Abstract
Background
It is now apparent that the complex microbial communities found on and in the human body vary across individuals. What has largely been missing from previous studies is an understanding of how these communities vary over time within individuals. To the extent to which it has been considered, it is often assumed that temporal variability is negligible for healthy adults. Here we address this gap in understanding by profiling the forehead, gut (fecal), palm, and tongue microbial communities in 85 adults, weekly over 3 months.
Results
We found that skin (forehead and palm) varied most in the number of taxa present, whereas gut and tongue communities varied more in the relative abundances of taxa. Within each body habitat, there was a wide range of temporal variability across the study population, with some individuals harboring more variable communities than others. The best predictor of these differences in variability across individuals was microbial diversity; individuals with more diverse gut or tongue communities were more stable in composition than individuals with less diverse communities.
Conclusions
Longitudinal sampling of a relatively large number of individuals allowed us to observe high levels of temporal variability in both diversity and community structure in all body habitats studied. These findings suggest that temporal dynamics may need to be considered when attempting to link changes in microbiome structure to changes in health status. Furthermore, our findings show that, not only is the composition of an individual’s microbiome highly personalized, but their degree of temporal variability is also a personalized feature.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-014-0531-y) contains supplementary material, which is available to authorized users.
Background
The increasing recognition that commensal and mutualistic microorganisms are necessary for many aspects of normal human physiology has altered the traditional pathogen-dominated view of human-bacterial interactions [1,2]. As a result of this paradigm shift, there is a tremendous amount of interest in understanding the factors that influence the diversity, composition, dynamics, and function of human-associated microbial communities. One of the primary objectives is to leverage this understanding in order to manage, restore, and/or exploit our microbial partners in ways that promote human health. However, our current understanding of how and why these communities vary through time is limited. Previous studies that have characterized human associated microbial communities over time have been based on relatively few individuals [3,4], intermittent sampling intervals [2,5,6], single body habitats [4,7-10], or focused on disease states [11], leaving us with an incomplete picture of the range of normal variability in the human microbiome. Only by conducting longitudinal studies of large cohorts of both healthy and diseased hosts can we begin to identify the ecological factors structuring the diversity, composition, and dynamics of the human microbiome.
Here, we investigated the temporal dynamics of forehead, gut (feces), palm, and tongue microbial communities of 85 college-age adults (Table 1) from three U.S. universities. Samples were self-collected weekly over a 3-month period beginning in January 2012. Bacterial and archaeal communities were characterized using high-throughput sequencing of the variable region 4 (V4) of the 16S rRNA gene [12]. In total, we generated 170,563,932 quality-filtered sequences from 3,655 samples, with all analyses conducted on samples rarefied to exactly 10,000 sequences per sample. To identify potential drivers of variability, we collected demographic, lifestyle, and hygiene data at the initiation of the sampling period using a standardized 49-question survey (Additional file 1). Weekly questionnaires were used to track changes in health status, medication use, menstrual cycle for women, and other dramatic changes in routine behavior (Additional file 2). De-identified responses to all questions are provided in Additional file 3.
Table 1.
Demographic summary of study participants
| Subject ID a | Age (years) | Gender | BMI | Ethnicity | Samples (forehead/gut/palm/tongue) (n) |
|---|---|---|---|---|---|
| A000 | 21 | Female | 19.94 | Caucasian | 9/9/10/10 |
| A003 | 21 | Female | 18.56 | Caucasian | 10/10/10/10 |
| A004 | 22 | Female | 25.85 | Caucasian | 8/10/10/10 |
| A007 | 22 | Female | 19.97 | Caucasian/Asian | 9/9/8/9 |
| A008 | 20 | Male | 20.67 | Caucasian | 10/10/9/- |
| A009 | 20 | Female | 22.31 | Caucasian | 9/9/9/10 |
| A010 | 20 | Female | ? | Caucasian | 10/10/8/10 |
| A011 | 29 | Female | 19.46 | Hispanic | 7/8/7/8 |
| A012 | 21 | Female | 26.61 | Caucasian | 10/9/10/10 |
| A015 | 20 | Female | 24.13 | Caucasian | 9/9/9/9 |
| A016 | 22 | Female | 25.75 | Caucasian | 8/-/7/9 |
| A017 | 21 | Female | 33.84 | Caucasian | 9/9/9/9 |
| A019 | 22 | Female | 18.29 | Caucasian | 9/9/9/9 |
| A026 | 21 | Male | 22.24 | Caucasian | 8/8/8/8 |
| A027 | 20 | Female | 21.79 | Other | 9/9/9/9 |
| A028 | 23 | Male | 23.06 | Asian/Pacific island | 8/8/7/8 |
| A029 | 21 | Male | 25.83 | Caucasian | 7/9/9/9 |
| A032 | 21 | Female | 18.88 | Caucasian | 8/8/-/8 |
