Abstract
Somatic embryogenesis receptor kinase (SERK) proteins play pivotal roles in regulation of plant development and immunity. The rice genome contains two SERK genes, OsSerk1 and OsSerk2. We previously demonstrated that OsSerk2 is required for rice Xa21-mediated resistance to Xanthomonas oryzae pv. oryzae (Xoo) and for normal development. Here we report the molecular characterization of OsSerk1. Overexpression of OsSerk1 results in a semi-dwarf phenotype whereas silencing of OsSerk1 results in a reduced angle of the lamina joint. OsSerk1 is not required for rice resistance to Xoo or Magnaporthe oryzae (M. oryzae). Overexpression of OsSerk1 in OsSerk2-silenced lines complements phenotypes associated with brassinosteroid (BR) signaling defects, but not the disease resistance phenotype mediated by Xa21. In yeast, OsSERK1 interacts with itself forming homodimers, and also interacts with the kinase domains of OsSERK2 and BRI1, respectively. OsSERK1 is a functional protein kinase capable of auto-phosphorylation in vitro. We conclude that, whereas OsSERK2 regulates both rice development and immunity, OsSERK1 functions in rice development but not immunity to Xoo and M. oryzae.
Keywords: Oryza sativa, Somatic embryogenesis receptor kinase, OsSERK1, Xanthomonas oryzae pv. Oryzae, Magnaporthe oryzae
INTRODUCTION
Somatic embryogenesis receptor kinase (SERK) proteins were initially identified because of their roles in the transition from somatic cells to embryogenic cells (Schmidt et al. 1997; Li 2010). In Arabidopsis thaliana, five SERK protein members (AtSERKs) have been reported (Schmidt et al. 1997; Li 2010). SERK proteins typically include five extracellular leucine-rich repeats (LRRs), a proline-rich region, a single-pass transmembrane domain, and a cytoplasmic kinase domain transducing extracellular signals to intracellular processes via protein phosphorylation (Hecht et al. 2001; Li 2010).
Apart from the role in plant regeneration from somatic embryos, AtSERK genes are better known for their functions in regulating plant development and immunity (Chinchilla et al. 2009; Roux et al. 2011; Gou et al. 2012). AtSERK3 (At4g33430) was independently identified as a brassinosteroid insensitive 1 (BRI1)-associated kinase (BAK1) because of its role in mediating brassinosteroid (BR) signal transduction (Li et al. 2002; Nam and Li 2002). Atserk3/bak1 mutants display certain degrees of bri1 symptoms whereas over-expression of AtSERK3/BAK1 complements bri1-5 dwarf phenotype (Gou et al. 2012). AtSERK3/BAK1 was later found to be required for immunity triggered by pathogen-associated molecular patterns (PAMPs), such as bacterial flagellin and elongation factor Tu (EF-Tu) (Chinchilla et al. 2007; Heese et al. 2007; Schwessinger et al. 2011; Roux et al. 2011). AtSERK3/BAK1 physically associates with the Arabidopsis pattern recognition receptors (PRRs) Flagellin Sensitive 2 (FLS2) and EF-Tu receptor (EFR) (Chinchilla et al. 2007; Heese et al. 2007; Roux et al. 2011; Schwessinger and Ronald 2012). The AtSERK3/BAK1 and FLS2 ectodomains form heterodimeric complexes and both directly interact with flg22 (Sun et al. 2013). Similarly the ectodomains of AtSERK3/BAK1 and BRI1 interact with BL as part of a heterodimeric complex (Wang and Chory, 2006; Santiago et al. 2013; Sun et al. 2013). Thus, AtSERK3/BAK1 functions as a co-receptor for receptor kinases BRI1, FLS2, and EFR and plays a pivotal role in regulating both plant development and immunity (Santiago et al. 2013; Chinchilla et al. 2007; Sun et al. 2013; Schwessinger and Ronald 2012). Subsequent studies have identified redundant roles among AtSERK members. For example, AtSERK4 (At2g13790) functions similarly as AtSERK3/BAK1 (BKK1, BAK1-like 1). Both are required for perception of PAMPs and for BR signaling (He et al. 2007; Roux et al. 2011). The AtSERK1 (At1g71830) ortholog in tomato is required for immune receptor Ve1-mediated resistance to race 1 of Verticillium dahlia (Fradin et al. 2011). Transfer of tomato Ve1 into Arabidopsis revealed that AtSERK1 is required in addition to AtSERK3/BAK1 for Ve1-mediated resistance (Fradin et al. 2011). Over-expression ofAtSERK1, AtSERK2, or AtSERK4/BKK1 suppressed the bri1-5 phenotype (Gou et al. 2012). AtSERK5 (At2g13790) from the Arabidopsis Columbia ecotype is nonfunctional. Recent studies of AtSERK members have revealed the molecular mechanisms underlying the contributions of AtSERKs to plant development and immunity (Gou et al. 2012; Schwessinger and Ronald 2012)
In contrast to the five SERK members in Arabidopsis, the rice genome contains only two genes encoding predicted SERK proteins (OsSERK1/Loc_Os08g07760 and OsSERK2/Loc_Os04g38480) (~76% identity to AtSERK proteins) (Singla et al. 2009; Chen et al. 2014). OsSERK1 and OsSERK2 are clustered in the same group as AtSERK1 and AtSERK2, but not with AtSERK3/BAK1 and AtSERK4/BKK1 (Chen et al. 2014). Because of the high degree of similarity of OsSERK1 an OsSERK2 with all the AtSERK proteins, it had been difficult to identify the rice equivalent of AtSERK3/BAK1. In fact, Li et al. (2009) hypothesized that OsSERK1 serves as OsBAK1, mainly based on the ability of OsSERK1 to restore the dwarf phenotype of the Arabidopsis bri1-5 mutant. Down-regulation experiments of OsSerk2 (named OsSERK1 in Hu et al. 2005) expression showed that OsSerk2 was involved in embryogenic cell formation and in plant development; overexpression of OsSerk2 increased rice resistance to the hemi-necrotrophic fungus Magnaporthe oryzae (M. orzyae), the causal agent of the rice blast disease. Simultaneous silencing of OsSerk1, OsSerk2 and other OsSERK-like genes enhanced rice susceptibility to M. orzyae (Hu et al. 2005; Park et al. 2011). These experiments indicated the involvement of OsSERK2 in resistance to M. oryzae, but did not specifically address the role of OsSERK1. In Arabidopsis, AtSERK genes are mainly associated with plant immunity to biotrophic pathogens although they are also involved in regulation of host resistance to hemi-necrotrophic and necrotrophic pathogens (Kemmerling et al. 2007; Roux et al. 2011). Recently, we reported that down-regulation of OsSerk2 expression almost completely abolished immunity mediated by XA21 and XA26, two rice PRRs (pattern recognition receptors) (Chen et al. 2014). Both XA21 and XA26 are phylogenetically closely related to Arabidopsis FLS2 and EFR and belong to the same LRR-RLK subfamily XII (Chen et al. 2014). OsSERK2 functions as a regulatory co-receptor kinase of XA21 and also regulates BR-mediated signaling. Thus, OsSERK2 possesses dual roles in rice development and in PRR-mediated immunity (Chen et al. 2014).
Compared with OsSERK2, OsSERK1 has slightly higher identity to AtSERK3/BAK1 (Chen et al. 2014). It is unknown if OsSERK1 contributes to rice immunity. In this study, we show that like OsSERK2, OsSERK1 functions as rice development, but unlike OsSERK2, OsSERK1 is not required for rice XA21-mediated immunity and does not contribute to resistance to Xoo and M. oryzae in the absence of XA21. We also found that specific silencing of OsSerk1 results in reduction of the angle of the lamina joint, but not affect other agronomic traits, such as leaf length and width, plant height, and seed set.
RESULTS
Overexpression of OsSerk1 results in a semi-dwarf phenotype
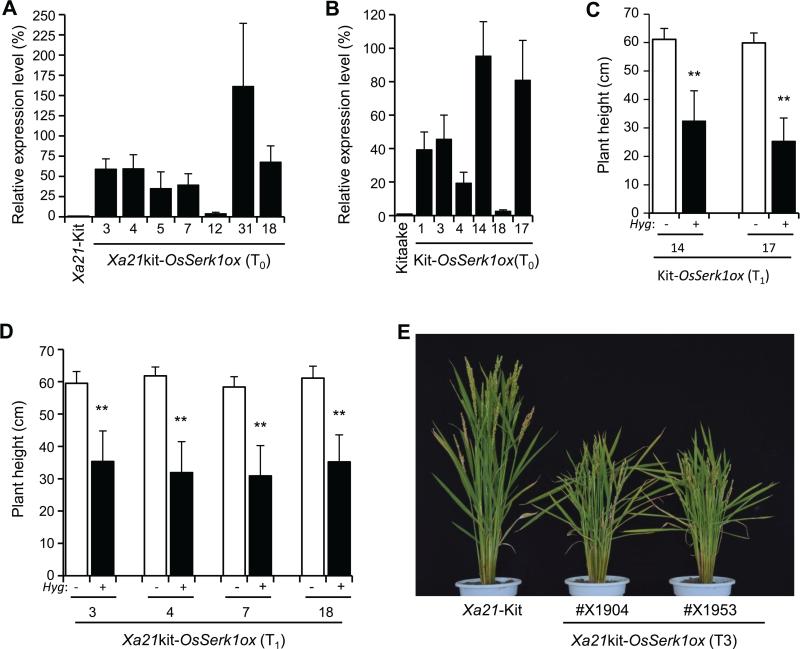
To investigate the function of OsSerk1, we isolated the full-length coding region of OsSerk1 and created an overexpression construct UbiC1300-OsSerk1 by using the maize ubiquitin 1 promoter to drive OsSerk1 expression. Using Agrobacterium-mediated transformation, we obtained 18 independent transgenicplants in the rice Kitaake genetic background (called Kit-OsSerk1ox) and 30 in the Xa21-Kitaake background (called Xa21kit-OsSerk1ox) (Figure S1). Nearly all T0 transgenic plants displayed semi-dwarf phenotypes compared to the wild type Kitaake control (Figure S1). The only exceptions were transgenic plants that did not overexpress OsSerk1. Two Kit-OsSerk1ox (#14 and #17) and four Xa21kit-OsSerk1ox (#3, #4, #7 and #18) T0 plants with high transcript levels of OsSerk1 were self-pollinated and used to analyze the correlation between the semi-dwarf phenotype and the transgene OsSerk1ox (Figure 1A, B). All plants carrying the OsSerk1ox transgene displayed significantly shorter than those lacking the OsSerk1ox transgene (Figure1C, D), suggesting that overexpression of OsSerk1 leads to the semi-dwarf phenotype. The semi-dwarf phenotype of homozygous OsSerk1ox plants included reduction of each internode length and increase of the angle of the lamina joint, compared with the control Xa21-Kitaake but did not affect seed size (Figure 1E, S2). These results suggest that OsSerk1 controls rice plant stature and the angle of the lamina joint.
