Abstract
Cholesterol 25-hydroxylase (CH25H) as an interferon-stimulated gene (ISG) has recently been shown to exert broad antiviral activity through the production of 25-hydroxycholesterol (25HC), which is believed to inhibit the virus-cell membrane fusion during viral entry. However, little is known about the function of CH25H on HCV infection and replication and whether antiviral function of CH25H is exclusively mediated by 25HC. In the present study, we have found that although 25HC produced by CH25H can inhibit HCV replication, CH25H mutants lacking the hydroxylase activity still carry the antiviral activity against HCV but not other viruses such as MHV-68. Further studies have revealed that CH25H can interact with the NS5A protein of HCV and inhibit its dimer formation, which is essential for HCV replication. Thus, our work has uncovered a novel mechanism by which CH25H restricts HCV replication, suggesting that CH25H inhibits viral infection through both 25HC-dependent and independent events.
Hepatitis C virus (HCV) has infected 170 million people worldwide1. Exposure to HCV after acute infection often leads to a chronic infection in the liver, eventually causing cirrhosis and hepatocellular carcinoma2,3. HCV possesses a 9.6-kb positive-sense RNA genome and its genome encodes a single polyprotein composed of approximately 3,000 amino acids which is processed by host and viral proteases, resulting in 10 viral proteins4. The nonstructural protein 5A (NS5A) is a 56-59–kDa phosphoprotein with an N-terminal amphipathic alpha helix (amino acids 5–25), and 3 structural domains (I, II and III) that is absolutely required for both RNA replication and virus assembly5,6.
Type-I interferons (IFNs), which include IFNα, -β, and –ω, are rapidly induced during viral infection and play a central role in restricting virus replication through the induction of a wide array of anti-viral effectors7,8. Hundreds of interferon stimulated genes (ISGs) have been identified since their discovery more than 25 years ago, and multiple ISGs have been reported to interfere with various key steps of HCV lifecycle via different mechanisms9,10. For example, ISG56 primarily inhibits HCV replication11, while IFITM1 has been shown to suppress both entry and replication process of the virus11,12. On other hand ISG20 and PKR are reported to inhibit HCV RNA synthesis depending on their 3′-5′ exonuclease and protein kinase activities, respectively13, while ISG15 was reported to inhibit HCV replication by decreasing the NS5A stability14. However, function of many other ISGs on HCV infection and replication remain to be elucidated. Cholesterol-25-hydroxylase (CH25H) is a 31.6-kDa endoplasmic reticulum-associated enzyme that catalyzes oxidation of cholesterol to 25-hydroxycholesterol (25HC), which serves as a corepressor of cholesterol biosynthetic enzymes by blocking sterol regulatory element binding protein processing15,16. CH25H is reported to be a conserved ISG, which is rapidly induced in many tissues including the liver, heart, brain, muscle, kidney and lung upon in vivo exposure to various toll-like receptor (TLR) ligands and IFN molecules17. Recently, CH25H has been identified as a major antiviral factor through producing 25HC, which is shown to inhibit a diverse array of viruses, including enveloped viruses (VSV, HSV, HIV, and MHV68) and acutely pathogenic EBOV, RVFV, RSSEV, and Nipah viruses by blocking membrane fusion between virus and cell18. Another study using quantitative metabolomic profiling also demonstrated that 25HC is the only secreted oxysterol synthesized by macrophages to act as a potent paracrine inhibitor of viral infection for a broad range of viruses at multiple levels19. Although 25HC has also been reported to possess anti-HCV activity20,21, the function of CH25H on HCV replication, and whether antiviral function of CH25H is exclusively mediated by 25HC are currently unknown.
In the present study, we show that CH25H has novel antiviral effects on HCV replication not only through its enzyme activity to produce 25HC but also by targeting NS5A leading to the selective inhibition of NS5A dimer formation.
Results
CH25H and its products suppress HCV infection
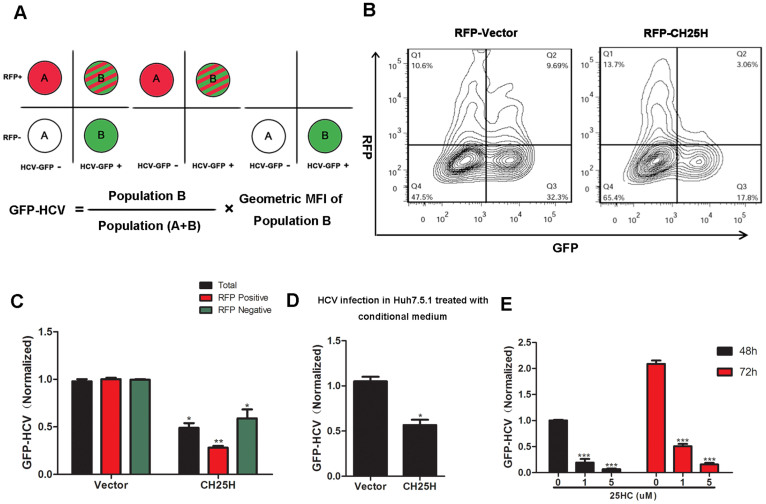