| A033 | 21 | Female | 27.44 | Caucasian | 10/9/10/10 |
| A036 | 20 | Female | 21.29 | Caucasian | 8/8/-/8 |
| A037 | 20 | Female | 20.80 | Hispanic | 9/8/9/10 |
| A038 | 21 | Female | ? | Hispanic | 8/8/8/9 |
| A040 | 22 | Male | 23.09 | Caucasian | 10/9/10/10 |
| A042 | 36 | Male | 25.40 | Caucasian | 9/10/10/10 |
| A044 | 21 | Male | 26.58 | Caucasian | 10/10/9/9 |
| A048 | 22 | Female | 22.14 | Caucasian | 10/10/10/10 |
| A049 | 20 | Female | ? | Caucasian | 7/8/-/10 |
| A050 | 41 | Female | 22.86 | Caucasian | 9/9/9/9 |
| A051 | 20 | Male | 22.47 | Caucasian | 7/-/7/- |
| A052 | 32 | Male | 31.84 | Hispanic | 10/10/10/10 |
| A053 | 23 | Female | 21.14 | Caucasian | 10/10/9/10 |
| A056 | 23 | Male | 23.73 | Caucasian | 8/8/8/8 |
| A061 | ? | Male | ? | ? | 7/-/-/7 |
| B101 | 24 | Male | 18.31 | Caucasian | 9/9/9/9 |
| B102 | 32 | ? | ? | caucasian | 7/-/-/7 |
| B105 | ? | ? | ? | caucasian | 7/7/7/7 |
| B106 | 21 | Female | 21.95 | Caucasian | 8/9/8/9 |
| B107 | 19 | Female | 20.37 | Caucasian | 9/8/-/9 |
| B108 | 30 | Female | 24.80 | Asian/Pacific island | 7/7/-/7 |
| B109 | 24 | Male | 23.67 | Caucasian | 9/9/8/9 |
| B110 | 20 | Female | 20.36 | Caucasian | 9/9/9/9 |
| B114 | 21 | Female | 17.54 | Caucasian | 8/9/-/9 |
| B117 | ? | Female | ? | Caucasian | 8/7/-/8 |
| B119 | 20 | Male | ? | Caucasian | -/8/-/8 |
| B121 | 20 | Female | 22.86 | Caucasian | 9/9/8/9 |
| B123 | 20 | Male | 25.07 | Caucasian | 9/8/9/9 |
| B124 | 21 | Female | 22.15 | Caucasian | 9/9/-/9 |
| B129 | 21 | Female | 18.40 | Caucasian | 9/8/-/9 |
| B130 | 22 | Female | 22.67 | Caucasian | 8/9/-/9 |
| B132 | 22 | Female | 16.82 | Hispanic | 9/9/9/9 |
| B133 | 22 | Male | 27.89 | Caucasian | 9/9/8/7 |
| B134 | ? | Male | 21.91 | Caucasian | 7/-/7/7 |
| B136 | 22 | Female | 19.22 | Caucasian | 8/9/7/8 |
| B137 | 22 | Female | ? | Hispanic | -/7/-/7 |
| B139 | 20 | Female | 17.75 | Caucasian | 9/9/-/9 |
| B144 | 33 | Male | 25.10 | Caucasian | 8/8/-/8 |
| B146 | 26 | Female | 20.60 | Caucasian | -/7/-/- |
| B147 | 51 | Female | ? | Caucasian | 7/7/7/- |
| B148 | 37 | Male | 20.08 | ? | 8/8/7/8 |
| B149 | 55 | Male | 25.10 | Caucasian | 9/9/9/9 |
| B150 | 32 | Female | 20.05 | Caucasian/Hispanic | 8/9/9/9 |
| B153 | 21 | Female | 21.93 | Caucasian | 8/8/-/8 |
| B154 | 21 | Female | 23.40 | Caucasian | 9/9/7/9 |
| B155 | 30 | Female | 23.62 | Caucasian | 9/9/9/9 |
| B156 | ? | Female | ? | Hispanic | -/7/-/7 |
| B157 | 25 | Male | 21.86 | Caucasian | 10/9/9/9 |
| B159 | 21 | Male | 25.10 | Caucasian | 7/8/-/9 |
| B160 | 22 | Female | 17.75 | Asian/Pacific island | 8/8/7/8 |
| B161 | 21 | Male | 26.58 | Caucasian | 7/-/-/7 |
| B164 | 22 | Male | 22.96 | Caucasian | 7/7/7/7 |
| C203 | 23 | Female | 21.74 | Caucasian | 9/7/9/9 |
| C204 | 25 | Male | 22.31 | Caucasian | 9/9/7/9 |
| C210 | 20 | Female | 24.03 | Caucasian | 8/-/8/8 |
| C212 | 22 | Female | 24.30 | Caucasian | 8/8/8/8 |
| C213 | 18 | Female | 22.71 | Caucasian | 8/8/7/8 |
| C214 | 27 | Male | 27.71 | Caucasian | 7/7/8/8 |
| C233 | 19 | Male | 22.96 | Caucasian/Hispanic | 9/7/9/8 |
| C234 | 18 | Female | 32.10 | Caucasian | 9/-/8/9 |
| C236 | 18 | Female | 18.09 | Caucasian | -/-/-/9 |
| C237 | 19 | Male | 18.65 | Caucasian | 7/7/-/- |
| C243 | 23 | Male | 23.63 | Caucasian | 8/-/8/8 |
| C248 | 21 | Male | 20.71 | Caucasian | 9/8/9/8 |
| C253 | 27 | Female | 23.21 | Caucasian | 8/9/9/9 |
| C255 | 22 | Female | 27.46 | Caucasian | 8/9/-/9 |
| C263 | 20 | Male | 20.09 | Caucasian | 7/8/8/7 |
aSingle-letter prefix represents the university of attendance.
Question marks denote data not provided by study participants. The last column shows the number of samples used in the time series analysis for each body habitat from each subject. Dashes indicate that samples from that individual were not including in the analysis for that particular body habitat. For full metadata, the reader is referred to Additional file 3.
BMI = body mass index.
Results and discussion
To quantify the amount of temporal variability in diversity of each body habitat, we calculated the coefficient of variation (CV = standard deviation/mean) for three alpha diversity metrics (phylogenetic diversity, phylotype richness, and Shannon index [13]) for each individual [14]. Low CV values indicate that an individual had relatively stable alpha diversity levels, whereas high CV values indicate than an individual had variable levels of alpha diversity over the 3-month study period. As evident in Figure 1a, there was a wide range of variability within each body habitat indicating that some individuals varied more than others. When we compare values across body habitats, we see that skin surfaces, particularly the palm, exhibited higher levels of temporal variability in diversity than gut or tongue (Figure 1a). These patterns were generally consistent regardless of the alpha diversity metric used. Skin surfaces also hosted the most diverse communities we surveyed (Additional file 4), as theory would predict for open, exposed environments that have a larger species pool of potential colonizers [15]. Ecological theory also predicts that habitats with large species pools should vary more through time [15], which is what we observed here.
Figure 1.
