Abstract
Limb development requires the coordinated growth of several tissues and structures including long bones, joints and tendons, but the underlying mechanisms are not wholly clear. Recently, we identified a small drug-like molecule -we named Kartogenin (KGN)- that greatly stimulates chondrogenesis in marrow-derived mesenchymal stem cells (MSCs) and enhances cartilage repair in mouse osteoarthritis (OA) models. To determine whether limb developmental processes are regulated by KGN, we tested its activity on committed preskeletal mesenchymal cells from mouse embryo limb buds and whole limb explants. KGN did stimulate cartilage nodule formation and more strikingly, boosted digit cartilaginous anlaga elongation, synovial joint formation and interzone compaction, tendon maturation as monitored by ScxGFP, and interdigit invagination. To identify mechanisms, we carried out gene expression analyses and found that several genes, including those encoding key signaling proteins, were up-regulated by KGN. Amongst highly up-regulated genes were those encoding hedgehog and TGFβ superfamily members, particularly TFGβ1. The former response was verified by increases in Gli1-LacZ activity and Gli1 mRNA expression. Exogenous TGFβ1 stimulated cartilage nodule formation to levels similar to KGN, and KGN and TGFβ1 both greatly enhanced expression of lubricin/Prg4 in articular superficial zone cells. KGN also strongly increased the cellular levels of phospho-Smads that mediate canonical TGFβ and BMP signaling. Thus, limb development is potently and harmoniously stimulated by KGN. The growth effects of KGN appear to result from its ability to boost several key signaling pathways and in particular TGFβ signaling, working in addition to and/or in concert with the filamin A/CBFβ/RUNX1 pathway we identified previously to orchestrate overall limb development. KGN may thus represent a very powerful tool not only for OA therapy, but also limb regeneration and tissue repair strategies.
Keywords: Limb development, Kartogenin, Skeletogenesis, Synovial joint formation, Joint interzone, TGFβ, BMP and hedgehog signaling, Lubricin
Introduction
The developing limb has long served as a favorable experimental system to unravel the mechanisms of skeletal development and growth, formation of synovial joints, evolution of the skeletal primordia into complex three dimensional functional entities, and regulation of related developmental steps and processes (Zeller et al., 2009). In the early limb, skeletal development initiates with the formation of mesenchymal condensations at appropriate sites and stages that prefigure the location and size of future skeletal elements, including the Y-shaped condensation that corresponds to the future single stylopod element (humerus or femur) and dual zeugopod elements (tibia/fibula and radius/ulna) (Pitsillides and Ashhurst, 2008). Following condensation and elimination of blood vessels (Yin and Pacifici, 2001), the cells undergo chondrogenic differentiation and produce readily recognizable cartilaginous primordia. The chondrocytes within each element then become organized into growth plates and are distinguishable into typical maturation zones that include the resting, proliferative, prehypertrophic and hypertrophic zones (Mackie et al., 2011). The hypertrophic chondrocytes mineralize their surrounding matrix, promote invasion of blood vessels and osteoprogenitors from neighboring perichondrial tissues, and pave the way for formation of endochondral bone and marrow. Such skeletal developmental processes are all intimately coordinated with concurrent processes occurring in surrounding tissues and structures that permit formation of anatomically appropriate muscles, tendons, ligaments and other locally specialized tissues. Given the complexity of these processes, it is not surprising that many signaling proteins, transcription factors, hormones and extracellular matrix macromolecules participate in their regulation (Capdevila and Belmonte, 2001). These include members of the transforming growth factor β (TGFβ), hedgehog and Wnt superfamilies of secreted signaling proteins, members of the Sox, Runx and Ets families of transcription factors, and a number of matrix collagens and proteoglycans (Karsenty et al., 2009; Pacifici et al., 2005; Song et al., 2009; Zeller et al., 2009). Despite undeniable progress, however, much remains unclear about limb skeletal development, particularly with regard to what drives limb elongation and growth at different stages, how the development of distinct tissues and structures is coordinated within each limb anatomical segment, and whether and which limb developmental event(s) is/are amenable to exogenous stimulation.
An essential limb developmental process for which even less is currently understood is synovial joint formation. The joints display remarkably distinct shapes, structure and organization –be it an elbow or an ankle- and contain unique tissues and components including articular cartilage, synovial capsule and lining, and a fluid rich in lubricin and other anti-adhesive molecules (Pitsillides and Ashhurst, 2008). Developmental studies originally suggested that highly condensed and flat mesenchymal cells emerging at each prospective joint site - collectively called the interzone- were important for joint formation (Holder, 1977). However, interzone cell fate, roles and modus operandi had long remained obscure (Khan et al., 2007). Using genetic tracing and tracking approaches in mouse, we showed that the interzone cells are not transient, actively participate in joint formation, and give rise to all joint tissues, including articular cartilage, synovial lining and intrajoint ligaments (Koyama et al., 2008). The interzone cells all share expression of the growth and differentiation factor 5 (Gdf5), a TGFβ superfamily member (Storm et al., 1994). Using Gdf5Cre mice to ablate floxed target genes, we and others showed that the behavior and function of interzone cells involve multiple mechanisms including Wnt/β-catenin signaling and cell surface/matrix macromolecule interactions (Koyama et al., 2008; Mundy et al., 2011). Recent studies have shown that joint formation also requires skeletal muscle function and contraction and signaling by β-catenin (Kahn et al., 2009; Pazin et al., 2012). The increasing research attention surrounding joint formation reflects the fact that many aspects of it remain stubbornly unclear and difficult to decipher, and that a much better understanding in this area could lead to the conception and creation of regenerative and repair tools for common and currently unsolved joint pathologies, including osteoarthritis (OA), severe joint injury, and congenital joint dysplasias (Onyekwelu et al., 2009; Sandell, 2012; Umlauf et al., 2010).
With these and other facts in mind, we recently carried out a high-throughput image-based screen to identify drugs with possible chondrogenic and chondroprotective capacity (Johnson et al., 2012). Amongst the over 22,000 structurally diverse, heterocyclic and drug-like molecules screened, we identified a molecule –we named Kartogenin (KGN)- that was able to stimulate chondrogenic differentiation of bone marrow mesenchymal stem cells (MSCs) in culture. The drug also displayed chondroprotective effects when injected into the operated joint in two mouse models of surgery and non-surgery induced OA (Johnson et al., 2012). Because of its remarkable properties, KGN attracted much attention (Marini and Forlino, 2012; Ray, 2012; Xu et al., 2013), but its overall biological properties, its mechanisms of action on developing skeletal cells, and its full therapeutic applications and potentials remain to be uncovered, understood and tested. We show here that KGN is in fact a potent stimulator of limb skeletal growth, facilitates joint formation by promoting interzone compaction and lubricin/Prg4 expression, and stimulates the activity of key signaling factors, including TGFβ superfamily members in particular. The data provide insights into the regulation and overall coordination of limb growth and development, also pointing to the possibility that KGN could be a powerful tool for limb repair and regenerative strategies.
Materials and Methods
Mouse lines, mating and genotyping
Female Gdf5-Cre transgenic mice (Rountree et al., 2004) were mated with male ROSA-mTomato/mGFP reporter mice which express constitutive red fluorescence prior to, and conditional green fluorescence following, Cre-mediated recombination (Muzumdar et al., 2007). Scleraxis-GFP (ScxGFP) mice were a kind gift from Dr. Ronen Schweitzer (Pryce et al., 2007). Gli1+/nLacZ mice are widely used as a functional readout of hedgehog signaling activity and range, and were obtained from Jackson laboratory (Ahn and Joyner, 2005). PTHrP-lacZ knockin mice were generously provided by Dr. Arthur Broadus (Chen et al., 2006). Pregnant and postnatal mice were maintained and sacrificed according to IACUC approved protocols. Genotyping was carried out with DNA isolated from tail clips.