To study the function of CH25H on HCV infection, the plasmid co-expressing CH25H and the red fluorescent protein RFP (linked by IRES) was transfected into Huh7.5.1 cells and the cells were subsequently infected with HCV GFP reporter virus (HCV-GFP). Expression of CH25H and replication of HCV were analyzed and measured by FACS based on RFP and GFP signals. The RFP positive population (RFP+) identifies cells that highly express CH25H, whereas the RFP negative population (RFP-) represents cells that do not express CH25H (Fig. 1A). Interestingly, overexpression of CH25H inhibited HCV infection not only in the RFP+ population of cells, but also in RFP- cells, suggesting that CH25H produces a soluble factor that can confer a cell non-autonomous anti-viral activity onto other adjacent cells (Fig. 1B and C). It was reported that expression levels of CH25H at steady-state are low-to-undetectable in most tissues and cells22, we then detected the expression of CH25H in 293T, Huh7.5.1 and Replicon cells by western blotting and the result showed that the endogenous expression of CH25H is very weak (see Fig. S1 in the supplemental material). CH25H catalyzes oxidation of cholesterol to 25-hydroxycholesterol (25HC), which is a soluble oxysterol that modulate cellular functions in both autocine and paracrine fashions. To determine whether transfection of CH25H led to the production of a soluble antiviral factor, we performed conditioned medium experiment. HEK293T cells were transfected with CH25H or vector control, and the conditioned medium was transferred onto freshly plated Huh7.5.1 cells for 12 hr before infection with HCV-GFP reporter virus (0.2 MOI). The result showed that the conditioned medium from CH25H transfected HEK293T cells generated ~50% HCV inhibition (Fig. 1D). Consistently, treatment of Huh7.5.1 cells with 25-hydroxycholesterol molecule also led to significant inhibition of HCV infection dose-dependently (Fig. 1E). The effect of 25HC on cell viability was also assessed and the result showed that 25HC treatment at 5 uM did not affect cell viability (see Fig. S2 in the supplemental material). Taken together, these data suggest that CH25H and its products could inhibit HCV infection in liver cells.
Figure 1. CH25H and its products suppress HCV infection.
A: Schematic diagram of FACS analysis of HCV-GFP in total, RFP+, RFP- populations. Relative replication of HCV-GFP reporter virus is defined as GFP+% x the geometric mean fluorescence intensity (MFI). B: Contour maps of HCV-GFP replication in Huh7.5.1 cells transfected with the indicated RFP-ISG. C: Effect of overexpression of RFP-CH25H on HCV-GFP replication in total, RFP- positive or negative population normalized to vector control. HCV-GFP was quantified by the calculated value of the percentage % GFP-positive population and MFI of GFP signal. D: Medium was collected from HEK293T cells after 48 h transfection with indicated expression vectors. Freshly plated Huh7.5.1 cells were treated with the conditioned medium for 8 h and then infected with HCV-GFP at 0.2MOI. HCV-GFP was quantified by FACS at 48 hpi. E: Huh7.5.1 cells were pretreated with increasing amounts of 25HC (0, 1, 5 uM) for 8 h. HCV-GFP infection and quantification were measured at 48 and 72 hpi. All experiments were performed in triplicates and data shown are representative of three independent experiments with SEM indicated by error bars. *P < 0.05; **P < 0.01; ***P < 0.001.
CH25H and its products suppress HCV replication
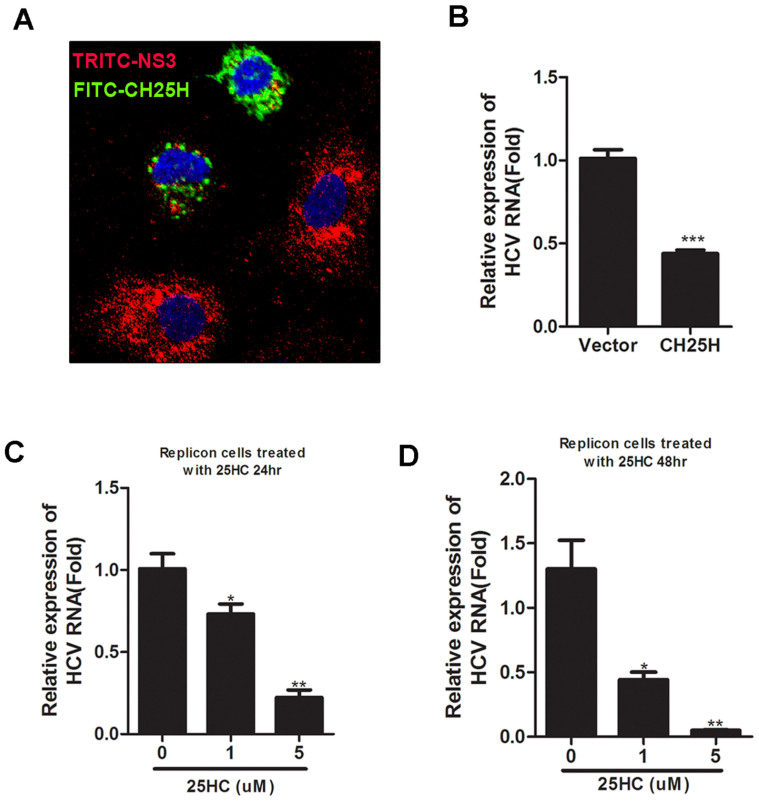
To further investigate how HCV replication was affected by CH25H, Huh-7 cells harboring an HCV subgenomic replicon mimicing the viral RNA replication23 was used for the study. Dual immunofluorcence staining for HCV antigen (NS3) and CH25H was performed to examine the expression of the replicon proteins and effect of CH25H. The result clearly showed that a much lower level of HCV NS3 expressed in those cells which were positive of CH25H expression (Fig. 2A). Transfection of CH25H also decreased HCV RNA levels in HCV replicon cells which demonstrated that enforced expression of CH25H could directly inhibit HCV RNA replication (Fig. 2B). We also examined the effect of 25-hydroxycholesterol on HCV RNA replication, and the result showed that HCV replicon activity was indeed inhibited by 25-hydroxycholesterol in a dose-dependent manner. (Fig. 2C and D). These results indicated that anti-HCV activity of CH25H can be mediated by its enzymatic product, 25-hydroxycholesterol.
