-dependent signaling demonstrates that low levels of LGP2 are synergistic with MDA5 [35, 47, 79, 82], a process that requires LGP2 ATP hydrolysis and RNA binding activities. The MDA5-stimulating activity of LGP2 is revealed in a narrow stoichiometric range, and further titration of LGP2 expression ultimately achieves an inhibitory concentration that disrupts RLR signaling activity [35, 76] [11, 66, 67].
Although many aspects of MDA5-LGP2 synergy remain to be addressed experimentally, our recent work has unraveled one element of this co-regulation (Figure 2C, [82]). It was observed that LGP2 catalytically increases the initial rate and stability of MDA5-dsRNA interactions. In addition, although LGP2 did not form dsRNA filaments by itself, the presence of LGP2 dramatically altered the quantity and quality of MDA5 filaments. LGP2 enabled MDA5 to generate a greater number of filaments, although the filaments were shorter in length. LGP2 enhances MDA5 activity by facilitating its initial RNA interaction, and has the effect of increasing the number and regulating the length of MDA5-RNA filaments. This MDA5 co-activation requires LGP2 ATP hydrolysis and RNA-binding activities, as well as an intact MDA5 oligomeric interface. This phenomenon apparently ensures MDA5 RNA recognition and optimizes signaling output, providing a plausible mechanistic basis for LGP2 co-activation.
Concluding Statements
In the decade since the recognition of RIG-I, MDA5, and LGP2 as a family of antiviral signaling proteins [20], our understanding of the mechanisms underlying intracellular RNA virus recognition and signal transduction has grown dramatically. Contemporary insights into antiviral signaling have rapidly moved from RLR identification to defining non-self RNA features and three-dimensional analysis of antiviral RNP filaments. Activation of beneficial MAVS prion-like assembly has drawn parallels with inflammasome processes, and RLRs have been independently implicated as mediators of immune defects of non-viral etiology. The next decade promises to expand upon these recent findings by revealing the secrets of each RLR protein, and exposing the mysteries of innate antiviral immune regulation.
Highlights.
RLRs share the ability to recognize nucleic acid signatures and activate antiviral signaling.
RLRs assemble dynamic multimeric ribonucleoprotein complexes.
RLRs differ in enzymatic properties, RNA recognition, and RNP assembly.
Acknowledgments
Research on RLRs in the Horvath lab was supported by NIH grants AI073919 and AI50707 to CMH. AMB was supported in part by a predoctoral fellowship from the NIH Cellular and Molecular Basis of Disease Training Grant T32GM008061. The authors apologize to any colleagues whose work was not mentioned due to space limitations or error.
Biographies

Curt Horvath is a Professor of Molecular Biosciences at Northwestern University, and co-directs the Signal Transduction in Cancer division of the Robert H. Lurie Comprehensive Cancer Center. His lab has uncovered diverse mechanisms of virus innate immune evasion aimed at RLR and JAK-STAT pathways, and current research on signal transduction and gene regulation includes investigation of virus-host interactions, protein-RNA interactions, and the molecular mechanisms underlying interferon production and cellular antiviral responses.
Annie Bruns received her bachelor’s degree at Luther College in Decorah Iowa, where she was awarded the prestigious McElroy Fellowship. She joined the Horvath lab as a Ph.D. student in the Northwestern University Interdisciplinary Biological Sciences graduate program and was supported by an NIH Cellular and Molecular Basis of Disease Training Program. Her thesis research focused on RNA recognition and signal transduction by the innate immune receptors in the RLR family, with emphasis on the regulatory roles for LGP2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunological reviews. 2011;243:91–8. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis ESC. Cytosolic sensing of viruses. Immunity. 2013;38:855–69. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs JL, Coyne CB. Mechanisms of MAVS regulation at the mitochondrial membrane. Journal of molecular biology. 2013;425:5009–19. doi: 10.1016/j.jmb.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rathinam VA, Fitzgerald KA. Cytosolic surveillance and antiviral immunity. Current opinion in virology. 2011;1:455–62. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–8. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 6.Ramos HJ, Gale M., Jr RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Current opinion in virology. 2011;1:167–76. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem. 2009;284:9700–12. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annual review of biophysics. 2008;37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Ranjith-Kumar CT, Brooks MT, Dharmaiah S, Herr AB, Kao C, et al. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009;284:13881–91. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:582–7. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–7. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Molecular cell. 2008;29:169–79. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Lu C, Stewart M, Xu H, Strong RK, Igumenova T, et al. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Archives of biochemistry and biophysics. 2009;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–74. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Molecular cell. 2008;29:428–40. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 17.Wang JP, Cerny A, Asher DR, Kurt-Jones EA, Bronson RT, Finberg RW. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J Virol. 2010;84:254–60. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. eLife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–61. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature immunology. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 21.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–22. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–35. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 23.Baum A, Garcia-Sastre A. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino acids. 2010;38:1283–99. doi: 10.1007/s00726-009-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 25.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–4. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang QX, Chen ZJ. Structural insights into the activation of RIG-I, a nanosensor for viral RNAs. EMBO reports. 2012;13:7–8. doi: 10.1038/embor.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohlway A, Luo D, Rawling DC, Ding SC, Pyle AM. Defining the functional determinants for RNA surveillance by RIG-I. EMBO reports. 2013;14:772–9. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO reports. 2013;14:780–7. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Molecular cell. 2013;51:573–83. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Oshiumi H, Matsumoto M, Seya T. Ubiquitin-mediated modulation of the cytoplasmic viral RNA sensor RIG-I. Journal of biochemistry. 2012;151:5–11. doi: 10.1093/jb/mvr111. [DOI] [PubMed] [Google Scholar]
