Highlights
-
•
The expression of lncRNAs in the immune system is cell type- and context-dependent.
-
•
Several lncRNAs identified to date regulate immune gene expression.
-
•
LncRNAs play crucial role in host–pathogen interactions.
-
•
The majority of disease-associated SNPs lie in regulatory regions of the genome.
Keywords: long non-coding RNA, lncRNA, lincRNA, innate immunity, gene expression, epigenetics
Abstract
All cells of the immune system rely on a highly integrated and dynamic gene expression program that is controlled by both transcriptional and post-transcriptional mechanisms. Recently, non-coding RNAs, including long non-coding RNAs (lncRNAs), have emerged as important regulators of gene expression in diverse biological contexts. lncRNAs control gene expression in the nucleus by modulating transcription or via post-transcriptional mechanisms targeting the splicing, stability, or translation of mRNAs. Our knowledge of lncRNA biogenesis, their cell type-specific expression, and their versatile molecular functions is rapidly progressing in all areas of biology. We discuss here these exciting new regulators and highlight an emerging paradigm of lncRNA-mediated control of gene expression in the immune system.
The immune system and non-coding RNAs
The immune system is equipped with an arsenal of strategies to combat infectious threats and maintain normal health. This is mediated by specialized immune cells dedicated to carrying out sophisticated and highly integrated functions of the innate and adaptive arms of the immune system. The development and activation state of immune cells is dependent on a tightly regulated and integrated gene-expression program controlled by well-established transcription factors and chromatin-modifying complexes. Our knowledge of the functional roles of proteins in the transcriptional and post-transcriptional regulation of gene expression is fairly well developed; however, we have only begun to appreciate the fundamental roles of regulatory RNAs in controlling all facets of gene expression.
Regulatory RNAs are non-protein-coding transcripts that mediate their functions strictly as RNA molecules [1]. Some of these, including small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA), have been known for decades. They are important players in essential biological processes including chromatin/nuclear organization (snRNA and snoRNA), mRNA splicing (snRNA), ribosome biogenesis and assembly (rRNA), and translation (tRNA) [2]. The discovery of RNA interference (RNAi) mediated by siRNA and miRNA led to a new paradigm in gene regulation by demonstrating that short complementary RNAs can directly regulate target mRNAs 3, 4, 5. More recently, a new class of regulatory RNAs known as lncRNAs has emerged as an additional layer of the circuitry controlling gene expression 6, 7. Our understanding of the functional roles of lncRNAs in controlling gene expression is evolving, and is redefining our basic understanding of biology. In the immune system, lncRNAs exhibit dynamic expression in cell type-, developmental stage-, and context-specific manners to coordinate several aspects of immune function. We describe here our current knowledge of these RNAs in the immune system.
lncRNAs: dark matter of the genome takes the center stage
One of the many unexpected surprises from the Encyclopedia of DNA Elements (ENCODE) consortium, founded in 2003, was that the vast majority of the mammalian genome is transcribed 8, 9. According to the latest GENCODE version 19 (http://www.gencodegenes.org), it is estimated that only ∼2% of the mammalian genome is composed of genes that encode proteins, while the vast majority (75–90%) is transcribed as non-coding RNA 10, 11. LncRNAs (13 870 in the human and 4074 in the mouse genome) account for most of this pervasive transcription (Figure 1 ). High-throughput transcriptome sequencing has led to the discovery of thousands of lncRNA genes, and these numbers are still growing 12, 13.
Figure 1.
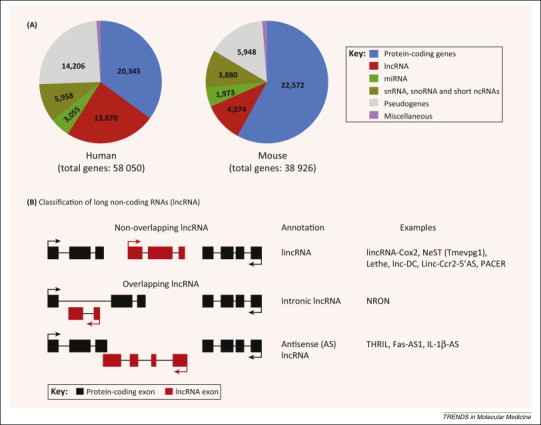
Long non-coding RNAs (lncRNAs) are the most abundant ncRNA species in the mammalian genome. (A) Pie charts showing the genome-wide distribution of protein- and non-coding genes in the human and mice genomes. Numbers shown are calculated from the GENCODE version 19 (July 2013 update; http://www.gencodegenes.org). (B) Classification of lncRNAs based on their genomic localization with respect to the neighboring protein-coding gene. LncRNAs in general are classified as non-overlapping (or the intergenic lncRNA; lincRNAs), or the overlapping lncRNAs, which include intronic and antisense (AS) lncRNAs. Intronic lncRNAs are transcribed within the intron of a protein-coding gene, and therefore do not contain sequences complementary to the mature, spliced mRNA of the protein-coding gene. Antisense lncRNA, however, contains region(s) of complementary sequences with the mature, spliced mRNA of the overlapping protein-coding gene. Examples of immune-related lncRNAs from the different lncRNA sub-classes are provided. Abbreviations: miRNA, micro-RNA; ncRNA, non-coding RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA.
