Abstract
The polyphosphate glucokinases can phosphorylate glucose to glucose 6-phosphate using polyphosphate as the substrate. ORF all1371 encodes a putative polyphosphate glucokinase in the filamentous heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. Here, ORF all1371 was heterologously expressed in Escherichia coli, and its purified product was characterized. Enzyme activity assays revealed that All1371 is an active polyphosphate glucokinase that can phosphorylate both glucose and mannose in the presence of divalent cations in vitro. Unlike many other polyphosphate glucokinases, for which nucleoside triphosphates (e.g. ATP or GTP) act as phosphoryl group donors, All1371 required polyphosphate to confer its enzymic activity. The enzymic reaction catalysed by All1371 followed classical Michaelis–Menten kinetics, with kcat = 48.2 s−1 at pH 7.5 and 28 °C and KM = 1.76 µM and 0.118 mM for polyphosphate and glucose, respectively. Its reaction mechanism was identified as a particular multi-substrate mechanism called the ‘bi-bi ping-pong mechanism’. Bioinformatic analyses revealed numerous polyphosphate-dependent glucokinases in heterocyst-forming cyanobacteria. Viability of an Anabaena sp. PCC 7120 mutant strain lacking all1371 was impaired under nitrogen-fixing conditions. GFP promoter studies indicate expression of all1371 under combined nitrogen deprivation. All1371 might play a substantial role in Anabaena sp. PCC 7120 under these conditions.
Introduction
Inorganic polyphosphate, which is a linear polymer of 10–1000 orthophosphates linked by phosphoanhydride bonds, has been found in all representative living cells, including bacteria, fungi, plants, animals and archaea (Achbergerová & Nahálka, 2011; Rao et al., 2009; Remonsellez et al., 2006; Scherer & Bochem, 1983). Polyphosphate is stored in the cytoplasm, where it can be visualized as metachromic inclusions (Meyer, 1902) or electron-dense granules (Jensen, 1968). Evolutionarily, polyphosphate stands as one of the earliest polymers produced in cells. Polyphosphate is considered to be the ancestor of ATP as an energy source (Resnick & Zehnder, 2000), given that hydrolysis of the phosphoanhydride bond between each orthophosphate yields free energy comparable to that generated by cleavage of ATP. In a bacterial cell, polyphosphate functions mainly as a dynamic storage compound for phosphate and energy (Harold, 1966; Kornberg et al., 1956). However, many other functions have been proposed for the polymer, including those in stress responses, complexation of heavy metals, biofilm formation and virulence (Kornberg, 1995; Rashid & Kornberg, 2000; Rashid et al., 2000; Tsutsumi et al., 2000). In bacteria, polyphosphate metabolism is driven by two kinds of enzymes: kinases and phosphatases. Polyphosphate is synthesized by polyphosphate kinase type 1 (Ahn & Kornberg, 1990; Kornberg et al., 1956), which catalyses the formation of the phosphoanhydride bonds between the growing polymer and the γ-phosphoryl residues of ATP or another nucleotide triphosphate. Conversely, polyphosphate is degraded mainly by exopolyphosphatases (Akiyama et al., 1993; Kornberg et al., 1999) and endopolyphosphatases (Lichko et al., 2010).
Polyphosphate glucokinase (PPGK; EC 2.7.1.63), a paralogue of the ATP-dependent glucokinase (Hsieh et al., 1993), catalyses the transfer of the terminal phosphoryl residue of polyphosphate to glucose in order to generate glucose 6-phosphate. The first ATP/polyphosphate-dependent glucokinase was discovered in Mycobacterium phlei (Szymona & Ostrowski, 1964). Since then, such enzymes have been found in many non-eukaryotic organisms (Liao et al., 2012; Lindner et al., 2010; Pepin & Wood, 1986; Phillips et al., 1993; Szymona & Widomski, 1974; Tanaka et al., 2003). All but one of the known PPGKs are bifunctional, in that they are able to utilize both ATP and polyphosphate as phosphoryl donors. The sole known exception to that is the PPGK of the polyphosphate-accumulating bacterium Microlunatus phosphovorus (Tanaka et al., 2003), which is a strictly polyphosphate-dependent enzyme.
The potential role of PPGKs in the complex metabolism of cyanobacteria has not yet been investigated. Cyanobacteria are a widespread group of oxygenic photosynthetic prokaryotes. During photosynthesis, energy is transiently stored in the energy-rich phosphoanhydride bonds of ATP molecules. Several genera of cyanobacteria perform both photosynthesis and N2 fixation; however, these two physiological processes are incompatible, because the oxygen-sensitive nitrogenase complex (Hill et al., 1981) is the key enzyme in N2 fixation. The diazotrophic cyanobacteria have developed special mechanisms to allow N2 fixation to take place under aerobic conditions (Berman-Frank et al., 2003). Some filamentous cyanobacteria, such as Anabaena sp. PCC 7120 (also called Nostoc sp. PCC 7120; hereafter Anabaena), form highly specialized cells called ‘heterocysts’, which fix N2 in a micro-oxic environment (Adams et al., 1981). The heterocysts are semi-regularly distributed along the filaments and rely on vegetative cells to supply them with photosynthetic products. In return, the heterocysts provide the filament with reduced nitrogen compounds (Flores & Herrero, 2010; Maldener & Muro-Pastor, 2010). In contrast, some unicellular diazotrophic cyanobacteria use a diurnal rhythm to separate N2 fixation and photosynthesis, protecting the nitrogenase from oxygen by employing it in the dark, when photosynthesis is quiescent (Mitsui et al., 1986; Toepel et al., 2008).
The purpose of this study was to characterize All1371, the PPGK from Anabaena, in vitro and to explore its biological function in vivo.
Methods
Sequence analysis.
A blastp search (Altschul et al., 1997) was performed against all cyanobacterial sequences available from the Integrated Microbial Genomes database (Markowitz et al., 2012) and against the sequences of Section V cyanobacteria identified by Dagan et al. (2013). The amino acid sequence of the PPGK from Anabaena (all1371: 637231738, gene ID Integrated Microbial Genomes database) was used as query. Similar amino acid sequences of proteins with known 3D structures were identified using the structure database PDBsum (http://www.ebi.ac.uk/pdbsum/). Sequences were aligned using clustalw2 (Larkin et al., 2007), and formatted with ESPript (Gouet et al., 1999). Sequence similarities were determined using the emboss needle software (http://www.ebi.ac.uk/Tools/psa/emboss_needle/).
Bacterial strains and culture conditions.
Anabaena was grown in fourfold diluted medium of Allen & Arnon (1955) (AA/4 medium) with or without 10 mM KNO3. Liquid cultures of Anabaena were grown under permanent illumination with white light of 70 µmol photons m−2 s−1 at 30 °C. Cultures were grown in air lift flasks (Ø 6 cm), bubbled with air enriched with 2 % (v/v) CO2. Mutants were grown in the presence of 50 µg neomycin ml−1 or 4 µg spectinomycin ml−1 and 1 µg streptomycin ml−1. Synechocystis sp. PCC 6803 (hereafter Synechocystis) was grown on BG11 agar plates (Rippka et al., 1979) additionally containing 20 mM HEPES. Liquid cultures were grown at 28 °C and under continuous illumination as described above. Liquid cultures of Mastigocladus laminosus SAG 4.84 and Fischerella muscicola PCC 7414 were grown in Castenholz medium D (8.24 mM NaNO3, 0.99 mM KNO3) or medium ND (without nitrate) (Castenholz, 1988) at 42 °C and under continuous illumination of 100 µmol photons m−2 s−1.
Chlorophyll a content was determined as described by de Marsac & Houmard (1988). For nitrogen starvation, exponentially grown cultures were harvested by centrifugation, washed twice with nitrate-free medium and resuspended to a final concentration of 7 µg chlorophyll ml−1 for further growth. Escherichia coli strains DH5α and BL21 (DE3) (Novagen; Merck Chemicals) were grown at 37 °C as batch culture in Erlenmeyer flasks with shaking at 300 r.p.m. in Luria–Bertani (LB) medium (Bertani, 1951) supplemented with 10 µg ampicillin ml−1, 150 µg neomycin ml−1 or 50 µg spectinomycin ml−1 when appropriate.
Construction of expression plasmid.