- 31.Maelfait J, Beyaert R. Emerging role of ubiquitination in antiviral RIG-I signaling. Microbiology and molecular biology reviews: MMBR. 2012;76:33–45. doi: 10.1128/MMBR.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–20. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 33.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–30. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–4. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruns AM, Pollpeter D, Hadizadeh N, Myong S, Marko JF, Horvath CM. ATP Hydrolysis Enhances RNA Recognition and Antiviral Signal Transduction by the Innate Immune Sensor, Laboratory of Genetics and Physiology 2 (LGP2) J Biol Chem. 2013;288:938–46. doi: 10.1074/jbc.M112.424416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–49. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiscott J, Ware C. Cytokines. Current opinion in immunology. 2011;23:561–3. doi: 10.1016/j.coi.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Current opinion in immunology. 2011;23:564–72. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T, et al. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell research. 2011;21:895–910. doi: 10.1038/cr.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8459–64. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. MDA-5 recognition of a murine norovirus. PLoS pathogens. 2008;4:e1000108. doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. The Journal of experimental medicine. 2008;205:1601–10. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–9. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nature immunology. 2011;12:137–43. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luthra P, Sun D, Silverman RH, He B. Activation of IFN expression by a viral mRNA through RNase L and MDA5. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2118–23. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. eLife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez KR, Bruns AM, Horvath CM. MDA5 and LGP2: Accomplices and Antagonists of Antiviral Signal Transduction. J Virol. 2014 doi: 10.1128/JVI.00640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berke IC, Li Y, Modis Y. Structural basis of innate immune recognition of viral RNA. Cellular microbiology. 2012 doi: 10.1111/cmi.12061. [DOI] [PubMed] [Google Scholar]
- 49.Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. The EMBO journal. 2012;31:1714–26. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18437–41. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, et al. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3340–9. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, et al. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–3. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- 53.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, et al. Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell. 2013;152:276–89. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 54.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, et al. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell reports. 2012;2:1187–96. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21010–5. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:637–42. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang DC, Gopalkrishnan RV, Lin L, Randolph A, Valerie K, Pestka S, et al. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene. 2004;23:1789–800. doi: 10.1038/sj.onc.1207300. [DOI] [PubMed] [Google Scholar]
- 58.Kovacsovics M, Martinon F, Micheau O, Bodmer JL, Hofmann K, Tschopp J. Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Current biology: CB. 2002;12:838–43. doi: 10.1016/s0960-9822(02)00842-4. [DOI] [PubMed] [Google Scholar]
- 59.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nature genetics. 2006;38:617–9. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 60.Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol. 2011;187:1298–303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Funabiki M, Kato H, Miyachi Y, Toki H, Motegi H, Inoue M, et al. Autoimmune Disorders Associated with Gain of Function of the Intracellular Sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 63.Miner JJ, Diamond MS. MDA5 and autoimmune disease. Nature genetics. 2014;46:418–9. doi: 10.1038/ng.2959. [DOI] [PubMed] [Google Scholar]
- 64.Kovacsovics M, Martinon F, Micheau O, Bodmer JL, Hofmann K, Tschopp J. Errata: Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Current biology: CB. 2002;12:1633. doi: 10.1016/s0960-9822(02)00842-4. [DOI] [PubMed] [Google Scholar]
- 65.Zhu Z, Zhang X, Wang G, Zheng H. The laboratory of genetics and physiology 2: emerging insights into the controversial functions of this RIG-I-like receptor. BioMed research international. 2014;2014:960190. doi: 10.1155/2014/960190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80:12332–42. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–8. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 68.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1512–7. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, et al. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity. 2012;37:235–48. doi: 10.1016/j.immuni.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–55. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 71.Pollpeter D, Komuro A, Barber GN, Horvath CM. Impaired cellular responses to cytosolic DNA or infection with Listeria monocytogenes and vaccinia virus in the absence of the murine LGP2 protein. PloS one. 2011;6:e18842. doi: 10.1371/journal.pone.0018842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Widau RC, Parekh AD, Ranck MC, Golden DW, Kumar KA, Sood RF, et al. RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E484–91. doi: 10.1073/pnas.1323253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liniger M, Summerfield A, Zimmer G, McCullough KC, Ruggli N. Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. J Virol. 2012;86:705–17. doi: 10.1128/JVI.00742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malur M, Gale M, Jr, Krug RM. LGP2 downregulates interferon production during infection with seasonal human influenza A viruses that activate interferon regulatory factor 3. J Virol. 2012;86:10733–8. doi: 10.1128/JVI.00510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Si-Tahar M, Blanc F, Furio L, Chopy D, Balloy V, Lafon M, et al. Protective Role of LGP2 in Influenza Virus Pathogenesis. The Journal of infectious diseases. 2014 doi: 10.1093/infdis/jiu076. [DOI] [PubMed] [Google Scholar]
- 76.Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, et al. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic acids research. 2009;37:2014–25. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Childs KS, Randall RE, Goodbourn S. LGP2 plays a critical role in sensitizing mda-5 to activation by double-stranded RNA. PloS one. 2013;8:e64202. doi: 10.1371/journal.pone.0064202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parisien JP, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, et al. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J Virol. 2009;83:7252–60. doi: 10.1128/JVI.00153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodriguez KR, Horvath CM. Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition. J Virol. 2014 doi: 10.1128/JVI.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, et al. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS pathogens. 2010;6:e1000734. doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bruns AM, Horvath CM. Activation of RIG-I-like receptor signal transduction. Critical reviews in biochemistry and molecular biology. 2012;47:194–206. doi: 10.3109/10409238.2011.630974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signal transduction by regulating MDA5-RNA interactions and filament assembly. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]