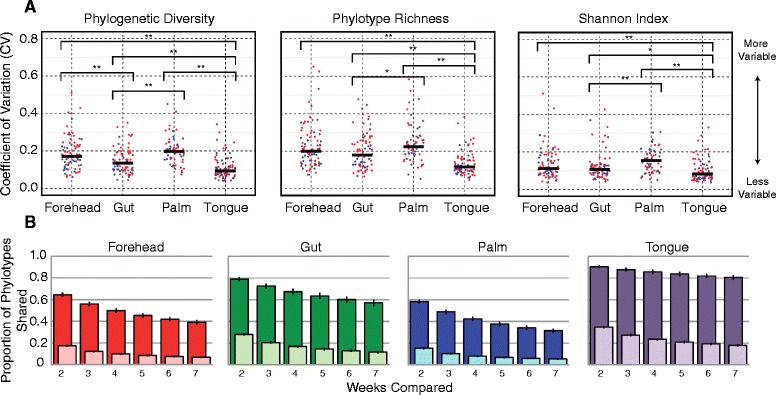
Body habitats exhibited different levels of temporal variability both in diversity (A) and membership (B). In (A), each point represents the temporal variability of a single individual colored by gender (red = female, blue = male) with black bars representing the median for a given body habitat and metric. Statistical differences were observed for each metric across body habitats (Kruskal-Wallis, P ≤0.01) and comparisons based on pairwise Mann-Whitney U-test are denoted by asterisks (* = corrected P ≤0.05, ** = corrected P ≤0.01). In (B), the smaller, lighter shaded bars in each plot are for all phylotypes except singletons and the larger, darker bars are only for the 100 most abundant phylotypes for each individual. Error bars in (B) are ±1 SEM.
This high degree of temporal variability in alpha diversity levels was matched by high variability (and hence instability) in community membership (Figure 1b). Comparing the proportion of phylotypes shared among time intervals within an individual shows that fewer phylotypes were shared through time in skin communities than in the gut or tongue communities. For example, on average only 15% of the phylotypes observed on the palm skin surface (excluding singletons on a per individual basis) were observed at any other point in time, whether samples were collected 1 or 6 weeks apart. A similar pattern was observed when we used median unweighted UniFrac values [16] (a phylogenetic metric of community membership) for each body habitat, where turnover was found to be greater for the skin than for the tongue and gut communities (Figure 2a and Additional file 5a). In contrast, variability in community structure, which accounts for phylotype abundance (median weighted UniFrac), was lower on the forehead than the other body habitats (Figure 2b and Additional file 5b), suggesting that the nature of variability differs depending on the body habitat in question. On the tongue and in the gut, changes in the relative abundance of persistent taxa (that is, those taxa that are consistently present over time) drive the temporal dynamics, whereas temporal variability in forehead communities appears to be driven more by the presence or absence of transient taxa on the skin surface. For the palm, both membership and structure appear highly dynamic, likely due to frequent hand washing and exchange of microbes with the numerous surfaces we touch on a daily basis, including our other body parts.
Figure 2.
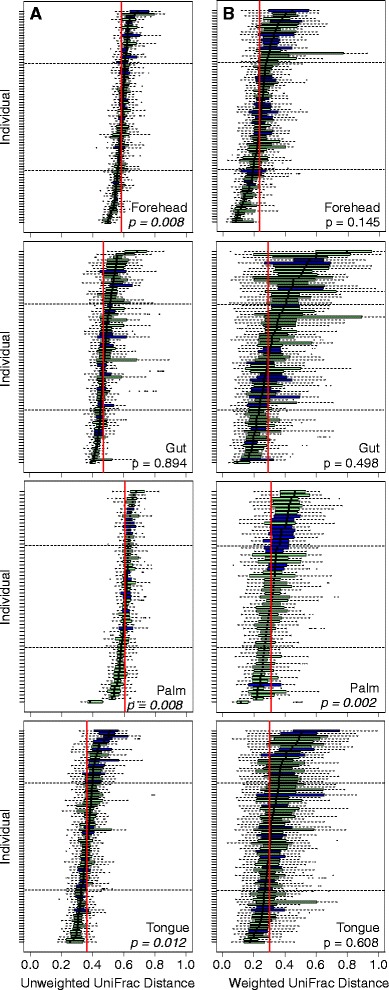
Boxplots of unweighted (A) and weighted (B) intra-individual UniFrac distances for each body habitat. A broad range of temporal variability in microbial community membership (A) and structure (B) was observed across body habitats and within body habitats across individuals. Individuals are sorted by median in each plot. Green bars depict individuals who did not report antibiotic use during the study period while blue bars indicate individuals who took antibiotics. The median values for each body habitat are shown with vertical red lines. Dotted horizontal lines in each plot divide the study population into first and fourth quartiles and depict ‘stable’ and ‘variable’ individuals, respectively. Non-parametric Mann-Whitney U-tests were used to determine the affect of antibiotic use on temporal variability within each body habitat. P values are shown in each panel. Note that statistical differences were observed for each metric across body habitats (Kruskal-Wallis, P ≤0.01).
Using median UniFrac values for each individual as our metric of temporal variability in community membership (unweighted) and structure (weighted), we found that individuals differed dramatically not only in the composition of their microbial communities (Additional file 6), as has been observed previously [2,5,17,18], but also in the degree of temporal variability observed in their microbial communities (Figure 2 and Additional file 7). This has been previously shown in vaginal communities [8], but we show here that this is a general characteristic of microbial communities across human body habitats. The variability of microbial communities in one body habitat, in general, did not predict the variability of microbial communities of other body habitats. The exception was the two skin habitats, where individuals that had more variable forehead communities also had more variable palm communities (Additional files 8 and 9). This finding suggests that the factors that contribute to intra-personal temporal variability in microbiome composition are shared across skin habitats, but not necessarily across other body habitats. Furthermore, relatively few individuals exhibited a significant time-decay relationship [19]; in general, samples collected closer together in time did not harbor more similar communities than those collected further apart in time (Additional file 10). These results highlight that attempts to predict what type of communities to expect in a given body habitat based on data collected during the previous week (or weeks) may be difficult for most individuals. However, it is important to note that if we had sampled more frequently (for example, on a daily basis [3]) or for a longer period of time, we may have been able to identify a stronger relationship between elapsed time and the composition of the communities within body habitats.