In general, lncRNAs are defined as non protein-coding transcripts larger than 200 nucleotides. Inclusion of the size criterion is primarily employed to distinguish lncRNAs from smaller ncRNA species such as siRNA, miRNA, and others. The majority of lncRNAs share features of protein-coding mRNAs because they are capped, polyadenylated, and spliced 14, 15. An important feature of lncRNAs is their lack of protein-coding capacity; however, as with any negative finding, it is difficult to prove the absence of encoded protein/peptide with absolute certainty. This aspect of lncRNA biology remains somewhat controversial 16, 17, 18. Therefore, it is crucial that lncRNA functionality is strictly attributed to the RNA (and not to a potentially encoded protein). Computational approaches (prediction of open reading frames, codon-substitution frequency, and evolutionary conservation) as well as experimental approaches (in vitro transcription and translation, polysome association, and ribosome footprinting followed by RNA-seq) are routinely employed to address the protein-coding potential of lncRNAs 14, 15. It is possible that some lncRNAs do produce small proteins (or peptides) which may mediate additional functions. Because the lncRNA field is rapidly evolving, a clear nomenclature for these RNAs is badly needed (Figure 1). Commonly used terms, such as long intergenic non-coding RNAs (lincRNA), refer to lncRNAs that are strictly intergenic and do not overlap with known protein-coding genes. All other forms of lncRNAs are either transcribed within (e.g., intronic lncRNA), or exhibit some overlap with a known protein-coding gene (e.g., antisense lncRNA). Antisense (AS) lncRNAs are particularly interesting because they appear to be the major lncRNA subtype: it is estimated that up to 72% of murine genomic loci show evidence of transcription [19].
A growing body of evidence indicates that lncRNAs play an important role in the transcriptional and post-transcriptional regulation of gene expression in a variety of biological processes such as X-chromosome inactivation (Xist/Tsix) 20, 21, genomic imprinting (H19 and Air) 22, 23, stem cell pluripotency [24], development [25], cancer metastasis (HOTAIR) [26], and atherosclerosis (Anril) [27]. A functional role of lncRNAs has also started to emerge in controlling gene expression in the immune system. lncRNAs are widely expressed in immune cells including monocytes, macrophages, dendritic cells (DC), neutrophils, T cells, and B cells during their development, differentiation, and/or activation. However, the functions of these RNAs are only beginning to be characterized.
Expression and the role of lncRNAs in immune cell development
An emerging theme in the biology of lncRNAs is their role in cell and tissue development [28]. This is highlighted by a large body of evidence indicating that lncRNAs play key roles during the development of erythrocytes 29, 30, adipocytes [31], cardiomyocytes [32], epidermal cells [33], and stem cells [24], among others. A recent study illustrates a similar functional role for lncRNAs in controlling the differentiation of human monocytes into dendritic cells (DC) [34]. DCs act as the primary antigen-presenting cells (APCs) for T cells and bridge the innate and adaptive arms of the immune system [35]. DCs come in two flavors: (i) conventional DCs (cDCs; also known as myeloid DCs), which function as APCs and producers of interleukin 12 (IL-12); and (ii) plasmacytoid DCs (pDCs) that produce abundant levels of type I interferon (IFN) following bacterial and viral infections [36]. To identify lncRNAs involved in DC maturation and function, the human transcriptome was profiled using RNA-seq at different stages of monocyte differentiation into DCs in vitro [34]. This approach identified a cohort of lncRNAs that were differentially expressed during the monocyte to DC transition. Detailed study of one of these RNAs, an intergenic lncRNA which the authors named lnc-DC, showed that it was expressed exclusively in DCs. Furthermore, lnc-DC was shown to be required for the differentiation of human monocytes into DCs in vitro, and played a similar role in mice. Expression of lnc-DC was driven by PU.1, a key transcription factor required for the DC maturation program [37]. In addition to controlling the maturation of DCs, lnc-DC also controls the expression of several DC genes including CD40, CD80, and HLA-DR. Therefore, lnc-DC controls the uptake of antigens by DCs, the subsequent induction of allogenic CD4+ T cell proliferation, and cytokine production [34]. Lnc-DC mediates these effects by binding the transcription factor STAT3 (signal transducer and activator of transcription 3) in the cytosol, preventing STAT3 binding to, and its dephosphorylation by, the tyrosine phosphatase SHP1 (Src homology region 2 domain-containing phosphatase-1). These events enhance STAT3 phosphorylation on tyrosine-705. The molecular action of lnc-DC in the cytosol appears to be analogous to another lncRNA, nuclear repressor of NFAT (NRON), that sequesters the transcription factor NFAT (nuclear factor of activated T cells) into an inactive RNA–protein scaffold in the cytosol of T cells [38]. Another interesting aspect of lnc-DC is its unique and abundant expression in cDC (around 100-fold higher than in pDCs or in any other immune cell type) suggesting that it can be utilized as a specific and sensitive marker of conventional DCs in the hematopoietic system.