Figure1. Identification of OsSerk1 overexpression transgenic plants.
(A) Transcript levels of OsSerk1 among wild-type control Xa21-Kitaake (Xa21-Kit) and independent Xa21kit-OsSerk1ox transgenic plants revealed by real-time RT-PCR.
(B) Transcript levels of OsSerk1 among the wild type Kitaake and independent Kit-OsSerk1ox T0 transgenic plants revealed by real-time RT-PCR. (C) and (D) Plant height of the transgenic T1 plants with or without the transgene OsSerk1ox. The primer pair (Hyg) specific to the hygromycin phosphotransferase gene was used to determine the plants with (represented by ‘+’) or without (represented by ‘-’) OsSerk1ox. Statistical significance comparison was conducted with ANOVA, where the mark ‘**’ on the column indicates difference with . (E) Stature of mature plants of wild-type Xa21 Kitaake, Xa21kit-OsSerk1ox #X1904 and #X1953. The #X1904 and #X1953 were the lines homozygous for OsSerk1ox that derived from Xa21kit-OsSerk1ox #3 and Xa21kit-OsSerkox #18 T0lines, respectively.
Overexpression of OsSerk1 does not affect rice resistance to Xoo
To test whether the overexpression of OsSerk1 enhanced rice resistance to the biotrophic pathogen Xoo, we inoculated Kit-OsSerk1ox T1 plants with Xoo strain PXO99 at two developmental stages (three or six weeks old). All progeny plants with or without the transgene displayed similar disease lesion lengths as the Kitaake control at both developmental stages (Figure S3A, B). Because Xa21 only shows partial resistance at the juvenile stage, we also inoculated Xa21kit-OsSerk1ox three week old plants to assess if overexpression of OsSerk1 could enhance Xa21 resistance at the seedling stage (Song et al. 1995; Park et al. 2010). We found no clear differences in lesion lengths among the plants with and without OsSerk1ox and the Xa21-Kitaake control (Figure S3B).
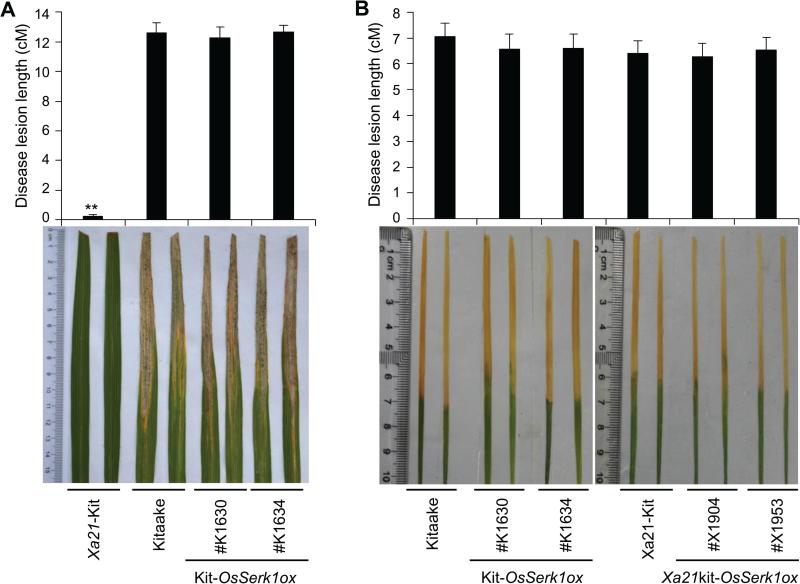
We further confirmed these results using lines homozygous for OsSerk1ox. Two Kit-OsSerk1ox lines, #K1630 and #K1634, and Kitaake were inoculated at the three and six weeks-old stages. We found that these plants displayed similar lesion lengths as the susceptible control Kitaake (Figure 2A, B). Similarly, the Xa21kit-OsSerk1ox lines homozygous for both Xa21 and OsSerk1ox,#X1904 and #X1953, displayed similar lesion lengths as the Xa21-Kitaake control at the seedling stage (Figure 2B). We also inoculated the two Xa21kit-OsSerk1ox lines and the Xa21-Kitaake control with the Xoo-4 strain, which is unable to activate the XA21-mediated immune response (Figure S4). As expected, the Xa21-Kitaake showed full susceptibility to Xoo-4. The Xa21kit-OsSerk1ox lines showed similar susceptibility to Xoo-4 as Xa21 Kitaake. Taken together, we conclude that overexpression of OsSerk1 does not affect rice resistance to Xoo.
Figure2. OsSerk1 overexpression does not affect rice basal resistance to Xoo.
(A) Lesion length of Xa21 Kitaake, Kitaake, and Kit-OsSerk1ox #K1603 and #K1634 plants inoculated with Xoo at the adult stage (six weeks old). The lines #K1603 and #K1634 were homozygous for OsSerk1ox in Kitaake background, which derived from independent T0 plants Kit-OsSerk1ox #14 and Kit-OsSerk1ox #17, respectively. All plants were inoculated with the PXO99 Xoo strain. Lesion lengths were measured at 15 days after inoculation (DAI) from 10 independent plants. Photographs depict representative symptom development in leaves at 15 DAI. (B) Lesion length of Xa21 Kitaake, Kitaake, Xa21kit-OsSerk1ox #X1904 and #X1953 plants inoculated at the seedling stage (three weeks old). All plants were inoculated with PXO99 at three weeks old. Lesion lengths were measured at 10 DAI from 10 independent plants. Photographs depict representative symptom development in leaves at 10DAI. Statistical significance comparison was conducted with ANOVA, where the mark‘**’ on the column indicates difference with .
Silencing of OsSerk1 mainly affects the angle of the lamina joint
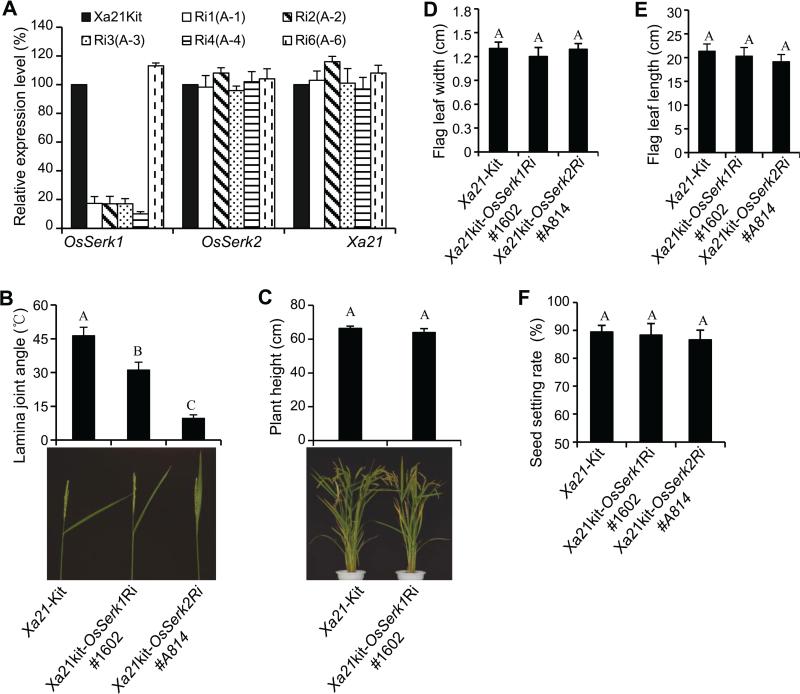
To further clarify the function of OsSerk1, we generated an RNAi constructpANDA-OsSerk1Ri and transformed it into the Xa21-Kitaake genetic background through Agrobacteria-mediated transformation. We obtained five independent Xa21kit-OsSerk1Ri plants. Through real-time RT-PCR, we found that the OsSerk1 transcript levels were significantly reduced in four of the five Xa21kit-OsSerk1Ri lines; while the OsSerk2 and Xa21 transcript levels in these lines showed no changes compared to the wild type Xa21-Kitaake used as the control (Figure 3A). This indicates that the four Xa21kit-OsSerk1Ri plants have specific down-regulation of OsSerk1 expression.
Figure3. Identification of Xa21kit-OsSerk1Ri transgenic lines with reduced expression specific to OsSerk1.
(A) Transcript levels of OsSerk1, OsSerk2, and Xa21 in wild-type Xa21-Kitaake and five independent Xa21kit-OsSerk1Ri lines as revealed by real-time RT-PCR. (B) Photographs and measured lamina joint angles of Xa21 Kitaake, Xa21kit-OsSerk1Ri #1602 and Xa21kit-OsSerk2Ri #A814 plants at 15 days after heading. (C) Photographs and measured plant heights of Xa21 Kitaake and Xa21kit-OsSerk1Ri #1602 at 25 days after heading. (D-F) Flag leaf width and length and seed set rates of Xa21-Kitaake and Xa21kit-OsSerk1Ri #1602 plants. Statistical significance comparison was conducted with ANOVA, where the different capital letters above the column indicate differences with , whereas the same letter indicates no significant differences.
