Abstract
Endothelin-converting enzyme-1 (Ece-1), a crucial component of the Endothelin signaling pathway, is required for embryonic development and is an important regulator of vascular tone, yet the transcriptional regulation of the ECE1 gene has remained largely unknown. Here, we define the activity and regulation of an enhancer from the human ECE1 locus in vivo. The enhancer identified here becomes active in endothelial progenitor cells shortly after their initial specification and is dependent on a conserved FOX:ETS motif, a composite binding site for Forkhead transcription factors and the Ets transcription factor Etv2, for activity in vivo. The ECE1 FOX:ETS motif is bound and cooperatively activated by FoxC2 and Etv2, but unlike other described FOX:ETS-dependent enhancers, ECE1 enhancer activity becomes restricted to arterial endothelium and endocardium by embryonic day 9.5 in transgenic mouse embryos. The ECE1 endothelial enhancer also contains an evolutionarily-conserved, consensus SOX binding site, which is required for activity in transgenic mouse embryos. Importantly, the ECE1 SOX site is bound and activated by Sox17, a transcription factor involved in endothelial cell differentiation and an important regulator of arterial identity. Moreover, the ECE1 enhancer is cooperatively activated by the combinatorial action of FoxC2, Etv2, and Sox17. Although Sox17 is required for arterial identity, few direct transcriptional targets have been identified in endothelial cells. Thus, this work has important implications for our understanding of endothelial specification and arterial subspecification.
Keywords: Etv2, FoxC2, Sox17, Ece-1, Endothelin, vasculogenesis, artery, vein, mouse, embryo
INTRODUCTION
Endothelin signaling is essential for embryonic development and is an important regulator of vascular tone postnatally (Barton and Yanagisawa, 2008; Kurihara et al., 1999). Endothelin signaling occurs via two G protein-coupled receptors, ETA and ETB, which are bound by Endothelin peptides and activate downstream intracellular signaling to control many diverse cellular outputs (Barton and Yanagisawa, 2008; Kedzierski and Yanagisawa, 2001). There are three distinct Endothelin ligands, ET-1, ET-2, and ET-3, which are encoded by three different genes and are initially produced as larger pro-proteins (Barton and Yanagisawa, 2008; Kedzierski and Yanagisawa, 2001). Preproendothelin proteins are cleaved by ubiquitously expressed Furin proteases to produce a precursor of Endothelin protein, referred to as Big Endothelin (Kido et al., 1997). Big Endothelins are subsequently processed into the mature 21 amino acid ligands in a regulated process by either of two highly specific proteases, Endothelin-converting enzyme (Ece)-1 or Ece-2 (Kedzierski and Yanagisawa, 2001). Inactivation of the Ece1 gene in mice results in perinatal lethality and essentially phenocopies the genetic loss of both Endothelin receptors in vivo (Yanagisawa et al., 1998). In contrast, mice lacking Ece-2 protein appear grossly normal, suggesting that Ece-2 plays a relatively more minor role in the pathway and that the majority of Endothelin converting activity in the embryo is mediated by Ece-1 (Yanagisawa et al., 2000).
Consistent with its critical role in Endothelin signaling, Ece-1 is widely expressed during development and in adulthood (Yanagisawa et al., 2000). In adults, Ece-1 is expressed in various organ systems but is particularly prominent in the vascular endothelium (Korth et al., 1999). In the developing mouse embryo, Ece1 is expressed in multiple tissues, including the cardiovascular system (Yanagisawa et al., 1998). Ece-1 function in the heart has been proposed to be important in the formation of the cardiac conduction system (Hall et al., 2004; Takebayashi-Suzuki et al., 2000). Four Ece-1 isoforms have been described (Schweizer et al., 1997; Valdenaire et al., 1999); the different ECE1 transcripts are initiated from distinct transcriptional start sites, which results in slight variations in the N-termini of the different proteins produced (Funke-Kaiser et al., 2000; Li et al., 2012; Orzechowski et al., 1997; Valdenaire et al., 1999). However, how ECE1 is controlled transcriptionally remains incompletely understood. Some analyses of proximal promoter elements in cell culture-based studies have been described (Funke-Kaiser et al., 2000; Li et al., 2012; Orzechowski et al., 1998; Orzechowski et al., 1997; Valdenaire et al., 1999), but the regulation of the ECE1 gene in vivo has not been addressed previously.
The Sox family of high mobility group (HMG) transcription factors is important for development (Kamachi and Kondoh, 2013). In particular, members of the SoxF subfamily, Sox7, Sox17, and Sox18, are key regulators of endothelial cell development and differentiation (Francois et al., 2010). Sox7 is an important regulator of vascular development (Costa et al., 2012; Wat and Wat, 2014). Sox18 is a key regulator of venous and lymphatic development and functions in a partially redundant manner with Sox7 (Francois et al., 2008; Tammela and Alitalo, 2010). Sox17, which is widely appreciated for its role in endoderm development (Hudson et al., 1997; Tam et al., 2003), is also a critical regulator of endothelial gene expression and arterial development (Choi et al., 2012; Corada et al., 2013; Sakamoto et al., 2007). Endothelial-specific deletion of Sox17 in the mouse results in embryonic demise with defects in vascular remodeling and an apparent failure to form major arteries (Corada et al., 2013; Kim et al., 2007; Lee et al., 2014). Though it is clear that SoxF factors are key regulators of endothelial development, few direct endothelial targets have been identified to date.
The Ets transcription factor Etv2 is a central regulator of endothelial cell specification (Lammerts van Bueren and Black, 2012; Meadows et al., 2011). Inactivation of the Etv2 gene in mice results in loss of the endothelial and hematopoietic lineages (Ferdous et al., 2009; Lee et al., 2008). Similarly, morpholino knock down of the Etv2 ortholog etsrp in zebrafish results in profound defects in vascular development (De Val et al., 2008; Sumanas and Lin, 2006). Co-expression of Etv2 and the Forkhead transcription factor FoxC2 results in dramatic cooperative activation of Etv2-dependent endothelial genes (De Val et al., 2008). Numerous endothelial genes may be direct transcriptional targets of Etv2 and FoxC2 via a composite DNA binding element, referred to as the FOX:ETS motif (De Val et al., 2008). Previously, we identified over 1500 evolutionarily-conserved FOX:ETS motifs associated with approximately 1200 endothelial genes in the human genome (De Val et al., 2008). Among the putative enhancers identified in that prior study was a small element from the human ECE1 gene (De Val et al., 2008). Indeed, a preliminary test of the ECE1 element in a transient transgenic mouse embryo assay found that it functioned as an endothelial-specific enhancer at E9.5 (De Val et al., 2008).
In the present study, we characterized the activity and regulation of the ECE1 endothelial-specific enhancer in detail. We found that the ECE1 enhancer is active in early endothelial progenitors and then rapidly restricted only to arterial endothelium and endocardium. We also found that enhancer activity was dependent on the previously predicted FOX:ETS motif, which we found was a bona fide element for cooperative activation by FoxC2 and Etv2. In addition, we show that the enhancer is dependent on an evolutionarily-conserved SOX site, which is bound and activated by the arterially-restricted Sox factor Sox17. Finally, we show that the Sox17, Etv2, and FoxC2 cooperatively activate the ECE1 endothelial enhancer. Thus, these studies define the first bona fide in vivo enhancer of ECE1 transcription, identify a direct arterial-specific target of Sox17, and establish cooperativity of Sox17 with other core endothelial transcription factors.