The all1371 gene was amplified by PCR using genomic Anabaena DNA as template and oligonucleotides 5′-AGGATCCTACTCAATGGTGGAAGATAACGG-3′ and 5′-GCGGCCGCTTCTATAGTGTTTTTTCATCTC-3′ (BamHI and NotI restriction sides highlighted in bold, stop codon underlined). The PCR product was ligated into the cloning vector pJET1.2 (Thermo Scientific) to ensure efficient restriction digests. After restriction digest of the resulting pJET-all1371 by BamHI and NotI, all1371 was inserted into the vector pGEX-6P-1 (GE Healthcare), leading to pGEX_all1371. The manipulations were checked by restriction analysis and DNA sequencing.
Protein expression and purification.
E. coli BL21(DE3) cells were transformed with pGEX_all1371. The recombinant strain was grown in LB medium containing 100 µg ampicillin ml−1 and 1 % (w/v) glucose. The expression was induced with 1 mM IPTG at an OD600 of 0.6. Cells were harvested 3 h after induction by centrifugation (15 min, 3800 g), resuspended in buffer containing 200 mM Tris/HCl (pH 8.5), 300 mM NaCl and 50 mM KCl, and disrupted by sonication. The debris was removed by centrifugation (15 min, 20 000 g). The glutathione-S-transferase (GST)–PPGK fusion was purified by affinity chromatography using Glutathion-Sepharose 4B (GE Healthcare) performed in a batch technique according to the manufacturer’s instructions. To elute All1371 the GST-tagged PPGK was cleaved on the column by PreScission protease (Walker et al., 1994) (GE Healthcare) overnight at 4 °C. The purity of the enzyme was verified by SDS-PAGE.
Preparation of cell-free cyanobacterial extract, electrophoresis and protein quantification.
Cyanobacterial cells were collected from liquid cultures (grown with or without nitrogen for 4 or 6 days) by centrifugation (6500 g). Sedimented cells were washed twice with 50 mM Tris/HCl buffer (pH 8.0) and stored at −20 °C. Thawed filaments of Mastigocladus laminosus and F. muscicola were pretreated by sonication. Cells were disrupted in a swing mill (Retsch MM 301) for 30 min at 30 Hz using glass beads (Ø 0.1 mm). Beads and crude extracts were separated by two sequential centrifugations at 10 000 g and 4 °C for 10 and 30 min. To remove small molecules the supernatants were purified using DextraSEC PRO2 columns (Applichem). The elution was performed by the original buffer. Protein concentrations were estimated according to Lowry et al. (1951) using BSA as reference. SDS-PAGE was performed on slab gels [15 % (w/v) acrylamide, 0.41 % (w/v) methylene-bisacrylamide] (Laemmli, 1970). The gels were stained with Coomassie brilliant blue R250.
Determination of molecular mass.
The molecular mass of native All1371 was determined by size exclusion chromatography on a Tricorn Superdex 200 10/300 GL column (GE Healthcare) calibrated with the gel filtration standards purchased from Bio-Rad (γ-globulin, 158 kDa; ovalbumin, 44 kDa; myoglobin, 17 kDa; cytochrome, ca. 12.4 kDa). As running buffer 100 mM Tris/HCl (pH 7.5), 200 mM NaCl, 6 mM MgCl2 (hereafter basic buffer) and 0.5 mM DTT were used at a flow rate of 0.8 ml min−1. Pure All1371 (100 µg) was loaded onto the column. The elution was monitored by measuring A280. Fractions of 0.5 ml were collected. Aliquots from these fractions were tested for PPGK activity. Precipitated fractions (Bensadoun & Weinstein, 1976) were analysed by SDS-PAGE. Two biological replicates were performed.
Activity assays and kinetic analyses.
Glucokinase activity and kinetics of the isolated All1371 were determined in vitro by coupling glucose 6-phosphate formation to the glucose-6-phosphate dehydrogenase reaction (Hsieh et al., 1993). Glucose 6-phosphate formation was monitored indirectly by measuring NADH development spectrophotometrically at 340 nm (ϵ340 = 6220 M−1 cm−1). Measurements were done in basic buffer with 0.6 mM NAD, 0.8 mM glucose, 0.01 mM polyP45 (phosphate glass type 45; Sigma-Aldrich; hereafter polyphosphate), 5.7 units glucose-6-phosphate dehydrogenase ml−1 and 0.26 µg All1371 ml−1 at 28 °C. To verify the cation dependency, MgCl2 was replaced by MnCl2 or water. One unit of PPGK activity was defined as the amount of enzyme which catalyses the formation of 1 µmol glucose 6-phosphate min−1. The reaction was started with All1371. All rates were determined from the linear region of the curves. To check the substrate specificity, the following substrates were applied to the assay at final concentrations of 10 µM, 100 µM and 1 mM: polyP45, ATP, ADP, AMP, GTP, UTP, CTP and pyrophosphate. Additionally ATP was tested as a substrate at final concentrations of 5 and 10 mM. Polyphosphate was used to check enzyme activity if no activity was detected in vitro. To determine the kinetics of All1371, both substrates (glucose, polyphosphate) were varied. The values of KM and kcat were calculated from the initial rate. Three biological replicates were performed. Kinetic parameters were analysed by Sigma Plot 2006 Enzyme Kinetics Module 1.3 (Systat software). The initial rate was measured for several glucose concentrations at different non-saturating polyP45 concentrations. Measurements were also taken for several polyphosphate concentrations at different non-saturating glucose concentrations.
Mannokinase activity was determined by monitoring the formation of NADH spectrophotometrically at 340 nm. Activity was measured in basic buffer including 0.6 mM NAD, 5–70 mM mannose, 0.02 mM polyP45, 5.7 units glucose-6-phosphate dehydrogenase ml−1, 1.0 units mannose-6-phosphate isomerase ml−1, 4.0 units glucose-6-phosphate isomerase ml−1 and 23–42 µg All1371 ml−1 at 28–30 °C.
When measuring PPGK activities in cell-free extracts, the extracts instead of the pure enzyme were used in standard assays as described above (glucokinase activity). The reaction was started with glucose. To confirm the linearity of the reaction different amounts of the extracts with 23–480 µg protein were added to the reaction mixture. All1371 was applied to the assay as a positive control.
Deletion of all1371 in Anabaena.
An Anabaena Δall1371 mutant was generated by replacing 771 nt including all1371 [720 nt, genomic region 1625 095–1625 814 (Nakao et al., 2010)] with an antibiotic resistance cassette, not affecting other ORFs.
Upstream and downstream regions of all1371 were amplified by PCR using genomic Anabaena DNA as template. Restriction sites introduced by the primers below are highlighted in the sequence in bold type and termed in parentheses. The upstream region (position 1625 815–1626 817) was amplified using the primers 5′-ATTGAGCTCAAGGACGGAAAAAATTACAC-3′ (SacI) and 5′-GAGTATTTACCTTTTTTCTAGAGACTGG-3′ (XbaI) yielding a PCR product of 973 nt after restriction. The downstream region was amplified using the primer pair 5′-CCCGAAACTCTAGATGTGACTGGGTATGGGG-3′ (XbaI) and 5′-AATGCTCGAGAACCAACCTATACCTGTGC-3′ (XhoI). The restricted product yielded a 943 nt fragment. The fragments were successively inserted into the pBluescript KSII+ vector (Stratagene) resulting in pKSII+_up_down. The resistance cassette C.K3 containing the neomycin phosphotransferase II gene was received from pRL448 (Elhai & Wolk, 1988a) and inserted into pKSII+_up_down via the XbaI site, yielding pKSII+_up-C.K3-down. The C.K3 cassette was inserted in the same direction as all1371. The correctness of the sequence was validated by DNA sequencing. To construct pRL271_up-C.K3-down used for deletion, the SacI/XhoI fragment excised from the prior plasmid was cloned into plasmid pRL271 (Black et al., 1993). This plasmid was conjugationally transferred to Anabaena by triparental mating using E. coli strain J53[RP4] and cargo strain E. coli HB101[pRL528] (Elhai & Wolk, 1988b). Neomycin-resistant double recombinants were identified by PCR and sacB selection (Cai & Wolk, 1990).
Viability tests.
Viability tests of Anabaena and the Δall1371 mutant were carried out as a spot assay on AA-plates (Allen & Arnon, 1955) with or without 10 mM KNO3 as a nitrogen source. A 10 µl volume of liquid cultures was applied per spot. These agar plates were exposed to continuous light of 60–70 µmol photons m−2 s−1 for 6 days. Three biological replicates were tested separately.
Generation of a GFP promoter fusion strain.