HOX antisense intergenic RNA myeloid 1 (HOTAIRM1) is encoded in the human HOXA gene cluster and is linked to the maturation of granulocytes [39]. HOTAIRM1 expression is restricted to myeloid cells, and is upregulated during retinoic acid (RA)-induced granulocyte differentiation of promyelocytic NB4 leukemia, and normal human hematopoietic stem cells. Knockdown of HOTAIRM1 blunts RA-induced expression of neighboring genes HOXA1 and HOXA2 (but not distal HOXA genes) as well as myeloid differentiation-associated genes CD11b and CD18. Because HOXA genes are involved in the transcriptional regulation of normal hematopoiesis [40] and acute myeloid leukemia (AML) 41, 42, it has been proposed that HOTAIRM1 plays an important role in myelopoiesis by regulating HOXA gene expression in cis. Together, these studies illustrate the importance of lncRNAs in controlling the development of immune cells. It remains to be seen whether other immune cell types such as monocyte–macrophages, innate lymphoid cells (ILC), B cells, and T cell subsets (Th1, Th2, Th17, and Treg) also express lncRNAs that control their development.
Functional lncRNAs in the innate immune system
Innate immunity is the first line of defense against microbial pathogens. Recent studies collectively indicate that lncRNAs play important functional roles in innate immune cells such as phagocytes. Notably, the discovery of lincRNA-Cox2 [43], Lethe [44], PACER [45] and THRIL (TNFα regulating hnRNPL interacting lncRNA) [46] represent exciting examples of the growing list of lncRNAs that have been implicated in controlling gene expression in immune cells.
Transcriptome profiling (RNA-seq) in mouse bone marrow-derived macrophages (BMDM) led to the identification of 72 lncRNAs that were significantly upregulated in macrophages exposed to the synthetic bacterial lipoprotein Pam3CSK4, which signals via Toll-like receptor 2 (TLR2) [43]. One of most robustly induced lncRNAs, lincRNA-Cox2, was shown to act as a regulator of the TLR induced transcriptional program in macrophages. The gene for lincRNA-Cox2, earlier identified as an intergenic transcript in the catalog of lincRNAs reported by Guttman et al. [6], is located ∼51 kb from the 3′ end of the prostaglandin-endoperoxide synthase 2 (Ptgs2; Cox2) gene on the opposite strand of chromosome 1. LincRNA-Cox2 was highly induced by multiple inflammatory triggers, including TLR ligands [lipopolysaccharide (LPS) and Pam3CSK4] and microbial pathogens (Listeria monocytogenes and Sendai virus) by a pathway involving MyD88 (myeloid differentiation primary response gene 88) and the transcription factor NF-κB (nuclear factor κ light-chain-enhancer of activated B cells) [43]. Functionally, lincRNA-Cox2 appears to activate and repress expression of distinct classes of immune genes. In resting macrophages, lincRNA-Cox2 represses the expression of ∼700 genes including chemokines (Ccl5 and Cx3cl1) and interferon-stimulated genes (ISG) (Irf7, Isg15, Ifi204, and Oas2), whereas it was required for the inducible expression of several other genes (IL6, Tlr1, and IL23a) turned on by the TLR2 pathway. lincRNA-Cox2 physically interacts with RNA-binding proteins (RBP) hnRNP-A2/B1 and hnRNP-A/B to mediate its repressive functions. Interestingly, these two hnRNPs are not involved in mediating the activating functions of this lincRNA, suggesting that additional lincRNA-Cox2 interacting protein(s) remain to be identified. How lincRNA-Cox2 contributes to TLR2-induced expression of IL6 and other genes is currently unknown. Future in-depth studies in animals lacking lincRNA-Cox2 will shed light on the in vivo immune functions of this RNA.
The genomic locus containing Ptgs2 (Cox2) contains two known lncRNA genes (lincRNA-Cox2, and Cox2-divergent (Ptgs2 opposite strand; Ptgs2os)) in mice 43, 44, and the recently described lncRNA gene PACER in humans [45]. The Cox2-divergent lncRNA is located at the 5′-end of Ptgs2 (non-overlapping), and is transcribed from the opposite (negative) DNA strand [44]. Although functions of Cox2-divergent are unclear, it is highly induced in mouse embryonic fibroblasts (MEF) exposed to tumor necrosis factor α (TNFα), and LPS, in an NF-κB-independent manner [44]. The lncRNA PACER (p50-associated COX-2 extragenic RNA) is exclusively involved in controlling the expression of COX-2 in cis in human monocyte–macrophage cell lines, and primary human mammary epithelial cells [45]. PACER expression is regulated by the chromatin-boundary/insulator factor CCCTC-binding factor (CTCF), which establishes an open chromatin domain in the upstream region of COX2 to promote PACER expression. In turn, PACER binds to the NF-κB homodimer p50/p50 (a transcriptional repressor complex) and titrates it away from the COX2 promoter. These events then favor the recruitment of the active NF-κB heterodimer p65/p50, which promotes the assembly of the transcription preinitiation complex containing histone acetyltransferase p300 and RNA polymerase II at the COX2 promoter. Therefore, PACER appears to function by occluding the repressor complex (p50/p50) to facilitate the expression of Ptgs2/COX2. This is remarkably similar to the lincRNA Jpx, which activates the expression of Xist during the initiation of X-chromosome inactivation by evicting CTCF from the Xist promoter [47].