Having established that the degree and nature of variability was specific to each body site and was in itself an important parameter that distinguished individuals from one another, we next sought to identify factors associated with this variation across individuals. Based on previous work [4,20], we expected that antibiotic usage would lead to profound shifts in the structure of an individual’s microbiome. Indeed, within a given individual, the largest shifts observed in community membership coincided with the time points that the individual reported having taken oral antibiotics (P <0.001 for both unweighted and weighted UniFrac, Monte Carlo t-test with 1,000 iterations). Across the study population, however, with the exception of palm communities, we did not find a significant effect of antibiotics on variability in community membership and structure; individuals who took antibiotics did not, on average, have more variable communities than those that did not take antibiotics over the time period of this experiment (Figure 2). Our observation that antibiotic use was not associated with increased temporal variability in microbial communities across the study population could be due to the fact that we did not control for the timing of sampling relative to antibiotic use, dosage, or type of antibiotics used by the individuals sampled here, or it may be because microbial community responses to antibiotics are highly individualized, as suggested by recent work [4,21].
We next used generalized linear models (GLMs) to identify which other factors or combination of factors best predicted why some individuals harbored more variable microbial communities than others. For these models, we again used median weighted or unweighted UniFrac values of each individual as our response variables for each body habitat. Potential predictive factors were compiled from the initial survey responses (Additional file 2) and we only included factors for which we had sufficient replication in survey responses (Additional file 3). Presented models included factors with a significance value <0.05. As shown in Table 2, our models were often able to explain much of the variability in the temporal stability of microbial communities across individuals, but the strength of the models was dependent on the body habitat in question or the distance metric used. Common predictive factors observed in multiple body habitats included median alpha diversity values (Shannon Index), university affiliation, and antibiotic use (Table 2). However, the strongest predictive variable for most body habitats was median diversity, measured using the Shannon index, suggesting an overall relationship between diversity and variability. Other factors appeared to have a body site-specific affect. For example, the number of roommates helped explain a significant amount of variability in the structure (weighted) of forehead microbial communities, a pattern that may driven by the exchange of skin bacteria between individuals sharing a common living area.
Table 2.
Measured factors that influenced the temporal variability of the human microbiome
| Parameter estimate | Sum of squares | F statistic | P value | BIC | R 2 | |
|---|---|---|---|---|---|---|
| Forehead - unweighted | ||||||
| Antibiotic use | 0.015 | 0.010 | 8.76 | 0.004 | -262.21 | 0.175 |
| University | 0.119 | 0.006 | 5.41 | 0.023 | -263.38 | |
| Forehead - weighted | ||||||
| Median Shannon | 0.038 | 0.090 | 32.2 | 3.61 e -7 | -190.67 | 0.580 |
| Gender | 0.023 | 0.027 | 9.54 | 0.003 | -193.54 | |
| Roommates (n) | 0.039 | 0.016 | 5.70 | 0.02 | -196.16 | |
| Gut - unweighted | ||||||
| Median Shannon | 0.063 | 0.081 | 73.24 | 4.3 e -12 | -240.90 | 0.570 |
| Over-the-counter acne product | 0.014 | 0.013 | 11.18 | 0.001 | -249.97 | |
| University | 0.014 | 0.007 | 6.64 | 0.012 | -254.34 | |
| Gut - weighted | ||||||
| Median Shannon | 0.107 | 0.238 | 20.64 | 2.61 e -5 | -85.83 | 0.319 |
| Over-the-counter acne product | 0.034 | 0.065 | 5.65 | 0.021 | -90.53 | |
| University | 0.028 | 0.047 | 4.08 | 0.047 | -90.55 | |
| Palm - unweighted | ||||||
| Exercise frequency | 0.033 | 0.022 | 15.74 | 2.00 e -4 | -188.6 | 0.310 |
| Lives with dogs | 0.014 | 0.010 | 7.18 | 0.009 | -189.9 | |
| Roommates (n) | 0.016 | 0.008 | 5.77 | 0.019 | -191.8 | |
| Palm - weighted | ||||||
| Antibiotic use | 0.026 | 0.024 | 4.97 | 0.029 | -129.8 | 0.080 |
| Tongue - unweighted | ||||||
| Antibiotic use | 0.018 | 0.015 | 7.75 | 0.007 | -217.82 | 0.215 |
| Median Shannon | 0.038 | 0.010 | 5.5 | 0.022 | -220.12 | |
| Tongue - weighted | ||||||
| No good model |
Generalized linear models (GLMs) were used to determine which of the measured factors or combination of factors best predicted variability in microbiome membership (unweighted UniFrac) and structure (weighted UniFrac). Unweighted UniFrac distances are a metric of the phylogenetic dissimilarity of samples through time. Weighted UniFrac distances weight dissimilarity both as a function of the phylogenetic dissimilarity and the relative abundance of taxa (such that two samples with the same phylogenetic dissimilarity are considered more different if one is dominated by a particular taxon).
BIC = Bayesian Information Criterion.
To explore the relationship between diversity and temporal variability in greater detail, we generated single-factor linear models using median Shannon index values as our metric of diversity and either median weighted or unweighted UniFrac values as our metric of stability (Figure 3). With these models, we observed statistically significant negative correlations between diversity and compositional variability for the gut and tongue communities; individuals with more diverse communities were less variable (more stable) than individuals with less diverse communities. In contrast, a positive relationship was observed between forehead community diversity and structural variability while no relationship was evident for palm communities. Similar directional patterns were observed with the other diversity metrics (Additional file 11). Our finding that microbial communities which likely experience lower rates of colonization from external environments (the gut and tongue) exhibit a positive diversity-stability relationship parallels patterns observed in many plant and animal communities where increases in species diversity often result in more stable communities and communities that are more resistant to invasions (that is, the portfolio effect) [22]. Although the health implications of the diversity-stability relationships observed here remain undetermined, recent work has shown that gut communities of lower diversity are often associated with disease phenotypes in humans [23].
Figure 3.