Figure 2. CH25H and its products suppress HCV subgenomic replicon replication.
A: Effect of CH25H on HCV replication. JFH HCV Replicon cells were transfected with HA-CH25H expression plasmid and immunofluorescence staining performed 48 hours later using a rabbit polyclonal anti-HA antibody and a mouse monoclonal anti-NS3 antibody, followed by an FITC-conjugated goat anti-rabbit and a TRITC-conjugated goat anti-mouse secondary antibodies. Red fluorescence shows NS3 staining, while green fluorescence detects CH25H. B: Effect of CH25H on HCV RNA expression. The CH25H expression plasmid or vector control plasmid was transfected into JFH replicon cells. Total RNA was extracted 48 hours after transfection and quantified by real-time RT-PCR. C: Effect of 25HC on HCV replication. Replicon cells were treated with increasing amounts of 25HC (0, 1, 5 uM) for 24 h (C) or 48 h (D). HCV Replicon RNA was quantified by real-time RT-PCR. All experiments were performed in triplicates and data shown are representative of three independent experiments with SEM indicated by error bars. *P < 0.05; **P < 0.01; ***P < 0.001.
Mutant forms of CH25H can inhibit HCV replication independent of its enzyme activity
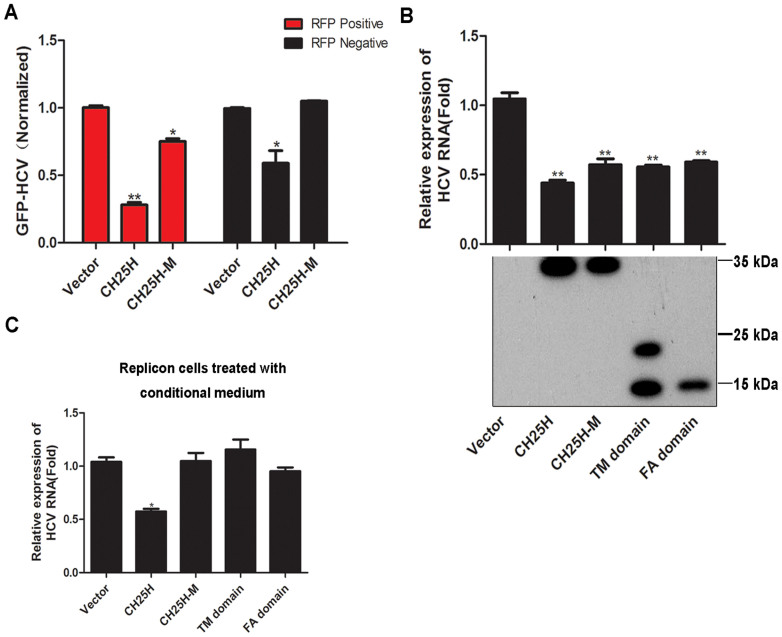
CH25H is a member of a small family of molecules that uses diiron cofactors to catalyze the hydroxylation of hydrophobic substrates. Clustered histidine residues in CH25H were reported to be important for its enzyme activity24. To delineate the anti-HCV mechanism of CH25H, a pair of histidine codons at positions 242 and 243 in the human CH25H were mutated to glutamine codons by site-directed mutagenesis. Liquid Chromatography Mass Spectrometry (LC-MS/MS) experiment was performed to determine whether the mutant form of CH25H (CH25H-M) has lost the ability to produce 25HC. LC-MS/MS results showed that while abundant 25HC was present in the conditioned medium from cells expressing wildtype CH25H, none was detected in the CH25H-M expressing conditioned medium (see Fig. S3 in the supplemental material), confirming that the enzyme activity is indeed disrupted in CH25H-M. To examine the HCV inhibitory activity of CH25H-M, we performed the similar cell autonomous and cell non-autonomous test experiment as described in Fig 1. We found that while expression of CH25H from RFP-linked expressing vector resulted in the inhibition of HCV infection in the RFP negative population of cells, CH25H-M failed to do so (Fig. 3A) indicating that cell non-autonomous inhibition of HCV infection in RFP negative cells requires the enzymatic activity of CH25H. Interestingly, however, the CH25H-M still inhibited HCV infection in RFP positive population of cells suggesting that CH25H can also inhibit HCV infection in cell autonomous fashion via hydroxylase independent mechanism. To further investigate whether the 25HC-independent antiviral mechanism of CH25H operates selectively for HCV infection, we conducted viral inhibition assays with another virus MHV-68. The result showed that in contrast to HCV, CH25H-M failed to inhibit MHV-68 infection (see Fig. S4 in the supplemental material). Thus the alternative hydroxylase-independent antiviral mechanism of CH25H acts selectively in the context of HCV infection. Next, we investigated which domains of CH25H, including Transmembrane (TM) domain and FAase (FA) domain, are responsible for its inhibitory effect on HCV replication. We found that both TM and FA domains could inhibit HCV replication (Fig. 3C). The conditioned medium from the cells transfected with CH25H or the truncated mutants was transferred onto freshly plated HCV Replicon cells to test their effect on HCV RNA replication. The result showed that while the conditioned medium from CH25H generated ~50% HCV RNA inhibition, the medium from cells expressing the mutants had no antiviral function on HCV replication, thus further proved that indeed the enzymatic function of CH25H is not sufficient for its anti-HCV activity (Fig. 3D). Taken together, our results demonstrated that CH25H can inhibit HCV infection and replication independent of its enzyme activity.