MATERIALS AND METHODS
Plasmids, cloning, and mutagenesis
FoxC2 and Etv2 expression vectors have been described previously (De Val et al., 2008). The mouse Sox17 cDNA was subcloned from the previously described plasmid pCIG-Sox17 (Sinner et al., 2007) into the pRK5 expression vector (Clontech) using the XhoI restriction site to create plasmid pRK5-Sox17. The reporter plasmid pTK-E-gal has been briefly described previously (Schachterle et al., 2012; Verzi et al., 2007) and contains a minimal promoter from the Herpes Simple virus thymidine kinase gene, sufficient to activate expression in cell culture and competent to respond to neighboring enhancer sequences when inserted into the vector, upstream of a lacZ reporter gene. ECE1[623]-lacZ has also been briefly described previously: a fragment from the human ECE1 locus was PCR amplified using the following primers: 5c-cataatcccggggcaaaaacacgcga-3c and 5c-tgactgacccgggccagacatcacc-3c, and the resulting 623 bp product was inserted into the transgenic reporter plasmid hsp68-lacZ vector after XmaI digest (De Val et al., 2008). For cell culture experiments, a shorter, 164 bp region of the ECE1 enhancer encompassing the core enhancer was PCR amplified using the following primers: 5ccataatcccggggcaaaaacacgcga-3c and 5c-attgctgctggcagatctgtccct-3c. The resulting amplicon was digested with BglII and was then ligated into the BglII and BamHI restriction sites in the pTK-E-gal reporter plasmid to create plasmid ECE1-TK-E-gal. Mutations were introduced into the ECE1 enhancer sequence by PCR as described (Dodou et al., 2003) to create the following mutant sequences: mutant FOX:ETS motif (mFEM), 5c-tagctaatgaggcagggaggcacaat-3c; mSOX, 5c-ggaagggaggcaggtagagaggaagt-3c. For the SOX site/FOX:ETS motif double mutant (mF/mS), the SOX site was mutated in the mFEM context using the same mutagenic primers.
Generation of transgenic mice
Transgene fragments were gel purified away from the parental plasmid backbone, prepared, and injected into oocytes by pronuclear injection as previously described (Dodou et al., 2003). The presence of a lacZ transgene in embryo yolk sacs or tail biopsies was determined by Southern blot or by PCR using the following primers: 5c-ctgttccaagagatgcttcctg-3c and 5c-ctcagtttggatgttcctggag-3c. All experiments using animals were reviewed and approved by the UCSF Institutional Animal Care and Use Committee and complied with all institutional and federal guidelines.
X-gal staining and immunofluorescence
X-gal staining for detection of β-galactosidase and counterstaining with Nuclear Fast Red were conducted as described previously (Anderson et al., 2004). For section immunofluorescence, embryos were sectioned, dewaxed, boiled in antigen retrieval solution (Biogenex), and blocked in PBS containing 10% sheep serum and 0.1% Triton X-100. The following primary antibodies were used at 1:100 dilutions in blocking serum: goat anti-ECE1 (R&D AF1784), goat anti-Nrp1 (R&D AF566), goat-anti Nrp2 (R&D AF2215), goat anti-connexin40 (Santa Cruz sc-20466), mouse anti-COUP-TFII (Perseus Proteomics PP-H7147-00), and chicken anti-β-galactosidase (Abcam ab9361). The following secondary antibodies were used: rabbit anti-goat 594 (Invitrogen A11080), rabbit anti-chicken 488 (Jackson ImmunoResearch 303-545-003), and rabbit anti-mouse 594 (Invitrogen A11062). Slides were mounted and photographed as described previously (Rojas et al., 2009).
Cell culture, transfection, and reporter assays
Cos-1 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin-streptomycin. Transfections were performed with Fugene6 (Roche) according to manufacturers instructions; transfection mixes contained 350ng of reporter construct and 450ng of total expression vectors. The total amount of transfected plasmid DNA was held constant in all cases. Cells were harvested 48 h after transfection, and β-galactosidase activity was determined using the Luminescent β-gal Detection Kit II (Clontech). Relative light units were normalized to total protein as measured by Bradford Assay (Bio-Rad).
Electrophoretic mobility shift assay (EMSA)
DNA binding assays were performed using modified 1× binding buffer (40mM KCl, 15mM HEPES pH7.9, 1mM EDTA, 0.5mM DTT, 50% glycerol) as described previously (Dodou et al., 2003). Recombinant proteins were made using the Promega TNT Coupled Reticulocyte System according to manufacturer's instructions. Full length FoxC2, Etv2, and Sox17 were expressed from their respective pRK5 vectors using SP6 polymerase. Control and mutated control probe sequences for FOX:ETS motif from the Mef2c gene and for the SOX site from the mouse laminin 1α gene have each been described previously (De Val et al., 2008; Niimi et al., 2004). ECE1 probes were designed with guanine overhangs for labeling with Klenow and 32P-dCTP. The sense strand sequences for the probes were: ECE1_FOX:ETS, 5’-gggagtctagctaaacaggaagggaggcacaatgagagga-3’; ECE1_mFEM, ’-gggagtctagctaatgaggcagggaggcacaatgagagga-3’; ECE1_SOX, 5’-ggaagggaggcacaatgagaggaagt-3’-; ECE1_mSOX, 5’-ggaagggaggcaggtagagaggaagt-3’.
RESULTS
ECE1 enhancer activity is restricted to arterial endothelium
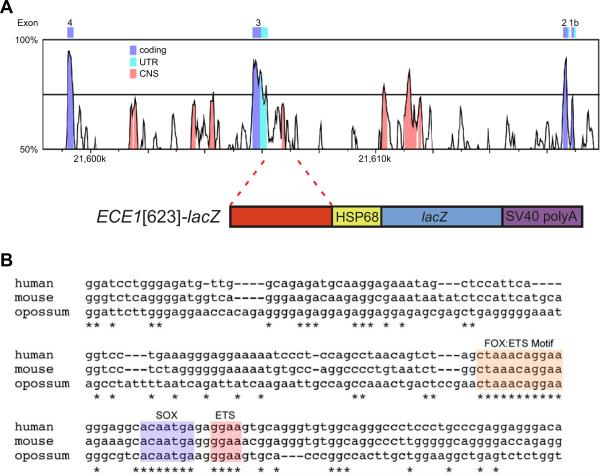
Previously, we cloned a 623-bp fragment of human genomic sequence (ECE1[623]) located 384 bp upstream of one of the three transcriptional start sites for the ECE1 gene (Fig. 1A; and De Val et al., 2008). This fragment contains the predicted FOX:ETS motif, and we previously established that this element functioned as an endothelial-specific transcriptional enhancer in transgenic mouse embryos at a single time point (De Val et al., 2008). To facilitate a more complete analysis of this regulatory element, we generated stable transgenic lines and analyzed enhancer activity in embryos from E7.5 to E18.5 (Fig. 2).
FIG. 1.