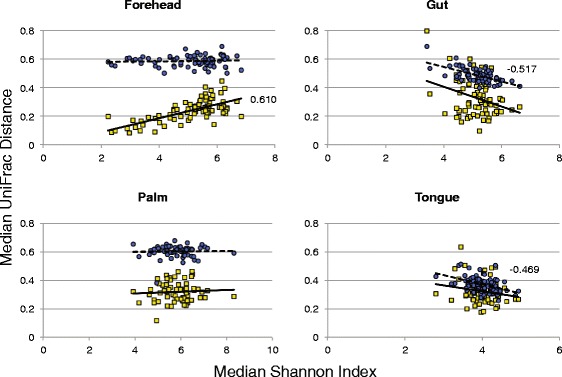
Relationship between diversity and variability of microbial communities associated with each body habitat. Diversity was measured as the median Shannon Diversity Index for each individual over the 3-month sampling period. Variability was measured as intra-individual median weighted (white boxes) and unweighted (gray circles) UniFrac distance. Each point represents values of the time-series data for one individual. Spearman rank correlation coefficients are presented for statistically significant relationships (P ≤0.01). Note that similar patterns were observed with other alpha diversity metrics (Additional file 11).
Individuals that had more stable communities harbored taxonomically distinct communities compared with those found in more variable individuals (Figure 4). For example, individuals with stable forehead communities had a greater relative abundance of Staphylococcaceae and Corynebacteriaceae, whereas individuals with highly variable forehead communities were enriched in Streptococcaceae and Lactobacillaceae (Figure 4a). The trade-off between Staphylococcaceae and Lactobacillaceae is intriguing because several Lactobacillaceae species inhibit attachment of Staphylococcaceae to epithelial cells [24,25]. In the gut, two of the dominant groups of Firmicutes, Clostridiaceae and Lactobacillaceae, were more abundant in variable individuals, whereas the Bacteroidaceae (the dominant family within the Bacteroidetes phylum) were most abundant in stable individuals (Figure 4b). A higher Firmicutes:Bacteriodetes ratio has been observed in guts of obese individuals [26,27], but we did not have enough diversity in body mass index (BMI) to formally test if temporal variability may also be associated with obesity. Although the mechanisms underlying these patterns remain unclear, these observations highlight the likely importance of bacterial interactions in determining the stability of human-associated microbial communities.
Figure 4.
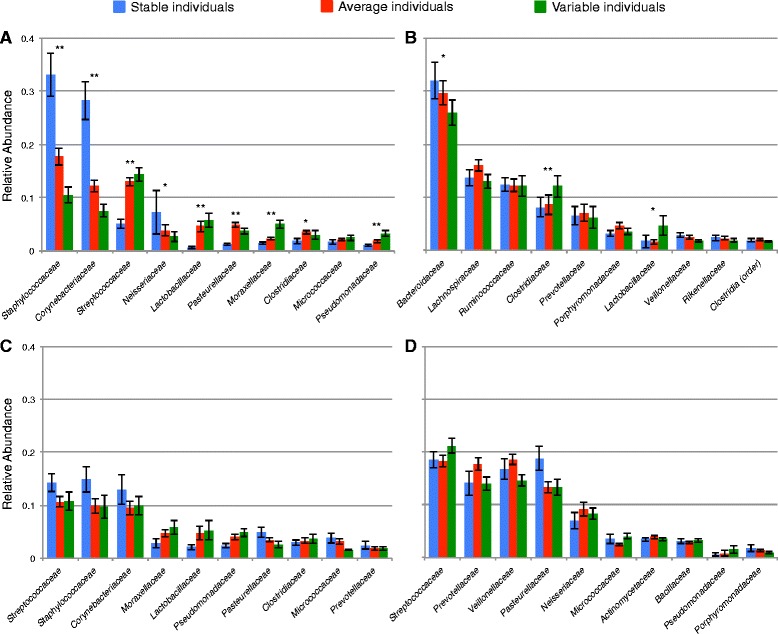
Average taxonomic composition was different among stability classes across individuals. Individuals were assigned to stability classes based on quartiles (first = stable (blue), second and third = average (red), fourth = variable (green)) of median weighted UniFrac distances for each body habitat. Significant differences were observed across forehead (A) and gut (B) communities but not in palm (C) or tongue (D) communities as determined by rank transforming the most abundant bacterial families (>1% in any group) for each body habitat and testing for differences between stability classes using the nonparametric Kruskal-Wallis analysis of variance. Significance is denoted with asterisks (* = corrected P ≤0.05, ** = corrected P ≤0.01).
Conclusions
Our findings suggest that the high degree of temporal variability in alpha diversity levels, community membership, and community structure observed across the sampled body habitats and across study participants is important to consider when designing studies to assess linkages between the human microbiome and health. Although the variability in community composition among healthy individuals exceeds the variability within individuals over time (Additional file 6, [3,5,8]), the intra-individual temporal variability is considerable and the degree of variability that an individual experiences over time may be a factor in determining disease state or differential treatment success. Further, because variability through time can be high, samples collected at one point in time may not adequately characterize an individual's microbiome, even if focusing on only the more abundant phylotypes (Figure 1b, dark shades). If the effect size of a change in disease state on the human microbiome is sufficiently large (for example, the loss of a major lineage), this intra-individual temporal variability may be irrelevant. However, if changes in disease state are associated with more subtle shifts in microbial community composition, it would be important to control for this temporal variability before one could establish causal linkages between changes in the microbiome and changes in health status. It is now well established that there is considerable inter-individual variability in the composition of the human microbiome [5,18], leading to the concept of a ‘personal microbiome’, and we are beginning to establish causal relationships between composition of the microbiome and disease [28]. Here we show that there is also a high-degree of inter-individual variability in the stability of the human gut, tongue, forehead, and palm microbiome. As a result, we suggest that the ‘personal microbiome’ concept should be extended to incorporate the rate of change of an individual’s microbiome, in addition to its composition (a feature which distinguishes the ‘personal microbiome’ from the ‘personal genome’) and that future investigations into associations between features of the microbiome and host phenotype may want to consider temporal variability as a potential explanatory factor.