We found that all the four Xa21kit-OsSerk1Ri lines displayed reduced angles if the lamina joint compared with the wild type Xa21-Kitaake. The homozygous OsSerk1Ri line (#1602) derived from the T0 line Ri4 (A-4) that expressed the lowest OsSerk1 transcript level (Figure 3A) was used in subsequent morphological analysis. The OsSerk1Ri #1602 plants displayed significantly smaller lamina joint angles, but showed almost the same plant height and leaf width and length as the wild type Xa21-Kitaake (Figure 3B-3E). Because OsSerk1 has higher transcript levels in the rice flowers (Chen et al. 2014), we reasoned that it might regulate seed development. We measured the seed set of OsSerk1Ri #1602. We did not find significant differences between the seed sets of OsSerk1Ri #1602 and the wild type Xa21-Kitaake (Figure 3F). These results indicate that OsSerk1 is mainly involved in the development of the angle of the lamina joint but does not affect traits controlling plant stature or seed set.
Silencing of OsSerk1 does not affect XA21-mediated immunity or rice basal resistance to Xoo
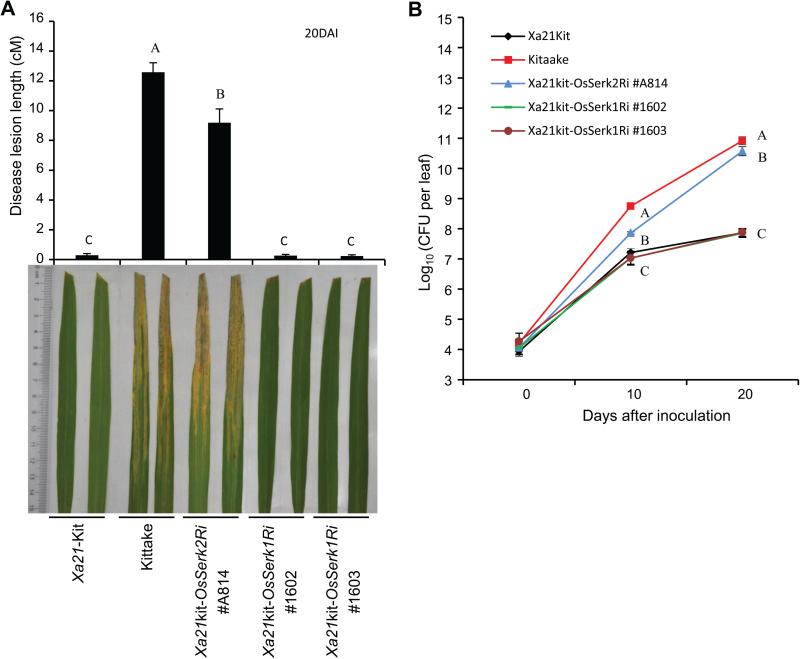
To test whether OsSerk1 is involved in XA21-mediated immunity to Xoo, we inoculated T1 plants from each of the four Xa21kit-OsSerk1Ri lines with PXO99 at six weeks old, and found all were resistant to PXO99, showing no significant differences on lesion lengths between the plants carrying or lacking the OsSerk1Ri transgene (Figure S5). To further confirm this result, we inoculated two OsSerk1Ri homozygous lines (#1602 and #1603) at six weeks old (Figure 4A). These two lines showed similar resistance levels as the Xa21-Kitaake plants. Both Xa21kit-OsSerk1Ri and Xa21-Kitaake plants had significantly shorter lesions than the Xa21kit-OsSerk2Ri #A814 and Kitaake plants. Bacterial growth curve analysis revealed that Xa21kit-OsSerk1Ri plants harbored similar Xoo bacterial populations as the Xa21-Kitaake plants at 0, 10 and 20 days after inoculation (Figure 4B).
Figure4. Silencing of OsSerk1 does not affect XA21-mediated immunity to Xoo.
(A) Disease lesion lengths of Xa21 Kitaake, Kitaake, Xa21kit-OsSerk1Ri #1602 and #1603 plants at 20 DAI. Lines #1602 and #1603, derived from independent T0 plants Ri1(A-1) and Ri4(A-4), respectively, are homozygous for the transgene OsSerk1Ri. All plants were inoculated at six weeks old in the field. Lesion lengths were measured at 20 DAI for 10 independent plants. The photograph depicts representative symptom development in leaves at 20 DAI. (B) Bacterial populations of Xa21 Kitaake, Kitaake, Xa21kit-OsSerk2Ri #A814, Xa21kit-OsSerk1Ri #1602 and #1603 lines at 0, 10 and 20 DAI. Each data point represents the average ± SD of 6 leaves from 3 independent plants. Statistical significance comparison was conducted with ANOVA, where the different capital letters above the columns and around the point indicate differences with .
To test if OsSerk1 is involved in rice basal resistance to Xoo, we inoculated the transgenic plants with the Xoo-4 strain, which is virulent on Xa21 plants. We found that the two lines (#1602 and #1603) homozygous for OsSerk1Ri showed similar susceptibility to Xoo-4 as the Xa21-Kitaake control (Figure S4), indicating that OsSerk1 is not involved in rice basal resistance to Xoo. Taken together, we conclude that unlike OsSerk2, specific silencing of OsSerk1 affects neither XA21-mediated immunity nor rice basal resistance to Xoo.
OsSerk1 is not involved in rice resistance to M. oryzae
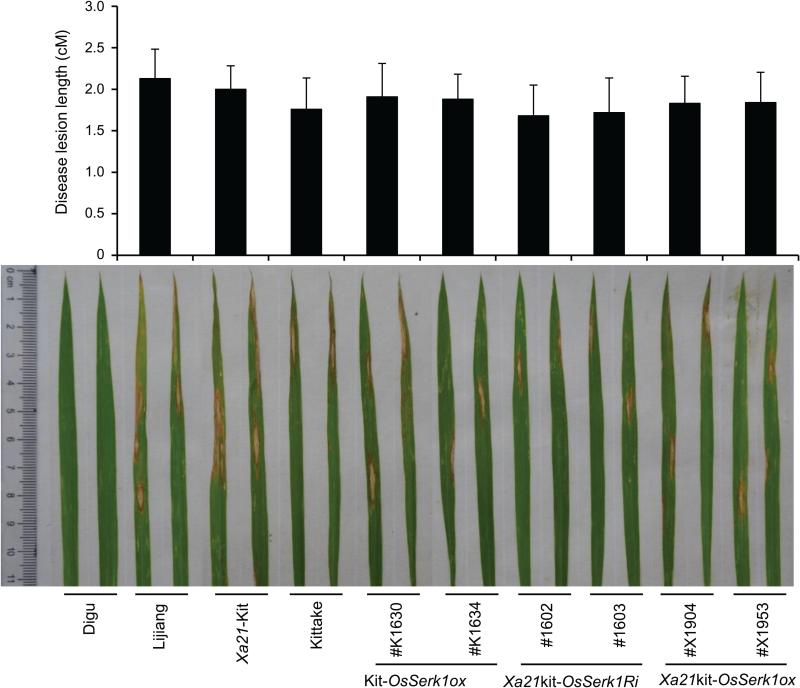
OsSerk1 does not function in rice immunity to biotrophic pathogen Xoo. We then tested whether it regulates rice resistance to hemi-necrotrophic pathogen M. oryzae. We inoculated the two OsSerk1Ri lines (#1602 and #1603), the two Xa21kit-OsSerk1ox lines (#X1904 and #X1953), and controls with M. oryzae strains, ZB13 and ZB25 (Figure 5, S6). Both Kitaake and Xa21-Kitaake are susceptible to ZB25 but resistant to ZB13. We found that all lines tested showed similar susceptibility to ZB25, except for the resistant control Digu (Figure 5). On the contrary, all lines showed resistance to ZB13, except for the susceptible control Lijiang (Figure S6). These results demonstrate that OsSerk1 does not regulate rice resistance to M. oryzae.
Figure 5. Altered expression of OsSerk1 does not affect rice resistance to Magnaporthe oryzae.
Digu and Lijiang are cultivars carrying broad-spectrum resistance and susceptibility to blast strain ZB25, respectively. Two-week old rice plants were used for inoculation with ZB25. The lesion length was measured and pictures were taken at 7 days after inoculation.
OsSerk1 cannot restore XA21-mediated immunity to Xoo in the OsSerk2 silenced line
OsSERK2 is a regulatory co-receptor kinase of XA21. Silencing of OsSerk2 in the Xa21-Kitaake genetic background severely compromises XA21-mediated immunity to Xoo strain PXO99 (Chen et al. 2014). Because OsSerk1 is expressed in rice leaves at very low levels, we tested if overexpression of OsSerk1 in the Oserk2-silenced line (Xa21kit-OsSerk2Ri #A814) would complement the mutant and restore XA21-mediated resistance. For this purpose, we generated several hybrid F1 plants by crossing three independent Xa21kit-OsSerk1ox lines (#3, #7, and #18, as pollen donor) with Xa21kit-OsSerk2Ri #A814 line (as recipient) to obtain OsSerk1oxOsSerk2Ri plants in Xa21-Kitaake background (called Xa21kit-OsSerk1oxOsSerk2Ri). F1 plants were inoculated at six weeks old. We found no significant differences in lesion length between the F1 plants carrying bothOsSerk1ox and OsSerk2Ri and those carrying only OsSerk2Ri (Figure S7A, B).