Lethe, a functional pseudogene (Rps15a-ps4) lncRNA, is also highly inducible in MEFs treated with the NF-κB-activating inflammatory triggers TNFα and IL-1β [44]. Lethe expression is also induced in response to the anti-inflammatory glucocorticoid receptor agonist, dexamethasone [44]. Lethe attenuates the NF-κB-dependent inflammatory response in MEFs by physically binding to p65 (RelA), which inhibits RelA occupancy at the promoter of target genes, including IL6, IL8, and superoxide dismutase 2 (SOD2) [44]. Consistent with this model, Lethe is primarily localized to chromatin. Interestingly, Lethe expression in the spleen of aged mice is markedly lower (20–50-fold) than in young mice, suggesting a functional link between decreased Lethe expression and enhanced NF-κB signaling associated with ageing [44]. Lethe therefore functions as a feedback regulator of the NF-κB signaling pathway to control the inflammatory response. In addition, Lethe has provided the first evidence for the existence of a functional pseudogene lncRNA, raising the possibility that many more currently annotated pseudogenes in the mammalian genome could in fact be functional (lncRNA) genes.
THRIL is another immune-related lncRNA, which primarily controls the expression of TNFα in the human monocyte-like THP-1 cell line [46]. THRIL was identified as one of 159 lincRNAs that were differentially expressed (80% down, 20% up) in Pam3CSK4-treated THP-1 cells [46]. Lentiviral short hairpin RNA (shRNA)-mediated knockdown of a pool of nine lincRNAs including THRIL resulted in impaired TNFα and/or IL-6 production, suggesting that several TLR2-induced lincRNAs could cooperate to regulate inflammatory responses in human monocytes [46]. RNA-seq following shRNA-silencing of THRIL expression in THP-1 cells revealed differential expression of 319 genes in response to Pam3CSK4, indicating that THRIL regulates a broad panel of immune genes [46]. THRIL interacts with hnRNPL to control TNFα expression, and both THRIL and hnRNPL localize to the TNFA promoter [46]. Because the expression of THRIL is inhibited by TNFα, it appears to act as a negative feedback regulator of TNFα expression in human monocytes. THRIL expression is significantly reduced during the acute stage of the inflammation-associated Kawasaki disease [46]; the biological significance of this disease association however remains unclear.
More recently, the nuclear enriched abundant transcript 1 (NEAT1), a key player controlling the formation of heterochromatin structures known as ‘paraspeckles’ [48], has been linked to the expression of the cytokine IL-8 in human cell lines following viral infections (HSV-1 and influenza A virus), or TLR3-activation by dsRNA [49]. NEAT1 relocates the repressor SFPQ (splicing factor proline glutamine-rich), a NEAT1-binding paraspeckle protein, from the IL8 promoter to the nuclear paraspeckle body, to mediate IL8 transcription [49]. In addition, NEAT1 also regulates HIV-1 by promoting the export of HIV-1 mRNA from the nucleus to the cytoplasm [50]. Collectively, these studies highlight the emergence of lncRNAs as an important regulatory layer in controlling gene expression in innate immunity.
lncRNAs in adaptive immunity
Lymphocytes (T and B cells) are the primary cellular mediators of the adaptive immune system. There is now clear evidence that lymphocytes express many lncRNAs, and that these play crucial roles in development, lineage-specific differentiation, and activation. Two lncRNAs expressed in T cells, NTT (non-coding transcript in CD4+ T cells) [51] and NRON [52], represent the earliest lncRNA genes identified in immune cells. NTT is a 17 kb polyadenylated, unspliced intergenic transcript localized to the nucleus. NTT is primarily expressed in activated human CD4+ T cells; however, its function remains to be defined. A potential functional link between NTT and the IFN-γ receptor (IFNGR) gene is one aspect that warrants further investigation because these genes share the same genomic locus (6q23–q24), and they exhibit similar spatiotemporal expression during T cell activation [51].
NRON, an intronic lncRNA, was identified in human T cells as a result of an shRNA screen exploring the role of 512 evolutionarily conserved ncRNAs that were known at the time [52]. NRON was found to regulate NFAT, a Ca2+-activated transcription factor, and modulate the expression of IL-2 in activated T cells. NRON interferes with the nuclear transport of NFAT (and not the transcriptional activity) by interacting with members of the importin-β superfamily, (including the nuclear transport factor KPNB1) [52] which promote the nucleocytoplasmic transport of cargos such as NFAT [53]. Subsequent studies however have shown that NRON acts as a structural scaffold lncRNA to sequester inactive, phosphorylated NFAT into a large cytosolic protein–RNA complex containing NFAT, IQGAP (IQ motif-containing GTPase activating protein), and several NFAT kinases [38]. Additional studies are needed to reconcile these seemingly disparate modes of NRON-mediated control of NFAT responses. Nonetheless, NRON appears to play a crucial function in controlling NFAT-dependent IL-2 expression in T cells. NRON expression is enriched in lymphoid tissue, consistent with its roles in modulating NFAT activity in T cells [52].