Generation of Prg4-mCherry reporter mice
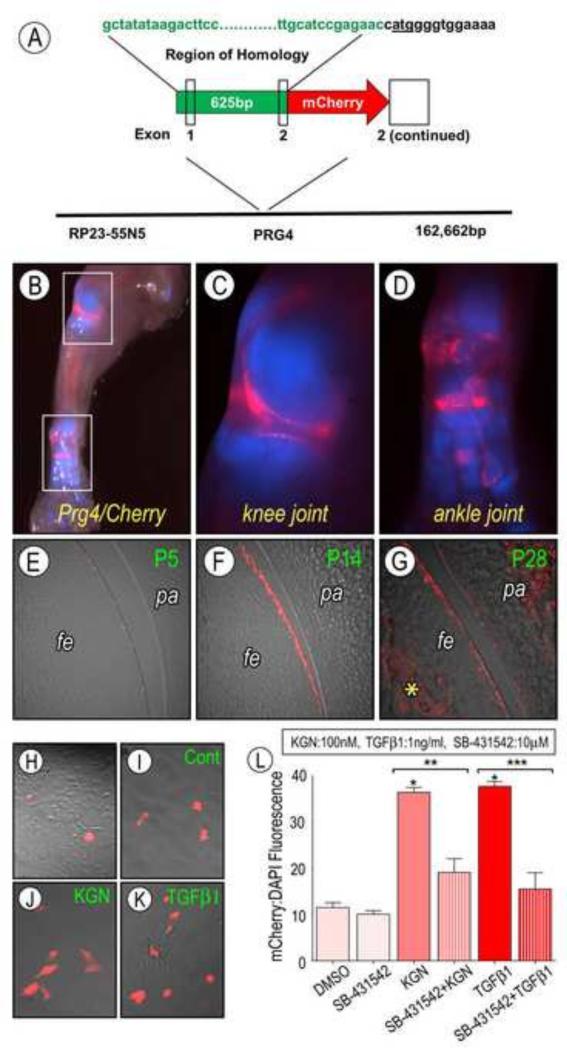
BAC clone RP23-55N5 was obtained from the Children's Hospital Oakland Research Institute (CHORI). The pLD53-SC2 and pSV1.RecA recombination vectors were generously provided by Shiaoching Gong (Gong et al., 2002). Transgenic animals were housed in a clean barrier facility and humanely treated in accordance with University of Connecticut Health Center institutional guidelines. A 625bp homology arm was PCR amplified from purified RP23-55N5 using the following primers: Prg4 (Sense) 5’-CTCTGCGGCCGCGCTATATAAGACTTCCAGCACACTGGAGA and Prg4 (antisense) 5’-CTCTGGATCCGTTCTCGGATGCAACGCCCTTGCTTGAGA-3’. The PCR amplicon was cloned into Not1 and BamH1 sites of the pLD53.sc2-mCherry shuttle vector using standard cloning methods. Recombinase A was introduced into RP23-55N5 host bacteria by transformation with pSV1.RecA vector (100 ng) and selected for on chloramphenicol (12.5 μg/ml)/tetracycline (10 μg/ml) LB agar plates. RP23-55N5 host bacteria containing RecA were then transformed by electroporation with 1 μg (1–2 μl) of the pLD53.sc2-mCherry containing the Prg4 homology arm. SOC medium (1 ml) was added and transformed bacteria were incubated with shaking at 200 rpm for one hour at 30°C. Recombinants were first selected by adding 5 ml of LB medium containing chloramphenicol (12.5 μg/ml), ampicillin (50 μg/ml), and tetracycline (10 μg/ml) to bacteria and grown overnight at 30°C with shaking at 200 rpm. Further selection for recombinant clones was carried out by plating 100 μl of overnight culture on to chloramphenicol (12.5 μg/ml), ampicillin (50 μg/ml) LB agar plates and incubated overnight at 42°C. Chloramphenicol/ampicillin resistant colonies were screened by colony PCR using primers: Prg4 5’ Recom (sense) 5’-GGACTAATTGGTTCATCCCAGTCCA-3’ and mCherry (antisense) 5’-GCACCTTGAAGCGCATGAACTCCTTGATGA-3’. Colony PCR-identified candidate recombinants were further verified by diagnostic restriction enzyme digestion and field inversion gel electrophoresis.
A verified Prg4-Cherry clone was grown and purified from 200 ml of bacterial culture using a Maxi kit (Qiagen, Valencia, CA) with minor modifications detailed here. After alkaline lysis 2 M potassium acetate was used in place of the standard 3 M solution. QF buffer was heated to 65°C for elution and the column eluate was further purified with a 1:1 phenol:chloroform extraction followed by a chloroform extraction. A 10 μg aliquot of the purified Prg4-Cherry BAC was linearized with PI-SceI and further purified on a Sepharose CL-4B column (Sigma, St. Louis, MO ) that had been equilibrated with injection buffer (10 mM Tris pH7.5, 0.1 mM EDTA, 100 mM NaCl). Ten 200 μl fractions were collected and the DNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific, Hudson, NH). A 30 μl aliquot of each fraction was run on a pulse field gel to assess DNA quality. Pronuclear injection was carried out at the UCONN Health Center Gene Targeting and Transgenic Facility (GTTF). To visualize overlap between Prg4 and Col2a1 expressing cells, Prg4-mCherry reporter mice were crossed with Col2a1-ECFP Reporter mice described previously (Maye et al., 2011).
Limb bud cell micromass cultures
Micromass cultures were prepared with mesenchymal cells isolated from E11.5 mouse embryo limb buds as previously described (Huegel et al., 2013). Briefly, cells were isolated by enzymatic digestion from freshly-dissected limb buds, suspended at a concentration of 5×106 cells/ml in DMEM containing 3% fetal bovine serum (FBS) and antibiotics, spotted onto tissue culture plates, allowed to adhere for 2 hrs, and finally flooded and maintained in the same medium. At 12 h of culture, medium was replaced and supplemented with DMSO solution (10 μl/ml, serving as control), 10 nM to 1μM KGN, 2.5 ng/ml TGFβ1 (Sigma Cat. # T7039) and/or Cyclopamine (CPN, 5μM). KGN stock solution was maintained at 100 μM in DMSO and diluted in culture medium as indicated. Where CPN treatment was applied, cells were pre-treated with CPN alone for 60 minutes prior to addition of TGFβ1 or KGN. Medium was replaced daily with fresh components. It should be noted that KGN is not stable over time and thus, new batches of KGN stock were used every 2 to 3 weeks. Cultures were stained with Alcian blue (Huegel et al., 2013) and staining quantified (Gutiérrez et al., 2012) as previously described.
Limb organ cultures
Whole forelimb and hindlimb buds were harvested from E12.5 or E13.5 CD1 mouse embryos. Limbs were placed on fine Nitex nylon mesh (03-20/14, Sefar, Buffalo, NY) and partially submerged in DMEM with 1% FBS. Limbs were maintained as matched pairs from each embryo, whereas limbs from one side of each embryo were treated with KGN (1μM) while limbs from the opposite side were treated with DMSO solution (10μl/mL) as controls. Culture medium was changed daily, and limbs were photographed at 0, 24, 48, 72 and 96 hrs with bright light and/or florescent microscopy. Digit growth was quantified with ImageJ software (Abràmoff et al., 2004) as the increase in distance from the metatarsal/proximal phalangeal joint to the distal tip of digit 3 between 0 and 96 hrs (Figure 2, J). Whole mount Alcian blue and β-galactosidase (LacZ) staining were completed using standard protocols.
Fig. 2.
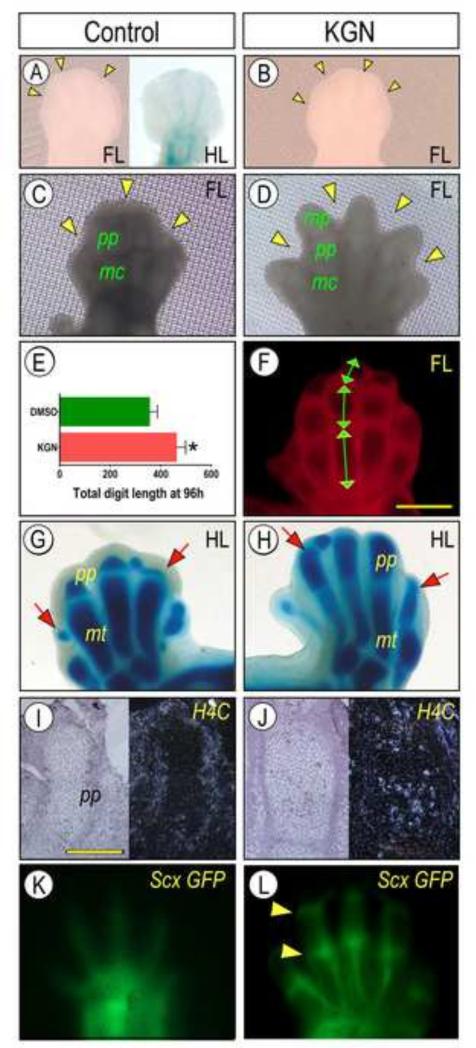
KGN promotes multiple limb developmental processes. E12.5 Forelimb (FL) and E13.5 hindlimb (HL) pairs isolated from mouse embryos were maintained in explant culture in control medium containing DMSO solution (10μl/mL) or 1 μM KGN. (A-D) Microscopic images of limbs at time of isolation (A-B) and at 96 hrs of culture in control (C) or KGN (D) medium. Note that KGN markedly increased overall limb growth, phalangeal elongation and interdigit mesenchymal invagination (arrowheads). Extent of growth stimulation was calculated by measuring length of each phalangeal element (E) and summation (F) using ImageJ software. Data are shown as average total digit length (pixels) + S.E.M. of 5 limbs at 96h (*p<0.05). (A, G-H) Images of E13.5 hindlimb explants at time of isolation (A) and maintained in control (G) or KGN (H) medium for 96 hrs and stained with Alcian blue. Note the significant growth of proximal phalanges (pp, arrows) caused by KGN. (I-J) Bright and dark field images of sections from 96 hrs forelimb specimens processed for analysis of proliferation marker H4C by in situ hybridization. Note the clearly higher hybridization signal over chondrocytes in KGN sample (J) versus control (I). (K-L) Fluorescence images of live forelimbs from E12.5 ScxGFP embryos at 96 hrs in control (K) or KGN (L) conditions. Tendon development and maturation were stimulated by KGN as indicated by stronger and sharper fluorescence signal and intense concentrated signal at joint locations (arrowheads). Bar for panels I-J, 100 μm. mc, metacarpal; mt, matarsal; pp, proximal phalange; and mp, medial phalange.
Joint superficial cell cultures
Superficial cells were isolated from the knee joints of Prg4-mCherry reporter mice at P5 as previously described (Yasuhara et al., 2011). After harvest, cells were expanded to 80% confluence in monolayer culture with DMEM and 10% FBS prior to passage and seeding into 96 well culture plates (Greiner Bio One, Cat#655090). Cells were serum-starved in DMEM with 2% FBS overnight prior to treatment with control 10μl/mL DMSO solution, 100 nM KGN, 1ng/mL TGFβ1 and/or the selective TGFβ signaling inhibitor SB-431542 (10uM, Sigma Cat. #S4317). Where applicable, cells were pretreated with SB-431542 for 90 min prior addition of KGN or TGFβ1. After 18 hrs, cells were fixed with 4% paraformaldehyde and stained with the nuclear stain DAPI. Total Prg4-mCherry reporter and DAPI fluorescence for each well were measured using a fluorescent microplate reader (Synergy HT, Biotek, Winooski, VT).