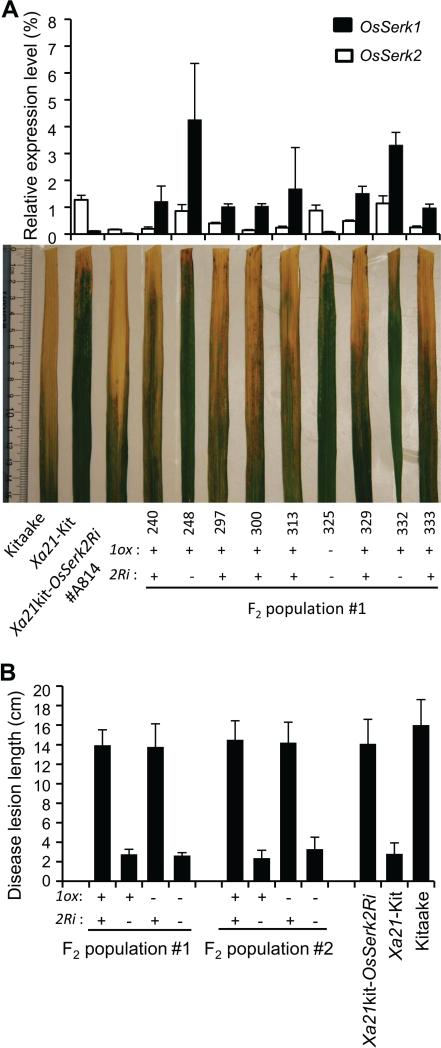
We next tested the resistance of two F2 populations, including 57 and 48 individual plants derived from the crosses of Xa21kit-OsSerk2Ri #A814/Xa21kit-OsSerk1ox #7 and #A814/Xa21kit-OsSerk1ox #18, respectively. The two transgenes (OsSerk1ox and OsSerk2Ri) segregated in this population based on the genotyping results (Figure S8A, B). Nine F2 plants with different combinations of OsSerk1ox and OsSerk2Ri were chosen to detect the expression levels of OsSerk1 and OsSerk2 by real time RT-PCR (Figure 6A). We found that OsSerk2 expression was significantly reduced in plants with the transgene OsSerk2Ri. The OsSerk1 transcription levels in the plants with only OsSerk1ox were higher than in those carrying both OsSerk2Ri and OsSerk1ox transgenes, indicating that the transgene OsSerk2Ri affects the overexpression level of OsSerk1 to a certain extent. However, even in the presence of OsSerk2Ri, the expression levels of OsSerk1 increased at least 10 fold compared with the wild type, and reached or slightly exceeded the OsSerk2 transcript levels in the wild type. This result indicates that the transcript level of OsSerk1 is strongly enhanced in the OsSerk2Ri background. All F2 plants were inoculated with PXO99 at six weeks old. We found that the Xa21kit-OsSerk1oxOsSerk2Ri plants showed similar susceptible phenotype (showing an average lesion length of 14 cm) as the Xa21kit-OsSerk2Ri plants, while the Xa21kit-OsSerk1ox plants displayed similar resistant phenotype (average lesion length 2.5 cm) as the Xa21-Kitaake control (Figure 6B, S8A-8B). These results demonstrate that overexpression of OsSerk1 is not able to complement the function of OsSerk2 in the XA21-mediated immune response. Taken together, we conclude that unlike OsSerk2, OsSerk1 is not involved in XA21-mediated immunity.
Figure 6. OsSerk1 overexpression cannot suppress the Xoo susceptibility of OsSerk2-knockdown plants in Xa21-Kitaake background.
(A) F2 population #1 was derived from the cross of Xa21kit-OsSerk2Ri #A814/Xa21kit-OsSerk1ox #18. Transcript levels of OsSerk1 and OsSerk2 revealed by real-time RT-PCR in plants carrying both transgenes OsSerk1ox and OsSerk2Ri, either of them, or none of them, which were genotyped by specific PCR primers 1ox for OsSerk1ox and 2Ri for OsSerk2Ri. The photograph depicts representative symptom development in leaves at 15 DAI. (B) Lesion lengths of Xa21kit-OsSerk2Ri #A814, Xa21 Kitaake, Kitaake, and F2 plants with both transgenes OsSerk1ox and OsSerk2Ri, either of them, or none of them. The F2 plants were derived from two F1 crosses, #A814/Xa21kit-OsSerk1ox #7 (F2 population #2) and #A814/Xkit-OsSerk1ox #18 (F2 population #1). All plants were inoculated with PXO99 at six weeks old, and lesion lengths were measured at 15 DAI for at least five leaves from three or more independent plants.
Overexpression of OsSerk1 is able to suppress the bri1-like phenotype caused by the OsSerk2 knockdown
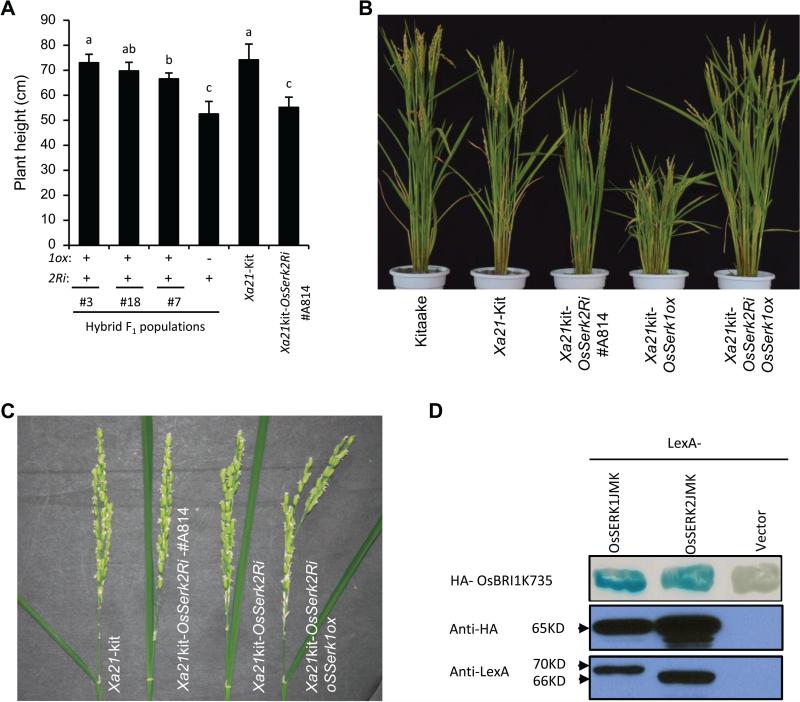
In previous studies, OsSerk2 was shown to be required for OsBRI1-mediated signaling (Hu et al. 2005; Chen et al. 2014). The Xa21kit-OsSerk2Ri #A814 plants (with reduced expression of OsSerk2) show a typical bri1-like phenotype, including erect leaves and semi-dwarfism (Chen et al. 2014). We measured the plant height of the Xa21kit-OsSerk1oxOsSerk2Ri plants to investigate whether overexpression ofOsSerk1 is able to suppress the bri1-like phenotype of Xa21kit-OsSerk2Ri. We found that all Xa21kit-OsSerk1oxOsSerk2Ri plants were significantly taller (with a range from 66.5±2.4cm to 73±3.4cm) than those carrying only OsSerk2Ri (52.5±5.1cm) and the Xa21kit-OsSerk2Ri plants (55.2±4.1cm) (Figure 7A). Compared with the Xa21-Kitaake control (74.2±6.3cm), the Xa21kit-OsSerk1oxOsSerk2Ri plants from two crosses (#A814/Xa21kit-OsSerk1ox #3 and #A814/ Xa21kit-OsSerk1ox #18) almost restored the semi-dwarf phenotype of #A814 (Figure 7A and 7B) to the normal level of Xa21-Kitaake. Furthermore, we observed that the angles of the lamina joints of all Xa21kit-OsSerk1oxOsSerk2Ri plants (ranging from 22.06±7.02° to 26.52±6.35°) increased by at least 17° compared with the angles of the Xa21kit-OsSerk2Ri (5.95±1.78°) and OsSerk2Ri plants (Table S1). However, complementation with OsSerk1 did not fully restore the lamina joint angles to the level of the wild type Xa21-Kitaake plants (38.07±10.01°) (Figure 7C; Table S1). These results indicate that OsSerk1 overexpression can suppress the semi-dwarf phenotype of the Xa21kit-OsSerk2Ri #A814 line and partially complements its erect-leaf phenotype.
Figure 7. OsSerk1 overexpression can suppress the bri1-like phenotype of OsSerk2-knockdown plants.
(A) Plant heights of F1 plants with or without the OsSerk1ox transgene and the control plants Xa21-Kitaake and Xa21kit-OsSerk2Ri #A814. Three hybrid F1 populations, generated by crossing #A814 (pollen recipient) and each of three Xa21kit-OsSerk1ox independent lines (#3, #7 and #18), were included. (B) The photograph depicts plant height of different genotypes at 20 days after heading. The Xa21kit-OsSerk2Ri and Xa21kit-OsSerk2RiOsSerk1ox plants were selected from the progeny of the #A814/Xa21kit-OsSerk1ox-#18 cross, that contain either only the OsSerk2Ri transgene or both transgenes OsSerk2Ri and OsSerk1ox (same for panel C). (C) The photograph depicts lamina joint angles of plants with different genotypes at 15 days after heading. (D) The OsSERK1 intracellular domain interacts with OsBRI1 in yeast-two hybrid system. The blue color indicates interaction between the two co-expressed proteins. The OsSERK1JMK and OsSERK2JMK were fused to the LexA tag, respectively, and OsBRI1K735 was fused to B42AD with HA tag. The expression of LexA and HA fusion proteins were confirmed by Western blot analyses using anti-LexA and anti-HA antibodies, respectively. This experiment was repeated three times with same results.
We next investigated whether OsSERK1 directly interacts with OsBRI1 by using a yeast two-hybrid assay. The truncated versions of OsSERK1 (OsSERK1JMK) and OsBRI1 (OsBRI1K735), both containing the whole intra-cellular domain and the entire juxtamembrane (JM) domain, were used as bait and prey, respectively. OsSERK2JMK was included as a positive control because it can directly interact with OsBRI1K735 in yeast (Chen et al. 2014). Indeed OsSERK1JMK and OsBRI1K735 directly interact in the yeast-two hybrid assays as indicated by the blue colony coloration specific for this combination and its absence in the respective control reactions (Figure 7D).
Taken together, we suggest that OsSerk1 encodes a similar function as OsSerk2 with regards to regulation of rice development and that this function is most likely exerted via its direct interaction with OsBRI1.
OsSERK1 interacts with itself and with OsSERK2 in vitro and is a functional protein kinase
Because OsSERK1 and OsSERK2 both can interact with OsBRI1, we tested if the two OsSERKs can directly interact with each other in the yeast two-hybrid assay (Figure S9). We found that yeast cells containing both OsSERK2JMK and OsSERK1JMK in either orientations display a light blue coloration. This indicates OsSERK1 weakly interacts with OsSERK2, suggesting they may form heterodimer in vitro. While BD-OsSERK1JMK and AD-OsSERK1JMK interact with each other in the yeast two-hybrid system, BD-OsSERK2JMK and AD-OsSERK2JMK do not (Figure S9). This indicates that OsSERK1 is capable of homodimerization in vitro, but OsSERK2 cannot.