A conserved, endothelial-specific enhancer in the ECE1 locus. (A) VISTA plot comparing human and mouse sequence within the ECE1 locus (upper schematic). Human sequence from hg19 (Build GRCh37.p13, Chr1: 21606465-21606628) is used as the reference sequence. CNS, conserved, non-coding sequence; UTR, untranslated exon sequence. A 623-bp fragment encompassing the CNS containing the FOX:ETS motif was cloned upstream of the hsp68 minimal promoter for expression of E-galactosidase (lower panel). (B) ClustalW multiple sequence alignment of the core 164-bp ECE1 enhancer region from human, mouse, and opossum. Conserved nucleotides are indicated with asterisks. The conserved FOX:ETS motif, a second conserved ETS site, and the conserved SOX site are indicated.
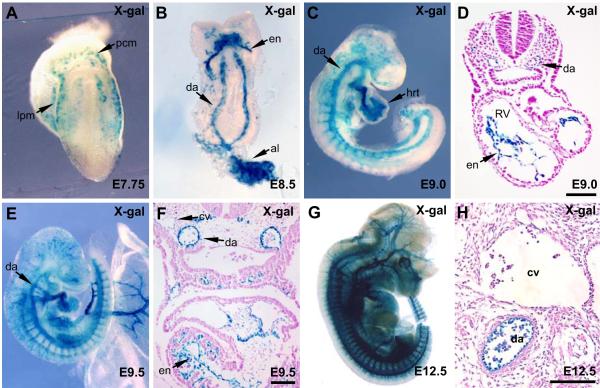
FIG. 2.
The ECE1 enhancer is endothelial specific and restricts to arterial endothelium. ECE1[623]-lacZ transgenic embryos were collected at E7.75 (A), E8.5 (B), E9.0 (C,D), E9.5 (E,F) and E12.5 (G,H) and were X-gal stained and then analyzed by whole mount (A,B,C,E,G) or were sectioned and counterstained (D,F,H) to analyze ECE1 enhancer activity. al, allantois; cv, cardinal vein; da, dorsal aorta; en, endocardium; hrt, heart; lpm, lateral plate mesoderm; pcm, precardiac mesoderm; RV, right ventricle. Scale bars, 100 μm.
The ECE1 endothelial enhancer first demonstrated activity in vivo beginning at E7.75 (Fig. 2A). At this stage, X-gal staining was apparent in cells within the lateral plate mesoderm (Fig. 2A), where vascular endothelial progenitors in the embryo proper originate (Patan, 2004). β-galactosidase activity was also evident more anteriorly in pre-cardiac mesoderm (Fig. 2A). By E8.5, X-gal staining was clearly visible in the paired dorsal aortae in the trunk and within the nascent endocardium in the heart (Fig. 2B). Following the onset of sprouting angiogenesis at approximately E9.0, ECE1 enhancer activity could be readily observed throughout the vasculature, including in intersegmental vessels (Fig. 2C). Within the heart, reporter activity was restricted to the endocardium at E9.0 (Fig. 2C,D). At E9.5, activity of the ECE1 enhancer was widely apparent in the vascular endothelium (Fig. 2E), but activity appeared to be restricting to arterial endothelium and endocardium, as X-gal staining was robust in arteries and very weak in developing veins (Fig. 2F). Enhancer activity remained robust in arterial endothelium throughout the remainder of mouse embryonic development. At E12.5, β-galactosidase expression remained completely restricted to arterial endothelium and endocardium (Fig. 2G, H).
To examine the activity of the ECE1 enhancer in more detail, we compared β-galactosidase expression from the ECE1 reporter to the expression of endogenous Ece-1 protein in arteries and veins and to artery and vein-specific markers by co-immunofluorescence at E12.5 (Fig. 3). We found that endogenous Ece-1 was expressed throughout the endothelium in both arteries and veins (Fig. 3A). In contrast, β-galactosidase expression was detectable exclusively in arteries (Fig. 3A’). The overlap of β-galactosidase and endogenous Ece-1 only in arteries (Fig. 3A″) suggests that the ECE1 enhancer described here controls only a subset of the endogenous expression pattern. Expression of the arterial markers Neuropilin-1 and Connexin40 (Buschmann et al., 2010; Herzog et al., 2001) overlapped completely with β-galactosidase expression (Fig. 3B,C), confirming the activity of the ECE1 enhancer in arterial endothelium. Likewise, comparison of E-galactosidase expression with the expression of the venous markers Neuropilin-2 and COUP-TFII (Chong et al., 2011; Herzog et al., 2001; You et al., 2005) also confirmed the arterial restriction of ECE1 enhancer activity since β-galactosidase expression did not overlap with expression of the venous markers (Fig. 3D,E). Taken together, the results presented in Figures 2 and 3 establish that ECE1[623] is an arterial-restricted enhancer in vivo.
FIG. 3.
ECE1 enhancer activity is restricted to arterial endothelium at E12.5. Immunostaining of transverse sections through the trunk region of ECE[623]-lacZ transgenic embryos collected at E12.5. Endogenous Ece-1 protein expression (A) is present in arteries and veins; β-galactosidase protein expression from the ECE1[623]-lacZ transgene (Ac) is restricted to arteries. β-galactosidase protein expression (Bc,Cc) overlaps with expression of the arterial markers Nrp1 (B) and Cx40 (C). β-galactosidase expression (D’,E’) is found only in arteries and is mutually exclusive from the expression of the venous markers Nrp2 (D) and COUP-TFII (E). Respective merged images for each of the markers are shown in A″-E″. A, artery, V, vein. Scale bars, 100 μm.
FoxC2 and Etv2 regulate the ECE1 arterial enhancer
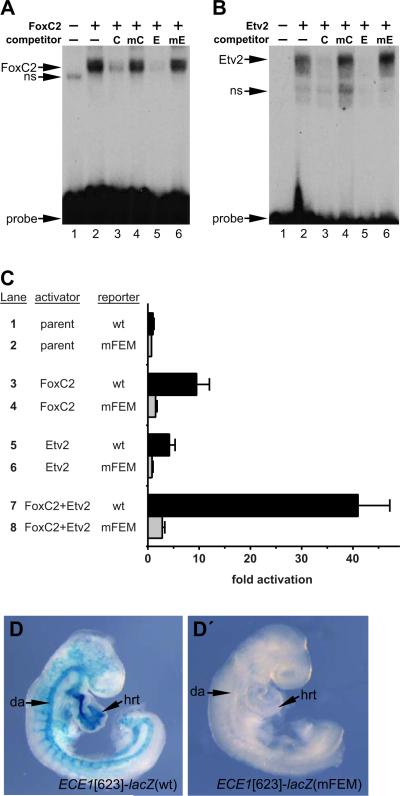
Although the ECE1 enhancer was originally identified based on the presence of a conserved FOX:ETS motif (De Val et al., 2008), the predicted FOX:ETS motif in the enhancer has not been previously validated as a bona fide binding site for FoxC2 and Etv2. Therefore, we performed electrophoretic mobility shift assays (EMSA) with recombinant FoxC2 and Etv2 proteins (Fig. 4A and B, respectively). Both factors efficiently bound to the ECE1 FOX:ETS motif (Fig. 4A,B, lane 2 in each panel). Unlabeled oligonucleotide probes representing a control FOX:ETS motif from an endothelium-specific enhancer of the mouse Mef2c gene and the ECE1 FOX:ETS motif itself efficiently competed for binding (Fig. 4A,B, lanes 3 and 5 in each panel). In contrast, mutant versions of each FOX:ETS motif failed to compete for binding even when present in ~100-fold excess (Fig. 4A,B, lanes 4 and 6 in each panel). Taken together, the results of the EMSA establish that the FOX:ETS motif in the ECE1 enhancer is a bona fide in vitro binding element recognized by FoxC2 and Etv2.