The gfp gene was amplified by PCR using the primer pair 5′-GATGGCTCTCTAGAATGAGTAAAGGAGAAG-3′ and 5′-CTTCTAGATTAATGTTTGTATAGTTCATC-3′ (XbaI in bold type, stop codon underlined) and plasmid pJET1.2-GFP as template (Baier, 2013). The gfp gene was obtained by XbaI digest and inserted in plasmid pKSII+_up_down (see above) via the XbaI site, leading to pKSII+_up-gfp-down. By this means, the gfp gene was integrated in this plasmid into the upstream region of all1371 32 nt after the transcription start site. This plasmid was used as template in a PCR performed with oligonucleotides 5′-CTATAGGGCGAATTCGAGCTCAAGGACGG-3′ and 5′-GTGTTCTTCTCCGAATTCCCATAC-3′ (EcoRI sites in bold type). Finally, the PCR product was inserted into pRL1049 (Black & Wolk, 1994) via the EcoRI sites, resulting in vector pRL1049-up-gfp-down, which was validated by DNA sequencing. To generate Anabaena all1371 GFP promoter fusion strains, pRL1049-up-gfp-down_all1371 was introduced in the Anabaena wild-type and Δall1371. Conjugation was performed as described above. Positive exconjugants selected on AA-agar plates (Allen & Arnon, 1955) containing 4 µg spectinomycin ml−1 and 1 µg streptomycin ml−1 were checked by PCR using primers 5′-GCCTGCATTTGGTGCTGGACTGG-3′ and 5′-GGTCTGCTAGTTGAACGCTTCC-3′. The plasmid pRL1049-up-gfp-down_all1371 was self-replicating in these exconjugants.
Confocal microscopy.
For confocal microscopy Anabaena and mutant strains (Δall1371, promoter fusion) were grown as liquid cultures with and without nitrate for 4 days. Fluorescence in cells of the Anabaena Δall1371 promoter fusion strain was visualized with a laser-scanning confocal microscope (Olympus FV-1000MPE). GFP was excited by an argon ion laser (488 nm irradiation). Fluorescence emission was recorded at 500–545 nm (for GFP) and 570–670 nm (for chlorophyll fluorescence) using a 60× water-immersion objective (Olympus IX-81 60×/1.2 Water UPlanSApo, DIC, fourfold zoom). All confocal images for each experiment were acquired using identical adjustments. The GFP fluorescence was quantified using Olympus Fluoview version 3.1. The fluorescence of a heterocyst was compared with that of the two adjacent vegetative cells. The Δall1371 mutant strain without GFP was used as control. Background fluorescence was subtracted.
Results
All1371 as putative PPGK
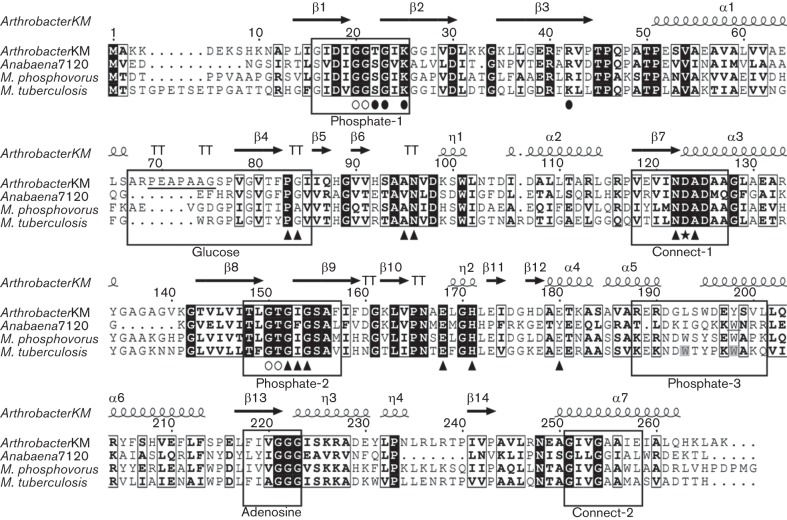
The ORF all1371 from Anabaena was assumed to encode a putative PPGK (EC 2.7.1.63), as its amino acid sequence has sequence similarity to several well-characterized bacterial PPGKs, including the polyphosphate/ATP-glucomannokinase from Arthrobacter sp. strain KM (Mukai et al., 2003, 2004) (53.1 % similarity), the polyphosphate-dependent PPGK from Microlunatus phosphovorus (Tanaka et al., 2003) (46.0 % similarity) and the polyphosphate/ATP-dependent glucokinase from Mycobacterium tuberculosis (Hsieh et al., 1993, 1996a, b) (47.4 % similarity). Comparison of these sequences, including secondary structures extrapolated from the crystal structure of the polyphosphate/ATP-glucomannokinase of Arthrobacter sp. strain KM (1WOQ) (Mukai et al., 2003, 2004), revealed the presence of some highly conserved common motifs (Fig. 1). Seven structural and functional motifs (Fig. 1, boxed) were found in all of the sequences including All1371.
Fig. 1.
Primary structural alignments of different PPGKs. Aligned primary structures from Arthrobacter sp. strain KM, Microlunatus phosphovorus (Tanaka et al., 2003), Mycobacterium tuberculosis (Hsieh et al., 1993; Phillips et al., 1999) and Anabaena sp. PCC 7120. Strictly conserved residues are shaded in black; similar residues are framed in black. Putative structural and functional domains are enclosed in boxes. Secondary structural elements [e.g. α helices, β sheets, turn-turns (TT)] of the polyP/ATP-glucomannokinase from Arthrobacter sp. strain KM are depicted above the alignment. Residues associated with β-d-glucose binding are marked with filled triangles. The heptapeptide is underlined. The catalytic aspartate (D) is highlighted with a star. Residues involved in the binding of both phosphate molecules used as ligands (open, phosphate A; filled, phosphate B) are marked with ovals (Mukai et al., 2004).
Purification of All1371 and molecular mass determination
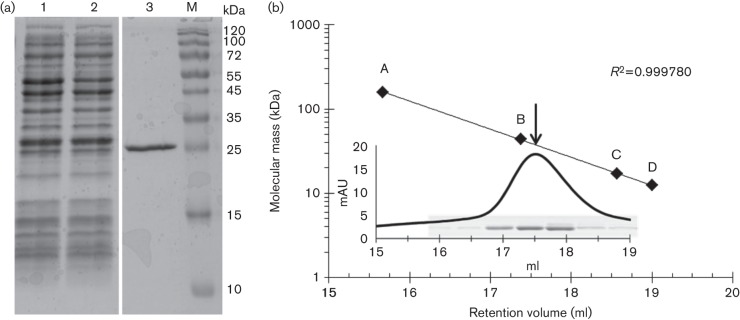
The N-terminal GST-fusion protein of All1371 was expressed for 3 h in E. coli BL21(DE3) carrying pGEX_all1371. After on-column cleavage with the PreScission protease, the 246 aa enzyme was eluted and analysed by SDS-PAGE (Fig. 2a). The enzyme appeared as a single band of 26 kDa (Fig. 2a, lane 3; apparent molecular mass). To investigate the oligomeric state of All1371, the recombinant PPGK was analysed by size-exclusion chromatography. A single symmetrical peak (Fig. 2b, inset) of approximately 39.0 kDa was obtained. Fractions corresponding to the protein elution peak showed a single protein band of 26 kDa on SDS-PAGE (Fig. 2b, inset), and exhibited polyphosphate-dependent activity in vitro (data not shown). The biochemical and kinetic properties of All1371 are summarized in Table 1.
Fig. 2.
Purification of recombinant All1371 and gel filtration. (a) SDS-PAGE (15 %) analysis of samples from the various purification steps performed after affinity chromatography. Lane 1, cell-free extract of recombinant E. coli, 15 µg protein; lane 2, flow-through, 15 µg protein; lane 3, All1371 after elution, 2 µg; lane M, protein standard. (b) All1371 was subjected to gel filtration calibrated with protein standards (filled diamonds; see Methods for details). The elution profile (indicating the elution position of All1371, arrow) and SDS-PAGE results of the obtained fractions are shown in the inset.
Table 1. Biochemical and kinetic properties of All1371.