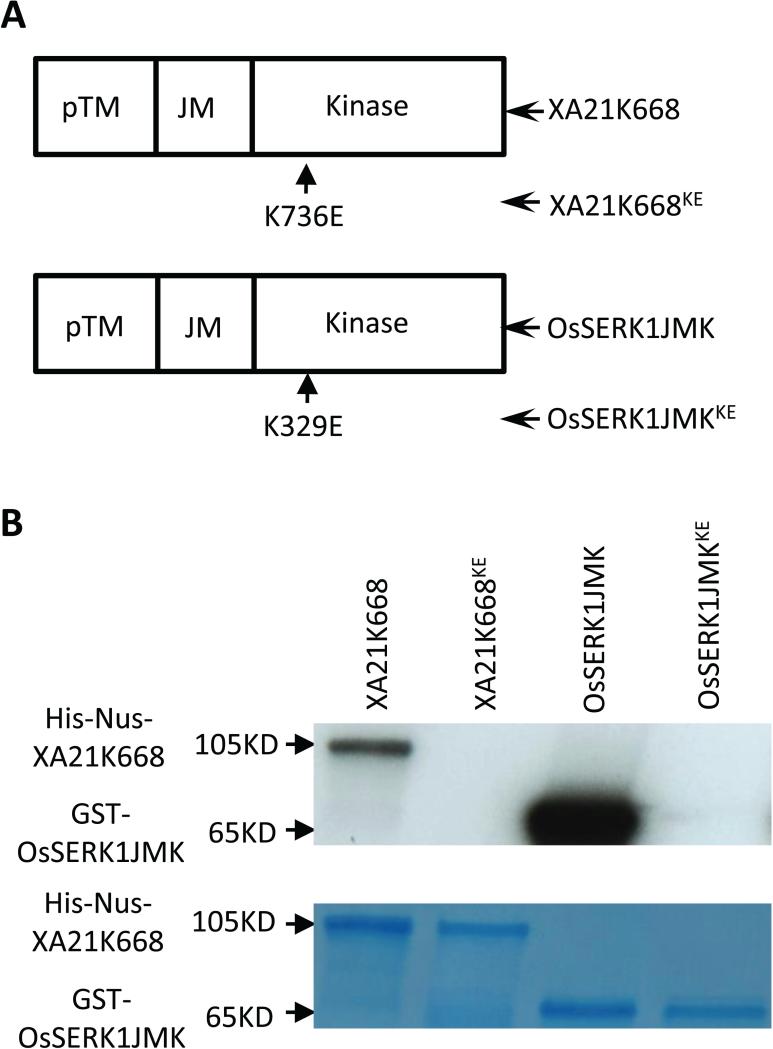
Because OsSerk1 encodes a predicted protein kinase, we next tested whether it possesses kinase activity. We expressed and purified a GST-OsSERK1JMK fusion protein and its catalytically inactive kinase variant GST-OsSERK1JMKKE, generated by mutating lysine (K) 329, which is conserved in all plant active kinase and required for ATP binding and the kinase catalytic activity, to glutamic acid (E). The E.coli-expressed XA21 kinase His-Nus-XA21K668 and its kinase inactive variant His-Nus-XA21K668KE (Chen et al. 2014) were used as the positive and negative controls, respectively, in the kinase assays. All four proteins contain the part of their transmembrane domains (TM) and full JM and kinase domains, as depicted in Figure 8A. These proteins were subjected to in vitro kinase assays using [32P]-γ-ATP. We found that the GST-OsSERK1JMK and His-Nus-XA21K668 fusion proteins were capable of auto-phosphorylation, whereas their respective kinase-inactive proteins failed to be autophosphorylated (Figure 8B). Notably, the OsSERK1 fusion protein showed much stronger kinase activity than the XA21 fusion protein (Figure 8B). We conclude that OsSERK1 is a functional protein kinase capable of auto-phosphorylation.
Figure 8. OsSERK1 is a functional protein kinase.
(A) The truncated protein of each of OsSERK1 and XA21 contains part of the transmembrane (pTM) domain, full justxamembrane (JM) and kinase domains. The mutation site was labeled under the sketch of each truncated protein. (B) The in vitro kinase assay was performed by incubating with [32P]-γ-ATP and each of the proteins, GST-OsSERK1JMK, GST-OsSERK1JMKKE, His-Nus-XA21K668, and His-Nus-XA21K668KE. Proteins were separated with SDS/PAGE and analyzed by autoradiography in the top panel and stained by Coomassie blue (CBB) in the bottom panel, respectively.
DISCUSSION
Orthologs of Arabidopsis SERK proteins in rice
In Arabidopsis, there are five SERK proteins that have evolved into two groups (Schmidt et al. 1997; Hecht et al. 2001). Group I consists of AtSERK1 and AtSERK2 that play redundant roles in regulation of plant development, while group II includes AtSERK3/BAK1 and AtSERK4/BKK1 that function redundantly in regulation of both plant immunity and development (Colcombet et al. 2005; Roux et al. 2011; Schwessinger and Ronald 2012). In contrast to the multiple SERK proteins in Arabidopsis, the cotton genome has only evolved three SERK orthologs. One of these is the ortholog of AtSERK1/SERK2 and the other two are the counterparts to AtSERK3/BAK1 (Gao et al. 2013). These studies illustrate the divergent evolution of SERK genes between species (Gao et al. 2013).
Through phylogenetic analysis, we identified two rice genes (OsSerk1 and OsSerk2) that encode proteins with typical structural characteristics of SERK proteins (Schmidt et al. 1997; Hecht et al. 2001; Chen et al. 2014). OsSERK1 and OsSERK2cluster with AtSERK1 and AtSERK2 but not AtSERK3/BAK1 and AtSERK4/BKK1 (Chen et al. 2014). OsSERK1 shows slightly higher identity (69.1%) with AtSERK3/BAK1 than OsSERK2 (61.2% identity), and can partially rescue the Arabidopsis bri1-5 mutant phenotype. For this reason, OsSERK1 was hypothesized, to be OsBAK1 by Li et al. (2009). Consistent with these observations, we found that OsSERK1 interacts with OsBRI1 (Figure 7D) and overexpression of OsSerk1 can suppress the bri1-like phenotype of transgenic OsSerk2Ri plants (Figure7A-7C). Based on these results, we hypothesize that OsSERK1 possesses a similar function in rice development as AtSERKs proteins, including AtSERK3/BAK1. Previous studies has demonstrated that simultaneous silencing of two OsSerk genes and others OsSerk-like genes increased expression levels of pathogenesis-related gene and enhance susceptibility to M. oryzae, suggesting the involvement of OsSerk or OsSerk-like genes in rice immunity (Park et al. 2011). However, these reports did not provide evidence that OsSerk1 was involved in regulation of rice immunity. Our study reveals that neither overexpression nor silencing of OsSerk1 affects rice resistance or susceptibility to M. oryzae (Figure 5, S6).
In our previous study, we showed that silencing of OsSerk2 disrupts XA21-mediated immunity to Xoo and that OsSERK2 physically associated with XA21 in vivo and served as a regulatory receptor kinase of XA21 (Chen et al. 2014). In addition, OsSerk2 also plays a pivotal role in regulating rice development through BR signaling. In summary, OsSerk2 has a dual function in rice development and immunity, similar to AtSERK3/BAK1 and AtSERK4/BKK1 in Arabidopsis (He et al. 2007; Roux et al. 2011; Gou et al. 2012). Compared with OsSerk2, OsSerk1 is expressed at much lower level in leaves (Chen et al. 2014). To investigate if OsSerk1 has similar function as OsSerk2 in regulating rice immunity to Xoo, we overexpressed OsSerk1 in Kitaake and Xa21-Kitaake genetic backgrounds and found that overexpression of OsSerk1 did not alter rice resistance to Xoo in either of the two genetic backgrounds (Figure2, S3). Altered OsSerk1 expression also did not influence the susceptibility to a Xoo strain that is able to evade XA21-mediated immunity (Figure S4). In addition, overexpression of OsSerk1 in Xa21kit-OsSerk2Ri lines could not restore the compromised XA21-mediated immunity caused by OsSerk2-silencing (Figure 6, S8). These results clearly demonstrate that OsSerk1 is not required for XA21-mediated immunity or for basal resistance to Xoo.
We found that overexpression of OsSerk1 can suppress the erect leaf and semi-dwarf phenotype resulting from OsSerk2-silencing (Figure 7). This suggests that OsSERK1 possesses a similar function as OsSERK2 in regulation of plant development. Both OsSerk1ox plants and OsSerk2Ri plants displayed semi-dwarf phenotype, while their hybrid F1 plants (harboring both OsSerk1ox and OsSerk2Ri transgenes) regained plant height similar to the wild type (Figure7A, B). In wild type plants, OsSerk1 is expressed at significantly lower levels than OsSerk2 in leaf tissues. In OsSerk1oxOsSerk2Ri lines, the OsSerk1 expression level reaches a level very similar to the OsSerk2 level in the wild type plants (Figure 6A). This may explain why OsSerk1ox can complement the semi-dwarf phenotype in OsSerk2Ri plants. To develop normal plant height, the optimum OsSERK protein (including both OsSERK1 and OsSERK2) level may be critical in order to properly regulate BRI1 function and maintain BR-signaling. This hypothesis is consistent with the observation that both OsSERK1 and OsSERK2 are able to directly interact with OsBRI1 in the yeast-two hybrid assay, suggesting that OsSERK1 and OsSERK2 may function interchangeably in modulation of BR signaling (Figure 7D). In the event that this optimum OsSERK protein level is shifted, either higher or lower, plants become dwarf or semi-dwarf due to inappropriate BR-signaling. Consequently, we conclude that the function of OsSERK1 in rice closely resembles the role of AtSERK1 and AtSERK2 in Arabidopsis, which function mainly in development, while OsSERK2 appears to be the true functional ortholog of AtSERK3/BAK1 and AtSERK4/BKK1, playing a major role in both immunity and development.