In situ hybridization
In situ hybridization was carried out limb sections as described (Koyama et al., 1999Limbs maintained in organ culture were fixed overnight in 4% paraformaldehyde before dehydration and paraffin embedding. Sequential 5 μM sections from control and KGN-treated limbs were mounted on the same slide so that data could be directly compared. Sections were treated for 15 min with a freshly prepared solution of 0.25% acetic anhydride in triethanolamine buffer and were hybridized with antisense or sense 35S-labeled riboprobes (approximately 1×106 DPM/section) at 50°C for 16 hrs. Whole-mount in situ hybridization was carried out as previously described with minor modifications (Koyama et al., 1996). After fixation in 4% paraformaldehyde, limbs were dehydrated and rehydrated in EtOH. Limbs were then washed in PBS+1% Tween-20 prior to Proteinase K digestion (20ug/mL) at 37°C for 1hr. Limbs were refixed in 0.2% glutaraldehyde and 4% paraformaldehyde, bleached in 6%H2O2, and prehybridized for 1 hr at 68°C. DIG-labeled probes were synthesized according to manufacturer's instructions (Roche, Cat. #1277073) and hybridized overnight at 68°C. Samples were washed with 50% formamide, 4× SSC, 1% Tween, then 0.5 M NaCl, 0.1 M Tris pH 7, 0.1% Tween, and finally 50% formamide, 4×SSC. Embryos were washed with TBST (0.1 M Tris, 0.15 M NaCl, 1% Tween) and blocked with 1.5% blocking reagent (Roche, Cat. #1099176) in TBST. Probes were detected using an anti-DIG Fab fragment antibody conjugated to alkaline phosphatase, preabsorbed with mouse embryonic powder. Excess antibody was washed with TBST, then NTMT (0.1 M NaCl, 0.1M Tris pH 9.5, 0.05 M MgCl2, 1% Tween), and finally NTMT with 5% polyvinyl alcohol. Antibody was visualized with NBT/BCIP substrate (Roche). cDNA clones used as templates for probes included: a 254 bp mouse histone 4C (H4C) (546–799; AY158963), Gfd5 (1321-1871; NM_008109) and Fgf10 probe included 694-1216 (NM_008002)
Immunoblots
Micromass cultures were harvested in 8M urea solution, disrupted via sonication and electrophoresed on 4–12% SDS-Bis-Tris gels (10 μg total proteins per lane). Transfer to PVDF membranes was completed using an iBlot Dry Blotting system (Invitrogen, Carlsbad, CA). Membranes were blocked for 30 min in 1% BSA (Sigma) and incubated overnight at 4°C with dilutions of antibodies against phospho-Smad1/5/8 (1:1000, Cat. 9511, Cell Signaling Technology, Beverly, MA) or phospho-Smad2/3 (1:1000, Cat. 8828, Cell Signaling Technology). Following incubation with primary antibody, membranes were rinsed in TBST and then incubated with biotin donkey anti-rabbit IgG (1: 10,000, Cat. RL611-7602, Rockland, Gilbertsville, PA) for 1 hr and strepavidin-HRP (1:15,000, Cat.170-6528, BioRad, Hercules, PA) for 1 hr. An enhanced chemiluminescent immunoblotting detection system (Thermo Fisher, Rockland, IL) was used to detect the antigen-antibody complexes. Membranes were re-blotted with antibodies to α-tubulin (1:10000, Cat. T-5168, Sigma) for normalization.
RNA isolation and RT-PCR
For limb organ culture experiments, tissue from the presumptive metacarpophalangeal joint regions of digits 2-4 was isolated after 48 or 96 hrs of culture. Pooled tissue from 4 limbs was used for each replicate sample (3 replicates, 12 limbs total) and disrupted using microcentrifuge grinder pestles (Bel-Art Products, Wayne, NJ). Total RNA was extracted using the RNeasy Micro Kit protocol (Qiagen). For micromass culture experiments, total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to manufacturer's protocols. RNA samples were reverse transcribed using the Quantiscript Reverse transcription system (Qiagen) according to manufacturer's protocol. For the RT2 Profiler BMP/TGFβ signaling array (Qiagen), 0.5 μg of cDNA from each pool of 4 limbs was used per plate. Primer sets used for RT-PCR are outlined in Supplemental Table 1.
Imaging
Bright and dark-field images of in situ hybridization were taken with a SPOT insight camera (Diagnostic Instruments, Inc.) operated with SPOT 5.1 software. Live cell images were taken at room temperature using an inverted ECLIPSE TE2000-U Nikon microscope with Image-Pro Plus 7.0 software (Media Cybernetics, Inc.). RT-PCR gels and immunoblot membranes were photographed using a GE Imagequant Luminescent Image Analysis system with accompanying Control Software version 1.2. Live fluorescent and bright field limb organ culture images were obtained at room temperature using an Olympus MVX10 MacroView microscope and Olympus cellSense Standard imaging software version1.3. A Leica TCS LSI confocal microscope with accompanying software was used for imaging of fluorescently labeled frozen tissue sections.
Results
Chondrogenesis and limb skeletal growth
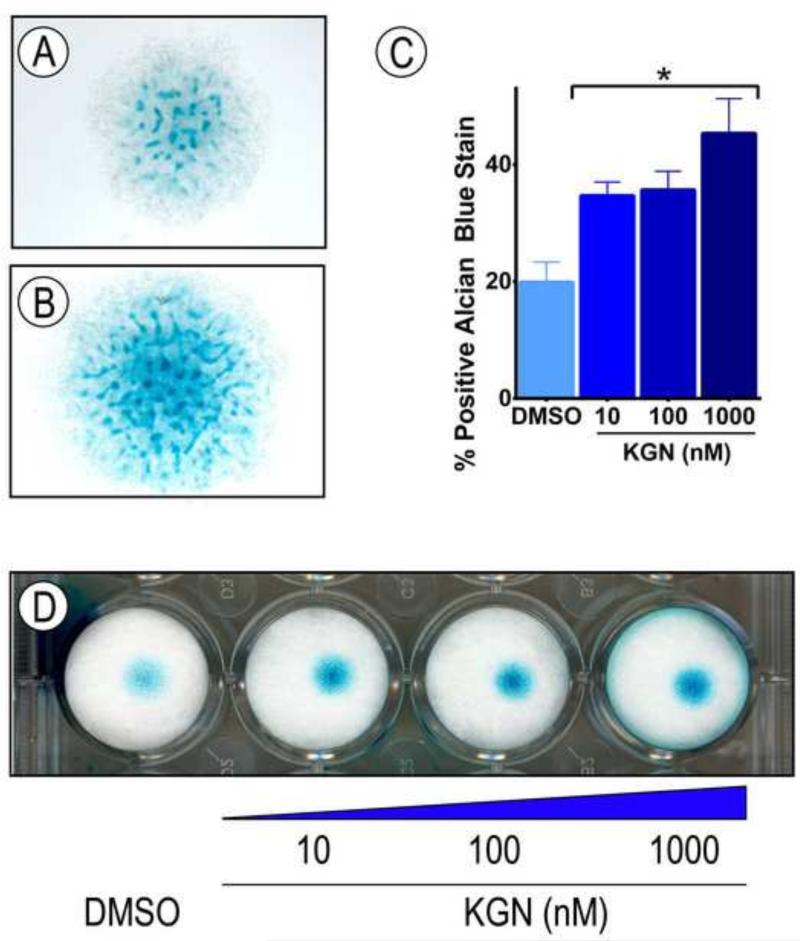
In our recent study we used human bone marrow-derived MSCs to identify KGN and test its chondrogenic capacity (Johnson et al., 2012). To extend these studies, we asked whether the drug would also stimulate chondrogenesis in committed preskeletal mesenchymal cells isolated from the E11.5 mouse embryo limb buds. After isolation, the cells were seeded in micromass culture, a widely used system that allows the cells to reestablish condensation and resume their chondrogenic differentiation (Ahrens et al., 1977). As expected, control micromass cultures displayed several cartilaginous cell nodules by day 5 that strongly stained with alcian blue (Fig. 1A). Treatment of companion cultures with KGN did in fact stimulate chondrogenesis and nodule formation (Fig. 1B) in a dose-dependent manner (Fig. 1D), with over a 100% increase in Alcian blue staining at a 1 μM concentration (Fig.1C).
Fig. 1.
Differentiation of preskeletal mesenchymal cells is stimulated by KGN. Cells were isolated from E11.5 mouse embryo limb buds, spotted in micromass cultures and reared in: (A) control medium containing vehicle solution (DMSO, 10μl/mL); or (B) medium containing 1 μM KGN for 5 days. Cultures were stained with Alcian blue to reveal cartilage nodule formation. (C-D) Similar cultures were treated with different concentrations of KGN (D), and ImageJ was used to calculate fraction of positive Alcian blue staining. Data are shown as averages of four independent experiments + SEM. * p<0.05, as determined by one-way ANOVA.