Figure 3. CH25H can inhibit HCV replication independent of its enzyme activity.
A: Effect of enforced expression of RFP-CH25H or RFP-CH25H-M plasmid on HCV-GFP in RFP-positive or negative population normalized to vector control. HCV-GFP was quantified as in Fig 1A. B: Effect of enforced expression of CH25H and its truncation mutants on HCV replication. The indicated CH25H-expression constructs were transfected into JFH replicon cells. Total RNA was extracted 48 hours after transfection and quantified by real-time RT-PCR. C: The conditioned medium was collected from HEK293T cells after 48 h transfection with indicated expression vector. Freshly plated Replicon cells were treated with conditioned medium for 24 h and total RNA were extracted and quantified by real-time RT-PCR. All experiments were performed in triplicates and data shown are representative of three independent experiments with SEM indicated by error bars. *P < 0.05; **P < 0.01. All the gels were run under the same experimental conditions as detailed in the Methods section, and full-length blots were cropped for final display.
CH25H selectively interacts with HCV NS5A
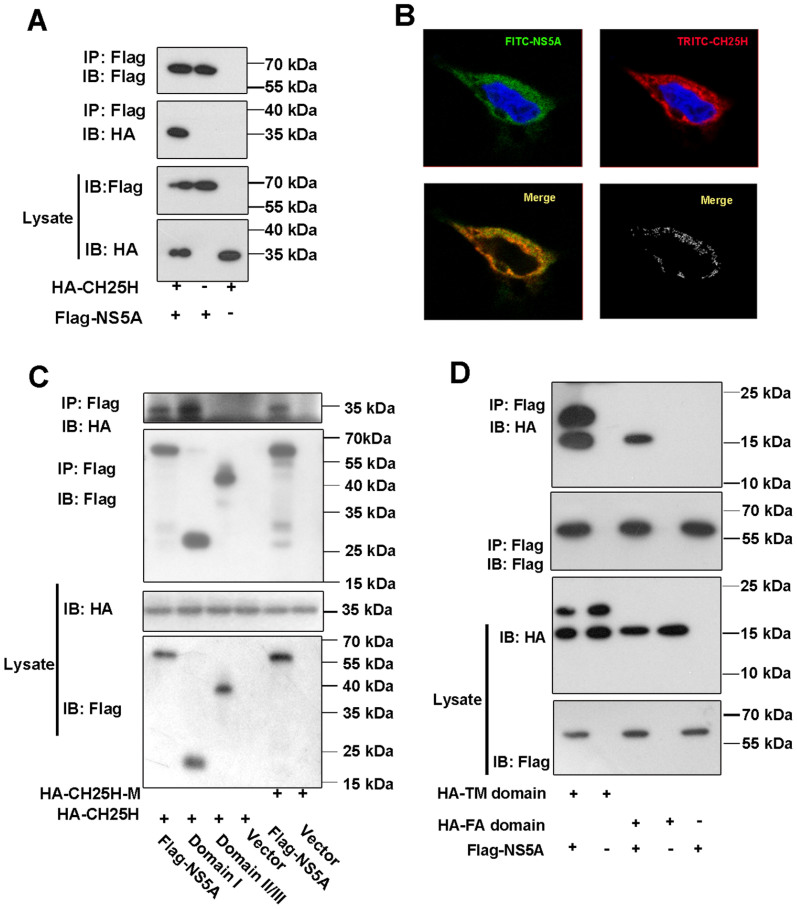
To study how CH25H inhibits HCV replication independent of its enzyme activity, we studied the interactions between CH25H and virus encoded proteins using Bi-molecular fluorescence complementation (BiFC) assay and found that CH25H interacted with NS5A (see Fig. S5 in the supplemental material). To further examine the interaction between NS5A and CH25H, Coimmunoprecipitation (Co-IP) experiments were performed by co-transfection of HA-tagged CH25H and Flag-tagged NS5A expression plasmids. The results showed that NS5A indeed interacted with CH25H (Fig. 4A). To further confirm their interaction inside cells, immunofluorescent microscopy analysis was conducted. Two-color immunofluorescent staining showed that CH25H and NS5A were indeed largely colocalized in the distinct cytoplasmic compartment (Fig. 4B). Having confirmed the molecular interaction between NS5A and CH25H, we subsequently carried out protein truncation assays to map the interaction regions of CH25H and NS5A. We constructed expression plasmids for the truncated proteins according to the reported domain structure of NS5A and CH25H, which were the domain I and domain II/III for NS5A, and the Transmembrane (TM) and FAase (FA) for CH25H. Coimmunoprecipitation experiments showed that the domain I but not domain II/III of NS5A interacted selectively with both TM and FA domains of CH25H (Fig. 4C and D), indicating the domain I of NS5A is distinctly targeted by CH25H.
Figure 4. CH25H interacts with HCV NS5A.