The first systematic study aimed at profiling the lncRNA transcriptome in CD8+ T cells was performed by Pang et al. and led to the identification of hundreds of lncRNAs in the mouse genome, many of which were lymphoid cell specific and differentially expressed in naïve, memory, or effector CD8+ T cells [1]. More recently, genome-wide transcriptional profiling of murine T cells has identified 1524 lincRNA gene clusters across a panel of T cell subsets, from early progenitors to terminally differentiated helper T cells [54]. These lincRNAs exhibited dynamic, cell- and activation state-specific expression. During CD4+ T cell differentiation into Th1 and Th2 cells, expression of lincRNAs in these T cell subsets was driven by the T cell lineage-specific transcription factors, T-bet and Stat4 for Th1 cells, and Stat6 and Gata3 for Th2 cells. One Th2-specific lincRNA, lincR-Ccr2-5′AS, is located upstream of the chemokine receptor Ccr2 gene, and is transcribed in the antisense (AS) direction [54]. LincR-Ccr2-5′AS, together with Gata3, controls the expression of immune genes in Th2 cells. This lincRNA also controls the migration of Th2 cells to the lungs in vivo, presumably by controlling the expression of several chemokine receptor genes (Ccr1, Ccr3, Ccr2, and Ccr5), which are all located in the same genomic locus as lincR-Ccr2-5′AS [54]. The molecular details of how lincR-Ccr2-5′AS mediates the expression of these genes remains unclear. In addition, many other lincRNAs are also specifically expressed in each of the CD4+ T cell subsets: naïve cells (79), Th1 (101), Th2 (63), Th17 (27), and induced regulatory T cells (iTreg) (37) [54]. However, what fraction of these lincRNAs are functionally linked to T cell development, as well as their effector functions, remain to be investigated. Another lincRNA expressed in human T cells, GAS5 (growth arrest-specific transcript 5), has been linked to cell cycle arrest in response to either nutrient deprivation or exposure to the mammalian target of rapamycin (mTOR) antagonist 55, 56, 57.
B cells, mediators of the antibody-dependent humoral arm of the adaptive immunity, also express lncRNAs. The antisense lncRNA Fas-AS1 (FAS antisense transcript 1) tightly controls the production of soluble Fas receptor (sFas), which binds Fas ligand to regulate Fas-induced apoptosis in B cell lymphomas [58]. Fas-AS1 binds to the splicing factor RBM5 to inhibit RBM5-mediated alternative skipping of the exon 6 of Fas (also known as CD95; TNFRSF6), which is required to generate the sFas mRNA. Because serum sFas levels are associated with poor prognosis in non-Hodgkin's lymphoma [59], the Fas-AS1 lncRNA is a potential therapeutic target in this setting. In addition, widespread antisense intergenic transcription has been shown to occur in the variable (V) region of the immunoglobulin heavy chain (Igh) locus in B cells, which is potentially linked to chromatin remodeling associated with V(D)J recombination involved in the production of the diverse repertoire of antigenic receptors in developing B cells 60, 61. Whether lncRNAs also play a role in the maturation and effector function of B cells remains an open question. Collectively, however, these studies demonstrate that immune cells express a vast repertoire of lncRNAs, many of which are expected to play key roles in the host immune response.
Role of lncRNAs in host defense against microbial infection
A functional role for lncRNAs in controlling the host immune response during microbial infection has also emerged. This is best highlighted by the discovery of a lincRNA termed NeST [62] (originally identified as Tmevpg1 [63]), a candidate gene controlling the persistence of Theiler's virus in the central nervous system in mice. In a recent study employing inter-crosses between susceptible SJL/J mice (these mice express NeST, develop persistent Theiler's virus infection, and clear Salmonella infection), and the resistant B10.S strain (lack NeST expression, clear Theiler's virus infection, and succumb to Salmonella infection), as well as through the generation of B10.S mice expressing a NeST transgene, Gomez et al. have provided compelling genetic evidence that NeST is the host factor responsible for the persistence of Theiler's virus as well as for the clearance of Salmonella infection in mice [62]. Nest is positioned near, and convergently transcribed to the Ifng gene. NeST is selectively expressed in CD4+ Th1 (but not Th2) cells, CD8+ T cells and natural killer (NK) cells 62, 63, 64. The transcription factors T-bet and Stat4, which are known to drive naive CD4+ T cell differentiation into Th1 cells, control the expression of NeST [64]. NeST binds to WDR5 (WD repeat-containing protein 5), a component of the histone methyltransferase complex, to mediate histone 3 lysine 4 trimethylation (H3K4me3) at the Ifng promoter to promote IFN-γ expression in CD8+ T cells [62]. Because Nest and Ifng are located in the same genomic locus, NeST is thought to act in cis as an enhancer RNA to promote Ifng expression. NeST alone, however, is not sufficient to drive Ifng expression because it works cooperatively with the transcription factor T-bet [64]. It is noteworthy that NeST, which is expressed at very low levels (∼0.15 copies per cell) in CD8+ T cells, mediates such profound effects upon IFN-γ production. The crucial role of NeST in determining the host susceptibility to an infectious disease further highlights the importance of lncRNA genes in the immune system. Hundreds of lncRNAs are also expressed in vivo following infection with coronavirus (the causative agent of acute respiratory syndrome) or influenza virus [65]. The functional importance of these virus-induced lncRNAs, however, is presently unknown.