FIG. 4.
The ECE1 enhancer contains a bona fide FOX:ETS motif. (A,B) EMSA using a radiolabeled double-stranded oligonucleotide probe encompassing the ECE1 FOX:ETS motif. Recombinant FoxC2 (A) and recombinant Etv2 (B) each bound the site (lane 2). Addition of excess unlabeled control FOX:ETS motif probe [C] or the ECE1 FOX:ETS motif probe [E] abolished binding of FoxC2 and Etv2 (lanes 3, 5). Competitor oligonucleotides in which the FOX:ETS motifs were mutated [mC, mE] did not compete for binding, even when added at ~100-fold excess (lanes 4, 6). Unprogrammed reticulocyte lysate is shown in lane 1 in each panel. Non-specific [ns] bands and free probe are denoted. (C) Cos-1 cells were co-transfected with pRK5 expression vector (lanes 1,2), pRK5-FoxC2 (lanes 3,4), pRK5-Etv2 (lanes 5,6), or pRK5-FoxC2 and pRK5-Etv2 (lanes7,8) and the wild type [wt] ECE1-TK-β-gal reporter (lanes 1,3,5,7) or a version of the reporter with a mutated FOX:ETS motif [mFEM] (lanes 2,4,6,8) and assayed for β-galactosidase activity. FoxC2 and Etv2 each activated the reporter (lanes 3,5). Cooperative activation was observed when FoxC2 and Etv2 were co-expressed (lane7). In all cases, activation was dependent on an intact FOX:ETS motif. Data are expressed as the mean fold activation plus SEM for 3 independent transfections and analyses. (D, D’) Wild type [wt] ECE1[623]-lacZ (D) or ECE1[623]-lacZ with a mutated FOX:ETS motif [mFEM] (D’) transgenic embryos were collected at E9.0 and were X-gal stained to detect β-galactosidase activity. The wild type enhancer was active throughout the arterial vasculature (7/9 independent transgenic lines; 2 lines showed little or no expression). Mutation of the FOX:ETS motif site completely abolished all activity in 3/3 independent transgenic lines examined. Representative transgenic embryos are shown. da, dorsal aorta; hrt, heart.
To further test whether the FOX:ETS motif in the ECE1 arterial enhancer is a functional element for FoxC2 and Etv2, we tested the ability of FoxC2 and Etv2 to transactivate the core region of the ECE1 enhancer (Fig. 1C). FoxC2 alone significantly activated the ECE1-TK-E-gal reporter (Fig. 4C, compare lanes 1 and 3; p=0.0043). This activation was dependent on an intact FOX:ETS motif since mutation completely abolished FoxC2-dependent transactivation (Fig. 4C, compare lanes 3 and 4). Similarly, Etv2 also significantly activated the ECE1 reporter (Fig. 4C, compare lanes 1 and 5; p=0.0100) and this activation was also dependent on an intact FOX:ETS motif (Fig. 4C, compare lanes 5 and 6).
Importantly, and consistent with previous studies of other FoxC2/Etv2-dependent enhancers (De Val et al., 2008), co-transfection of FoxC2 and Etv2 expression constructs with the ECE1-TK-β-gal reporter resulted in strong (>40-fold) activation of the ECE1 enhancer compared to control (Fig. 4C, compare lanes 1 and 7; p<0.001). Mutation of the FOX:ETS motif nearly abolished reporter activation (Fig. 4C, compare lanes 7 and 8). The cooperative activation of the reporter by both factors together represents a significant increase over the transactivation by either factor alone (p<0.001 versus either FoxC2 or Etv2 alone). These data demonstrate that the ECE1 endothelial enhancer is cooperatively regulated by FoxC2 and Etv2.
To determine if the ECE1 FOX:ETS motif was necessary for reporter activity in vivo, we generated embryos with either wild type ECE1[623]-lacZ or a mutant version with a disrupted FOX:ETS motif and examined enhancer activity at E9.0 (Fig. 4D, Dc). As expected, X-gal staining demonstrated robust β-galactosidase activity in embryos harboring the wild transgene (Fig. 4D). In contrast, embryos transgenic for the mutant transgene (mFEM) exhibited no detectable X-gal staining (Fig. 4D’). These data demonstrate that the ECE1 FOX:ETS is required for enhancer activity in vivo. Taken together, the results of Fig. 4 support the idea that the ECE1 FOX:ETS motif is a bona fide binding element for FoxC2 and Etv2 and is required for enhancer activation in vitro and in vivo.
A SOX binding site is essential for ECE1 enhancer activity
Comparison of ECE1 enhancer sequences from human, mouse, and opossum in the core region of human:mouse conservation revealed that only three short stretches of sequence are perfectly conserved between the two placental mammals and the marsupial opossum (Fig. 1B). Two of those elements are the FOX:ETS motif and a neighboring ETS site. The only other perfectly conserved sequence in the core enhancer is a consensus SOX site (Fig. 1B; and Hosking et al., 1995; Niimi et al., 2004). This candidate site attracted our attention given the essential role of the SoxF subfamily in vascular endothelial development (Francois et al., 2010). In particular, Sox17 expression is restricted to arteries and has recently been described to be a key regulator of arterial development (Corada et al., 2013). Therefore, we reasoned that Sox17 might be a regulator of the ECE1 arterial-restricted enhancer via the consensus SOX site.
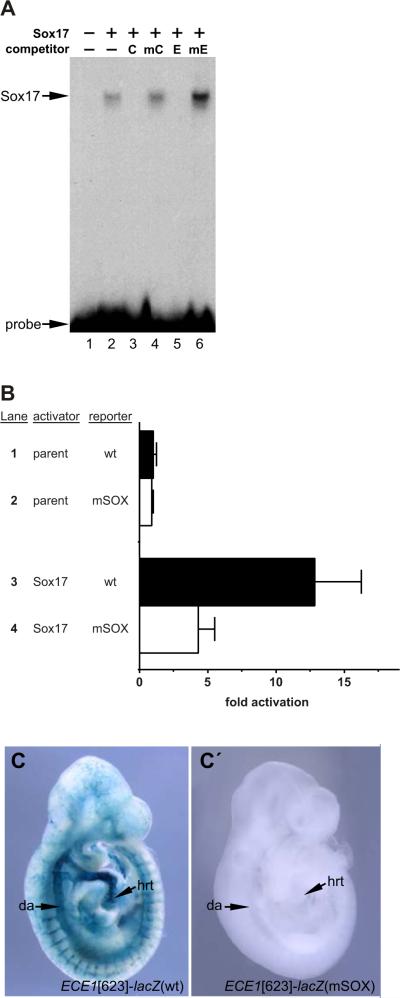
To determine whether the SOX motif in the ECE1 enhancer might function as a Sox17 binding site, we performed EMSA with recombinant Sox17 protein (Fig. 5A). Sox17 bound efficiently to the ECE1 SOX motif (Fig. 5A, lane 2). Binding to the SOX site by Sox17 was specific since addition of excess unlabeled oligonucleotide probe from a control Sox17 site from the laminin 1D gene competed with the radiolabeled probe and prevented the shift from occurring (Fig. 5A, lane 3). Similarly, binding of Sox17 to the ECE1 SOX site was abolished by addition of an unlabeled double-stranded oligonucleotide with the exact ECE1 SOX site sequence (Fig. 5A, lane 5). Mutant versions of the control SOX site and the ECE1 SOX site were unable to compete for binding even when added at ~100-fold excess (Fig. 5A, lanes 4 and 6, respectively). These results establish that the consensus SOX motif in the ECE1 enhancer is bona fide binding site recognized by Sox17 in vitro.