A: NS5A interacts with CH25H. Expression vectors for Flag-tagged NS5A and HA-tagged CH25H were transfected into HEK293T cells. After 36 h, the cells were harvested and lysed, and then the whole cell extracts were subjected to Co-IP assay using an anti-Flag M2-agarose. B: Colocalization of CH25H with NS5A. HEK293T cells were co-transfected with Flag-tagged NS5A and HA-tagged CH25H. After 36 h, the cells were fixed and stained with anti-Flag or anti-HA antibody, followed by TRITC- or FITC-conjugated anti-mouse or anti-rabbit secondary antibody, then analyzed by confocal microscopy and the Pearson's Coefficient is 0.86473. C: CH25H and CH25H-M interact with domain I of NS5A. Expression plasmids for HA-tagged CH25H or CH25H-M were co-transfected with Flag-tagged NS5A truncation mutant constructs. Co-IP assays were performed using cell lysates from cells expressing the indicated constructs. D: Both of TM and FA domains of CH25H interact with NS5A. Expression plasmids for HA-tagged CH25H TM or FA domain were co-transfected with Flag-tagged NS5A. Co-IP assays were done as in C. All results were confirmed by three independent experiments. All the gels were run under the same experimental conditions as detailed in the Methods section, and full-length blots were cropped for final display. Full-length blots are included in the supplementary information.
CH25H specifically disrupts NS5A dimer formation
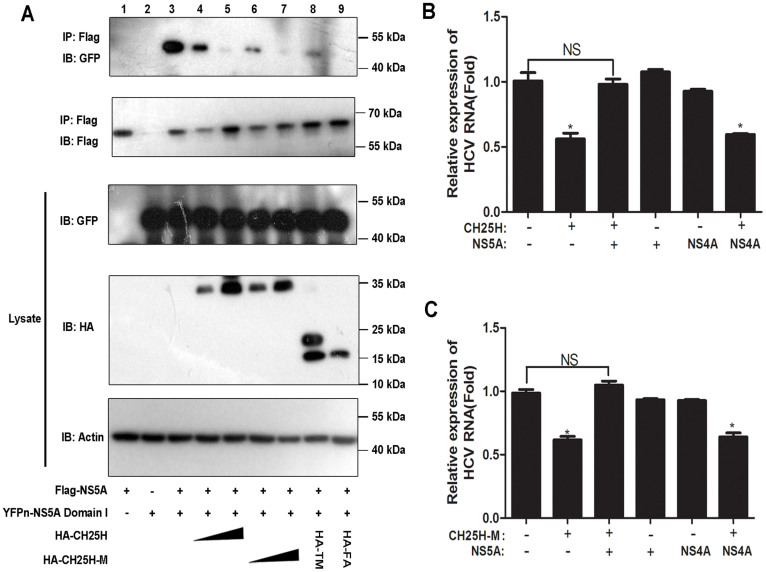
The domain I of NS5A has been reported to be responsible for the formation of NS5A homo-dimer, which is crucial for HCV viral RNA synthesis and replication25,26. We thus hypothesized that CH25H binding to NS5A domain I interferes NS5A dimer formation. To test this hypothesis, we performed competitive coimmunoprecipitation experiments. The results demonstrated that CH25H could indeed disrupt the formation of NS5A dimer in a dose-dependent manner (Fig. 5A lane 4 and 5). To further establish the relationship of NS5A dimer disruption and inhibition of HCV replication, we carried out extended experiment to examine the effect of the CH25H mutants on NS5A dimer formation. The results showed that consistently CH25H-M and the truncated mutants all could disrupt NS5A dimer formationas potent as was the wildtype CH25H (Fig. 5A lane 6-lane 9). Collectively, our results showed that NS5A dimer formation could be inhibited by CH25H and its TM and FA domains suggesting that disruption of NS5A dimer is a critical mechanism how CH25H inhibits HCV replication.
Figure 5. CH25H disrupts NS5A dimer formation.
A: Co-IP assays using Flag control vector or Flag-tagged full-length NS5A as bait to capture YFPn-tagged NS5A domain I were performed in the presence of an increasing amount of HA-tagged CH25H, HA-tagged CH25H-M, or HA-tagged CH25H truncation mutant constructs. Captured proteins were analyzed by Westernblot using anti-GFP and anti-Flag antibodies. All results were confirmed by three independent experiments. All the gels were run under the same experimental conditions as detailed in the Methods section, and full-length blots were cropped for final display. Full-length blots are included in the supplementary information. Effects of NS5A or NS4A on antiviral activities of CH25H (B) or CH25H-M (C). The indicated plasmids expressing control, NS5A or NS4A expression plasmids were cotransfected with CH25H wt or mutant CH25H expression plasmids into Replicon cells. HCV RNA level was measured by real-time RT-PCR 48 hours after transfection. All experiments were performed in triplicates and data shown are representative of three independent experiments with SEM indicated by error bars. *P < 0.05.
To further validate the notion that competitive binding of CH25H to NS5A is indeed responsible for the inhibition of HCV replication, we co-transfected NS5A and CH25H or CH25H-M plasmids into HCV replicon cells and measured the replicon RNA levels. We found that overexpression of NS5A rescued HCV replication inhibited by CH25H or CH25H-M, while another HCV encoded protein NS4A which does not bind to CH25H failed to do so (Fig. 5B and C). Therefore, these data collectively demonstrated that CH25H inhibits HCV replication by binding to NS5A selectively.
Discussion
The current combination therapy for chronic hepatitis C virus (HCV) infection includes Type 1 interferon (IFN) which induces a wide array of interferon-stimulated genes (ISGs) to inhibit virus infection27. ISGs work to restrict viral infection through a wide variety of mechanisms, however, only a few ISGs have so far been defined to have anti-HCV properties. An understanding of the mechanisms by which ISGs exert their inhibitory activities against HCV would enrich the treatment modalities to improve the effectiveness of the current anti-HCV therapeutics while reducing unwanted side effect.