When limb buds are isolated from the embryo and placed as intact explants in organ culture, they remain viable for several days, but grow and develop slowly (Neubert et al., 1974; Smith et al., 2013). We exploited this explant culture system to further interrogate the developmental properties and action of KGN. Forelimb bud pairs were isolated from the left and right sides of E12.5 mouse embryos. To minimize possible developmental variability from embryo to embryo, one limb bud was reared in control conditions and the contralateral limb bud from the same embryo was reared in KGN-containing medium. At the moment of isolation, the limb buds displayed a typical round morphology at their distal end, with minimal visible skeletal development, limited Alcian blue staining along the digital rays, and minimal tissue invagination at each prospective interdigit location (Fig. 2A-B, arrowheads). By 96 hrs in organ culture, the control forelimb buds had grown moderately and their metacarpal (MC) and middle phalangeal (MP) cartilaginous elements were now appreciable by microscopy on live specimens (Fig. 2C); however, interdigit invagination was still minimal (Fig. 2C, arrowheads). In sharp contrast, the digits of companion limb buds treated with KGN had elongated considerably, with well-defined phalangeal elements identifiable by Alcian blue staining (Supplemental Fig. 1). The interdigit invagination process had also advanced (Fig. 2D, arrowheads), easily revealing each digit under live microscopy (Fig. 2D). Imaging-assisted quantification of digit/phalangeal size (Fig. 2E-F) showed that the total digit length of KGN-treated limbs at 96 hrs was significantly greater than that of control limbs (Fig. 2G, p<0.05).
To verify that similar responses would occur posteriorly hindlimb pairs were harvested from E13.5 embryos, which correspond to the E12.5 forelimbs used above given the approximate half-day developmental delay in hindlimbs versus forelimbs (Towers and Tickle, 2009). E13.5 hindlimb buds were maintained in organ culture for 96 hrs and were whole mount-stained with Alcian blue. In controls, the cartilaginous metatarsal (mt) elements were well developed and strongly stained, but the proximal phalanges (pp) were still rudimentary (Fig. 2G, arrows). Indeed, KGN treatment of companion specimens greatly stimulated overall growth, and this was clearly revealed by the presence of conspicuous Alcian blue-stainable proximal phalanges (pp) (Fig. 2H, arrows). It should be noted that limb buds harvested at younger or later developmental stages were less responsive to KGN.
Development of the most distal forelimb phalanges was further assessed by Safranin-O staining. A loosely-organized mesenchymal cell population was found distal to the middle phalange (mp) in control forelimbs after 96h in organ culture (Supplemental Fig.2A,C). In striking contrast, chondrocytes had condensed to form rudimentary distal phalange in KGN treated digits (Supplemental Fig.2 B, D. Development of the penultimate phalange and digit tip is mediated in part by Fgf signaling (Sanz-Ezquerro and Tickle, 2003). We performed whole-mount in situ hybridization to evaluate Fgf10 mRNA expression in limb explants. After 96h, enhanced staining of KGN treated limbs indicated elevated Fgf signaling as compared to controls (Supplemental Fig. 2E-F).
Skeletal elongation and overall limb outgrowth at different stages are propelled by a number of processes that include cell proliferation. Thus, we examined whether KGN would promote limb cell proliferation by evaluating mRNA expression of proliferation marker histone 4 C (H4C). In control forelimb bud specimens at 96 hrs, there was clear H4C expression in several cell types including perichondrial cells, but expression was lower within the cartilaginous elements themselves (Fig. 2I). H4C expression was, however, much stronger in the chondrocytes of KGN-treated companions (Fig. 2J).
Limb skeletal growth and elongation are coordinated with multiple differentiation, growth and maturation processes that lead to formation of muscles, tendons and other surrounding structures. To test whether KGN stimulatory effects would extend to such processes and structures, we examined tendon development as a representative example. Tendons and ligaments arise from progenitor cell populations expressing the transcription factor Scleraxis (Scx) (Schweitzer et al., 2001). Studies with transgenic ScxGFP reporter mice have revealed that changes in Scx expression patterns coincide with distinct phases of tendon morphogenesis, including initial Scx expression in the superficial proximomedial domain of limb buds at E10.5 and proceeding to development of more distinct tendon fibers by E14.5 (Schweitzer et al., 2001, Pryce et al., 2007). In control E12.5 forelimb buds after 96 hrs of organ culture, ScxGFP expression remained poorly defined and limited to superficial fibrous-like structures along each digit (Fig. 2K). These patterns had much advanced in companion limb buds reared in KGN-containing medium (Fig. 2L). ScxGFP expression was not only strong in the well-defined and elongated tendon structures along the digits, but also at the sites of the metacarpophalangeal and proximal phalangeal joints (Fig. 2L, arrowheads), confirming an increase in localized ScxGFP expression surrounding joint digits as reported by others (Li et al., 2010).
Limb joint development
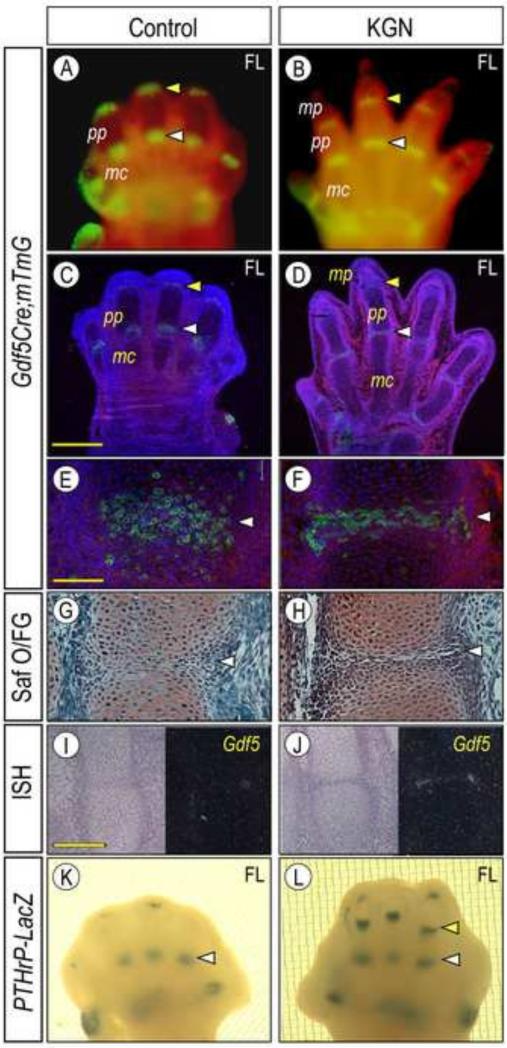
We previously showed that Gdf5-expressing mesenchymal interzone cells emerge at presumptive limb joint sites and eventually give rise to most joint tissues over time, including articular cartilage, ligaments and synovial lining (Koyama et al., 2008). To determine whether KGN would affect these processes, forelimb bud pairs were isolated from E12.5 compound transgenic Gdf5-Cre; ROSA-mTomato/mGFP reporter mice. As shown previously, Gdf5 transcripts first appear at putative metacarpophalangeal joint sites along the digital rays of E12.5 limbs and appear at more distal sites over developmental time (Storm and Kingsley, 1996). The ROSA-mTomato/mGFP reporter mice express constitutive Red Tomato fluorescence prior to Cre-mediated recombination and conditional mGFP fluorescence following recombination (Muzumdar et al., 2007). Accordingly, compound transgenic limb buds reared in organ culture were examined live by fluorescence microscopy over time. Strong Gdf5-Cre driven mGFP reporter signal was appreciable in all 5 incipient metacarpal and phalangeal joints in controls (Fig. 3A, white arrows). Similar overall patterns were seen in companion specimens reared in KGN (Fig. 3B, white arrows), though the digits had elongated considerably and the proximal phalanges (pp) were now clearly appreciable (Fig. 3B). To examine the joint formation process more closely, we prepared histological sections from 96 hr specimens and focused on the incipient joint between metacarpal (mc) and proximal phalange (pp). In controls, the joint was barely visible histologically (Figs. 3C, arrow) and the prospective mGFP-positive interzone cells were scattered and not well compacted (Figs. 3E, 3G). In the KGN-treated companions, however, the joint was clear and exhibited an obvious interzone between the flanking cartilaginous elements and, most importantly, its interzone cells were now compacted, flat-shaped and all transversely oriented with respect to the digit long axis (Fig. 3F, 3H, arrow). KGN also appreciably stimulated Gdf5 expression at the mRNA level as revealed by in situ hybridization (Figs. 3I-3J).
Fig. 3.
Synovial joint development and interzone compaction are promoted by KGN. (A-D) Forelimb bud pairs from E12.5 Gdf5Cre; ROSA-mTomato/mGFP mouse embryos were maintained in control and KGN medium for 96 hrs. (A-B)Whole mount and (C-D) tissue section fluorescence images showing strong mGFP signal at joint sites (arrowheads) in both control (A, C) and treated (B, D) specimens. (E-J) Higher magnification images of serial sections of metacarpal-phalangeal joints (white arrowhead) showing that the interzone had undergone compaction and maturation (F, H) and expressed higher Gdf5 (J) after KGN treatment compared to control specimens (E, G, I). E-F are fluorescence images of mGFP signal; G-H are bright field images of sections after Safranin O/fast green (Saf O/FG) staining; and I-J are bright and dark field images of Gdf5 in situ hybridization. (K-L) Whole mount images of E12.5 PTHrP-LacZ forelimbs pairs that were reared for 96 hrs in control (K) and KGN (L) explant conditions and processed for β-galactosidase detection. Note that the PTHrP-LacZ signal was much stronger in KGN specimens and was also visible in more distal joints (arrowheads) including the metacarpal-phalangeal joint (yellow arrowhead). Bar for panels E-H, 100 μm; bar for panels I-J, 200 μm. mc, metacarpal; pp, proximal phalange; and mp, medial phalange.