FIG. 5.
The ECE1 enhancer contains a bona fide SOX site. (A) EMSA using a radiolabeled double stranded oligonucleotide probe corresponding to the ECE1 SOX site. Recombinant Sox17 bound to the probe (lane 2). Addition of excess unlabeled control SOX site [C] or the ECE1 SOX site [E] abolished binding (lanes 3,5). Unlabeled mutant versions of the SOX sites [mC, mE] did not compete for Sox17 binding at ~100-fold excess (lanes 4,6). Unprogrammed reticulocyte lysate is shown in lane 1. Free probe is denoted. (B) Cos-1 cells were co-transfected with pRK5 expression vector (lanes 1,2) or with pRK5-Sox17 (lanes 3,4) and the wild type [wt] ECE1-TK-β-gal reporter (lanes 1,3) or a version of the reporter with a mutated SOX site [mSOX] (lanes 2,4) and assayed for β-galactosidase activity. Sox17 strongly activated the reporter (lane 3) and this activation was largely dependent on an intact SOX site (lane 4). Data are expressed as the mean fold activation plus SEM for 3 independent transfections and analyses. (C, C’) Wild type [wt] ECE1[623]-lacZ (C) or ECE1[623]-lacZ with a mutated SOX site [mSOX] (Cc) transgenic embryos were collected at E9.5 and were X-gal stained to detect E-galactosidase activity. The wild type enhancer was active throughout the arterial vasculature (7/9 independent transgenic lines; 2 lines showed little or no expression). Mutation of the SOX site completely abolished all activity in all 3/3 independent transgenic lines examined. Representative transgenic embryos are shown. da, dorsal aorta; hrt, heart.
We also tested the ability of Sox17 to transactivate the ECE1 enhancer (Fig. 5B). Co-transfection of a Sox17 expression vector with the ECE1-TK-β-gal reporter resulted in a significant increase in reporter activity compared to the activation obtained with the parent expression vector (Fig. 5B, compare lanes 1 and 3; ~13-fold activation, p= 0.0038). Mutation of the SOX site in the enhancer significantly reduced activity, although some residual reporter activity remained (Fig. 5B, compare lanes 3 and 4; p= 0.0330). The slight residual activity of the enhancer even with the mutant SOX site may be due to the existence of nonconsensus binding sites in the ECE1[164] core enhancer that allow for some activation of the reporter to occur, or it may be due to indirect activation by Sox17. Overall, however, the results presented in Fig. 5B demonstrate that Sox17 is sufficient for activation of the ECE1 enhancer, and that this activation is mediated primarily by binding to the conserved SOX site in the core enhancer.
We next asked if the SOX site was required for ECE1 enhancer activity in vivo. To test this, we generated transgenic mouse embryos harboring wild type ECE1[623]-lacZ and a mutant version with a disrupted SOX site (Fig. 5C,Cc). As described above, embryos transgenic for the wild type ECE1[623]-lacZ construct showed strong X-gal staining in the vascular endothelium at E9.5 (Fig. 5C). In contrast, we did not observe any staining in embryos carrying the mutant version of the reporter construct (Fig. 5Cc). Thus, the activity of the ECE1 enhancer in vivo is dependent on the SOX binding site.
Sox17, FoxC2, and Etv2 synergistically activate the ECE1 enhancer
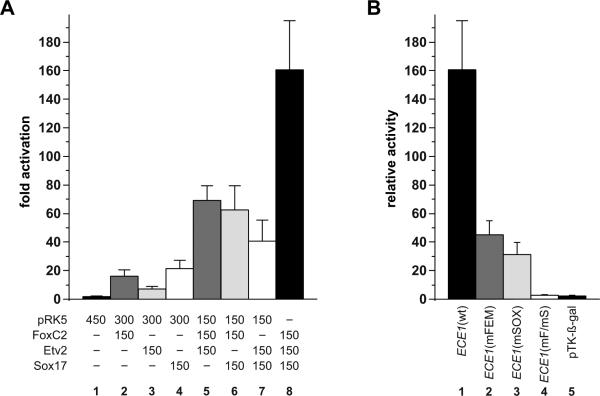
The proximity of the FOX:ETS and SOX binding sites in the core enhancer suggested the possibility that FoxC2, Etv2, and Sox17 might function collaboratively to activate the ECE1 enhancer. To test this possibility, we examined the combined effects of these three transcription factors on ECE1 enhancer transactivation (Fig. 6A). As expected, FoxC2 and Etv2 exhibited strong cooperativity on the ECE1 enhancer (Fig. 6A, lane 5). Importantly, co-expression of Sox17 with either FoxC2 or Etv2 also resulted in synergistic activation of the reporter (Fig. 6A, lanes 6 and 7). Co-expression of all three factors resulted in even more robust activation of the ECE1 reporter (>160-fold) than the activation observed with any combination of two factors (Fig. 6A, lane 8). Taken together, these observations suggest that Sox17 can function cooperatively with Etv2 and FoxC2 separately or combinatorially to activate endothelial-specific regulatory elements.
FIG. 6.
Synergistic activation of the ECE1 enhancer by combinatorial expression of Sox17, FoxC2, and Etv2 requires binding to the cognate binding site for each factor. (A) Cos-1 cells were transfected with the wild type ECE1-TK-β-gal reporter plasmid and various combinations of pRK5 expression plasmids for FoxC2, Etv2, and Sox17 and were assayed for E-galactosidase activity. The amount (ng) of each transfected expression plasmid is indicated. Data are expressed as the mean fold activation of reporter co-transfected with parental expression plasmid only plus SEM for 3 independent transfections and analyses. (B) Cos-1 cells were transfected with expression plasmids for FoxC2, Etv2, and Sox17 (150 ng each plasmid) and various ECE1-TK-β-gal reporter plasmids, including the wild type (wt, lane 1), mutated FOX:ETS motif (mFEM, lane 2), mutated SOX site (mSOX, lane 3), both FOX:ETS motif and SOX sites mutated (mF/mS, lane 4), and the parental pTK-E-gal reporter lacking ECE1 elements (lane 5). Maximal activation by the three transactivators was only observed when both cis-acting elements were intact (lane 1). Data are expressed as the mean relative β-galactosidase activity plus SEM for 3 independent transfections and analyses.