Measurements were performed at 28 °C, pH 7.5; n≥3; 100 % = 107.1 U mg−1.
| Property | Value (mean±sd) | |
| Molecular mass (kDa) | Native complex* | 39 |
| Monomer, apparent† | 26.0 | |
| Monomer, calculated‡ | 26.6 | |
| Oligomeric structure | Monomer or homodimer | |
| KM (mM) | Glucose | 0.118±0.01 |
| KM (µM) | Polyphosphate | 1.76±0.26 |
| vmax (U mg−1) | Glucose, polyphosphate | 107.1±15.3 |
| kcat (s−1) | Glucose, polyphosphate | 48.2±6.9 |
| K0.5 (mM) | Mannose | 24.3±2.36 |
| vmax (U mg−1) | Mannose | 0.43±0.04 |
| kcat (s−1) | Mannose | 0.21±0.017 |
| Phosphoryl donor specificity (% vmax) | Polyphosphate | 100 |
| ATP, GTP, UTP, CTP, ADP, AMP | 0 | |
| Pyrophosphate | 0 | |
| Without polyphosphate | 0 |
Determined by size exclusion chromatography.
Determined by SDS-PAGE.
Calculated according to the primary structure, including the linker peptide of the GST tag.
Biochemical properties of All1371
Our enzyme activity assays indicated that purified All1371 uses polyphosphate to phosphorylate glucose and mannose, with a higher preference for glucose (Table 1). All1371 activity was strictly dependent on the presence of Mg2+ or Mn2+ (data not shown). All1371 had high substrate specificity and acted as a strict polyphosphate-dependent enzyme. No other phosphoryl group donor was accepted. Kinetic analysis indicated that the reactions of All1371 with polyphosphate and glucose followed Michaelis–Menten kinetics. The KM values for polyphosphate and glucose were 1.76 µM and 0.118 mM, respectively (at 28 °C and pH 7.5). The maximum rate of All1371-mediated catalysis was 107 U mg −1, yielding a kcat of 48.2 s −1 (Table 1). Furthermore, our kinetic analysis revealed that All1371 had a kcat of 0.19 s−1 and a K0.5 of 24.3 mM for mannose (Table 1).
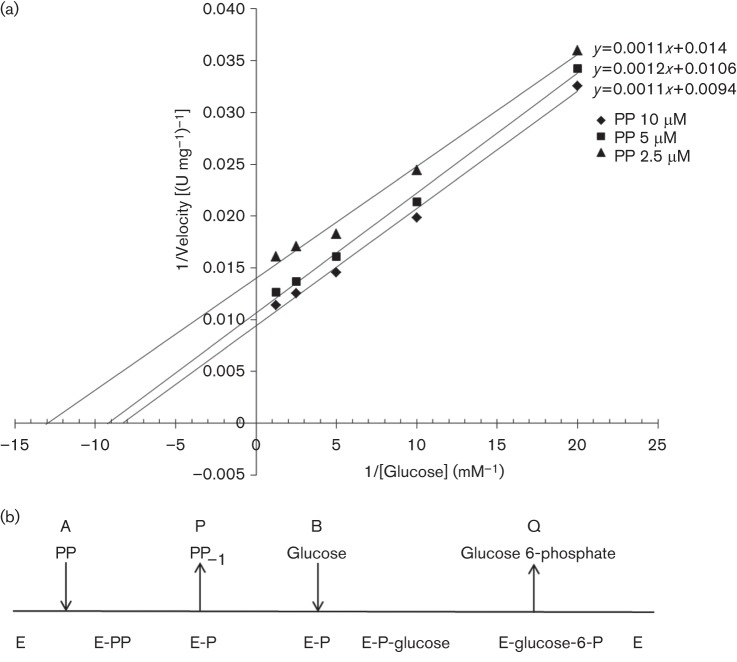
To characterize the enzymic mechanism of All1371, additional kinetic analyses with glucose and polyphosphate were performed. The initial rate of All1371 activity was determined with varying concentrations of glucose and fixed concentrations of polyphosphate. We obtained a linear double reciprocal plot with parallel lines (Fig. 3a). When polyphosphate was varied, we obtained a similar graph with parallel lines (data not shown).
Fig. 3.
Kinetics of the All1371 reaction. (a) Activity of All1371. Primary double-reciprocal plot of initial velocity with glucose as the variable substrate and different concentrations of polyP45 (PP) as the fixed substrate (pH 7.5; n = 3). (b) Schematic of the bi-bi ping-pong mechanism. Polyphosphate (PP) acts as the first substrate (A) by covalently binding to PPGK (E). The first product (P), polyphosphate reduced at one phosphate (PP–1), is released, and binding of the second substrate, glucose (B), occurs on the phosphorylated enzyme (E-P). Finally, the second product, glucose 6-phosphate (Q), is released and the enzyme is restored (E).
Distribution of putative PPGKs in cyanobacteria and PPGK activity in cell-free cyanobacterial extracts
To examine the distribution of PPGKs among cyanobacteria, we performed a blastp search (Altschul et al., 1997) against all sequenced cyanobacterial genomes (September 2013; 141 genomes) using the amino acid sequence of All1371 as the query. Our analysis revealed that in 34 % of all sequenced cyanobacteria a putative PPGK is present. PPGKs were found in all five sections of cyanobacteria (Table 2) with the highest frequency in the heterocyst-forming species of Section IV (85 %) and Section V (54.5 %), followed by the non-heterocystous species of Section III (52.9 %), Section II (50 %) and Section I (5.7 %). More complete data are presented in Table S1 (available in the online Supplementary Material). Cell-free extracts of some of these cyanobacteria were tested for specific PPGK activity under different nitrogen conditions, including Anabaena (Section V), Mastigocladus laminosus (Section V) (Nürnberg et al., 2014), F. muscicola (Section V) and Synechocystis (Section I), the last lacking a predicted PPGK (negative control). As expected, cell-free extracts of Synechocystis did not show any PPGK activity, whereas those of Anabaena, Mastigocladus laminosus and F. muscicola exhibited detectable PPGK activity (Table 3). In Anabaena, PPGK activity was increased slightly under nitrogen depletion. PPGK activity in cell-free extracts from the two diazotrophic, branched filamentous cyanobacterial strains of Section V was three- to fourfold higher than in non-branched Anabaena cells (Table 3). Interestingly, we found a decrease of PPGK activity in extracts of Section V cells grown without combined nitrogen (Table 3).
Table 2. Distribution of PPGKs among cyanobacteria.
| Section | |||||
| I | II | III | IV | V | |
| No. of PPGKs | 4 | 3 | 18 | 17 | 6 |
| No. of cyanobacteria | 70 | 6 | 34 | 20 | 11 |
| Percentage | 5.7 | 50.0 | 52.9 | 85.0 | 54.5 |
Table 3. PPGK activities in cell-free extracts of cyanobacteria.
Measurements were performed at 28–30 °C and pH 7.5; n≥3; nd, not detectable (≤1 nmol min−1 mg−1).
| Strain | Section | Growth conditions | PPGK activity (nmol min−1 mg−1) (mean±sd) |
| Anabaena | IV | +N | 4.3±1.3 |
| −N | 5.8±1.3 | ||
| Anabaena Δall1371 | IV | +N | nd |
| −N | nd | ||
| F. muscicola | V | +N | 15.3±2.3 |
| −N | 13.1±2.1 | ||
| Mastigocladus laminosus | V | +N | 23.6±0.7 |
| −N | 13.9±3.2 | ||
| Synechocystis | I | +N | nd |
| −N | nd |
Viability of Δall1371
To confirm a function of All1371 in vivo, we generated a mutant strain in which ORF all1371 was replaced with the neomycin-resistance cassette, C.K3 (Fig. 4a). Complete segregation of the mutant was validated by PCR (Fig. S1), and no PPGK activity was detected in cell-free extracts of Δall1371 (Table 3). Using light and fluorescence microscopy, we monitored the shape and autofluorescence of cells. When the Δall1371 mutant was deprived of combined nitrogen, the filaments showed both vegetative cells and morphologically mature heterocysts. Analysis of Δall1371 mutant and WT cells under different light and nitrogen conditions revealed that viability of the mutant was distinctly decreased under combined nitrogen-limiting conditions (Fig. 4b, Fig. S2). This effect was increased under light–dark cycle conditions, but in this case we also noted a reduced viability of the mutant in the presence of nitrate (Fig. S3b).
Fig. 4.