Potential use of OsSerk1 in developing rice varieties with improved plant architecture
Plant architecture is a major factor affecting grain yield (Reinhardt and Kuhlemeier 2002; Jiao et al. 2010). Yield-related plant architecture includes plant height, tillering pattern, and leaf angle (Yang et al. 2008; Zhang et al. 2012). By using the semi-dwarf gene sd-1, the rice yield has experienced a remarkable increase, which was known as “The Green Revolution” (Spielmeyer et al. 2002). Recently, the rice ideotype approach has been used in breeding programs at the International Rice Research Institute (IRRI) and in China to further improve rice yield potential (Peng et al. 2008; Sharma et al. 2013). One of the most important characters for rice ideotype is erect leaves (or small leaf angles) (Peng et al. 2008; Jiao et al. 2010; Zhang et al. 2012), which can improve light penetration and canopy net photosynthesis rate and ultimately improve grain-yield (Zhang et al. 2012; Sharma et al. 2013). In addition, researchers may also increase yield by increasing the density of plants with erect leaves in the field (Sakamoto 2006). In the present study, we found that the transgenic plants carrying OsSerk1Ri exhibited reduced lamina joint angles (Figure 3B). Notably, these plants do not show any obvious difference in other agronomic traits, grain-yield associated components, or resistance to Xoo compared with the parental Xa21-Kitaake plants (Figure 3C-3F, 4). These results indicate that down-regulating the expression of OsSerk1 is able to improve the plant architecture without observable negative effects, which are consistent with the report of Li et al. (2005). Thus, modulating the expression level of OsSerk1 may serve as a useful strategy to develop rice varieties with enhanced yield.
MATERIALS AND METHODS
Plant materials, growth and pathogens inoculation conditions
Rice (Oryza sativa L.) lines employed in this work included japonica cultivar Kitaake, transgenic Xa21 line in Kitaake genetic background (hereafter called Xa21 Kitaake), and the transgenic line Xa21kit-OsSerk2Ri #A814 (Chen et al. 2014) with knock-down of OsSerk2 in Xa21-Kitaake genetic background. The Xa21-Kitaake plants show robust resistance to Xoo due to the Xa21 transgene, while the Xa21kit-OsSerk2Ri plants are fully susceptible to Xoo due to the reduced expression of OsSerk2 (Chen et al. 2014). For adult rice plant inoculation, the plants were grown in the greenhouse till six weeks of age and transferred to the growth chamber before inoculation with the Xoo strain PXO99 or Xoo-4. PXO99 carries a genetic factor that triggers XA21-mediated immunity while Xoo-4 lacks this genetic factor and can evade XA21-mediated immunity (Xoo-4 was kindly provided by Dr. Zhihui Xia from Hainan University, China). For seedling rice inoculation, the plants were grown in the greenhouse till 2.5 weeks of age before transferred to the growth chamber for inoculation (Park et al. 2010). Growth chambers were set on 14 hours light/10 hours dark photoperiod, 28/26°C temperature cycle, and 85/90% humidity. Xoo bacterial suspension (OD600 of 0.5) was used to inoculate rice by the scissors-dip method. The disease lesion length and bacterial population accumulated in rice leaf were evaluated as reported before (Chern et al. 2005). The ANOVA (analysis of variance) program packaged in SPSS16.0 software was adopted to assess significance in statistics.
For rice blast inoculation, plants were grown in the growth chamber at 28°C in 12h light/12h dark photoperiod with 75% humidity. Two-week old rice plants were used for inoculation with M. oryzae strains (ZB13 and ZB25) that were collected in Sichuan of China. The Digu and Lijiang rice varieties were used as the resistant and susceptible controls, respectively, to the two strains. The concentration of spore was 5×105/ml with 0.2% Tween-20. The fungal- and mock-inoculated rice seedlings were kept in dark inoculation chambers with 95% humidity at 28°C. The lesion length was measured and pictures were taken at 7 days after inoculation.
Plasmid constructs
For RNAi construct, a 432bp unique cDNA fragement of OsSerk1 (amplified by primer pair OsSerk1Ri-1/-2: 5’-CACCATCCGTGCACTTGGTTTCAT -3’/5’-AAGGGTTGTTGGCAAAACTG-3’) from japonica variety Nipponbare was cloned into the pENTR™/D-TOPO® (Invitrogen) vector and then put into pANDA (Kindly provided by Professor Ko Shimamoto, Nara Institute of Science and Technology, Japan) vector through LR recombination to generate OsSerk1Ri construct.
For overexpression construct, a 1875 bp full-length cDNA fragment of OsSerk1 (amplified by primer pair 07760cDNA-F/07760cDNA-R(Stop) (5’-CACCATGGCGGCGCATCGGTGGGCGGTG-3’/5’-TCACCTCGGCCCTGATA GCTCAACC-3’) from japonica cultivar Nipponbare was cloned into the pENTR™/D-TOPO® (Invitrogen) vector and then put into the Ubi-NC1300RFCA vector through LR recombination to generate UbiC1300-OsSerk1 construct. The Ubi-NC1300RFCA vector was developed by introducing the 1711bp RFCA (reading frame cassette A) fragment into Ubi/NC1300 that has been reported previously (Chern et al, 2005). In the UbiC1300-OsSerk1 construct, the OsSerk1 gene is under control of the maize ubiquitin promoter.
For constructs used in yeast two-hybrid assay, the partial cDNA sequence of OsSerk1 (named OsSERK1JMK), containing juxtamembrane and kinase domains (JMK) with stop codon, was amplified by primer pair OsSerk1G257-F/OsSerk1G257-R(w/stop) (5’-CACCATGGGTTTTGCATGGTATCGGCGC-3’/5’-TTATCATCTCGGCCCTGA TAGCTCAACCG-3) and cloned into pENTR™/D-TOPO® (Invitrogen) to create pENTR-OsSERK1JMK. The pENTR-OsSERK2JMK and pENTR-OsBRIK735 constructs were generated previously (Chen et al 2014). The pENTR-OsBRIK735 was recombined with the pB42AD vector to yield HA-tagged fusion protein. The pENTR--OsSERK1JMK and pENTR-OsSERK2JMK plasmids were recombined with the pLexA vector to produce LexA fusion proteins.
Development of rice transgenic lines and crossing
Through Agrobacterium-mediated transformation described previously (Chern et al. 2005), the overexpression construct of OsSerk1 was introduced into Xa21-Kitaake and Kitaake plants, respectively. The RNAi construct of OsSerk1 was introduced into Xa21-Kitaake plant. Because the transgenic Xa21-Kitaake plant is mannose resistant, transgenes OsSerk1Ri and OsSerk1ox were selected with hygromycin in the present study. The Xa21kit-OsSerk2Ri #A814 plants carrying reduced OsSerk2 expression was used as the pollen recipient to cross with transgenic OsSerk1ox plants in Xa21-Kitaake background to obtain OsSerk1oxOsSerk2Ri plants. PCR-based genotyping was performed to determine the transgenic plants with or without the transgene(s) according to the description of previous study (Chen et al. 2010). The PCR specific primer pairs used for genotyping transgenes OsSerk1Ri, OsSerk1ox and OsSerk2Ri were Ubi-pro-F(5′-CATACGCTATTTATTTGCTTGG)/OsSERK1Ri-2(5′-AAGGGTTGTT GGCAAAACTG-3′), Ubi-pro-F/OsSerk1oX-genotype-R(5′-GTATCGTTCCGCTTATGTTATT-3′), and Ubi-pro-F/ OsSerk1Ri-R(5′-CCAATCGAGCAACATCACAT-3′), respectively.
RNA extraction and real time RT-PCR analyses
Total RNA was isolated from rice plant tissue using Invitrogen RNA isolation kit, TRIzol (Invitrogen), following the manufacture's manual. Total RNA was treated with DNase I and used for the first strand cDNA synthesis using the Invitrogen reverse transcription kit (Invitrogen) following the provided manual. Quantitative real time PCR (qRT-PCR) was performed on a Bio-Rad CFX96 Real-Time System coupled to a C1000 Thermal Cycler (Bio-Rad). For qRT-PCR reactions, the Bio-Rad SsoFast Eva Green Supermix was used. qRT-PCR primer pairs used were as follows: OsSerk1-Q1/-Q2 (5’-TGCATTGCATAGCTTGAGGA-3’/5’- GCAGCATTCCCAAGATCAAC-3’) for the OsSerk1 gene, Xak1-Q1/Q2 (5’-TAGTCTGCGCCAAAGTCTGA-3’/5’- GCACCTGACAGTTGTGCATT -3’) for the OsSerk2 gene, Xa21-Q1/-Q2 (5’-TGACACGAAGCTCATTTTGG-3’/5’- TTGATGGCATTCAGTTCGTC-3’) for the Xa21 gene, and actin-Q1/-Q2 (5’-TCGGCTCTGAATGTACCTCCTA-3’/5’-CACTTGAGTAAAGACTGTCACTT G-3’) for the reference gene Actin. qRT-PCR reactions were run for 40 cycles with annealing at 56°C for 12 sec and denaturation at 95°C for 8 sec. The expression levels of OsSerk1, OsSerk2 and Xa21 were normalized to the Actin gene expression level.
Yeast Two-Hybrid Assays
The Matchmaker LexA two-hybrid system (Clontech) was used for yeast two hybrid assays. Yeast pEGY48/p8op-lacZ (Clontech) was co-transformed with the BD and AD vectors by using the Frozen-EZ yeast transformation II kit (Zymo Research) and spread on an appropriate medium following the procedures described previously (Chen et al. 2010).
Immune-blotting
Total protein extraction from yeast cell and immuno-blotting (Western blotting) was performed as previously described (Chen et al. 2010). The anti-LexA antibody (Clontech) was used to detect LexA-fused protein and the anti-HA antibody (Covance) used to detect HA-fused protein.
Purification of recombinant proteins and in vitro protein kinase assay
Recombinant fusion proteins were produced in E.coli BL21 (Novagen). GST-tagged fusion proteins (GST-OsSERK1JMK, GST-OsSERK1JMKKE) were enriched using Glutathione Sepharose Fast Flow (GE Healthcare) according to the manufacturer's protocol. His-Nus-tagged fusion proteins (His-Nus-XA21K668, His-Nus-XA21K668KE) were enriched using His-Bind Resin (Novagen) according to the manufacturer's protocol (Chen et al. 2014). After elusion, the fusion proteins were adjusted to the same concentration in 10% glycerol solution and stored at -70°C until usage.