As interzone formation progresses, local gene expression patterns change as well (Koyama et al., 2008), and one gene expressed in late and more mature interzones is parathyroid hormone-related protein (PTHrP) as shown recently in PTHrP-LacZ knock-in reporter mice (Chen et al., 2008). In control E12.5 PTHrP-LacZ forelimb explants, PTHrP-LacZ activity became clear in metacarpophalangeal joints by 96 hrs (Fig. 3K, arrows), but remained barely visible in the more distal proximal phalangeal joints (Fig. 3K, arrowheads). KGN treatment of companion limbs increased overall reporter signal that was also strong in more distal joints (Fig. 3L, arrowheads), reiterating KGN's ability to advance joint formation and maturation.
By the end of embryogenesis the joints begin to cavitate, forming two distinct articular surfaces separated by a synovial fluid that contains lubricin/Prg4 and other anti-adhesive macromolecules produced by surface lining cells (Koyama et al., 2008). Cavitation occurs in a proximal to distal fashion as limb joints develop, with the most distal phalangeal joints initiating cavitation by late embryogenesis, and is completed in early postnatal life (Mitrovic, 1978). While the well organized, compacted appearance of KGN treated joints indicated that they had progressed further towards cavitation than controls, there was no evidence that joints had yet formed two distinct articular surfaces. When we assessed cavitation by onset of lubricin/Prg4 expression, we did not observe overt signs of it in control or KGN-treated explants even after prolonged culture. This is likely due to the very young age of the initial E12.5 limb buds and as importantly, the absence of muscle-driven movement needed to drive cavitation (Kahn et al., 2009). To test whether KGN could enhance any aspect of that process, we isolated superficial articular cells from the knee joints of neonatal transgenic Prg4-mCherry reporter mice and reared them in control or KGN-containing medium. We would like to emphasize that Prg4-mCherry is a novel transgenic mouse line. Its construction is described in Materials and Methods, and detailed overall phenotypic characteristics will be detailed elsewhere. Briefly, whole mount fluorescence analysis at P2 showed that there was very strong mCherry reporter activity in limb joints (Fig. 4B-D, Supplemental Fig. 3), and frozen serial sections showed that reporter activity did in fact characterize surface cells of the knee at postnatal stages (Fig. 4E-G). Surface cell populations were isolated from the knee joints of P5 Prg4-mCherry mice by sequential enzymatic digestion as we described previously (Yasuhara et al., 2011) and seeded in monolayer culture. The cells exhibited the expected flat and fibroblastic cytoarchitecture soon after plating (Fig. 4H), and about 10% of them displayed strong Prg4-mCherry reporter activity at later days (Fig. 4I). Treatment of parallel cultures with KGN for 18hr led to a significant increase in the fraction of reporter positive cells, more than doubling it at 1 μM concentration (Fig. 4J and 4L, p<0.05) compared to control (Fig 4I).
Fig. 4.
Analysis of Prg4/lubricin expression. (A) Schematic showing the region of homology used to insert a mCherry encoding transgene into mouse Prg4 locus. (C-D) Whole mount fluorescent images of Prg4/mCherry (red signal) × Col2a1/CFP (blue signal) at postnatal day 2. White squares shown in B are presented at higher magnification in C and D. Note the strong intensity of the signal and its restricted presence at the knee and ankle joint sites. (E-F) Sections of the knee joint harvested (E) P5, (F) P14 and (G) P28 showing that positive signal was restricted to articular cartilage surface of the femur (fe) and patella (pa) and increased with age. (G) By P28, the Prg4/mCherry signal was present and still strong on the joint surface. Prg4 expression is also seen in the underlying bone (*) as reported previously (Ikegawa 2000). (H-K) Fluorescent images of live surface cells isolated from P5 Prg4/mCherry knee joints and viewed immediately after plating (H) or 18 hr treatment with 100nM KGN (J) or 1 ng/ml TGFβ1 (K). Note the increased Prg4/mCherry positive cells after either treatment in comparison to control (I). (L) Percentage Prg4/mCherry fluorescence as compared to DAPI for cultures treated with KGN, TGFβ1 and/or the TGFβ signaling inhibitor SB-431542 plotted versus control. KGN and TGFβ1 Prg4/mCherry fluorescence as compared to DMSO (*p<0.05). Pretreatment with SB-431542 prevented this response in KGN (**p<0.001) and TGFβ1 (***p<0.001) treated samples. Data are expressed as averages of twelve independent wells + S.E.M.
Mechanisms of action
Several signaling pathways and complex interplays amongst them participate in, and promote, skeletal growth and elongation and joint formation, maturation and postnatal consolidation. Thus, we sought to clarify whether and which pathway(s) may account for the strong stimulatory bioactivity of KGN on those processes uncovered by our data above.
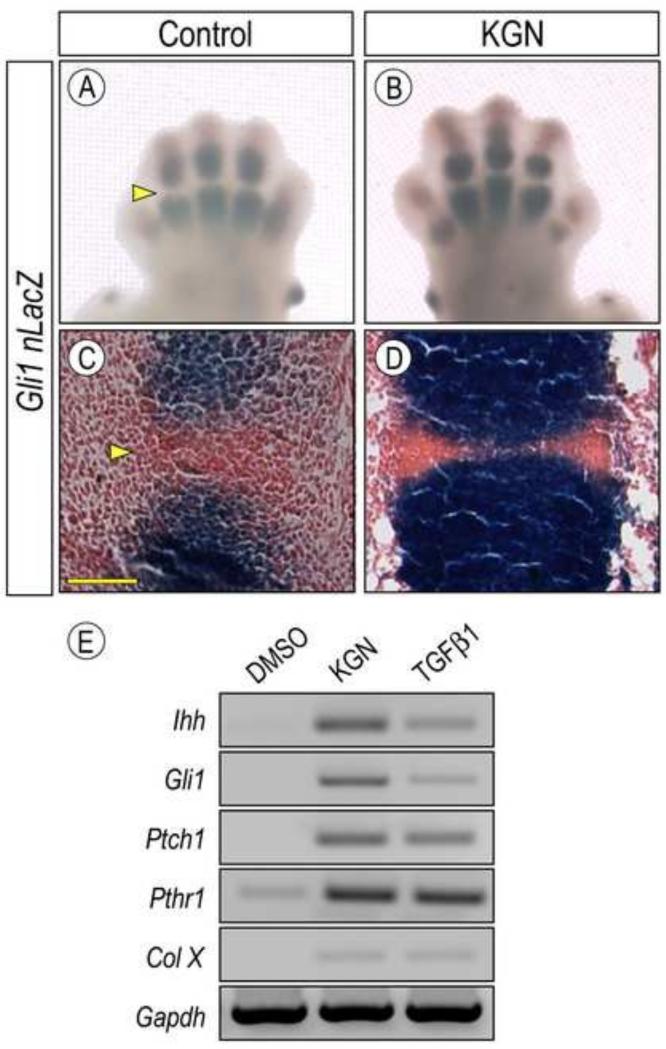
Indian hedgehog (Ihh) is a key signaling protein secreted by prehypertrophic chondrocytes in the growth plate that is needed to sustain chondrocyte proliferation, long bone elongation and bone formation; its genetic ablation also prevents formation of synovial joints, leading to fusion of adjacent cartilaginous anlaga (Koyama et al., 2008; St-Jacques et al., 1999). To monitor hedgehog signaling, we used Gli +/nLacZ knock-in reporter mice that have been widely used as an effective read-out of hedgehog protein distribution and activity on target cells in a variety of developmental systems and processes (Bai et al., 2002). Thus, we isolated forelimb bud pairs from E12.5 Gli +/nLacZ mouse embryos and reared them in absence or presence of 1 μM KGN in organ culture. Whole mount staining at 48 hrs of culture showed that LacZ activity was readily appreciable in the developing cartilaginous phalangeal elements in control limbs (Fig. 5A) and was much lower and nearly absent in incipient joints as expected (Fig. 5A, arrowhead). These patterns were more obvious after tissue sectioning (Fig. 5C). Treatment with KGN boosted LacZ activity dramatically, and this was quite clear by both whole mount and section staining (Fig. 5B, 5D). Interestingly, stimulation of Ihh signaling has previously been shown to increase chondrocyte proliferation within putative hypertrophic zones of developing limbs (Long et. al., 2001). As described above, KGN treatment greatly stimulated mRNA expression of the cell proliferation marker H4C within these regions (Fig. 2 I-J), suggesting a link between increased digit length and upregulation of Ihh signaling by KGN. To verify these observations at the gene expression level, we isolated preskeletal limb bud cells from E11.5 embryos and reared them in micromass culture in control or KGN-containing medium as above. Whole cell RNAs were isolated at 24 hrs of culture so that immediate early responses could be measured. Semi-quantitative RT-PCR analysis showed that KGN treatment had induced expression of Ihh as well as hedgehog target genes Gli-1 and Ptch1 (Fig. 5E), whose expression was barely appreciable in controls (Fig. 5E). This stimulation did depend on active hedgehog signaling since it was wholly prevented by co-treatment of KGN and cyclopamine (CPN), a powerful hedgehog chemical inhibitor (Supplemental Fig. 4, lane 4).
Fig. 5.
Hedgehog signaling is stimulated by KGN. (A-D) Forelimb pairs from E12.5 Gli 1+/nLacZ mouse embryos were kept for 96 hrs in control or KGN-containing medium and processed for whole mount (A-B) or tissue section (C-D) detection of β-galactosidase activity. Note that KGN increased Gli1nLacZ activity in the cartilaginous phalangeal elements (B and D) but remained low at joint sites as expected (A and C, arrowhead). However, the signal-free space had considerably narrowed after treatment (D). (E) RT-PCR analysis of hedgehog signaling and maturation related molecules in E12.5 limb bud mesenchymal cells maintained in micromass culture and treated with 100nM KGN or 2.5 ng/ml TGFβ1 for 24h. Note that similar responses were achieved by treatment with either KGN or TGFβ1.