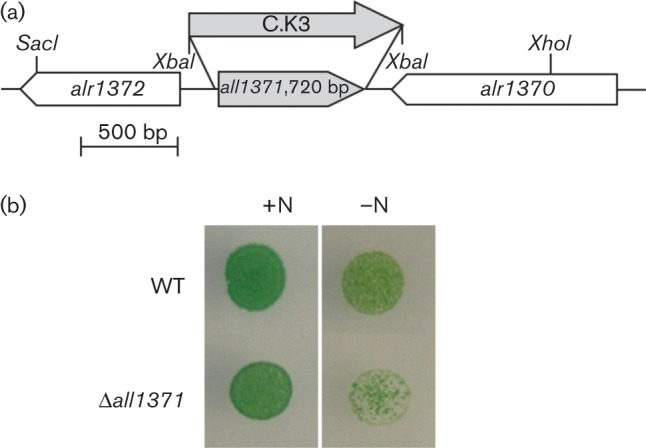
The Δall1371 mutant. (a) Schematic of the chromosomal region surrounding all1371 and the gene inactivation strategy, in which all1371 was replaced with the C.K3 cassette. (b) Viability analyses of Anabaena wild-type (WT) and the all1371 knockout mutant strain (Δall1371) on AA-agar plates containing 10 mM KNO3 (+N) or lacking combined nitrogen (–N). In total, 4.8 ng chlorophyll a per spot was plated. The plates were incubated under continuous light for 6 days.
Expression analysis using a GFP promoter fusion
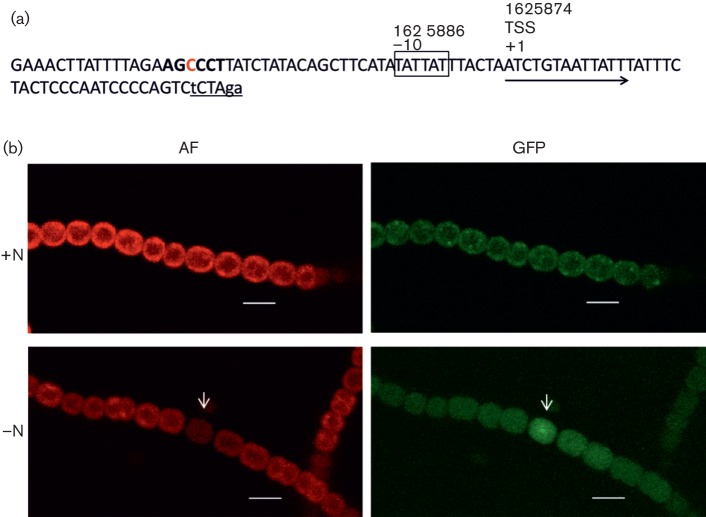
The transcriptional changes experienced by Anabaena during nitrogen-depletion-induced cell differentiation were recently analysed by Mitschke et al. (2011). They identified numerous transcription start sites (TSSs) in Anabaena, including a TSS of all1371 at position 1625 874. The organization of the predicted all1371 promoter region is depicted in Fig. 5(a). We identified a putative palindromic motif that is likely to be a (DIF)+ motif (Mitschke et al., 2011). This putative (DIF)+ motif displays an inverse orientation and one mismatch [Fig. 5a; (DIF)+ in bold type, mismatch in red, AGCCCT].
Fig. 5.
All1371 promoter activity in heterocysts. (a) The promoter region of all1371, including the TSS and the −10 region (boxed) (Mitschke et al., 2011), was integrated into a self-replicating plasmid. A promoter-less gfp gene was integrated via the underlined XbaI site (altered bases are depicted in lower case); the presumed DIF+ domain (AGCCCT) is shown in bold. (b) Fluorescence in the Anabaena Δall1371 promoter fusion strain grown under N2-fixing conditions (–N) and grown with combined nitrogen (+N). A heterocyst is indicated with an arrow. Bars, 5 µm. AF, auto-fluorescence (red); GFP, GFP fluorescence (green).
To investigate the expression of all1371 along the filaments, the gfp gene was transcriptionally fused to the all1371 promoter and transformed into the Δall1371 mutant strain. In the absence of combined nitrogen, we noted distinct GFP fluorescence in the mature heterocysts of 4-day-old filaments of the promoter fusion strain (Fig. 5b). An overview of fluorescence (GFP, autofluorescence) of the promoter fusion strain is given in Fig. S4. No distinct GFP fluorescence was observed in the filaments of the promoter fusion strain when combined nitrogen was supplied (Fig. 5b) compared with Δall1371 (Fig. S5). GFP fluorescence in heterocysts was first detected 24 h after nitrogen step down, and persisted until the filaments were harvested 4 days later (data not shown). Quantification of GFP fluorescence of numerous cells in the promoter–gfp fusion strain in comparison with cells of the parent strain Δall1371 confirmed our microscopic observations: the fluorescence intensity in heterocysts (n = 42) of the promoter fusion strain was ~2.1-fold higher than in vegetative cells (n = 71). The ratio of fluorescence intensity from heterocysts (n = 18) to vegetative cells (n = 34) in filaments of the parent strain Δall1371 was 1 : 0.9 (Table S2).
Discussion
Identification of the putative PPGK, All1371
The structural and functional motifs found in all the aligned amino acid sequences (Fig. 1, boxed) are predicted to interact with the substrates glucose, mannose, ATP and polyphosphate (Liao et al., 2012; Mukai et al., 2003). Five of the identified domains have been proposed to interact with the ATP molecule: phosphate-1 and phosphate-2 are involved in binding β-phosphates and γ-phosphates, while connect-1, connect-2 and the adenosine motif interact with the adenine ring of ATP (Mukai et al., 2003). The conserved aspartate residue in the connect-1 region (Fig. 1, star) is believed to be essential for catalytic activity (Arora et al., 1991; Mukai et al., 2004). The glucose motif has been suggested to participate in glucose binding. The heptapeptide PEAPAAG (Fig. 1, underlined) was proposed to be responsible for mannose phosphorylation in the sequence of Arthrobacter sp. strain KM (Mukai et al., 2003). The phosphate-3 motif was predicted to be a binding region for polyphosphate (Liao et al., 2012). Residues Trp193 and Trp198 (Fig. 1, grey shading) were proposed to be essential for catalytic activity in Mycobacterium tuberculosis (Phillips et al., 1999). A residue equivalent to Trp198 is present in the amino acid sequence of Anabaena (Fig. 1, underlined W). Furthermore, the phosphate-1 domain is likely to contain a putative polyphosphate-binding site, as both the anionic phosphates used as ligands in a crystallographic study (Mukai et al., 2004) bound at highly conserved amino acid residues similar to Lys25. It was proposed that there may be shared ATP- and polyphosphate-binding sites in the phosphate-1 and phosphate-2 regions (Mukai et al., 2004). Thus, the present amino acid sequence analysis of All1371 (Fig. 1) and the previous findings in similar proteins collectively suggest that All1371 functions as a PPGK.
Purification of All1371, and molecular mass determination
The purified All1371 appeared as a single protein band of 26 kDa in SDS-PAGE analysis (Fig. 2a, lane 3). This result is consistent with the expected molecular mass of 26.6 kDa calculated with the ProtParam tool (http://web.expasy.org/protparam/) (Wilkins et al., 1999) for one monomer of the recombinant All1371.
The protein peak of 39 kDa obtained in size-exclusion chromatography (Fig. 2b) indicates that the native enzyme may exist as either a monomer or a homodimer. PPGK homodimers have also been reported in Mycobacterium tuberculosis, Propionibacterium shermanii and Propionibacterium arabinosum (Phillips et al., 1999), whereas the polyphosphate/ATP-dependent glucomannokinase of Arthrobacter sp. strain KM was determined to exist as a monomer (Mukai et al., 2003).
Biochemical properties of All1371
All1371 uses polyphosphate exclusively to phosphorylate glucose and mannose (Table 1) and is strictly dependent on the presence of divalent cations. This requirement for divalent cations is shared with the PPGKs of Microlunatus phosphovorus (Tanaka et al., 2003), Arthrobacter sp. (Mukai et al., 2003), Mycobacterium tuberculosis and Mycobacterium phlei (Szymona & Ostrowski, 1964; Szymona & Widomski, 1974). Recently, Mg2+ was found to be an indispensable cofactor for the PPGK of Thermobifida fusca (Liao et al., 2012). Here, we report that All1371 showed high substrate specificity and acted as a strict polyphosphate-dependent enzyme. This result is a notable feature, as most of the previously described glucokinases utilized either ATP alone, or ATP and polyphosphate. The previous in vitro studies on PPGKs revealed that these enzymes were often bi-functional and not restricted to polyphosphate. For example, the PPGKs from Mycobacterium tuberculosis (Hsieh et al., 1996a), Propionibacterium shermanii (Phillips et al., 1993) and Corynebacterium glutamicum (Lindner et al., 2010) were also able to use ATP or GTP. The present work showed that, along with the PPGK of Microlunatus phosphovorus (Tanaka et al., 2003), All1371 is one of only two known PPGKs that uses only polyphosphate as its phosphate donor.