In this work, we define CH25H as an anti-HCV effector ISG that can potently inhibit HCV infection and replication (Fig. 1 and 2). CH25H is an enzyme that catalyzes oxidation of cholesterol to 25-hydroxycholesterol (25HC)24. However, little is known about the functional aspects of CH25H that contribute to its antiviral activity. Recent work has started to reveal that CH25H inhibits VSV and HIV infection at the level of viral entry, specifically by inhibiting the virus-cell membrane fusion through the production of 25HC18. 25HC was also previously reported to inhibit HCV replication20 which is consistent with our data that CH25H inhibits HCV infection and replication partly dependent on the enzyme activity to produce 25HC. 25HC belongs to a diverse class of endogenous oxysterols, the oxidation products of cholesterol, which is well characterized as a soluble factor that control fatty acid biosynthesis through regulation sterol-responsive element binding proteins (SREBP)15. It was also identified to bind and activate the nuclear hormone receptor liver X receptors (LXR)28. LXRs play an important role on lipid metabolism and were reported to control the low-density lipoprotein (LDL) receptor (LDLR) expression29,30, suggesting that 25HC could inhibit LDLR expression. Recent studies demonstrated that hepatocytes treated with 25HC exhibited decreased LDLR mRNA levels31. LDLR is a membrane glycoprotein which has been reported to be essential for replication of the HCV genome32. Our preliminary study result also showed that CH25H could inhibit LDLR expression which might provide an explanation for 25HC mediated antiviral function on HCV.
In an attempt to delineate the anti-HCV mechanism of CH25H, we generated a mutant CH25H construct lacking enzymatic activity to produce 25HC. Although the enzymatic dead mutant CH25H-M failed to produce 25HC and was unable to inhibit HCV infection in trans by the paracrine mechanism, however, the mutant CH25H still inhibits HCV infection in RFP positive population (Fig. 3A), and overexpression of CH25H but not CH25H-M inhibits MHV-68 growth. All truncations of CH25H not the conditional medium inhibit HCV replication (Fig. 3B and C) confirming that an alternative antiviral mechanism for the protein specifically in the context of HCV is independent on its enzyme activity.
NS5A is a pleiotropic virus-encoded protein required for HCV RNA replication33. The structure of Domain I suggests that it is dimeric in nature which forms a positively charged groove involved in RNA binding25. Recently several studies have shown that the NS5A dimer formation mediated by domain I is essential for HCV replication26,34. Therefore it is an attractive target for direct antiviral therapy of HCV35. The remarkably potent RNA replication inhibitor, BMS-790052, has been implicated to target NS5A by inhibiting its dimerization36. Viperin, one conserved type I ISG, was also reported to inhibit HCV replication by interacting with NS5A37,38. Using BiFC screening system we demonstrated that CH25H specifically interacts with HCV NS5A, not other viral proteins including Core, NS3, NS4A or NS5B. Co-IP data demonstrated that both TM and FA domains of CH25H interact with the domain I of NS5A (Fig. 4). Thus CH25H binds to the same domain of NS5A responsible for dimer formation, suggesting that competition for dimer formation may serve as a strategy for host to inhibit HCV replication. Indeed, we showed that NS5A dimerization was strongly disrupted in the presence of CH25H or the mutant forms of CH25H which remain activity in HCV inhibition, even though lacking hydroxylase activity (Fig. 5A). At the same time, our results also confirmed the importance of the dimer formation of NS5A in HCV replication. We have further substantiated this finding by showing that overexpression of NS5A but not other HCV viral proteins, such as NS4A could rescue the inhibition of HCV replication imposed by CH25H or CH25H-M (Fig. 5B and C), confirming that CH25H inhibits HCV replication by competitive binding with NS5A. In addition to its essential function in HCV RNA synthesis and replication, NS5A also attracts considerable interest because of its potential role in evading host defense by modulating the response to IFN-α therapy. One of the critical genetic determinant for IFN resistant of various HCV genotypes is mapped to the region in NS5A called the interferon-a sensitivity-determining region (ISDR). It will be interesting to test in future work whether the adaptive mutations found in this region affect the interaction between CH25H and NS5A.
In conclusion, the work presented herein has extended our understanding of the newly identified antiviral protein CH25H. The findings described here as well as reported by others clearly show that CH25H can broadly suppress virus infection by producing 25HC18,19. Nevertheless, we have identified another novel antiviral mechanism to HCV independent of its enzyme activity by specifically binding to NS5A and inhibiting its dimer formation, suggesting that CH25H is a multiple functional antiviral protein which is able to inhibit viral infection and replication through both 25HC-dependent and independent pathways. Taken together, our work sheds interesting light on the mechanisms by which CH25H restricts HCV replication and also points out a potential new strategy to improve the existing anti-HCV therapies by effectively inducing CH25H expression and function.
Methods
Reagents and Antibodies
Mouse monoclonal and rabbit polyclonal antibodies against FLAG (Sigma), HA (Sigma), mouse monoclonal antibodies against β-actin (Sigma), NS3 (Abcam) and rabbit polyclonal antibody against GFP (Abcam), 25-hydroxycholesterol (Sigma) were purchased from the indicated companies.
HCV Replicon Cells and HCV-JFH1 Cell Culture
HCV replicon cells (JFH strain) from Dr. Jin Zhong were maintained in complete DMEM supplemented with 0.5 g/L G418 (Invitrogen). The human hepatocellular carcinoma cell line Huh7.5.1 and HCVJFH1-infected Huh7.5.1 cells were maintained in complete DMEM. The Huh7.5.1 cells were infected with GFP-HCV (JFH1 strain) as previously described39.