In addition to host-encoded lncRNAs, several microbial species also express lncRNAs, which in some cases subvert host immunity [66]. Several studies have highlighted a functional role for a non-coding polyadenylated nuclear (PAN) RNA encoded in the Kaposi's sarcoma-associated herpesvirus (KSHV) genome [67]. The KSHV PAN lncRNA facilitates the conversion of latent to lytic (active) infection, presumably by regulating the dissociation of LANA (latency associated nuclear antigen) from the KSHV genome [68]. In addition, the PAN lncRNA recruits the demethylase JMJD3 and UTX to epigenetically repressed regions of the KSHV genome to enhance viral genome expression [69]. The KSHV PAN lncRNA also suppresses antiviral host factors including IFN-α, IFN-γ, and RNase L through its interaction with the polycomb repressive complex 2 (PRC2) to subvert host immunity and promote viral replication in infected cells 67, 70. Similarly, the herpes virus encodes HSUR1 and HSUR2 (herpesvirus saimiri U-rich ncRNAs) that target host miRNAs miR-27 and miR-16, respectively, to alter the expression of miRNA-regulated genes [71]. Another example of a viral lncRNA is the HIV-1 encoded antisense lncRNA (localizing to the viral 5′-long terminal repeat promoter), which suppresses viral transcription through the endogenous RNA-directed epigenetic pathway involving DNA methyltransferase 3a (Dnmt3a), histone deacetylase 1 (HDAC1), and the PRC2 complex protein EZH2 [72]. Plasmodium falciparum, the causative agent of fatal cerebral malaria, also encodes ∼60 putative lncRNAs including the lncRNA-TARE (telomere-associated repeat elements) family of 22 telomere-associated lncRNAs [73]. It remains to be determined whether P. falciparum-encoded lncRNA-TAREs are involved in telomere maintenance, and/or gene expression in the parasite. Collectively, these studies indicate that lncRNAs lie at the heart of host–pathogen interactions.
How do lncRNAs work?
Our knowledge of the molecular functions of lncRNAs is expanding rapidly. A detailed review of how lncRNAs act is provided in several excellent recent reviews 7, 15. A fascinating aspect of lncRNA biology is their versatility; work on several fronts provides compelling evidence that lncRNAs regulate gene expression by targeting either the production, splicing, decay, or translation of target mRNAs 14, 15. Biochemically, lncRNAs employ RNA–RNA, RNA–DNA, and/or RNA–protein interactions to exert their activities (Figure 2 ). Such interactions have been known for decades for RNA–protein (RNP) complexes, wherein snRNAs interact with RNA-binding proteins (RBP) to assemble the functional RNP complexes involved in pre-mRNA processing and chromatin organization [74]. The mechanism(s) employed by lncRNAs differ depending on their cytosolic or the nuclear location (Figure 2). In the cytosol, lncRNAs directly interact with target mRNAs (or miRNAs) to control their expression or to regulate mRNA translation. For example, an antisense lncRNA to Uchl1, a gene involved in brain function and neurodegenerative diseases [75], enhances Uchl1 mRNA translation by promoting its association with polysomes [76], whereas lincRNA-p21 represses the translation of complementary mRNAs [77]. The muscle-specific lincRNA, linc-MD1, governs muscle differentiation by acting as a competing endogenous lncRNA (ceRNA) in mouse and human myoblasts to ‘sponge’ miR-133, thereby regulating the expression of genes involved in muscle differentiation [78]. In addition, cytosolic lncRNAs can interact with specific signaling proteins. For example, lnc-DC and NRON interact with Stat3 and NFAT, respectively, to regulate pathway-specific gene expression programs 34, 52.
Figure 2.
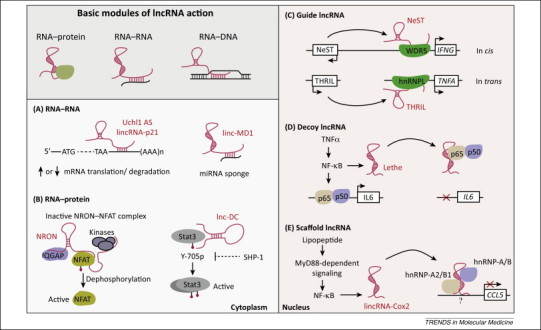
Long non-coding RNAs (lncRNAs) are versatile regulators of gene expression. lncRNAs employ three basic modules, RNA–RNA, RNA–protein and RNA–DNA interactions, to exert their functions either in the cytosol (A,B) or the nucleus (C–E). (A) In the cytosol lncRNAs interact with target mRNAs through base-pairing to either enhance translation (e.g., Uchl1 AS), or repress translation (e.g., lincRNA-p21). In addition, cytosolic lncRNAs are also known to regulate mRNA stability. For example, the lncRNA termed ½-Staufen binding site (sbs) RNA forms an RNA duplex with mRNAs containing partial complementarity in the 3′-untranslated region (UTR) to promote Staufen 1 binding, which drives Staufen-mediated mRNA degradation. (B) Cytosolic lncRNAs are also known to regulate immune signaling pathways through RNA–protein interactions. NRON and lnc-DC act as a scaffold for the transcription factors NFAT and Stat3, respectively, regulating phosphorylation status and thereby the expression of target genes in immune cells. (C–E) In the nucleus, lncRNAs function by acting as a guide (C, NeST; THRIL), decoy (D, Lethe), or scaffold (E, lincRNA-Cox2) to interact with specific protein(s) to silence gene expression. Abbreviations: hnRNP, heterogeneous ribonucleoprotein; IQGAP, IQ motif-containing GTPase activating protein; MD1, muscle differentiation 1; NFAT, nuclear factor of activated T cells; NRON, non-coding repressor of NFAT; THRIL, TNFα and hnRNPL related immunoregulatory lincRNA; Uchl1, ubiquitin carboxy-terminal hydrolase L1; WDR5, WD repeat-containing protein 5.