Methods
Subject recruitment and sample collection
Volunteers were recruited from three Universities (University of Colorado, Boulder (UCB), Northern Arizona University (NAU), and North Carolina State University (NCSU)) in January/February of 2012 and asked to donate weekly self-collected samples for a minimum of 10 weeks using sterile, pre-labeled, double-tipped swabs (Becton, Dickinson and Company, Sparks, MD, USA.). Participants were instructed to sample two skin habitats (foreheads and palms) and the surface of their tongue by swabbing for 10 to 15 s. Gut (fecal) samples were collected by touching cotton swabs to used toilet paper so that a small amount of fecal material was transferred to each pair of swabs. Volunteers were asked to collect samples before showering and as close to drop-off times as possible without placing samples in freezers to avoid freeze-thaw cycles. One representative at each University collected samples from students and placed them in a -20°C freezer until shipping on dry ice to the UCB where all sample processing occurred. Volunteers were also asked to provide a variety of demographic and behavioral metadata at the initiation of the project using a scantron-based survey (Additional file 1). Weekly questionnaires (Additional file 2) were also provided with sampling kits to collect information on changes in health status, medication use, and menstruation for women. At the conclusion of the study prior to publication, study participants were provided their personalized results via a password-protected website [29]. All volunteers were made aware of the nature of this project and gave written consent in accordance with protocols approved by each University's Institutional Review Board (IRB) (UCB 409.13; NAU 12.0169; NCSU 2443). Per IRB regulations, volunteers were able to drop out of the study at any time and were not required to answer any or all survey questions.
Sample processing
Samples from NAU and NCSU were shipped on dry ice to UCB at the conclusion of sampling. Upon arrival, individual swabs were linked with Personal IDs using digital barcodes and logged into an Excel worksheet. Swabs were then sorted by body habitat and the tip of one duplicate swab was aseptically cut into single wells in 2 mL 96-well deep-well plates (Axygen Inc., Union City, CA, USA). Plates were sealed with silicone Axymat sealing mats (Axygen Inc., Union City, CA, USA). Each plate contained negative control samples that included swab blanks (sterile swabs), extraction blanks (reagents), and a PCR control. Forehead, gut, and tongue plates also included positive controls that were collected from one individual at the initiation of the project and stored/shipped with samples at each university. No differences were observed in community membership or structure in positive control samples.
DNA extraction, PCR amplification, and sequencing
DNA extraction and PCR amplification of the variable region 4 (V4) of the 16S rRNA gene using Illumina adapted universal primers 515 F/806R [12,30] was conducted using the direct PCR protocol as previously described [31].
Aliquots (4 μL) from the fecal and tongue extracts were transferred into 384-well plates for triplicate PCR reactions, while skin aliquots (forehead and palm, 4 μL) were transferred into 96-well plates. PCRs were conducted in triplicate 20 μL reactions and thermal cycling conditions for the 384-well plates were: initial denaturation for 3 min at 94°C; 35 cycles (94°C, 60 s; 50°C, 60 s; 72°C, 105 s) followed by a final elongation for 10 min at 72°C. Conditions for the 96-well plates were identical except for shorter denaturation (94°C, 45 s) and elongation (72°C, 90 s) steps. PCR products from triplicate reactions of each sample were pooled, visualized on an agarose gel, and quantified using the PicoGreen dsDNA assay (Invitrogen, Carlsbad, CA, USA). Positive amplicons from each body habitat (forehead, gut, palm, and tongue) were then pooled in equimolar concentrations into composite samples that were cleaned using a single-tube MoBio Ultraclean PCR Clean-up Kit (MoBio Laboratories, Carlsbad, CA, USA). Each body habitat was sequenced on an individual lane (4 lanes total) of an Illumina HiSeq2000 instrument at the University of Colorado BioFrontiers Institute Advanced Genomics Facility.
Data processing
All data processing was performed using QIIME 1.6.0-dev unless otherwise noted. The specific processing steps were as follows. Raw fastq data were demultiplexed and quality filtered as described previously [32]. Sequences that passed quality filtering were clustered into phylotypes (Operational Taxonomic Units, OTUs) at 97% sequence identity using a uclust-based [33] closed-reference protocol against the 12_10 revision of the Greengenes database [34], where reads that did not match a sequence in the reference set at least 97% identity were excluded from subsequent analyses. The taxonomy of each phylotype was assigned as the taxonomy associated with the Greengenes sequence defining that OTU. The Greengenes phylogenetic tree was used for phylogenetic diversity calculations. A median of 49,242.0 sequences was collected per sample. After removing phylotypes appearing in negative controls at high abundance (≥0.5% across all controls) [31], all samples were rarefied to 10,000 sequences for all downstream analyses unless otherwise noted.
Potentially mislabeled samples were detected using the random forest classification approach described previously [35]. Briefly, the full sample-by-phylotype abundance matrix (that is, OTU table) was filtered to exclude phylotypes that were observed in fewer than 10 samples. The OTU table was then randomly subsampled to exactly 1,000 sequences per sample. Three samples achieved a probability of being mislabeled greater than 90%, and were excluded from all downstream analyses.
Alpha diversity metrics (phylogenetic diversity (PD), phylotype richness, and Shannon Index [13]) were computed as implemented in QIIME. Comparisons of alpha diversity presented in this study are computed at exactly 10,000 sequences per sample. Beta diversity was computed using the weighted and unweighted UniFrac metrics [16] at exactly 10,000 sequences per sample.
The time series samples were defined as the set of samples that came from an individual’s body site where at least seven samples were collected and successfully sequenced from that individual’s body site over a 10-week sampling period. For example, if six fecal samples and seven forehead samples were sequenced from an individual, their fecal samples would not be included in any time series analyses, but their forehead samples would be included. This resulted in 80 individuals for which we had a forehead time series (48 women, 30 men, 2 unknown), 75 individuals with a gut time series (48 women, 26 men, 1 unknown), 61 individuals with a palm time series (35 women, 25 men, 1 unknown), and 80 individuals with a tongue time series (50 women, 28 men, 2 unknown) from 85 subjects (Table 1).