Kinetic analyses of All1371 (Table 1) revealed a relatively low KM value obtained for polyphosphate (1.76 µM), suggesting that All1371 has a high affinity for its sole substrate. In comparison with the KM values for polyphosphate and glucose (Table 1), the polyphosphate- and ATP-dependent PPGK from Corynebacterium glutamicum yielded KM values of 0.2 mM for polyP45 and 1 mM for glucose (Lindner et al., 2010); the PPGK of Propionibacterium shermanii yielded a KM value of 1.2 µM for polyP35 (Phillips et al., 1993); the PPGK of Mycobacterium tuberculosis yielded a KM value of 4.6 µM for polyP35 (Phillips et al., 1999); and the PPGK of Microlunatus phosphovorus yielded a KM of 3.8 mM for polyP30 (Tanaka et al., 2003). The turnover number of All1371 of 48.2 s −1 (Table 1) is comparable with the kcat value of 57.0 s−1 determined for the PPGK from Propionibacterium shermanii against polyP35 (Phillips et al., 1999).
According to our analyses, All1371 is a polyphosphate-dependent glucomannokinase. Interestingly, the heptapeptide in the sequence of Arthrobacter sp. strain KM (Fig. 1, underlined), which is assumed to be responsible for mannose phosphorylation (Mukai et al., 2004), is not present in the corresponding Anabaena sequence. The results of additional kinetic analyses (Fig. 3a) were consistent with the so-called ‘bi-bi ping-pong’ mechanism (Cleland, 1963). As illustrated in Fig. 3(b), this mechanism is a particular multi-substrate reaction that includes two substrates and two products (bi-bi) and is characterized by alternating processes of substrate binding and product release (ping-pong) for the two substrates. In a first step, polyphosphate is covalently bound to All1371, which is then phosphorylated. Approximately one orthophosphate-reduced polyphosphate is released from the enzyme as the first product. In a second step, glucose is bound to the phosphorylated enzyme, and the second substrate is phosphorylated. Glucose 6-phosphate is released as a second product, and the enzyme returns to its initial state (Fig. 3b). In contrast, the ATP/polyphosphate-dependent PPGK of Mycobacterium tuberculosis (Hsieh et al., 1996a) and the ATP-dependent glucokinase from Streptomyces coelicolor (Imriskova et al., 2005) were both found to display ordered bi-bi sequential mechanisms. The ordered bi-bi sequential mechanism differs from the bi-bi ping-pong mechanism in that both substrates (glucose and ATP or polyphosphate) bind to the enzyme first before the two products are released.
Putative PPGKs in cyanobacteria
Cyanobacteria may be grouped into five sections according to their morphology (Rippka et al., 1979). While species of Sections I and II are unicellular forms, those of Sections III, IV and V show filamentous forms. Cyanobacteria of Sections IV and V are additionally able to form heterocysts. The highest level of complexity is seen among Section V strains, which form true branches within their filaments (Golubic et al., 1996). Diazotrophic growth has been observed in both unicellular and filamentous strains (reviewed by Stal, 1995). Our blastp search revealed that PPGKs were found very frequently in cyanobacteria of Sections IV and V, which are all diazotrophic strains forming heterocysts. All genomes of the analysed Section IV and Section V strains contained PPGK genes. In about half of the analysed genomes of Section III we found putative PPGK genes (52.9 %). In 11 of these 18 PPGK gene-containing genomes (61 %) we also found nifH genes encoding the key enzyme of N2 fixation (Table S1). Some of these Section III organisms are known to fix N2 under micro-oxic conditions, such as Pseudanabaena sp. ATCC 27183 (Rippka & Waterbury, 1977) (synonymous with Pseudanabaena sp. PCC 6802). Among cyanobacteria of Section II, 50 % of sequenced unicellular strains were also predicted to contain a PPGK. All are known to fix N2 under anaerobic conditions (Rippka et al., 1979) or to have a nitrogenase complex (Rippka & Waterbury, 1977), or a putative dinitrogenase has been annotated in the genome (Markowitz et al., 2012). Furthermore, strains belonging to Chroococcidiopsis are closely related to the heterocyst-forming cyanobacteria (Fewer et al., 2002). These facts may suggest a possible correlation between PPGK appearance and the ability to fix N2 under anoxic/micro-oxic conditions provided by either heterocysts or the environment. This presumption is supported by the results obtained by analysing Section I organisms. Only four (5.7 %) of the unicellular strains of Section I were found to contain a putative PPGK. Synechococcus sp. PCC 7335 and Synechococcus sp. PCC 7502 arose through morphological transition events (Robertson et al., 2001; Shih et al., 2013). Interestingly, putative PPGKs were not found in the genomes of Cyanothece strains that are able to grow diazotrophically in diurnal rhythm. Based on the present findings, we hypothesized that the presence of a PPGK in cyanobacterial genomes is strongly related to the organism’s ability to fix N2 in heterocysts. A correlation between PPGK appearance and an organism’s ability to fix N2 under anoxic conditions is possible but has to be analysed further, especially from a phylogenetic point of view.
To determine whether PPGK activity was present in vivo, PPGK activity in cell-free extracts of some heterocyst-forming cyanobacteria with putative PPGKs was determined. As summarized in Table 3, PPGK activity in Anabaena is increased after 4 days of nitrogen depletion. An increase of PPGK activity under this condition is in line with the results of Flaherty et al. (2011). Using deep sequencing analyses performed 21 h after nitrogen deprivation, they found a 4.8-fold increase in the mRNA expression level of all1371. Furthermore, the increased PPGK activity is in line with a previous report (Thompson et al., 1994), showing that in Anabaena flos-aquae, phosphate is stored as sugar phosphate under N2-fixing conditions, but as polyphosphate in the presence of combined nitrogen. We found that PPGK activity was higher in cell-free extracts from thermophilic Section V strains of F. muscicola and Mastigocladus laminosus than in Anabaena (Table 3). A higher in vitro activity might be the result of a higher robustness of the PPGK due to its thermophilic origin (Beadle et al., 1999). Interestingly, we observed an increased PPGK activity in cell-free extracts obtained from these strains grown in the presence of nitrate than under diazotrophic conditions. The higher complexity of Section V strains differing in the regulation of diazotrophic growth (Nürnberg et al., 2014) might explain this observation.
Viability of Δall1371
The impaired viability of the mutant implies that All1371 plays an important role in providing glucose-6-phosphate in Anabaena, supporting the canonical hexokinase. This is supported by the PPGK activity measured in cell-free extracts of Anabaena obtained from nitrate-supplemented cultures (Table 3). Under diazotrophic conditions heterocysts are not able to fix carbon dioxide. Carbon compounds, probably in the form of sucrose (Curatti et al., 2002), are imported from vegetative cells. NAD(P)H, needed as a reducing equivalent, is generated in heterocysts (Maldener & Muro-Pastor, 2010). There, glucose 6-phosphate is used as substrate for glucose-6-phosphate dehydrogenase, a main enzyme of the oxidative pentose phosphate pathway. Because of the decreased viability of the mutant observed under diazotrophic conditions, we conclude that All1371 may represent an alternative enzyme completing the hexokinase under ATP-consuming (diazotrophic) growth conditions.
Expression analysis using a GFP promoter fusion
In cyanobacteria, the nitrogen-regulated genes are mainly controlled by the transcriptional regulators NtcA and HetR (Kumar et al., 2010). Recently, chromatin immunoprecipitation analysis followed by high-throughput sequencing was used to identify all of the NtcA-binding sites of Anabaena at 3 h after a nitrogen step down (Picossi et al., 2014). Interestingly, they detected an internal NtcA-binding site in all1371 whereas the impact of binding of NtcA remains unclear. Further HetR-controlled promoters characterized by an differentiation-related change (DIF)+ motif (TCCGGA, a palindrome at or close to position −35) were identified by comparing results found in Anabaena with a ΔhetR mutant 8 h after a nitrogen step down (Mitschke et al., 2011). The putative (DIF)+ motif with an inverse orientation located in the promoter region of all1371 (Fig. 5 a) additionally indicates that the promoter might be HetR-dependent. In fact, the promoter of all1371 responded to nitrogen depletion in WT but not in the ΔhetR mutant, indicating a HetR dependency (W. Hess, personal communication). Our results obtained with a GFP promoter fusion strain show that the all1371 promoter activity is particularly enhanced under nitrogen starvation in mature heterocysts. N2 fixation in heterocysts is an energy-intensive process requiring 16 molecules of ATP to reduce one molecule of N2 (Hill et al., 1981; Howard & Rees, 1996). The ability of PPGKs to utilize polyphosphate instead of ATP for glucose phosphorylation might allow the heterocysts to save ATP for the essential process of N2 fixation.