By contrast, nucleus-localized lncRNAs largely function by modulating epigenetic processes to alter gene expression by acting as a guide, decoy, or scaffold (Figure 2). Many nuclear lncRNAs function by localizing to their genomic targets through either RNA:DNA interactions or by employing specific protein(s). The list of lncRNA-interacting proteins is ever growing, and includes hnRNPs such as hnRNP-K for lincRNA-p21 [79], hnRNP-A/B and -A2/B1 for lincRNA-Cox2 [43], and hnRNPL for THRIL [46]; transcription factors CTCF for Jpx [47], NF-κB p65 (RelA) for Lethe [44], and NF-κB p50 for PACER [45]; and components of the epigenetic machinery including WDR5 for NeST [62], and PRC2 for HOTAIR 26, 80. The PRC2 complex is particularly interesting because it binds to many lncRNAs [81] and mediates the repressive histone mark H3K27me3. Whether PRC2–lncRNA interactions exhibit specificity is somewhat unclear because recent studies have shown that PRC2 binding to RNAs is promiscuous, and shows no specificity towards either the type (mRNAs or ncRNAs) or the RNA length (mature versus nascent transcripts) 82, 83. Therefore, the precise role of PRC2 in lncRNA function needs to be examined more closely. Together, these studies highlight the ability of lncRNAs to control gene expression through diverse mechanisms. The discovery of these RNAs is leading to a paradigm shift in our understanding of epigenetics and gene regulation.
Role of lncRNAs in human diseases
It has recently become clear that only ∼7% of disease-associated single-nucleotide polymorphisms (SNPs) identified in genome-wide association studies (GWAS) lie in protein-coding genes [8]. The majority of GWAS SNPs, however, are localized to regulatory regions of the genome, including regulatory elements (e.g., enhancers) and intergenic regions of the genome (which are rich in lncRNAs) [84]. Thus, genetic variations could affect the expression and/or function of lncRNAs, and have far-reaching implications for human diseases. Altered expression of lncRNAs has been noted for several immune-related diseases including inflammatory bowel disease (IBD), diabetes, and multiple sclerosis [85]. An immediate challenge, however, is to define the functional roles of these lncRNAs in the context of disease pathophysiology. Whether aberrant expression of lncRNAs plays a causal role or whether they are a mere consequence of disease pathology remains an open question.
Concluding remarks and future perspectives
Tremendous progress in recent years has provided clear evidence that the majority of the mammalian genome is transcribed. Although we are beginning to appreciate the functional roles of these RNAs, it remains to be seen what fraction of the mammalian transcriptome is functional. The emergence of lncRNAs as key regulators of gene expression has provided an additional layer of regulation in immunity. Studies on lncRNA biology in the past few years have vastly changed our very basic knowledge about how genes are regulated. In the immune system, lncRNAs play important roles in immune cell development, lineage differentiation, and effector function. Future studies will undoubtedly unveil additional and exciting new insights into lncRNA biology (Box 1 ). Defining the in vivo functions of immune-related lncRNAs in animal models is one exciting area for future studies. The application of recently developed genome-wide, high-resolution technologies such as chromatin isolation by RNA purification (ChiRP) [86] and RNA antisense purification (RAP) [87] will uncover mechanistic details of how lncRNAs regulate chromatin structure and gene expression. Given the rapid pace of research in the field of lncRNAs, we are certain that many more surprises and novel concepts will emerge in the coming years.
Box 1. Outstanding questions.
-
•
What roles do lncRNAs play in the development of the immune system?
-
•
What are the in vivo functions of immune-related lncRNAs?
-
•
What are the rules governing specificity of lncRNA–protein complexes?
-
•
What are the structural determinants of lncRNA function?
-
•
Are lncRNA functions evolutionarily conserved?
-
•
What factors drive the (low) expression of lncRNAs? Do lncRNAs exhibit heterogeneous expression?
-
•
What are the functional consequences of disease-associated SNPs localized to lncRNAs?
Acknowledgments
We apologize to all colleagues whose work could not be discussed owing to space limitations. This work is supported by the American Heart Association post-doctoral fellowship (14POST18930001) to M.K.A, and by grants from the National Institute of Health (AI067497) to K.A.F.