Two micrograms of each fusion protein was incubated in 30 ul kinase buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, 10 mM MnCl2, 1mM DTT) in the presence of 0.5 ul (5 mCui) [32P]-γ-ATP for 30 min at 30°C with shaking at 1200rpm. The reaction was stopped by adding 10ul 4xLDS loading dye (Invitrogen) and immediately transferred to 80°C for 10 min. The reaction mixture was separated by SDS-PAGE. Post electrophoresis, proteins were transferred onto PVDF membranes followed by staining with 0.2% w/v ponceau S in TCA (3% v/v). The membranes were dried at room temperature for 20 min and then followed by autoradiograph analysis as described previously (Chen et al. 2010).
Supplementary Material
Figure S1. Plant heights of transgenic T0 plants with OsSerk1 overexpression
(A) Plant heights of the transgenic Kit-OsSerk1ox T0 plants in Kitaake background. Total of 18 independent Kit-OsSerk1ox T0 plants were obtained. Three of them were confirmed by PCR not containing the OsSerk1ox transgene, which was represented by ‘(-)’ and white columns (same for the bottom panel). One of them did not show overexpression of OsSerk1 by real-time RT-PCR, which was represented by grey columns (same for the bottom panel). (B) Plant heights of transgenic Xa21kit-OsSerk1ox T0 plants in Xa21 Kitaake background. Total of 30 independent Xa21kit-OsSerk1ox T0 plants were obtained. One (shown as white bar) of them was confirmed not containing OsSerk1ox, and two (grey bars) of them did not show overexpression of OsSerk1.
Figure S2. Morphological phenotypes of transgenic lines with OsSerk1 overexpression
(A) The internode length and lamina joint angle of each line were measured from 10 independent plants at 25 days after heading. The control lines Xa21 Kitaake (Xa21-Kit) and Kitaake, and two lines homozygous for OsSerk1ox from each background of Xa21-Kit and Kitaake. The two lines #X1904 and #X1953 homozygous for OsSerk1ox were derived from Xa21kit-OsSerk1ox-#3 and Xa21kit-OsSerkox #18 T0 lines, respectively. The lines #K1630 and #K1653 homozygous for OsSerk1ox were derived from Kit-OsSerk1ox #14 and Kit-OsSerk1ox #17, respectively. (B) and (C) Photographs for seed size and lamina joint angle were taken at 25 days after heading.
Figure S3. Disease resistance determination of Kittake- and Xa21 kittake carrying-OsSerk1ox transgene after inoculation with Xoo
(A) Disease lesion lengths of OsSerk1ox T1 plants in Kitaake background (Kit-OsSerk1ox) and the Xa21 Kitaake (Xa21-Kit) and Kitaake control plants at 15 days after inoculation at the adult stage. All plants were inoculated with Xoo strain PXO99, which can trigger XA21 mediated immune response, at 6 weeks old. Lesion lengths were measured at 15 DAI for at least 6 leaves from three or more independent plants. “+” and “-” indicate presence or absence of the OsSerk1ox transgene revealed by PCR with Hyg specific primers (same for below). (B) Disease lesion lengths of OsSerk1ox T1 plants and the control plants at 10 DAI at the seedling stage. All plants were inoculated with PXO99 at 3 weeks old. Lesion lengths were measured for at least 8 leaves from three or more independent plants at 10 DAI. Statistical significance comparison was conducted with ANOVA, where the different ‘**’ and ‘*’ marks above the columns indicate differences with and , respectively.
Figure S4. Disease resistance determination of Xa21kit-OsSerk1ox and Xa21kit-OsSerk1Ri lines after inoculation with a virulent Xoo strain to Xa21 gene The rice lines, Xa21 kitaake, Xa21kit-OsSerk1Ri (#1602 and #1603) and Xa21kit-OsSerk1ox (#1904 and #1953) were inoculated with the Xoo-4 strain, which is virulent on Xa21 plants, at 6 weeks old. Lesion lengths were measured at 15 DAI for at least 6 leaves from three or more independent plants. The photograph depicts representative symptom development in leaves at 15 DAI.
Figure S5. Disease resistance determination of Xa21 Kitaake carrying OsSerk1Ri transgeneto Xoo
Four independent OsSerk1Ri T1 plants and Xa21 Kitaake (Xa21-Kit), Kitaake, and Xa21kit-OsSerk2Ri were included in this experiment. All plants were inoculated at 6 weeks old. “+” and “-” indicate the presence and absence of the OsSerk1Ri transgene in T1 plants respectively. Lesion lengths were measured at 20 DAI.
Figure S6. Altered expression of OsSerk1 does not affect rice resistance to Magnaporthe oryzae
The Digu and Lijiang are the cultivars having broad-spectrum resistance and susceptibility to blast strain ZB13, respectively. Two-week old rice plants were used for inoculation. The lesion length was measured and pictures were taken at 7 days after inoculation. This experiment was repeated three times with same results.
Figure S7. Disease resistance determination of Xa21 kitaake plants carrying both OsSerk1ox and OsSerk2Ri transgenes after inoculation with Xoo
(A) Lesion lengths of the Xa21 Kitaake, Kitaake, and Xa21kit-OsSerk2Ri #A814 plants, and the three F1 hybrid lines (Xa21kit-OsSerk2RiOsSerk1ox #3, #7, #14). Three F1 hybrid populations were generated by crossing #A814 with each of the three independent OsSerk1ox lines, Xa21kit-OsSerk1ox #3, #7, and #14, respectively. The F1 population consisted of plants carrying (represented by ‘+’) OsSerk1ox and null segregants (represented by ‘-’) determined by specific PCR primers. The numbers of Xa21kit-OsSerk1oxOsSerk2Ri vs Xa21kit-OsSerk2Ri F1 plants obtained were 10/7, 29/34, and 29/24 from crosses, #A814/Xa21kit-OsSerk1ox #3, #A814/Xa21kit-OsSerk1ox #7 and #A814i/Xa21kit-OsSerk1ox #18, respectively. Inoculation with PXO99 was conducted at six weeks old. Lesion lengths were measured at 15 DAI for at least 14 leaves from ten or more independent plants. Statistical significance comparison was conducted with ANOVA, where the different capital letters above the columns indicate differences with . (B) The photograph depicts representative symptom development in leaves at 15 DAI. In each F1 population, only Xa21kit-OsSerk1oxOsSerk2Ri F1 plants were included in the photograph.
Figure S8. Long disease lesion lengths are correlated with the presence of OsSerk2Ri but not with OsSerk1ox
Two F2 populations (A and B) were included in this experiment that were derived from two F1 hybrids from crosses of Xa21kit-OsSerk2Ri #A814 with Xa21kit-OsSerk1ox #7 or Xa21kit-OsSerk1ox #18. All plants, including the Xa21 Kitaake (Xa21-Kit), #A814, and Kitaake controls, were inoculated with PXO99 at 6 weeks old. Lesion lengths were measured at 15 DAI. The genotyping primers specific to OsSerk2Ri and OsSerk1ox, respectively, were used to determine the plants with OsSerk2Ri and/or OsSerk1ox. In both populations, the plants containing OsSerk2Ri all showed longer lesion lengths than those without this transgene. The plants with and without OsSerk1ox showed no correlation with the disease lesion lengths.
Figure S9. Interaction of OsSERK1 and OsSERK2 in yeast
The OsSERK1 and OsSERK2 intracellular domains were fused to B42AD with HA tag and to LexA tag, respectively. The blue colors indicate interaction between the two co-expressed proteins. This experiment was repeated three times with same results.
ACKNOWLEDGEMENTS
We are grateful to Dr. Bo Ding and Professor Wende Liu from Institution of Plant Protection of Chinese Academic of Agricultural Science for their helpful discussions on this manuscript. We also thank Professor Zhihui Xia from Hainan University, China, for kindly providing the Xoo strain Xoo-4. This work was supported by a National Institutes of Health grant (GM59962) to Pamela C Ronald, Jiangsu Government scholarship for overseas study and the fund for short-term visit of foreign research fellows to Shimin Zuo, National Science Fund of China (31171622 and 31371705) and Sichuan ‘Hundred Talents Plan’ fund to Xuewei Chen, and an EMBO (European Molecular Biology Organization) long-term post-doctoral fellowship (ALTF 1290-2011) and a Human Frontiers Science Program long-term post-doctoral fellowship (LT000674/2012) to Benjamin Schwessinger.