All QIIME commands for performing these processing steps can be found in Additional file 12.
Statistical analysis
To assess the temporal variability of within sample diversity (alpha diversity), we calculated the coefficient of variation (CV) for three diversity metrics (phylogenetic diversity - PD, OTU richness, and Shannon index) for each body habitat of each individual through time. Individual values were used to determine the per body site median across the study population, with higher values indicative of more variable communities.
Variability in community composition (beta diversity) was determined per body habitat by calculating the median weighted and unweighted UniFrac distances for each individual over time. With this metric, communities with a higher median value are more variable whereas a lower value indicates more stable communities. (Note that because we summarize temporal data in a single measurement, we do not need to account for lack of independence of temporal samples from a single individual in evaluations based on this metric.) Differences across body sites for both alpha- and beta-diversity were assessed using the non-parametric Kruskal-Wallis one-way analysis of variance with pairwise comparisons made using the Mann-Whitney U-test, as implemented in R.
To determine the number of phylotypes shared by an individual over different windows of time, we converted the OTU tables of each body habitat to a presence/absence matrix, split it by individual, filtered out singletons, and determined the number of OTUs found in exactly two samples, three samples, four samples, and so on up to seven samples using a custom R script. Samples did not have to be from consecutive weeks. We repeated this analysis on only the top 10% most abundant OTUs per individual. The numbers of phylotypes shared per individual were then averaged across individuals for each window of time and each body habitat.
For each body habitat, the study population was divided into quartiles based on median intra-individual UniFrac values where the first quartile was defined as ‘stable’, the second and third quartiles as ‘average’, and the fourth quartile as ‘variable.’ To determine if certain taxa were more or less abundant in the different quartiles (that is, stability classes), we rank transformed the most abundant bacterial families (>1% across individuals) for each body habitat and tested for differences between the groups using the nonparametric Kruskal-Wallis analysis of variance.
adonis [36], ANOSIM [37], and PERMDISP [36] (using 999 permutations) were used to test for differences in community composition between individuals at each body site. The statistical methods were used to analyze both weighted and unweighted UniFrac distance matrices, with only the time series samples being included in the analyses.
To determine the affect of antibiotic use on community variability, we grouped individuals based on their usage (yes or no) and used the non-parametric Mann-Whitney U-test to test for differences between the two groups. Spearman rank correlations were used to determine if community variability as measured using median UniFrac distances was correlated across pairs of body habitats. To assess if patterns in community composition could be related to time between sampling events, Mantel tests (Spearman-rank correlations on 999 permutations) were conducted for each individual using both weighted and unweighted UniFrac values and Manhattan time-distance matrices calculated in R using the VEGAN package [35]. Using the mean of the different alpha diversity metrics (PD, phylotype richness, and Shannon index [13]) as our metrics of diversity and median UniFrac distances (both weighted and unweighted) as our metric of community variability on a per individual basis, we constructed linear models for each body habitat across individuals to examine the relationship between diversity and stability.
We identified key predictors of the variability in composition of bacterial assemblages using generalized linear models. We used a model simplification procedure, removing non-significant terms (α =0.05) in a stepwise fashion [38], to explore the relative contributions of the various terms included in the start model. Model simplification approaches have been criticized [39], but in the absence of strong a priori information on the drivers of variability of bacterial assemblages, this approach is a useful first step [40]. The final models we present are those that exclusively include variables that explain significant variation in our dependent variables. We also used model simplification in which final models were those in which Bayesian information criterion (BIC) was minimized. However, these ‘best’ models ended up including all variables we tested and so here we focus on those variables with significant explanatory power.
To determine if the weeks where individuals reported taking antibiotics were the weeks where they experience the largest changes in their gut community compositions, we ran per-body-site one-tailed, rank-based Monte Carlo t-tests. The adjacent-week UniFrac distances were compiled for each individual on a per-body-site basis (that is, the distance between their gut samples on week 1 and week 2, week 2 and week 3, and so on). Each individual’s UniFrac distances were ranked from smallest to largest, and assigned their rank value. Across individual ranks were grouped into distributions based on whether they occurred in a week where the individual reported taking antibiotics or not. Those distributions were then compared with a one-tailed Monte Carlo t-test with 1,000 iterations.
Data availability
Sequence data and accompanying de-identified metadata have been deposited in the EMBL under accession number (ERP005150-ERP005153).
Acknowledgments
We thank MJ Gebert, G Humphrey, and CL Lauber for logistical support throughout the project; N Rountree for sample collection; A Shade for providing custom R scripts; G Ackermann for IRB assistance; and M Watwood, E Schwartz, and J Wilder for help recruiting study participants. Computational support was provided by an Amazon Web Services research grant to JGC and RK Support for YVB, AG, and RK came from the National Institutes of Health, the Crohns and Colitis Foundation of America, and the Howard Hughes Medical Institute. RRD was supported by grant #52006933 to NC State University from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Program (http://www.hhmi.org/grants/office/undergrad/).
Abbreviations
- BIC
Bayesian information criterion
- BMI
body mass index
- CA
California
- CV
coefficient of variation
- dsDNA
double-stranded deoxyribonucleic acid
- GLM
generalized linear model
- IRB
Institutional Review Board
- MD
Maryland
- mL
milliliter
- NAU
Northern Arizona University
- NCSU
North Carolina State University
- OTU
operational taxonomic unit
- PCR
polymerase chain reaction
- PD
phylogenetic diversity
- QIIME
Quantitative Insights Into Microbial Ecology
- rRNA
ribosomal ribonucleic acid
- sec
seconds
- UCB
University of Colorado, Boulder
- μL
microliter
- U.S.
United States
- USA
United States of America
- V4
variable region 4
Additional files
The pre-study questionnaire used to collect demographic, lifestyle, and hygiene data on study participants.
The weekly questionnaire used to collect information about changes in health status, medication use, stage of menstrual cycle for women, and any other dramatic changes in the routine of study participants.
A list of all samples collected in this study with corresponding de-identified personal IDs of study subjects and answers to survey questions.