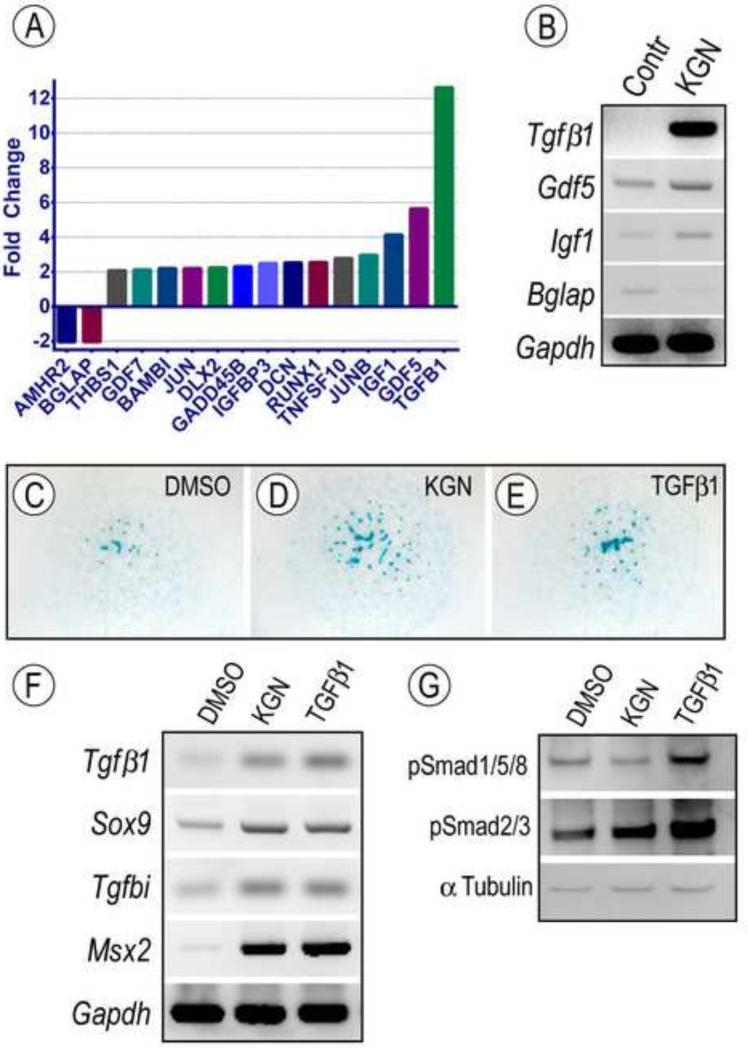
Members of the TGFβ signaling protein superfamily are also important regulators of skeletogenesis and joint development (Seo and Serra, 2007; Spagnoli et al., 2007) and TGFβ1 in particular is a stimulator of chondrogenesis and lubricin expression (Chimal-Monroy and Diaz de Leon, 1997). To obtain a global view of their possible involvement, we isolated total RNAs from the metacarpophalangeal region of E12.5 limb bud pairs reared in control or KGN-containing medium for 96 hrs, a region responsive to KGN treatment as shown above. RNAs were processed for quantitative evaluation using TGFβ/BMP Signaling Pathway RT2 Profiler PCR Arrays, which allow simultaneous analysis of 84 genes involved in TGFβ/BMP action and signal transduction. Using Gapdh expression set at value 1, we found that while several genes were minimally affected by KGN treatment, 16 genes were up-regulated by more than 2 fold (Fig. 6A; Supplemental Table 1). Of these, TGFβ1 expression was up-regulated by more than 10 fold, and Gdf5 and Igf1 expression was up-regulated more than 4 fold (Fig. 6A). The latter increases as well as concurrent decreases in Bglap expression were validated by RT-PCR (Fig. 6B).
Fig. 6.
Modulation of signaling protein activities and growth factor expression by KGN. (A-B) E12.5 limbs were maintained ex vivo in control conditions or medium containing 1 μM KGN or 10uL/mL DMSO as control. (A) Graphic representation of genes regulated more than two fold by KGN treatment in putative joint sites after 96h. Note the multi-fold induction of TGFβ1, Gdf5 and IGF1 expression with Gapdh set at 1; also included are two genes (Amhr2 and Bglap) that were down-regulated. (B) Image of RT-PCR analysis verifying the changes in expression of indicated genes by KGN. (C-G) E11.5 limbs bud mesenchymal cells were maintained in micromass cultures in control conditions or medium containing 100nM KGN or 2.5 ng/ml TGFβ1 for 24h (C-G). Bright field images of micromass cultures stained with alcian blue. Note that both KGN and TGFβ1 stimulated cartilage nodule formation. (F) RT-PCR analysis of changes in gene expression of TGFβ target genes in micromass cultures treated with KGN or TGFβ1 compared to control cultures (DMSO). (G) Immunoblot analysis of levels of phosphorylated Smads in control, KGN-treated and TGFβ1-treated micromass cultures. Both KGN and TGFβ1 strongly up-regulated pSmad2/3 levels, but only TGFβ1 increased pSmad1/5/8 levels. Protein content per lane was verified by immunoblot with α-Tubulin antibodies.
The dramatic increase in TGFβ1 expression led us to ask whether exogenous TGFβ1 would stimulate chondrogenesis in limb bud cell micromasses similar to KGN. A 24h treatment with 2.5 ng/ml recombinant human TGFβ1 or 100nM KGN did in fact elicit comparable stimulations in: (i) Alcian blue-positive nodule formation (Fig. 6C-E); (ii) expression of the master chondrogenic gene Sox9 and of TGFβ-target genes MSX2 and Tgfbi, including TGFβ1 itself (Fig. 6F); and (iii) hedgehog signaling associated genes Ihh, Gli1, Ptch and Pthr1 (Fig 5). The Gli1 increase was prevented by co-treatment with cyclopamine (Supplemental Fig. 4, lane 6). In a similar fashion, TGFβ1 treatment stimulated Prg4/lubricin expression as did KGN (Fig. 4K). Notably, these effects were attenuated by pretreatment of joint superficial cells with the TGFβ signaling inhibitor SB-431542 (P<0.001, Fig. 4L).
Lastly, we assessed activation of the TGFβ signaling cascade in micromass cultures after 24h of KGN treatment. Consistent with up-regulated TGFβ expression, KGN increased the phosphorylation and thus activation of Smad2 and Smad3, the putative TGFβ downstream transcription factors, to a comparable extent as did TGFβ (Fig. 6G). The phosphorylation of other Smad family members (Smad 1/5/8) downstream of BMP signaling cascade was also increased upon TGFβ stimulation, in agreement with previously demonstrated crosstalk between the two signaling pathways (Keller et al., 2011). Interestingly, KGN treatment exhibited no major effect on Smad 1/5/8 phosphorylation, implying potential difference between KGN and TGFβ in determining cell fate in developmental and adult tissues. The differential effects of KGN on Smad 2/3 and suppression of Smad 1/5/8 by KGN were further confirmed in MSCs by gene expression and Ingenuity pathway analyses (Supplemental Fig 5).