We next tested whether the FOX:ETS and SOX sites in the ECE1 enhancer were required for cooperative activation by FoxC2, Etv2, and Sox17 (Fig. 6B). Mutation of either the FOX:ETS motif or the SOX site dramatically reduced, but did not completely eliminate, transactivation of the reporter by the three factors (Fig. 6B, lanes 2 and 3). In contrast, simultaneous disruption of the FOX:ETS motif and the SOX site completely abolished all reporter activity (Fig. 6B, lane 4). These data establish that maximal synergy by FoxC2, Etv2, and Sox17 requires the presence of functional binding sites for each factor.
DISCUSSION
Modular regulation of ECE1 transcription
Ece-1 functions as an important regulatory node in Endothelin signaling by controlling the production of the mature Endothelin peptide (Turner and Murphy, 1996). In spite of its importance in embryonic development and in the control of vascular tone postnatally, the transcriptional regulation of ECE1 has not been extensively investigated. In the present study, we conducted the first analysis of ECE1 transcriptional regulation using an in vivo approach. Compared to endogenous Ece1, which is expressed broadly in the developing mouse embryo, including throughout the vasculature (Yanagisawa et al., 1998; and data not shown), the ECE1 enhancer described here only marks a subset of the endogenous gene expression pattern. This suggests that additional enhancers regulate ECE1 in other parts of its expression domain. Modular regulation by multiple, distinct enhancers is a recurrent mechanism for many developmental genes with complex expression patterns, such as ECE1, and is thought to help achieve precise temporal and spatial control of expression (Spitz and Furlong, 2012).
Restriction of ECE1 enhancer activity to arterial endothelium and endocardium
The ECE1 enhancer described here rapidly restricts to arterial endothelium. However, at early times, activity appears to be present more broadly in the endothelium, including in veins or possibly in their progenitors. This may be due to reporter expression in the endothelial progenitor population and the perdurance of β-galactosidase in veins once they have formed. Alternatively, it is possible that the enhancer remains active in presumptive veins until the venous character of the vessel has been fully established. This may suggest that a population of early endothelial cells may be neither arterial nor venous and that the ECE1 enhancer is active in these early cells, in addition to being active subsequently in arterial endothelium.
The three SoxF family members, Sox7, Sox17, and Sox18 are each expressed in endothelial cells, including in the arterial endothelium (Francois et al., 2010; Young et al., 2006), but only Sox17 is thought to restrict exclusively to arterial endothelium within the vasculature (Corada et al., 2013; Liao et al., 2009). Consistent with this notion, deletion of Sox17 specifically in vascular endothelium results in arteriovenous malformations (Corada et al., 2013). Furthermore, an arterial enhancer of Dll4 contains a SOX binding site that is crucial for activity in arterial endothelium (Sacilotto et al., 2013). Our findings are consistent with a role for Sox17 as a key regulator of arterial fate and arterial gene expression; the evidence presented here suggests that Sox17 is a bona fide regulator of the ECE1 arterial enhancer. However, we can't rule out that other SoxF factors or other transcription factors acting via the SOX consensus element may be involved in ECE1 enhancer activity and contribute to its arterial restriction during embryogenesis.
Notch signaling in the vascular endothelium is also restricted to arteries and is crucial for arterial identity (Lawson et al., 2001; Roca and Adams, 2007; Zhong et al., 2001). However, it is currently unknown how many arterial genes are direct Notch signaling targets. Recently, a binding site for RBPJ, a downstream effector of Notch signaling, has been identified in the Dll4 arterial enhancer (Sacilotto et al., 2013; Wythe et al., 2013), suggesting that Dll4 is a direct Notch target. In contrast, the ECE1 arterial-restricted enhancer described here does not contain any discernible RBPJ binding sites (Fig. 1B), suggesting that arterial restriction of this enhancer may occur independent of direct Notch activation. This finding supports the idea that regulatory pathways upstream or parallel to Notch may also be sufficient for artery-restricted expression and is consistent with some recent studies that suggest that Sox17 functions in arterial identity upstream or possibly independent of Notch signaling (Clarke et al., 2013; Corada et al., 2013). Conversely, other recent studies suggest that Notch functions upstream of Sox17 (Lee et al., 2014). Thus, it will be important for future studies to resolve the relationship between Notch signaling and Sox17 and their respective roles in arterial identity.
Transcription factor interactions in the endothelial cell gene regulatory network
Etv2 is essential for endothelial progenitor cell specification and is thought to be a central upstream regulator of the gene regulatory network that establishes the endothelial cell phenotype (De Val and Black, 2009; Lammerts van Bueren and Black, 2012). Previous work has shown that Etv2 activity is dramatically enhanced by cooperative interactions with other transcription factors (De Val et al., 2008; Shi et al., 2014). We show here that the ECE1 enhancer, like other previously described endothelial enhancers (De Val et al., 2008), is cooperatively activated by FoxC2 and Etv2 through its conserved FOX:ETS motif. Importantly, we found that Sox17 also functions cooperatively with Etv2 to activate the ECE1 enhancer and that transactivation by all three factors, FoxC2, Etv2, and Sox17, results in additional synergistic activation.
After the initial specification of the endothelial lineage, Etv2 expression is rapidly extinguished (Ferdous et al., 2009; Kataoka et al., 2011; Lee et al., 2008), whereas Sox17 becomes restricted to the arterial compartment within the vasculature (Corada et al., 2013; Liao et al., 2009). FoxC2 is expressed throughout the early vascular endothelium and its expression persists in the vascular endothelium (Seo et al., 2006). Our observation that Sox17 cooperates with FoxC2 without a requirement for Etv2 (Fig. 6A) is consistent with the possibility that the combined activities of FoxC2 and Sox17 may account for the continued activation of the ECE1 enhancer in arteries, following initial enhancer activation by Etv2, possibly in combination with Forkhead and SoxF factors. It is interesting to speculate that this may represent a general model for early endothelial gene activation in which Etv2 and its cofactors are involved in the initiation of gene expression at early times in the endothelial gene regulatory network. Subsequently, as Etv2 expression is rapidly switched off, other transcription factors function combinatorially to maintain and restrict endothelial gene expression to specific vascular compartments. It will be important to decipher these transcription factor relationships in order to understand the endothelial cell gene regulatory network more completely.
Highlights.