REFERENCES
- Chen X, Chern M, Canlas PE, Ruan D, Jiang C, Ronald PC. An ATPase promotes autophosphorylation of the pattern recognition receptor XA21 and inhibits XA21-mediated immunity. Proc Natl Acad Sci USA. 2010;107:8029–8034. doi: 10.1073/pnas.0912311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XW, Zuo SM, Schwessinger B, Chern MS, Canlas PE, Ruan DL, Zhou XG, Wang J, Daudi A, Petzold CJ, Heazlewood JL, Ronald PC. An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol Plant. 2014 doi: 10.1093/mp/ssu003. Doi: 10.1093/mp/ssu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern MS, Canlas PE, Fitzgerald H, Ronald PC. NRR, a negative regulator of disease resistance in rice that interacts with arabidopsis NPR1 and rice NH1. Plant J. 2005;43:623–635. doi: 10.1111/j.1365-313X.2005.02485.x. [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Shan LB, He P, Vries S, Kemmerling B. One for all: The receptor-associated kinase BAK1. Trends in Plant Sci. 2009;14:535–541. doi: 10.1016/j.tplants.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell. 2005;17:3350–3361. doi: 10.1105/tpc.105.036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Abd-EI-Haliem A, Masini L, van den Berg GC, Joosten MH, Thormma BP. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 2011;156:2255–2265. doi: 10.1104/pp.111.180067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XQ, Li FJ, Li MY, Kianinejad AS, Dever JK, Wheeler TA, Li ZH, He P, Shan LB. Cotton GhBAK1 mediates verticillium wilt resistance and cell death. J Integr Plant Biol. 2013;55:586–596. doi: 10.1111/jipb.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou XP, Yin HJ, He K, Du JB, Yi J, Xu SB, Lin HH, Clouse SD, Li J. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012;8:e1002452. doi: 10.1371/journal.pgen.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol. 2007;17:1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, de Vries SC. The Arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in cultures. Plant Physiol. 2001;127:803–816. [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Xiong L, Yang Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta. 2005;222:107–117. doi: 10.1007/s00425-005-1534-4. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Müssig C, Thomma BP, Albrecht C, de Vries SC, Hirt H, Nürnberger T. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor - like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J. 2009;7:791–806. doi: 10.1111/j.1467-7652.2009.00444.x. [DOI] [PubMed] [Google Scholar]
- Li J. Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr Opin in Plant Biol. 2010;13:509–514. doi: 10.1016/j.pbi.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Park CJ, Lee SW, Chern MS, Sharma R, Canlas PE, Song MY, Jeon JS, Ronald PC. Ectopic expression of rice Xa21 overcomes developmentally controlled resistance to Xanthomonas oryzae pv. oryzae. Plant Sci. 2010;9:66–71. doi: 10.1016/j.plantsci.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Ryu HY, Kim BH, Kim SY, Yoon IS, Nam KH. A subset of OsSERK genes, including OsBAK1, affects normal growth and leaf development of rice. Mol Cells. 2011;32:561–569. doi: 10.1007/s10059-011-0178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SB, Khush GS, Virk P, Tang QY, Zou YB. Progress in ideotype breeding to increase rice yield potential. Field Crops Res. 2008;108:32–38. [Google Scholar]
- Reinhardt D, Kuhlemeier C. Plant architecture. EMBO Rep. 2002;3:846–851. doi: 10.1093/embo-reports/kvf177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tor M, de Vries S, Zipfel C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23:2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol. 2006;24:105–109. doi: 10.1038/nbt1173. [DOI] [PubMed] [Google Scholar]
- Santiago J, Henzler C, Hothorn M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science. 2013;341:889–892. doi: 10.1126/science.1242468. [DOI] [PubMed] [Google Scholar]
- Schmidt EDL, Guzzo F, Toonen MAJ, de Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124:2049–2062. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C. Phosphorylation-Dependent Differential Regulation of Plant Growth, Cell Death, and Innate Immunity by the Regulatory Receptor-Like Kinase BAK1. PLoS Genet. 2011;7:e1002046. doi: 10.1371/journal.pgen.1002046. doi:10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC. Plant innate immunity: Perception of conserved microbial signatures. Annu Rev Plant Biol. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- Santiago J, Henzler C, Hothorn M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science. 2013;341:889–892. doi: 10.1126/science.1242468. [DOI] [PubMed] [Google Scholar]
- Sharma D, Sanghera GS, Sahu P, Sahu PS, Parikh M, Sharma B, Bhandarkar S, Chaudhari PR, Jena BK. Tailoring rice plants for sustainable yield through ideotype breeding and physiological interventions. African J Agric Res. 2013;8:5004–5019. [Google Scholar]
- Singla B, Khurana JP, Khurana P. Structural characterization and expression analysis of the SERK/SERL gene family in rice (Oryza sativa). Int J Plant Genomics. 2009 doi: 10.1155/2009/539402. Doi:10.1155/2009/539402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sun YD, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 2013;342:624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- Sun YD, Han ZF, Tang J, Hu ZH, Chai CL, Zhou B, Chai JJ. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Research. 2013;23:1326–1329. doi: 10.1038/cr.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- Yang XC, Hwa CM. Genetic modification of plant architecture and variety improvement in rice. Heredity. 2008;101:396–404. doi: 10.1038/hdy.2008.90. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xu YY, Guo SY, Zhu JY, Huan Q, Liu HH, Wang L, Luo GZ, Wang XJ, Chong K. Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 2012 doi: 10.1371/journal.pgen.1002686. Doi: 10.1371/journal.pgen.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Plant heights of transgenic T0 plants with OsSerk1 overexpression
(A) Plant heights of the transgenic Kit-OsSerk1ox T0 plants in Kitaake background. Total of 18 independent Kit-OsSerk1ox T0 plants were obtained. Three of them were confirmed by PCR not containing the OsSerk1ox transgene, which was represented by ‘(-)’ and white columns (same for the bottom panel). One of them did not show overexpression of OsSerk1 by real-time RT-PCR, which was represented by grey columns (same for the bottom panel). (B) Plant heights of transgenic Xa21kit-OsSerk1ox T0 plants in Xa21 Kitaake background. Total of 30 independent Xa21kit-OsSerk1ox T0 plants were obtained. One (shown as white bar) of them was confirmed not containing OsSerk1ox, and two (grey bars) of them did not show overexpression of OsSerk1.
Figure S2. Morphological phenotypes of transgenic lines with OsSerk1 overexpression
(A) The internode length and lamina joint angle of each line were measured from 10 independent plants at 25 days after heading. The control lines Xa21 Kitaake (Xa21-Kit) and Kitaake, and two lines homozygous for OsSerk1ox from each background of Xa21-Kit and Kitaake. The two lines #X1904 and #X1953 homozygous for OsSerk1ox were derived from Xa21kit-OsSerk1ox-#3 and Xa21kit-OsSerkox #18 T0 lines, respectively. The lines #K1630 and #K1653 homozygous for OsSerk1ox were derived from Kit-OsSerk1ox #14 and Kit-OsSerk1ox #17, respectively. (B) and (C) Photographs for seed size and lamina joint angle were taken at 25 days after heading.
Figure S3. Disease resistance determination of Kittake- and Xa21 kittake carrying-OsSerk1ox transgene after inoculation with Xoo
(A) Disease lesion lengths of OsSerk1ox T1 plants in Kitaake background (Kit-OsSerk1ox) and the Xa21 Kitaake (Xa21-Kit) and Kitaake control plants at 15 days after inoculation at the adult stage. All plants were inoculated with Xoo strain PXO99, which can trigger XA21 mediated immune response, at 6 weeks old. Lesion lengths were measured at 15 DAI for at least 6 leaves from three or more independent plants. “+” and “-” indicate presence or absence of the OsSerk1ox transgene revealed by PCR with Hyg specific primers (same for below). (B) Disease lesion lengths of OsSerk1ox T1 plants and the control plants at 10 DAI at the seedling stage. All plants were inoculated with PXO99 at 3 weeks old. Lesion lengths were measured for at least 8 leaves from three or more independent plants at 10 DAI. Statistical significance comparison was conducted with ANOVA, where the different ‘**’ and ‘*’ marks above the columns indicate differences with and , respectively.
Figure S4. Disease resistance determination of Xa21kit-OsSerk1ox and Xa21kit-OsSerk1Ri lines after inoculation with a virulent Xoo strain to Xa21 gene The rice lines, Xa21 kitaake, Xa21kit-OsSerk1Ri (#1602 and #1603) and Xa21kit-OsSerk1ox (#1904 and #1953) were inoculated with the Xoo-4 strain, which is virulent on Xa21 plants, at 6 weeks old. Lesion lengths were measured at 15 DAI for at least 6 leaves from three or more independent plants. The photograph depicts representative symptom development in leaves at 15 DAI.
Figure S5. Disease resistance determination of Xa21 Kitaake carrying OsSerk1Ri transgeneto Xoo
Four independent OsSerk1Ri T1 plants and Xa21 Kitaake (Xa21-Kit), Kitaake, and Xa21kit-OsSerk2Ri were included in this experiment. All plants were inoculated at 6 weeks old. “+” and “-” indicate the presence and absence of the OsSerk1Ri transgene in T1 plants respectively. Lesion lengths were measured at 20 DAI.
Figure S6. Altered expression of OsSerk1 does not affect rice resistance to Magnaporthe oryzae
The Digu and Lijiang are the cultivars having broad-spectrum resistance and susceptibility to blast strain ZB13, respectively. Two-week old rice plants were used for inoculation. The lesion length was measured and pictures were taken at 7 days after inoculation. This experiment was repeated three times with same results.
Figure S7. Disease resistance determination of Xa21 kitaake plants carrying both OsSerk1ox and OsSerk2Ri transgenes after inoculation with Xoo
(A) Lesion lengths of the Xa21 Kitaake, Kitaake, and Xa21kit-OsSerk2Ri #A814 plants, and the three F1 hybrid lines (Xa21kit-OsSerk2RiOsSerk1ox #3, #7, #14). Three F1 hybrid populations were generated by crossing #A814 with each of the three independent OsSerk1ox lines, Xa21kit-OsSerk1ox #3, #7, and #14, respectively. The F1 population consisted of plants carrying (represented by ‘+’) OsSerk1ox and null segregants (represented by ‘-’) determined by specific PCR primers. The numbers of Xa21kit-OsSerk1oxOsSerk2Ri vs Xa21kit-OsSerk2Ri F1 plants obtained were 10/7, 29/34, and 29/24 from crosses, #A814/Xa21kit-OsSerk1ox #3, #A814/Xa21kit-OsSerk1ox #7 and #A814i/Xa21kit-OsSerk1ox #18, respectively. Inoculation with PXO99 was conducted at six weeks old. Lesion lengths were measured at 15 DAI for at least 14 leaves from ten or more independent plants. Statistical significance comparison was conducted with ANOVA, where the different capital letters above the columns indicate differences with . (B) The photograph depicts representative symptom development in leaves at 15 DAI. In each F1 population, only Xa21kit-OsSerk1oxOsSerk2Ri F1 plants were included in the photograph.
Figure S8. Long disease lesion lengths are correlated with the presence of OsSerk2Ri but not with OsSerk1ox
Two F2 populations (A and B) were included in this experiment that were derived from two F1 hybrids from crosses of Xa21kit-OsSerk2Ri #A814 with Xa21kit-OsSerk1ox #7 or Xa21kit-OsSerk1ox #18. All plants, including the Xa21 Kitaake (Xa21-Kit), #A814, and Kitaake controls, were inoculated with PXO99 at 6 weeks old. Lesion lengths were measured at 15 DAI. The genotyping primers specific to OsSerk2Ri and OsSerk1ox, respectively, were used to determine the plants with OsSerk2Ri and/or OsSerk1ox. In both populations, the plants containing OsSerk2Ri all showed longer lesion lengths than those without this transgene. The plants with and without OsSerk1ox showed no correlation with the disease lesion lengths.
Figure S9. Interaction of OsSERK1 and OsSERK2 in yeast
The OsSERK1 and OsSERK2 intracellular domains were fused to B42AD with HA tag and to LexA tag, respectively. The blue colors indicate interaction between the two co-expressed proteins. This experiment was repeated three times with same results.