A figure showing the amount of microbial diversity observed in each sample.
A figure depicting the temporal variability observed in microbial community membership and structure for each body habitat of each individual.
A table showing that the composition of each individual’s microbiome is personalized through time.
A figure showing how the microbial communities of selected individuals vary through time.
A table showing the results of Spearman rank correlation of community membership across different body habitats.
A table showing the results of Spearman rank correlation of community structure across different body habitats.
A table of Mantel test results correlating microbial community membership and structure with time between samples (time distance-decay).
A table of results correlating microbial diversity with temporal variability in community membership and structure for each body habitat.
A list of all QIIME commands used in data processing.
Footnotes
Gilberto E Flores and J Gregory Caporaso contributed equally.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GEF conceived of and designed the study, collected samples, analyzed and interpreted the data, and wrote the paper. JGC conceived of and designed the study, analyzed and interpreted the data, and wrote the paper. JBH conducted all wet lab work. JRR analyzed and interpreted the data. DD collected samples and analyzed and interpreted the data. JC analyzed and interpreted the data. JWL analyzed and interpreted the data. YVB analyzed and interpreted the data. AG analyzed and interpreted the data. RK conceived of and designed the study, analyzed and interpreted the data, and wrote the paper. RRD conceived of and designed the study, analyzed and interpreted the data, and wrote the paper. NF conceived of and designed the study, analyzed and interpreted the data, and wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Gilberto E Flores, Email: gilberto.flores@csun.edu.
J Gregory Caporaso, Email: gregcaporaso@gmail.com.
Jessica B Henley, Email: jessicabhenley@gmail.com.
Jai Ram Rideout, Email: jai.rideout@gmail.com.
Daniel Domogala, Email: ddd38@nau.edu.
John Chase, Email: chase.john@gmail.com.
Jonathan W Leff, Email: leff.jonathan@gmail.com.
Yoshiki Vázquez-Baeza, Email: yoshiki89@gmail.com.
Antonio Gonzalez, Email: antgonza@gmail.com.
Rob Knight, Email: rob.knight@colorado.edu.
Robert R Dunn, Email: rroberdeaudunn@gmail.com.
Noah Fierer, Email: noahfierer@gmail.com.
References
- 1.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Munzy DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camarinha-Silva A, Jauregui R, Pieper DH, Wos-Oxley ML. The temporal dynamics of bacterial communities across human anterior nares. Environ Microbiol Rep. 2012;4:126–132. doi: 10.1111/j.1758-2229.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 8.Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra152. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Murray PR, Segre JA. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magurran A. Measuring Biological Diversity. Oxford: Blackwell Publishing; 2004. [Google Scholar]
- 14.Shade A, Caporaso JG, Handelsman J, Knight R, Fierer N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 2013;7:1493–1506. doi: 10.1038/ismej.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukami T. Community assembly along a species pool gradient: implications for multiple-scale patterns of species diversity. Popul Ecol. 2004;46:137–147. doi: 10.1007/s10144-004-0182-z. [DOI] [Google Scholar]
- 16.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. Forensic identification using skin bacterial communities. Proc Natl Acad Sci U S A. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White PE, Adler BP, Lauenroth KW, Gill AR, Greenberg D, Kaufman MD, Rassweiler A, Rusak AJ, Smith DM, Steinbeck RJ, Waide RB, Yao J. A comparison of the species–time relationship across ecosystems and taxonomic groups. Oikos. 2006;112:185–195. doi: 10.1111/j.0030-1299.2006.14223.x. [DOI] [Google Scholar]
- 20.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilman D. The ecological consequences of changes in biodiversity: A search for general principles. Ecology. 1999;80:1455–1474. [Google Scholar]
- 23.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jorgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tim S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 24.Prince T, McBain AJ, O’Neill CA. Lactobacillus reuteri protects epidermal keratinocytes from Staphylococcus aureus-induced cell death by competitive exclusion. Appl Environ Microbiol. 2012;78:5119–5126. doi: 10.1128/AEM.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarate G, Nader-Macias ME. Influence of probiotic vaginal lactobacilli on in vitro adhesion of urogenital pathogens to vaginal epithelial cells. Lett Appl Microbiol. 2006;43:174–180. doi: 10.1111/j.1472-765X.2006.01934.x. [DOI] [PubMed] [Google Scholar]
- 26.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 27.Bervoets L, Van Hoorenbeeck K, Kortleven I, Van Noten C, Hens N, Vael C, Goossens H, Desager KN, Vankerckhoven V. Differences in gut microbiota composition between obese and lean children: a cross-sectional study. Gut Pathogens. 2013;5:10. doi: 10.1186/1757-4749-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.My Microbes, Personalized Microbiome Results of the Student Microbiome Project. [http://my-microbes.qiime.org]
- 30.Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flores GE, Henley JB, Fierer N. A direct PCR approach to accelerate analyses of human-associated microbial communities. PLoS One. 2012;7:e44563. doi: 10.1371/journal.pone.0044563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knights D, Kuczynski J, Koren O, Ley RE, Field D, Knight R, DeSantis TZ, Kelley ST. Supervised classification of microbiota mitigates mislabeling errors. ISME J. 2011;5:570–573. doi: 10.1038/ismej.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. R package version 20–5. Wien: Institute for Statistics and Mathematics; 2012. vegan: Community Ecology Package. [Google Scholar]
- 37.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 38.Crawley MJ. Statistical Computing: An Introduction to Data Analysis Using S-Plus. Chichester: John Wiley & Sons, Ltd.; 2002. [Google Scholar]
- 39.Whittingham MJ, Stephens PA, Bradbury RB, Freckleton RP. Why do we still use stepwise modelling in ecology and behaviour? J Anim Ecol. 2006;75:1182–1189. doi: 10.1111/j.1365-2656.2006.01141.x. [DOI] [PubMed] [Google Scholar]
- 40.Anderson DR. Model Based Inference in the Life Sciences: A Primer on Evidence. New York: Springer Science+ Business Media, LLC; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data and accompanying de-identified metadata have been deposited in the EMBL under accession number (ERP005150-ERP005153).