Acknowledgements
We thank Thomas Zielke for his valuable support with fluorescence microscopy. We are also grateful to Faranak Fassihianifard for performing the initial enzymic analysis.
Abbreviations:
- Anabaena
Anabaena sp. PCC 7120
- GST
glutathione-S-transferase
- PPGK
polyphosphate glucokinase
- TSS
transcription start site
Footnotes
Two supplementary tables and five supplementary figures are available with the online Supplementary Material.
References
- Achbergerová L., Nahálka J. (2011). Polyphosphate – an ancient energy source and active metabolic regulator. Microb Cell Fact 10, 63. 10.1186/1475-2859-10-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. G., Carr N. G., Wilcox M. (1981). The developmental biology of heterocyst and akinete formation in cyanobacteria. Crit Rev Microbiol 9, 45–100. 10.3109/10408418109104486 [DOI] [PubMed] [Google Scholar]
- Ahn K., Kornberg A. (1990). Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem 265, 11734–11739. [PubMed] [Google Scholar]
- Akiyama M., Crooke E., Kornberg A. (1993). An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem 268, 633–639. [PubMed] [Google Scholar]
- Allen M. B., Arnon D. I. (1955). Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol 30, 366–372. 10.1104/pp.30.4.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora K. K., Filburn C. R., Pedersen P. L. (1991). Glucose phosphorylation. Site-directed mutations which impair the catalytic function of hexokinase. J Biol Chem 266, 5359–5362. [PubMed] [Google Scholar]
- Baier, A. (2013). Untersuchungen zum stickstoffinduzierten Phycobilisomenabbau - NblA, ein kleines Protein mit großer Wirkung. Doctoral thesis, Humboldt-Universität zu Berlin. [Google Scholar]
- Beadle B. M., Baase W. A., Wilson D. B., Gilkes N. R., Shoichet B. K. (1999). Comparing the thermodynamic stabilities of a related thermophilic and mesophilic enzyme. Biochemistry 38, 2570–2576. 10.1021/bi9824902 [DOI] [PubMed] [Google Scholar]
- Bensadoun A., Weinstein D. (1976). Assay of proteins in the presence of interfering materials. Anal Biochem 70, 241–250. 10.1016/S0003-2697(76)80064-4 [DOI] [PubMed] [Google Scholar]
- Berman-Frank I., Lundgren P., Falkowski P. (2003). Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res Microbiol 154, 157–164. 10.1016/S0923-2508(03)00029-9 [DOI] [PubMed] [Google Scholar]
- Bertani G. (1951). Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black T. A., Wolk C. P. (1994). Analysis of a Het- mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J Bacteriol 176, 2282–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black T. A., Cai Y., Wolk C. P. (1993). Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol 9, 77–84. 10.1111/j.1365-2958.1993.tb01670.x [DOI] [PubMed] [Google Scholar]
- Cai Y. P., Wolk C. P. (1990). Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172, 3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castenholz R. W. (1988). Culturing methods for cyanobacteria. Methods Enzymol 167, 68–93. 10.1016/0076-6879(88)67006-6 [DOI] [Google Scholar]
- Cleland W. W. (1963). The kinetics of enzyme-catalyzed reactions with two or more substrates or products. III. Prediction of initial velocity and inhibition patterns by inspection. Biochim Biophys Acta 67, 188–196. 10.1016/0926-6569(63)90227-X [DOI] [PubMed] [Google Scholar]
- Curatti L., Flores E., Salerno G. (2002). Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett 513, 175–178. 10.1016/S0014-5793(02)02283-4 [DOI] [PubMed] [Google Scholar]
- Dagan T., Roettger M., Stucken K., Landan G., Koch R., Major P., Gould S. B., Goremykin V. V., Rippka R. & other authors (2013). Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol 5, 31–44. 10.1093/gbe/evs117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Marsac N. T., Houmard J. (1988). Complementary chromatic adaptation: physiological conditions and action spectra. Methods Enzymol 167, 318–328. 10.1016/0076-6879(88)67037-6 [DOI] [Google Scholar]
- Elhai J., Wolk C. P. (1988a). Conjugal transfer of DNA to cyanobacteria. Methods Enzymol 167, 747–754. 10.1016/0076-6879(88)67086-8 [DOI] [PubMed] [Google Scholar]
- Elhai J., Wolk C. P. (1988b). A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68, 119–138. 10.1016/0378-1119(88)90605-1 [DOI] [PubMed] [Google Scholar]
- Fewer D., Friedl T., Büdel B. (2002). Chroococcidiopsis and heterocyst-differentiating cyanobacteria are each other’s closest living relatives. Mol Phylogenet Evol 23, 82–90. 10.1006/mpev.2001.1075 [DOI] [PubMed] [Google Scholar]
- Flaherty B. L., Van Nieuwerburgh F., Head S. R., Golden J. W. (2011). Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics 12, 332. 10.1186/1471-2164-12-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E., Herrero A. (2010). Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol 8, 39–50. 10.1038/nrmicro2242 [DOI] [PubMed] [Google Scholar]
- Golubic S., Hernandez-Marine M., Hoffmann L. (1996). Developmental aspects of branching in filamentous Cyanophyta/Cyanobacteria. Arch Hydrobiol Suppl Algol Stud 83, 303–329. [Google Scholar]
- Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999). ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308. 10.1093/bioinformatics/15.4.305 [DOI] [PubMed] [Google Scholar]
- Harold F. M. (1966). Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol Rev 30, 772–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S., Kennedy C., Kavanagh E., Goldberg R. B., Hanau R. (1981). Nitrogen fixation gene (nifL) involved in oxygen regulation of nitrogenase synthesis in K. pneumoniae. Nature 290, 424–426. 10.1038/290424a0 [DOI] [PubMed] [Google Scholar]
- Howard J. B., Rees D. C. (1996). Structural basis of biological nitrogen fixation. Chem Rev 96, 2965–2982. 10.1021/cr9500545 [DOI] [PubMed] [Google Scholar]
- Hsieh P. C., Shenoy B. C., Jentoft J. E., Phillips N. F. (1993). Purification of polyphosphate and ATP glucose phosphotransferase from Mycobacterium tuberculosis H37Ra: evidence that poly(P) and ATP glucokinase activities are catalyzed by the same enzyme. Protein Expr Purif 4, 76–84. 10.1006/prep.1993.1012 [DOI] [PubMed] [Google Scholar]
- Hsieh P. C., Kowalczyk T. H., Phillips N. F. (1996a). Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis. Biochemistry 35, 9772–9781. 10.1021/bi9528659 [DOI] [PubMed] [Google Scholar]
- Hsieh P. C., Shenoy B. C., Samols D., Phillips N. F. (1996b). Cloning, expression, and characterization of polyphosphate glucokinase from Mycobacterium tuberculosis. J Biol Chem 271, 4909–4915. 10.1074/jbc.271.9.4909 [DOI] [PubMed] [Google Scholar]
- Imriskova I., Arreguín-Espinosa R., Guzmán S., Rodriguez-Sanoja R., Langley E., Sanchez S. (2005). Biochemical characterization of the glucose kinase from Streptomyces coelicolor compared to Streptomyces peucetius var. caesius. Res Microbiol 156, 361–366. 10.1016/j.resmic.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Jensen T. E. (1968). Electron microscopy of polyphosphate bodies in a blue-green alga Nostoc pruniforme. Arch Mikrobiol 62, 144–152. 10.1007/BF00410400 [DOI] [Google Scholar]
- Kornberg A. (1995). Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J Bacteriol 177, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A., Kornberg S. R., Simms E. S. (1956). Metaphosphate synthesis by an enzyme from Escherichia coli. Biochim Biophys Acta 20, 215–227. 10.1016/0006-3002(56)90280-3 [DOI] [PubMed] [Google Scholar]
- Kornberg A., Rao N. N., Ault-Riché D. (1999). Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem 68, 89–125. 10.1146/annurev.biochem.68.1.89 [DOI] [PubMed] [Google Scholar]
- Kumar K., Mella-Herrera R. A., Golden J. W. (2010). Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol 2, a000315. 10.1101/cshperspect.a000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A. & other authors (2007). clustal w and clustal_x version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Liao H., Myung S., Zhang Y. H. (2012). One-step purification and immobilization of thermophilic polyphosphate glucokinase from Thermobifida fusca YX: glucose-6-phosphate generation without ATP. Appl Microbiol Biotechnol 93, 1109–1117. 10.1007/s00253-011-3458-1 [DOI] [PubMed] [Google Scholar]