Discussion
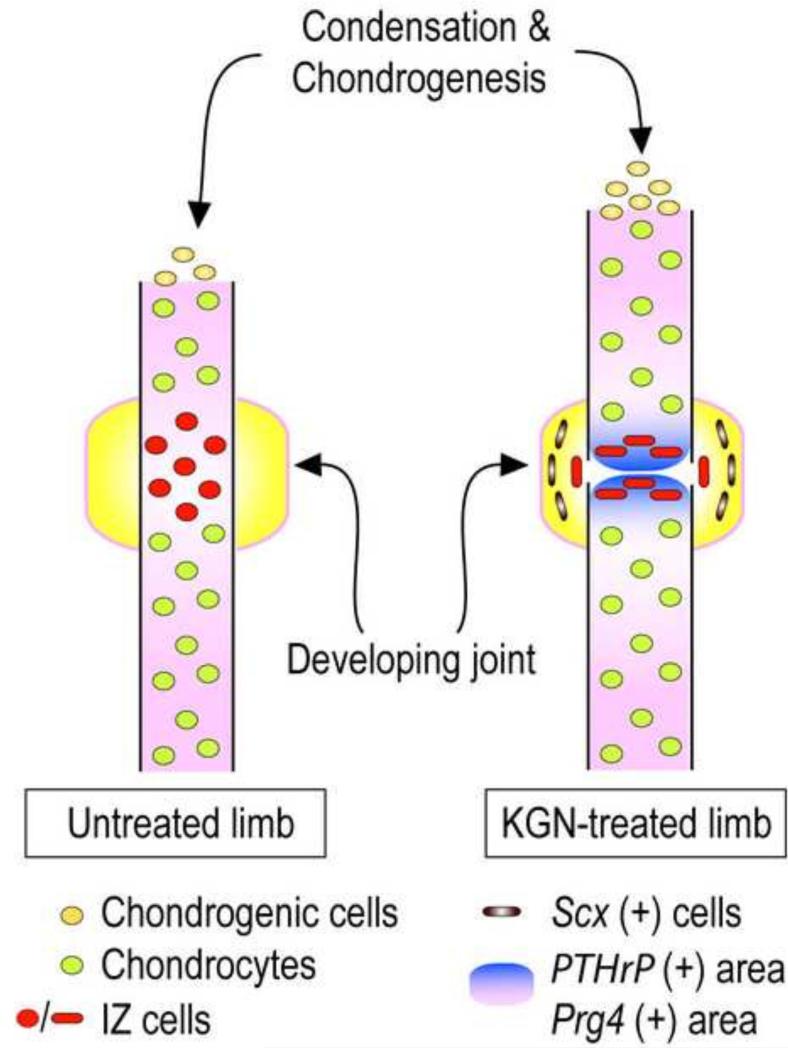
These data provide clear evidence that multiple and diverse limb developmental processes are harmoniously stimulated by KGN, including interzone and synovial joint formation, limb skeletal growth and elongation, interdigit invagination, and tendon maturation. Each of these processes is rather complex, requires programming, differentiation, migration and morphogenetic action of distinct cell types and progenitors, and proceeds through distinct spatio-temporal steps (Zeller et al., 2009b). The data indicate that KGN acted through central, comprehensive mechanisms that normally dictate, promote and orchestrate overall limb development. In particular, the interzone was originally recognized as the first discernible trait of location and impending formation of synovial joints in the developing limb (Holder, 1977). Early experiments found that microsurgical removal of the interzone in the developing chick limb prevented joint formation, establishing the interzone as an essential requirement for joint development (Holder, 1977). Our Gdf5-driven genetic cell tracking data have reiterated its importance and demonstrated the participation of its cells in joint tissue formation (Koyama et al., 2008). What has remained unclear is how the interzone forms to begin with, and how it acquires its compacted, avascular structure composed of flat cells. The future joint sites in the limb are initially cartilaginous and contiguous with the flanking long bone anlaga; in the autopod region, these uninterrupted cartilaginous rods are referred to as digit rays (Hamrick, 2001). The cartilaginous cells are all descendants of early Sox9-expressing progenitors (Akiyama et al., 2005) and it is possible that the interzone cells could be part of this lineage or may also include cells from adjacent perichondrial tissues (Soeda et al., 2010). Our data now provide further insights into the behavior of incipient Gdf5-expressing interzone cells (schematically summarized in Fig. 7). The cells initially appear dispersed and somewhat scattered though still restricted to the joint site, becoming compacted, flattened and oriented perpendicularly upon KGN treatment of the limb explants. How could KGN elicit such a striking and joint promoting response? It is possible that KGN acted on the interzone cells via its strong stimulation of hedgehog and TGFβ signaling pathways and PTHrP expression, all of which are required for joint formation (Koyama et al., 2007; Seo and Serra, 2007; Spagnoli et al., 2007). Thus, signaling could have stimulated interzone cell migration, cell-cell adhesion and cell flattening and compaction (Fig. 7). Alternatively, it could have stimulated growth of the flanking cartilaginous elements, narrowing the interzone space and aiding interzone compaction indirectly. Whatever scenario turns out to be true, the data raise the thorny issue of how these and other KGN-stimulated pathways could promote a mesenchymal character and a flat morphology in interzone cells, while they would promote a cartilaginous character and a round cell morphology in flanking cartilaginous cells. In our previous study we excluded the possibility that KGN stimulates chondrogenesis -and its dependence on a round cell shape (Benya and Shaffer, 1982)- by affecting G- or F-actin and thus cytoskeletal rearrangements (Johnson et al., 2012). These conundrums are not new, and one likely explanation is that a number of additional mechanisms operate within the interzone itself to create and maintain its mesenchymal character, including local expression of potent anti-chondrogenic Wnt proteins and hedgehog inhibitors such as Hip (Koyama et al., 2007; Gao et al., 2009). Thus, the multiple and distinct effects elicited by KGN would reflect not only its extraordinary biological potency, but also intervention of distinct local mechanisms operating in, and instructing, different limb cell populations.
Fig. 7.
Schematic depicting the multiple and concerted processes regulating the early phases of digit skeletal element growth and joint formation. Cartilaginous digit formation and growth involve condensation of mesenchymal cells at the distal end and chondrogenic differentiation. These processes were clearly stimulated by KGN. In the prospective joint site, the incipient interzone cells were initially dispersed and there was minimal cell condensation, compaction and alignment perpendicular to the limb long axis. KGN coordinately promoted these processes that led to interzone compaction and cell alignment, increases in PTHrP and lubricin expression, and consolidation of Scx expression. These complex processes as well the formation of an incipient synovial cavity, were likely promoted by the harmonious and multifaceted action of KGN on TGFβ and hedgehog signaling pathways.
The positive influence KGN exerts on these multiple events and cell types indicates that it acts directly or indirectly on a number of central mechanisms that normally coordinate limb development and growth and would also mediate KGN's comprehensive and multi-faceted effects. Our previous gene array analyses and gain- and loss-of-function studies on bone marrow MSCs suggested that KGN stimulates chondrogenesis by inducing the release of core binding factor β (CBFβ) from filamin A (FLNA) in cytoplasm, and subsequent CBFβ translocation to the nucleus and interaction with RUNX1 (Johnson et al., 2012). This mechanism also likely applies to the developmental processes described here given that these genes are expressed in limb preskeletal mesenchymal cells (Wang et al., 2005; Yoshida et al., 2002), and RUNX1 has been shown to stimulate their differentiation (Wang et al., 2005) (Supplemental Table 2). Our current data suggest that KGN enhances chondrogenesis, skeletal growth and other processes by also stimulating several signaling pathways, including those involving members of the hedgehog, FGF and IGF families, and the TGFβ superfamily. Indeed, hedgehog (Murtaugh et al., 1999), IGF-I (Coxam et al., 1995) and TGFβ1 (Guerne et al., 1994) stimulate chondrogenesis, with both hedgehog (Vortkamp et al., 1996) and TGFβ (Serra et al., 1999) stimulating chondrocyte proliferation via crosstalk with PTHrP. Likewise, TGFβ signaling is a major regulator and stimulator of tendon formation and maturation (Pryce et al., 2007).
The significant stimulation of TGFβ expression and signaling by KGN is in line with the fact that treatment of limb bud mesenchymal cells with KGN or TGFβ1 activates overlapping downstream pathways and effectors, including hedgehog signaling, Smad 2/3 phosphorylation and Sox9 expression. Recent studies have also shown that excess TGFβ signaling can cause impairment of joint function and early onset OA in mouse models, likely due to its deleterious effects on both subchondral bone and joint mechanical support (Zhen et al., 2013). When mesenchymal cells were treated with exogenous TGFβ1, the phosphorylation of Smad 1/5/8 (BMP Smads) increased in addition to the phosphorylation of Smad 2/3 (TGFβ Smads). Phosphorylation and subsequent activation of Smad 1/5/8 have been shown act upstream of RUNX2 (Kempf et al., 2007; Drissi et al., 2003), which is believed to be largely responsible for chondrocyte hypertrophy and terminal differentiation. Interestingly, and distinct from the effects of TGFβ1, KGN treatment specifically increases Smad 2/3 phosphorylation without affecting Smad 1/5/8 phosphorylation significantly. The concerted activation of Smad2/3 and suppression of Smad 1/5/8 have been proposed to inhibit chondrocyte terminal differentiation (van der Kraan et al., 2009), which is consistent with the chondroprotective effects of KGN and with gene expression data showing that KGN suppresses Smad 1/5/8 and RUNX2 expression in MSCs. These data indicate that KGN may promote embryonic joint development and adult chondrogenesis through RUNX1 mediated transcriptional effects, and partially through the activation of the TGFβ as well as hedgehog signaling pathway. The effects on other parallel pathways by KGN, for example down-regulation of BMP family growth factors, may concomitantly regulate the magnitude and duration of TGFβ signaling to suppress potential deleterious activities. Clearly, the detailed mechanisms underlying KGN's effects involving its direct target pathway (FLNA, CBFβ and RUNX1) and downstream effectors (TGFβ, BMPs and hedgehogs) and the potential interplays amongst these pathways warrant further investigation under both developmental and regenerative chondrogenesis conditions.
Finally, we also show that KGN stimulates lubricin/Prg4 expression in joint surface cells (Fig. 4). Lubricin is an essential component of the synovial fluid and its deficiency or mutations are associated with joint disease in patients (Marcelino et al., 1999). This mucin-rich protein is produced by the superficial layer of articular cartilage as well as cells that line the synovial tissue facing the synovial cavity (Jay et al., 2001). Cells from both sites were likely present in the populations we isolated from the P5 joints. The data are in line with our previous findings that KGN stimulated lubricin expression in bone marrow MSCs (Johnson et al., 2012), and indicate that KGN can promote not only early steps in joint formation, but also traits needed for long term joint function and friction-less movement. In conclusion the studies described here support the notion that KGN may provide a promising new approach to the treatment of developmental and degenerative joint disease, but also suggest that the detailed mechanism of action of KGN may involve a complex combination of effects on transcriptional and signaling pathways. The data also suggest that KGN could be a potent tool for limb regeneration and tissue repair strategies.
Supplementary Material
Supplemental Fig.1. Alcian blue staining of E12.5 forelimbs at time of harvest and after 96h in ex vivo organ culture with DMSO control (10ul/mL) or KGN (1μM).
Supplemental Fig.2. Development of forelimb digit tips in response to KGN. (A-E) Safranin-O staining of E12.5 forelimb digits after 96h in DMSO control (10ul/mL) or KGN (1μM). The middle phalange (mp) (A, C) was well developed in control samples, while cells condensed to form a distal phalange (dp) in response to KGN (B, D). (E-F) Whole mount in situ hybridization for Fgf10 mRNA
Supplemental Fig. 3. Expression Prg4/lubricin in control early postnatal Prg4-mCherry × Col2a1-ECFP Reporter mice. (A-C) mCherry fluorescence was strong in superficial cells of the knee and (D-E) wrist at postnatal day 2.