The human ECE1 locus contains an arterial-restricted enhancer
The ECE1 arterial enhancer is dependent on a conserved FOX:ETS motif
The ECE1 arterial enhancer is dependent on a conserved SOX binding site
Etv2, FoxC2, and Sox17 directly binding and activate the ECE1 arterial enhancer
Spx17, Etv2, and FoxC2 cooperatively activate the ECE1 arterial enhancer
ACKNOWLEDGEMENTS
We thank Jim Wells (Cincinnati) for providing the Sox17 cDNA clone, Will Schachterle for helpful advice, and Ann Zovein and Barbara Celona for helpful comments on the manuscript. S.C.M. is supported by CIRM TG2-01153. R.M.B. was supported by NIH T32 HL007544. This work was supported by NIH grant HL064658 and AHA grant 14ISA20880001 to B.L.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson JP, Dodou E, Heidt AB, De Val SJ, Jaehnig EJ, Greene SB, Olson EN, Black BL. HRC is a direct transcriptional target of MEF2 during cardiac, skeletal, and arterial smooth muscle development in vivo. Mol Cell Biol. 2004;24:3757–3768. doi: 10.1128/MCB.24.9.3757-3768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M, Yanagisawa M. Endothelin: 20 years from discovery to therapy. Can J Physiol Pharmacol. 2008;86:485–498. doi: 10.1139/Y08-059. [DOI] [PubMed] [Google Scholar]
- Buschmann I, Pries A, Styp-Rekowska B, Hillmeister P, Loufrani L, Henrion D, Shi Y, Duelsner A, Hoefer I, Gatzke N, Wang H, Lehmann K, Ulm L, Ritter Z, Hauff P, Hlushchuk R, Djonov V, van Veen T, le Noble F. Pulsatile shear and Gja5 modulate arterial identity and remodeling events during flow-driven arteriogenesis. Development. 2010;137:2187–2196. doi: 10.1242/dev.045351. [DOI] [PubMed] [Google Scholar]
- Choi E, Kraus MR, Lemaire LA, Yoshimoto M, Vemula S, Potter LA, Manduchi E, Stoeckert CJ, Jr., Grapin-Botton A, Magnuson MA. Dual lineage-specific expression of Sox17 during mouse embryogenesis. Stem Cells. 2012;30:2297–2308. doi: 10.1002/stem.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong DC, Koo Y, Xu K, Fu S, Cleaver O. Stepwise arteriovenous fate acquisition during mammalian vasculogenesis. Dev Dyn. 2011;240:2153–2165. doi: 10.1002/dvdy.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RL, Yzaguirre AD, Yashiro-Ohtani Y, Bondue A, Blanpain C, Pear WS, Speck NA, Keller G. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat Cell Biol. 2013;15:502–510. doi: 10.1038/ncb2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corada M, Orsenigo F, Morini MF, Pitulescu ME, Bhat G, Nyqvist D, Breviario F, Conti V, Briot A, Iruela-Arispe ML, Adams RH, Dejana E. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat Commun. 2013;4:2609. doi: 10.1038/ncomms3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa G, Mazan A, Gandillet A, Pearson S, Lacaud G, Kouskoff V. SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of haematopoietic development. Development. 2012;139:1587–1598. doi: 10.1242/dev.071282. [DOI] [PubMed] [Google Scholar]
- De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, Latif S, Hammer RE, Harvey RP, Olson EN, Kyba M, Garry DJ. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc Natl Acad Sci U S A. 2009;106:814–819. doi: 10.1073/pnas.0807583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois M, Caprini A, Hosking B, Orsenigo F, Wilhelm D, Browne C, Paavonen K, Karnezis T, Shayan R, Downes M, Davidson T, Tutt D, Cheah KS, Stacker SA, Muscat GE, Achen MG, Dejana E, Koopman P. Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008;456:643–647. doi: 10.1038/nature07391. [DOI] [PubMed] [Google Scholar]
- Francois M, Koopman P, Beltrame M. SoxF genes: Key players in the development of the cardio-vascular system. Int J Biochem Cell Biol. 2010;42:445–448. doi: 10.1016/j.biocel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Funke-Kaiser H, Bolbrinker J, Theis S, Lemmer J, Richter CM, Paul M, Orzechowski HD. Characterization of the c-specific promoter of the gene encoding human endothelin-converting enzyme-1 (ECE-1). FEBS Lett. 2000;466:310–316. doi: 10.1016/s0014-5793(00)01086-3. [DOI] [PubMed] [Google Scholar]
- Hall CE, Hurtado R, Hewett KW, Shulimovich M, Poma CP, Reckova M, Justus C, Pennisi DJ, Tobita K, Sedmera D, Gourdie RG, Mikawa T. Hemodynamic-dependent patterning of endothelin converting enzyme 1 expression and differentiation of impulse-conducting Purkinje fibers in the embryonic heart. Development. 2004;131:581–592. doi: 10.1242/dev.00947. [DOI] [PubMed] [Google Scholar]
- Herzog Y, Kalcheim C, Kahane N, Reshef R, Neufeld G. Differential expression of neuropilin-1 and neuropilin-2 in arteries and veins. Mech Dev. 2001;109:115–119. doi: 10.1016/s0925-4773(01)00518-4. [DOI] [PubMed] [Google Scholar]
- Hosking BM, Muscat GE, Koopman PA, Dowhan DH, Dunn TL. Trans-activation and DNA-binding properties of the transcription factor, Sox-18. Nucleic Acids Res. 1995;23:2626–2628. doi: 10.1093/nar/23.14.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, Jakt ML, Tarui H, Nishikawa S. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRalpha+ primitive mesoderm. Blood. 2011;118:6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- Kedzierski RM, Yanagisawa M. Endothelin system: the double-edged sword in health and disease. Annu Rev Pharmacol Toxicol. 2001;41:851–876. doi: 10.1146/annurev.pharmtox.41.1.851. [DOI] [PubMed] [Google Scholar]
- Kido T, Sawamura T, Hoshikawa H, D'Orleans-Juste P, Denault JB, Leduc R, Kimura J, Masaki T. Processing of proendothelin-1 at the C-terminus of big endothelin-1 is essential for proteolysis by endothelin-converting enzyme-1 in vivo. Eur J Biochem. 1997;244:520–526. doi: 10.1111/j.1432-1033.1997.00520.x. [DOI] [PubMed] [Google Scholar]
- Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth P, Bohle RM, Corvol P, Pinet F. Cellular distribution of endothelin-converting enzyme-1 in human tissues. J Histochem Cytochem. 1999;47:447–462. doi: 10.1177/002215549904700403. [DOI] [PubMed] [Google Scholar]
- Kurihara H, Kurihara Y, Nagai R, Yazaki Y. Endothelin and neural crest development. Cell Mol Biol (Noisy-le-grand) 1999;45:639–651. [PubMed] [Google Scholar]
- Lammerts van Bueren K, Black BL. Regulation of endothelial and hematopoietic development by the ETS transcription factor Etv2. Curr Opin Hematol. 2012;19:199–205. doi: 10.1097/MOH.0b013e3283523e07. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, Weinstein BM. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim DS, Choi K. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lee S, Yang H, Song S, Kim K, Saunders TL, Yoon JK, Koh GY, Kim I. Notch pathway targets proangiogenic regulator sox17 to restrict angiogenesis. Circ Res. 2014;115:215–226. doi: 10.1161/CIRCRESAHA.115.303142. [DOI] [PubMed] [Google Scholar]