Plasmids and Expression Vectors
Mammalian expression plasmids for full-length wildtype human CH25H (aa1-aa272), CH25H TM domain (aa1-aa135) and CH25H FAase domain (aa136-aa272) were PCR and cloned into pCMV-HA to generate HA tagged expression constructs by standard molecular cloning techniques. Site-directed mutagenesis was carried out to convert histidine codons at positions 242 and 243 of the wildtype CH25H to glutamine codons according to manufacturer instructions (TransGen Biotech) to create the mutant form (CH25H-M) deprived of catalytic activity. Sequences encoding full-length HCV NS5A (aa1978-aa2458), NS5A domain I (aa1978-aa2173), NS5A domain II and III (aa2174-aa2458) and NS4A (aa1683-aa1715) were amplified by PCR from a plasmid encoding the full-length genome of JFH-1 HCV and cloned into pENTR/D-TOPO vector (Invitrogen). Through Gateway recombination, the NS5A ORF fragments cloned in pENTR/D-TOPO were transferred individually into pL-EF1a-DEST-Flag or pBabe-CMV-YFPn-DEST-neo expression vectors described in40 to generate C-terminal Flag-tagged or YFPn-tagged full length or truncated NS5A constructs. Similarly, the ORFs of CH25H and CH25H-M were transferred into pL-EF1a-DEST-SFB-IRES/TagRFP expression vector to co-express target gene and IRES-linked RFP marker. Primer pairs used for PCR amplification of the genes are listed (see Table S1 in the supplemental material). The target genes in the resulting Gateway expression plasmids were all confirmed by DNA sequencing, diagnostic restriction enzyme digestion analyses, and further by westernblot.
Transfection
Plasmid transfections were performed using Lipofectamine 2000 following the manufacturer's instructions (Invitrogen).
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100/PBS before incubation with primary antibodies (anti-NS3, anti-HA or anti-Flag) for 1 hour at room temperature (RT). Cells were washed in PBS, followed by incubation with TRITC or FITC-conjugated anti-mouse or anti-rabbit immunoglobulin (Ig) secondary antibodies. Nuclei were stained with DAPI. Confocal microscopy was performed with Olympus FV1000 Confocal Microscope (Olympus).
Flow cytometry
Cells were collected by trypsinization, fixed in 2% paraformaldehyde and analyzed on BD FACS-Calibur flow cytometer (BD Biosciences).
Quantitative RT–PCR (qPCR) analysis
Total RNA was extracted from cultured cells with TRIzol reagent (Invitrogen). Real-time PCR was performed with Power SYBR Green PCR master mix (Applied Biosystems) in an ABI 7500 Real-time PCR system (Applied Biosystems). The primers for real-time PCR are as follows: JFH-HCV (Forward, 5′-TCTGCGGAACCGGTGAGTA-3′; Reverse, 5′-TCAGGCAGTACCACAAGGC-3′); GAPDH (Forward, 5′-GAACGGGAAGCTCACTGG-3′; Reverse, 5′-GCCTGCTTCACCACCTTCT-3′). All gene expression data were normalized to the expression of GAPDH.
Co-immunoprecipitation and Western Blot Analysis
Cells were lysed 36–48 h after transfection of expression plasmids using 50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100 containing cocktail. For immunoprecipitation, lysates were incubated with the appropriate antibodies for 2 h on ice, followed by precipitation with protein G Sepharose (Santa Cruz). Samples were separated by SDS–PAGE and transferred to PVDF membranes. After blocking in PBS containing 0.1% Tween-20 and 5% skim-milk, the blots were probed with indicated antibodies. Westernblot visualization was done with enhanced chemiluminescence.
Quantification of 25-Hydroxycholesterol
2 mL of cell culture medium was extracted with 5 mL of methanol and 1 g of sodium sulfate anhydrous (Na2SO4) twice. After centrifugation, the supernatant was evaporated to dryness with nitrogen gas flow. The residue was reconstituted with 0.5 mL of acetonitrile and 0.5 mL of water and analyzed by LC-MS/MS with ESI+ ion source (API 5000, Applied Biosystems, ON, Canada). The instrument procedure follows the description41.
MHV-68 infection
HEK293T cells were infected with MHV-68(M3FL)42 at MOI = 0.25. After 24hpi, the supernatant was tansferred to naïve BHK21 and luciferase activity was measured at 9hpi.
Statistical methods
GraphPad Prism 5 software (GraphPad Software, San Diego, CA) was used for data analysis using a two-tail unpaired t test. A p value <0.05 was considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Author Contributions
Y.C. designed and performed experiments, analyzed data and wrote the paper; S.W., Z.Y., H.T., R.A. performed experiments; Y.L., G.C. performed LC-MS experiment; P.L., P.D. and L.S. analyzed the data; J.Z. and X.C. provided critical regents; F.X.Q. organized, designed, and wrote the paper, H.D. and G.H.C. initiated the study, organized, designed, and wrote the paper.
Supplementary Material
Supplementary information
Acknowledgments
We are grateful to Dr. Liguo Zhang for advice and Dr. Chunli Jiang for technical help in confocal microscopy. This work was supported by Grant No. 81325012 from the National Natural Science Foundation of China, Grant No. 2015CB554302, No. 2009CB522505 and No. 2013CB911103 from the Ministry of Science and Technology of China and National Institutes of Health Grant (AI078389 and AI069120).