References
- 1.Pang K.C. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J. Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 2.Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee R.C. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart B.J. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Fire A. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Guttman M. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djebali S. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrien T. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrow J. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabili M.N. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hangauer M.J. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingolia N.T. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman M. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bazzini A.A. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werner A. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 20.Brown C.J. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.T. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 22.Brannan C.I. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sotomaru Y. Unregulated expression of the imprinted genes H19 and Igf2r in mouse uniparental fetuses. J. Biol. Chem. 2002;277:12474–12478. doi: 10.1074/jbc.M109212200. [DOI] [PubMed] [Google Scholar]
- 24.Guttman M. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauvageau M. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta R.A. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bochenek G. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 2013;22:4516–4527. doi: 10.1093/hmg/ddt299. [DOI] [PubMed] [Google Scholar]
- 28.Hu W. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu W. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Dominguez J.R. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123:570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun L. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grote P. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kretz M. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 35.Steinman R.M., Hemmi H. Dendritic cells: translating innate to adaptive immunity. Curr. Top. Microbiol. Immunol. 2006;311:17–58. doi: 10.1007/3-540-32636-7_2. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt S.V. Regulatory dendritic cells: there is more than just immune activation. Front. Immunol. 2012;3:274. doi: 10.3389/fimmu.2012.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy K.M. Transcriptional control of dendritic cell development. Adv. Immunol. 2013;120:239–267. doi: 10.1016/B978-0-12-417028-5.00009-0. [DOI] [PubMed] [Google Scholar]
- 38.Sharma S. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA–protein scaffold complex. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11381–11386. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bei L. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding beta3 integrin during myeloid differentiation. J. Biol. Chem. 2007;282:16846–16859. doi: 10.1074/jbc.M609744200. [DOI] [PubMed] [Google Scholar]
- 41.Eklund E.A. The role of HOX genes in myeloid leukemogenesis. Curr. Opin. Hematol. 2006;13:67–73. doi: 10.1097/01.moh.0000208467.63861.d6. [DOI] [PubMed] [Google Scholar]
- 42.Rice K.L., Licht J.D. HOX deregulation in acute myeloid leukemia. J. Clin. Invest. 2007;117:865–868. doi: 10.1172/JCI31861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carpenter S. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rapicavoli N.A. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krawczyk M., Emerson B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun S. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clemson C.M. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirose T. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4 doi: 10.1128/mBio.00596-12. e00596–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu A.Y. The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics. 1997;39:171–184. doi: 10.1006/geno.1996.4463. [DOI] [PubMed] [Google Scholar]
- 52.Willingham A.T. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 53.Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 54.Hu G. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013;14:1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kino T. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mourtada-Maarabouni M. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5) Mol. Pharmacol. 2010;78:19–28. doi: 10.1124/mol.110.064055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mourtada-Maarabouni M. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J. Cell Sci. 2008;121:939–946. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- 58.Sehgal L. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia. 2014 doi: 10.1038/leu.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niitsu N. A high serum soluble Fas/APO-1 level is associated with a poor outcome of aggressive non-Hodgkin's lymphoma. Leukemia. 1999;13:1434–1440. doi: 10.1038/sj.leu.2401502. [DOI] [PubMed] [Google Scholar]
- 60.Verma-Gaur J. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17004–17009. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolland D.J. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 62.Gomez J.A. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vigneau S. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 2003;77:5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Collier S.P. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J. Immunol. 2012;189:2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng X. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1 doi: 10.1128/mBio.00206-10. e00206–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scaria V., Pasha A. Long non-coding RNAs in infection biology. Front. Genet. 2012;3:308. doi: 10.3389/fgene.2012.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rossetto C.C., Pari G.S. Kaposi's sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J. Virol. 2011;85:13290–13297. doi: 10.1128/JVI.05886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campbell M. A lytic viral long noncoding RNA modulates the function of a latent protein. J. Virol. 2014;88:1843–1848. doi: 10.1128/JVI.03251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossetto C.C., Pari G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 2012;8:e1002680. doi: 10.1371/journal.ppat.1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rossetto C.C. Regulation of viral and cellular gene expression by Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013;87:5540–5553. doi: 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cazalla D., Steitz J.A. Down-regulation of a host microRNA by a viral noncoding RNA. Cold Spring Harb. Symp. Quant. Biol. 2010;75:321–324. doi: 10.1101/sqb.2010.75.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saayman S. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol. Ther. 2014;22:1164–1175. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Broadbent K.M. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 2011;12:R56. doi: 10.1186/gb-2011-12-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dreyfuss G. Ribonucleoprotein particles in cellular processes. J. Cell Biol. 1988;106:1419–1425. doi: 10.1083/jcb.106.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Setsuie R., Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem. Int. 2007;51:105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 76.Carrieri C. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 77.Yoon J.H. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cesana M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huarte M. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rinn J.L. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khalil A.M. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davidovich C. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaneko S. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ricano-Ponce I., Wijmenga C. Mapping of immune-mediated disease genes. Annu. Rev. Genomics Hum. Genet. 2013;14:325–353. doi: 10.1146/annurev-genom-091212-153450. [DOI] [PubMed] [Google Scholar]
- 85.Kumar V. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chu C. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Engreitz J.M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]