- Lichko L. P., Kulakovskaya T. V., Kulaev I. S. (2010). Properties of partially purified endopolyphosphatase of the yeast Saccharomyces cerevisiae. Biochemistry (Mosc) 75, 1404–1407. 10.1134/S0006297910110131 [DOI] [PubMed] [Google Scholar]
- Lindner S. N., Knebel S., Pallerla S. R., Schoberth S. M., Wendisch V. F. (2010). Cg2091 encodes a polyphosphate/ATP-dependent glucokinase of Corynebacterium glutamicum. Appl Microbiol Biotechnol 87, 703–713. 10.1007/s00253-010-2568-5 [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951). Protein measurement with the Folin phenol reagent. J Biol Chem 193, 265–275. [PubMed] [Google Scholar]
- Maldener I., Muro-Pastor A. M. (2010). Cyanobacterial heterocysts. In eLS. Chichester: Wiley. [10.1002/9780470015902.a0000306.pub2] 10.1002/9780470015902.a0000306.pub2 [DOI] [Google Scholar]
- Markowitz V. M., Chen I. M., Palaniappan K., Chu K., Szeto E., Grechkin Y., Ratner A., Jacob B., Huang J. & other authors (2012). IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40 (Database issue), D115–D122. 10.1093/nar/gkr1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. (1902). Orientierende Untersuchungen über Verbreitung, Morphologie, und Chemie des Volutins. Bot Zeitschr 62, 113–152. [Google Scholar]
- Mitschke J., Vioque A., Haas F., Hess W. R., Muro-Pastor A. M. (2011). Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci U S A 108, 20130–20135. 10.1073/pnas.1112724108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui A., Kumazawa S., Takahashi A., Ikemoto H., Cao S., Arai T. (1986). Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature 323, 720–722. 10.1038/323720a0 [DOI] [Google Scholar]
- Mukai T., Kawai S., Matsukawa H., Matuo Y., Murata K. (2003). Characterization and molecular cloning of a novel enzyme, inorganic polyphosphate/ATP-glucomannokinase, of Arthrobacter sp. strain KM. Appl Environ Microbiol 69, 3849–3857. 10.1128/AEM.69.7.3849-3857.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Kawai S., Mori S., Mikami B., Murata K. (2004). Crystal structure of bacterial inorganic polyphosphate/ATP-glucomannokinase. Insights into kinase evolution. J Biol Chem 279, 50591–50600. 10.1074/jbc.M408126200 [DOI] [PubMed] [Google Scholar]
- Nakao M., Okamoto S., Kohara M., Fujishiro T., Fujisawa T., Sato S., Tabata S., Kaneko T., Nakamura Y. (2010). CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res 38 (Database issue), D379–D381. 10.1093/nar/gkp915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberg D. J., Mariscal V., Parker J., Mastroianni G., Flores E., Mullineaux C. W. (2014). Branching and intercellular communication in the Section V cyanobacterium Mastigocladus laminosus, a complex multicellular prokaryote. Mol Microbiol 91, 935–949. 10.1111/mmi.12506 [DOI] [PubMed] [Google Scholar]
- Pepin C. A., Wood H. G. (1986). Polyphosphate glucokinase from Propionibacterium shermanii. Kinetics and demonstration that the mechanism involves both processive and nonprocessive type reactions. J Biol Chem 261, 4476–4480. [PubMed] [Google Scholar]
- Phillips N. F., Horn P. J., Wood H. G. (1993). The polyphosphate- and ATP-dependent glucokinase from Propionibacterium shermanii: both activities are catalyzed by the same protein. Arch Biochem Biophys 300, 309–319. 10.1006/abbi.1993.1043 [DOI] [PubMed] [Google Scholar]
- Phillips N. F., Hsieh P. C., Kowalczyk T. H. (1999). Polyphosphate glucokinase. Prog Mol Subcell Biol 23, 101–125. 10.1007/978-3-642-58444-2_6 [DOI] [PubMed] [Google Scholar]
- Picossi S., Flores E., Herrero A. (2014). ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genomics 15, 22. 10.1186/1471-2164-15-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N. N., Gómez-García M. R., Kornberg A. (2009). Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78, 605–647. 10.1146/annurev.biochem.77.083007.093039 [DOI] [PubMed] [Google Scholar]
- Rashid M. H., Kornberg A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97, 4885–4890. 10.1073/pnas.060030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M. H., Rao N. N., Kornberg A. (2000). Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol 182, 225–227. 10.1128/JB.182.1.225-227.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remonsellez F., Orell A., Jerez C. A. (2006). Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology 152, 59–66. 10.1099/mic.0.28241-0 [DOI] [PubMed] [Google Scholar]
- Resnick S. M., Zehnder A. J. (2000). In vitro ATP regeneration from polyphosphate and AMP by polyphosphate : AMP phosphotransferase and adenylate kinase from Acinetobacter johnsonii 210A. Appl Environ Microbiol 66, 2045–2051. 10.1128/AEM.66.5.2045-2051.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R., Waterbury J. B. (1977). Synthesis of nitrogenase by non-heterocystous cyanobacteria. FEMS Microbiol Lett 2, 83–86. 10.1111/j.1574-6968.1977.tb00913.x [DOI] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111, 1–61. 10.1099/00221287-111-1-1 [DOI] [Google Scholar]
- Robertson B. R., Tezuka N., Watanabe M. M. (2001). Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int J Syst Evol Microbiol 51, 861–871. 10.1099/00207713-51-3-861 [DOI] [PubMed] [Google Scholar]
- Scherer P. A., Bochem H. P. (1983). Ultrastructural investigation of 12 Methanosarcinae and related species grown on methanol for occurrence of polyphosphatelike inclusions. Can J Microbiol 29, 1190–1199. 10.1139/m83-182 [DOI] [Google Scholar]
- Shih P. M., Wu D., Latifi A., Axen S. D., Fewer D. P., Talla E., Calteau A., Cai F., Tandeau de Marsac N. & other authors (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci U S A 110, 1053–1058. 10.1073/pnas.1217107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stal L. J. (1995). Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol 131, 1–32. 10.1111/j.1469-8137.1995.tb03051.x [DOI] [PubMed] [Google Scholar]
- Szymona M., Ostrowski W. (1964). Inorganic polyphosphate glucokinase of Mycobacterium phlei. Biochim Biophys Acta 85, 283–295. [DOI] [PubMed] [Google Scholar]
- Szymona M., Widomski J. (1974). A kinetic study on inorganic polyphosphate glucokinase from Mycobacterium tuberculosis H37RA. Physiol Chem Phys 6, 393–404. [PubMed] [Google Scholar]
- Tanaka S., Lee S. O., Hamaoka K., Kato J., Takiguchi N., Nakamura K., Ohtake H., Kuroda A. (2003). Strictly polyphosphate-dependent glucokinase in a polyphosphate-accumulating bacterium, Microlunatus phosphovorus. J Bacteriol 185, 5654–5656. 10.1128/JB.185.18.5654-5656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P. A., Oh H. M., Rhee G. Y. (1994). Storage of phosphorus in nitrogen-fixing Anabaena flos-aquae (Cyanophyceae). J Phycol 30, 267–273. 10.1111/j.0022-3646.1994.00267.x [DOI] [Google Scholar]
- Toepel J., Welsh E., Summerfield T. C., Pakrasi H. B., Sherman L. A. (2008). Differential transcriptional analysis of the cyanobacterium Cyanothece sp. strain ATCC 51142 during light-dark and continuous-light growth. J Bacteriol 190, 3904–3913. 10.1128/JB.00206-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi K., Munekata M., Shiba T. (2000). Involvement of inorganic polyphosphate in expression of SOS genes. Biochim Biophys Acta 1493, 73–81. 10.1016/S0167-4781(00)00165-2 [DOI] [PubMed] [Google Scholar]
- Walker P. A., Leong L. E., Ng P. W., Tan S. H., Waller S., Murphy D., Porter A. G. (1994). Efficient and rapid affinity purification of proteins using recombinant fusion proteases. Biotechnology (N Y) 12, 601–605. 10.1038/nbt0694-601 [DOI] [PubMed] [Google Scholar]
- Wilkins M. R., Gasteiger E., Bairoch A., Sanchez J. C., Williams K. L., Appel R. D., Hochstrasser D. F. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112, 531–552. [DOI] [PubMed] [Google Scholar]