- Li Y, Seidel K, Marschall P, Klein M, Hope A, Schacherl J, Schmitz J, Menk M, Schefe JH, Reinemund J, Hugel R, Walden P, Schlosser A, Volkmer R, Schimkus J, Kolsch H, Maier W, Kornhuber J, Frolich L, Klare S, Kirsch S, Schmerbach K, Scheele S, Grittner U, Zollmann F, Goldin-Lang P, Peters O, Kintscher U, Unger T, Funke-Kaiser H. A polymorphic microsatellite repeat within the ECE-1c promoter is involved in transcriptional start site determination, human evolution, and Alzheimer's disease. J Neurosci. 2012;32:16807–16820. doi: 10.1523/JNEUROSCI.2636-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WP, Uetzmann L, Burtscher I, Lickert H. Generation of a mouse line expressing Sox17-driven Cre recombinase with specific activity in arteries. Genesis. 2009;47:476–483. doi: 10.1002/dvg.20520. [DOI] [PubMed] [Google Scholar]
- Meadows SM, Myers CT, Krieg PA. Regulation of endothelial cell development by ETS transcription factors. Semin Cell Dev Biol. 2011;22:976–984. doi: 10.1016/j.semcdb.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T, Hayashi Y, Futaki S, Sekiguchi K. SOX7 and SOX17 regulate the parietal endoderm-specific enhancer activity of mouse laminin alpha1 gene. J Biol Chem. 2004;279:38055–38061. doi: 10.1074/jbc.M403724200. [DOI] [PubMed] [Google Scholar]
- Orzechowski HD, Gunther A, Menzel S, Funke-Kaiser H, Richter M, Bohnemeier H, Paul M. Endothelial expression of endothelin-converting enzyme-1 beta mRNA is regulated by the transcription factor Ets-1. J Cardiovasc Pharmacol 31 Suppl. 1998;1:S55–57. doi: 10.1097/00005344-199800001-00018. [DOI] [PubMed] [Google Scholar]
- Orzechowski HD, Richter CM, Funke-Kaiser H, Kroger B, Schmidt M, Menzel S, Bohnemeier H, Paul M. Evidence of alternative promoters directing isoform-specific expression of human endothelin-converting enzyme-1 mRNA in cultured endothelial cells. J Mol Med (Berl) 1997;75:512–521. doi: 10.1007/s001090050136. [DOI] [PubMed] [Google Scholar]
- Patan S. Vasculogenesis and angiogenesis. Cancer Treat Res. 2004;117:3–32. doi: 10.1007/978-1-4419-8871-3_1. [DOI] [PubMed] [Google Scholar]
- Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007;21:2511–2524. doi: 10.1101/gad.1589207. [DOI] [PubMed] [Google Scholar]
- Rojas A, Schachterle W, Xu SM, Black BL. An endoderm-specific transcriptional enhancer from the mouse Gata4 gene requires GATA and homeodomain protein-binding sites for function in vivo. Dev Dyn. 2009;238:2588–2598. doi: 10.1002/dvdy.22091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacilotto N, Monteiro R, Fritzsche M, Becker PW, Sanchez-Del-Campo L, Liu K, Pinheiro P, Ratnayaka I, Davies B, Goding CR, Patient R, Bou-Gharios G, De Val S. Analysis of Dll4 regulation reveals a combinatorial role for Sox and Notch in arterial development. Proc Natl Acad Sci U S A. 2013;110:11893–11898. doi: 10.1073/pnas.1300805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y, Hara K, Kanai-Azuma M, Matsui T, Miura Y, Tsunekawa N, Kurohmaru M, Saijoh Y, Koopman P, Kanai Y. Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem Biophys Res Commun. 2007;360:539–544. doi: 10.1016/j.bbrc.2007.06.093. [DOI] [PubMed] [Google Scholar]
- Schachterle W, Rojas A, Xu SM, Black BL. ETS-dependent regulation of a distal Gata4 cardiac enhancer. Dev Biol. 2012;361:439–449. doi: 10.1016/j.ydbio.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Valdenaire O, Nelbock P, Deuschle U, Dumas Milne Edwards JB, Stumpf JG, Loffler BM. Human endothelin-converting enzyme (ECE-1): three isoforms with distinct subcellular localizations. Biochem J. 1997;328(Pt 3):871–877. doi: 10.1042/bj3280871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Fujita H, Nakano A, Kang M, Duarte A, Kume T. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev Biol. 2006;294:458–470. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Shi X, Richard J, Zirbes KM, Gong W, Lin G, Kyba M, Thomson JA, Koyano-Nakagawa N, Garry DJ. Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev Biol. 2014;389:208–218. doi: 10.1016/j.ydbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi-Suzuki K, Yanagisawa M, Gourdie RG, Kanzawa N, Mikawa T. In vivo induction of cardiac Purkinje fiber differentiation by coexpression of preproendothelin-1 and endothelin converting enzyme-1. Development. 2000;127:3523–3532. doi: 10.1242/dev.127.16.3523. [DOI] [PubMed] [Google Scholar]
- Tam PP, Kanai-Azuma M, Kanai Y. Early endoderm development in vertebrates: lineage differentiation and morphogenetic function. Curr Opin Genet Dev. 2003;13:393–400. doi: 10.1016/s0959-437x(03)00085-6. [DOI] [PubMed] [Google Scholar]
- Tammela T, Alitalo K. Lymphangiogenesis: Molecular mechanisms and future promise. Cell. 2010;140:460–476. doi: 10.1016/j.cell.2010.01.045. [DOI] [PubMed] [Google Scholar]
- Turner AJ, Murphy LJ. Molecular pharmacology of endothelin converting enzymes. Biochem Pharmacol. 1996;51:91–102. doi: 10.1016/0006-2952(95)02036-5. [DOI] [PubMed] [Google Scholar]
- Valdenaire O, Lepailleur-Enouf D, Egidy G, Thouard A, Barret A, Vranckx R, Tougard C, Michel JB. A fourth isoform of endothelin-converting enzyme (ECE-1) is generated from an additional promoter molecular cloning and characterization. Eur J Biochem. 1999;264:341–349. doi: 10.1046/j.1432-1327.1999.00602.x. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Agarwal P, Brown C, McCulley DJ, Schwarz JJ, Black BL. The transcription factor MEF2C is required for craniofacial development. Dev Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wat JJ, Wat MJ. Sox7 in vascular development: review, insights and potential mechanisms. Int J Dev Biol. 2014;58:1–8. doi: 10.1387/ijdb.130323mw. [DOI] [PubMed] [Google Scholar]
- Wythe JD, Dang LT, Devine WP, Boudreau E, Artap ST, He D, Schachterle W, Stainier DY, Oettgen P, Black BL, Bruneau BG, Fish JE. ETS factors regulate Vegf-dependent arterial specification. Dev Cell. 2013;26:45–58. doi: 10.1016/j.devcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Hammer RE, Richardson JA, Emoto N, Williams SC, Takeda S, Clouthier DE, Yanagisawa M. Disruption of ECE-1 and ECE-2 reveals a role for endothelin-converting enzyme-2 in murine cardiac development. J Clin Invest. 2000;105:1373–1382. doi: 10.1172/JCI7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Hammer RE, Richardson JA, Williams SC, Clouthier DE, Yanagisawa M. Role of Endothelin-1/Endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. J Clin Invest. 1998;102:22–33. doi: 10.1172/JCI2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- Young N, Hahn CN, Poh A, Dong C, Wilhelm D, Olsson J, Muscat GE, Parsons P, Gamble JR, Koopman P. Effect of disrupted SOX18 transcription factor function on tumor growth, vascularization, and endothelial development. J Natl Cancer Inst. 2006;98:1060–1067. doi: 10.1093/jnci/djj299. [DOI] [PubMed] [Google Scholar]
- Zhong TP, Childs S, Leu JP, Fishman MC. Gridlock signalling pathway fashions the first embryonic artery. Nature. 2001;414:216–220. doi: 10.1038/35102599. [DOI] [PubMed] [Google Scholar]