References
- Hajarizadeh B., Grebely J. & Dore G. J. Epidemiology and natural history of HCV infection. Nat. Rev. Gastroerterol. Hepatol. 10, 553–562 (2013). [DOI] [PubMed] [Google Scholar]
- Shlomai A., de Jong Y. P. & Rice C. M. Virus associated malignancies: The role of viral hepatitis in hepatocellular carcinoma. Semin. Cancer Biol. 26C, 78–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. Y. Molecular pathogenesis of hepatitis C virus-associated hepatocellular carcinoma. Front. Biosci. 12, 222–233 (2007). [DOI] [PubMed] [Google Scholar]
- Ashfaq U. A., Javed T., Rehman S., Nawaz Z. & Riazuddin S. An overview of HCV molecular biology, replication and immune responses. Virol. J. 8, 161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. et al. Hepatitis C Virus Nonstructural Protein 5A: Biochemical Characterization of a Novel Structural Class of RNA-Binding Proteins. J. Virol. 84, 12480–12491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B. D. & Rice C. M. Unravelling hepatitis C virus replication from genome to function. Nature 436, 933–938 (2005). [DOI] [PubMed] [Google Scholar]
- Liu S. Y., Sanchez D. J. & Cheng G. New developments in the induction and antiviral effectors of type I interferon. Curr. Opin. Immunol. 23, 57–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K., Onoguchi K., Takahasi K. & Fujita T. Type I interferon production induced by RIG-I-like receptors. J. Interferon Cytokine Res. 30, 875–881 (2010). [DOI] [PubMed] [Google Scholar]
- Rice C. M. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz P. et al. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology 56, 2082–2093 (2012). [DOI] [PubMed] [Google Scholar]
- Raychoudhuri A. et al. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 85, 12881–12889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C. et al. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 57, 461–469 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D. et al. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82, 1665–1678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J. & Yoo J. Y. Inhibition of hepatitis C virus replication by IFN-mediated ISGylation of HCV-NS5A. J. Immunol. 185, 4311–4318 (2010). [DOI] [PubMed] [Google Scholar]
- Adams C. M. et al. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and insigs. J. Biol. Chem. 279, 52772–52780 (2004). [DOI] [PubMed] [Google Scholar]
- Kandutsch A. A., Chen H. W. & Shown E. P. Binding of 25-hydroxycholesterol and cholesterol to different cytoplasmic proteins. Proc. Natl. Acad. Sci. U. S. A. 74, 2500–2503 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. & Scott A. L. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 88, 1081–1087 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. Y. et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38, 92–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M. et al. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 38, 106–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A. I. et al. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 99, 15669–15674 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. et al. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. U. S. A. 100, 15865–15870 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. & Scott A. L. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 88, 1081–1087 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V. et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285, 110–113 (1999). [DOI] [PubMed] [Google Scholar]
- Lund E. G., Kerr T. A., Sakai J., Li W. P. & Russell D. W. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J. Biol. Chem. 273, 34316–34327 (1998). [DOI] [PubMed] [Google Scholar]
- Tellinghuisen T. L., Marcotrigiano J. & Rice C. M. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435, 374–379 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P. J. et al. Correlation between NS5A dimerization and hepatitis C virus replication. J. Biol. Chem. 287, 30861–30873 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q. L., Zhang J. Y., Zhang Z., Wang L. F. & Wang F. S. Sofosbuvir and ABT-450: terminator of hepatitis C virus? World J. Gastroenterol. 19, 3199–3206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski B. A. et al. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. U. S. A. 96, 266–271 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto K. et al. Identification of human low-density lipoprotein receptor as a novel target gene regulated by liver X receptor alpha. FEBS Lett. 580, 4929–4933 (2006). [DOI] [PubMed] [Google Scholar]
- Zelcer N., Hong C., Boyadjian R. & Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 325, 100–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina S. et al. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J. Hepatol. 46, 411–419 (2007). [DOI] [PubMed] [Google Scholar]
- Albecka A. et al. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology 55, 998–1007 (2012). [DOI] [PubMed] [Google Scholar]
- Gu M. G. & Rice C. M. Structures of hepatitis C virus nonstructural proteins required for replicase assembly and function. Curr. Opin. Virol. 3, 129–136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. Y. et al. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280, 36417–36428 (2005). [DOI] [PubMed] [Google Scholar]
- Debes J. D. & Smith C. I. NS5A: a new target for antiviral drugs in the treatment of hepatitis C virus infection. Hepatology 56, 797–799 (2012). [DOI] [PubMed] [Google Scholar]
- Gao M. et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465, 96–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnweiss A. et al. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J. Immunol. 184, 5179–5185 (2010). [DOI] [PubMed] [Google Scholar]
- Helbig K. J. et al. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology 54, 1506–1517 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q. X. et al. Compensatory mutations in NS3 and NS5A proteins enhance the virus production capability of hepatitis C reporter virus. Virus Res. 145, 63–73 (2009). [DOI] [PubMed] [Google Scholar]
- Chen L. Y., Liu D. & Zhou S. Y. Telomere maintenance through spatial control of telomeric proteins. Mol. Cell. Biol. 27, 5898–5909 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. et al. Investigation of Testosterone, Androstenone, and Estradiol Metabolism in HepG2 Cells and Primary Culture Pig Hepatocytes and Their Effects on 17 beta HSD7 Gene Expression. PLoS One 7, e52255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. Y., Sanchez D. J., Aliyari R., Lu S. & Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U. S. A. 109, 4239–4244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information