Supplemental Fig. 4. RT-PCR analysis of Gli1 expression in E12.5 limb bud mesenchymal cells in micromass that were treated (+) or co-treated with 1 μM KGN, 2.5 ng/ml TGFβ1 and/or 5μM cyclopamine (CPN). Note that Gli1 was induced by treatment with KGN or TGFβ1 and was counteracted in either case by CPN co-treatment.
Supplemental Fig. 5. Kartogenin modulates multiple components of the TGFβ signaling pathway. hMSC were treated for 24h with 100nM of KGN in serum free DMEM. Ingenuity Pathway Analysis software was used to evaluate regulation of TGFβ signaling. Network components surrounded by a purple border are significantly disregulated by KGN (p<0.01).
Supplemental Table 1. Sequences of primer sets used for RT-PCR analyses.
Supplemental Table 2. Fold change in genes related to TGFβ/BMP Signaling after KGN treatment. RNA isolated from putative joint regions of E12.5 forelimbs after 96h of culture in DMSO or 1μM KGN was used to analyze gene expression on an RT-Profiler TGFβ/BMP Signaling quantitative RT-PCR array. Fold regulation is calculated as change in expression of KGN treated limbs relative to controls.
Highlights.
Embryonic limb development is potently and harmoniously stimulated by KGN treatment.
KGN stimulates several key developmental pathways, including TGF-beta signaling.
KGN may be a powerful tool for limb regeneration and tissue repair strategies.
Acknowledgements
We express our gratitude to: Ms. Hyo-bin Um and Dr. Paul Billings for advice and suggestions; Dr. Ronen Schweitzer for the ScxGFP mice; Dr. Arthur Broadus for the PTHrPLacZ mice; Dr. David Butler for the Col2a1-ECFP mice; and Dr. David Kingsley for the Gdf5Cre mice. We thank the Penn Center for Musculoskeletal Disease core facilities for help and advice with gene array data analysis and informatics. This work was supported by National Institutes of Health grants AR062908 (M.P. and M.E.I) and AR046000 (M.P.), and California Institute for Regenerative Medicine (CIRM) grant TR2-01829 (P.G.S.). R.S.D. is the recipient of a postdoctoral training grant (1F32AR064071) from the National Institutes of Health. The above funding sources had no role in study design, data collection or preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics Intern. 2004;11:36–43. [Google Scholar]
- Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Kim J-E, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- Benya PD, Shaffer JD. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell. 1982;30:215–224. doi: 10.1016/0092-8674(82)90027-7. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Belmonte JCI. Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell Dev. Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- Chen X, Macica CM, Dreyer BE, Hammond VE, Hens JR, Philbrick WM, Broadus AE. Initial Characterization of PTH-Related Protein Gene-Driven lacZ Expression in the Mouse. J. Bone Miner. Res. 2006;21:113–123. doi: 10.1359/JBMR.051005. [DOI] [PubMed] [Google Scholar]
- Chen X, Macica CM, Nasiri A, Broadus AE. Regulation of articular chondrocyte proliferation and differentiation by indian hedgehog and parathyroid hormone–related protein in mice. Arthritis Rheum. 2008;58:3788–3797. doi: 10.1002/art.23985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimal-Monroy J, Diaz de Leon L. Differential effects of transforming growth factors beta 1, beta 2, beta 3 and beta 5 on chondrogenesis in mouse limb bud mesenchymal cells. Int. J. Dev. Biol. 1997;41:91–102. [PubMed] [Google Scholar]
- Coxam V, Miller M, Bowman B, Qi D, Miller S. Insulin-like growth factor 1 and parathyroid hormone effects on the growth of fetal rat metatarsal bones cultured in serum-free medium. Neonatology. 1995;68:368–376. doi: 10.1159/000244257. [DOI] [PubMed] [Google Scholar]
- Drissi MH, Li X, Sheu TJ, Zuscik MJ, Schwarz EM, Puzas JE, Rosier RN, O'Keefe RJ. Runx2/Cbfa1 stimulation by retinoic acid is potentiated by BMP2 signaling through interaction with Smad1 on the collagen X promoter in chondrocytes. J. Cell. Biochem. 2003;90:1287–1298. doi: 10.1002/jcb.10677. [DOI] [PubMed] [Google Scholar]
- Gao B, Hu J, Stricker S, Cheung M, Ma G, Law KF, Witte F, Briscoe J, Mundlos S, He L. A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature. 2009;458:1196–1200. doi: 10.1038/nature07862. [DOI] [PubMed] [Google Scholar]
- Geng H, Lan R, Wang G, Siddiqi AR, Naski MC, Brooks AI, Barnes JL, Saikumar P, Weinberg JM, Venkatachalam MA. Inhibition of autoregulated TGFβ signaling simultaneously enhances proliferation and differentiation of kidney epithelium and promotes repair following renal ischemia. Am. J. Pathol. 2009;174:1291–1308. doi: 10.2353/ajpath.2009.080295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kγ origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerne PA, Sublet A, Lotz M. Growth factor responsiveness of human articular chondrocytes: distinct profiles in primary chondrocytes, subcultured chondrocytes, and fibroblasts. J. Cell. Physiol. 1994;158:476–484. doi: 10.1002/jcp.1041580312. [DOI] [PubMed] [Google Scholar]
- Gutiérrez ML, Guevara J, Barrera LA. Semi-automatic grading system in histologic and immunohistochemistry analysis to evaluate in vitro chondrogenesis. Univ. Scientiarum. 2012;17:167–178. [Google Scholar]
- Hamrick MW. Primate origins: evolutionary change in digital ray patterning and segmentation. J. Hum. Evol. 2001;40:339–351. doi: 10.1006/jhev.2001.0467. [DOI] [PubMed] [Google Scholar]
- Holder N. An experimental investigation into the early development of the chick elbow joint. J. Embryol. Exp. Morphol. 1977;39:115–127. [PubMed] [Google Scholar]
- Huegel J, Mundy C, Sgariglia F, Nygren P, Billings PC, Yamaguchi Y, Koyama E, Pacifici M. Perichondrium phenotype and border function are regulated by Ext1 and heparan sulfate in developing long bones: A mechanism likely deranged in Hereditary Multiple Exostoses. Dev. Biol. 2013;377:100–112. doi: 10.1016/j.ydbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegawa S, Sano M, Koshizuka Y, Nakamura Y. Isolation, characterization and mapping of the mouse and human PRG4 (proteoglycan 4) genes. Cytogenet. Cell Genet. 2000;90:291–297. doi: 10.1159/000056791. [DOI] [PubMed] [Google Scholar]
- Jay GD, Tantravahi U, Britt DE, Barrach HJ, Cha CJ. Homology of lubricin and superficial zone protein (SZP): products of megakaryocyte stimulating factor (MSF) gene expression by human synovial fibroblasts and articular chondrocytes localized to chromosome 1q25. J. Orthop. Res. 2001;19:677–687. doi: 10.1016/S0736-0266(00)00040-1. [DOI] [PubMed] [Google Scholar]
- Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X, Schultz PG. A stem cell-based approach to cartilage repair. Science. 2012;336:717–721. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- Kahn J, Shwartz Y, Blitz E, Krief S, Sharir A, Breitel DA, Rattenbach R, Relaix F, Maire P, Rountree RB, Kingsley DM, Zelzer E. Muscle Contraction Is Necessary to Maintain Joint Progenitor Cell Fate. Dev. Cell. 2009;16:734–743. doi: 10.1016/j.devcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu. Rev. Cell and Dev. Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- Keller B, Yang T, Chen Y, Munivez E, Bertin T, Zabel B, Lee B. Interaction of TGFβ and BMP signaling pathways during chondrogenesis. PloS one. 2011;6:e16421. doi: 10.1371/journal.pone.0016421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf H, Ionescu A, Udager AM, Lassar AB. Prochondrogenic signals induce a competence for Runx2 to activate hypertrophic chondrocyte gene expression. Dev. Dyn. 2007;236:1954–1962. doi: 10.1002/dvdy.21205. [DOI] [PubMed] [Google Scholar]
- Khan I, Redman S, Williams R, Dowthwaite G, Oldfield S, Archer C. The development of synovial joints. Curr. Top. Dev. Biol. 2007;79:1–36. doi: 10.1016/S0070-2153(06)79001-9. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Kronenberg HM. Overview of Skeletal Development, Skeletal Development and Repair. Springer; 2014. pp. 3–12. [DOI] [PubMed] [Google Scholar]
- Koyama E, Golden EB, Kirsch T, Adams SL, Chandraratna RA, Michaille J-J, Pacifici M. Retinoid signaling is required for chondrocyte maturation and endochondral bone formation during limb skeletogenesis. Dev. Biol. 1999;208:375–391. doi: 10.1006/dbio.1999.9207. [DOI] [PubMed] [Google Scholar]
- Koyama E, Ochiai T, Rountree RB, Kingsley DM, Enomoto-Iwamoto M, Iwamoto M, Pacifici M. Synovial Joint Formation during Mouse Limb Skeletogenesis. Ann. N. Y. Acad. Sci. 2007;1116:100–112. doi: 10.1196/annals.1402.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree RB, Kingsley DM, Iwamoto M, Enomoto-Iwamoto M, Pacifici M. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev. Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama E, Yamaai T, Iseki S, Ohuchi H, Nohno T, Yoshioka H, Hayashi Y, Leatherman JL, Golden EB, Noji S. Polarizing activity, Sonic hedgehog, and tooth development in embryonic and postnatal mouse. Dev. Dyn. 1996;206:59–72. doi: 10.1002/(SICI)1097-0177(199605)206:1<59::AID-AJA6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Li Y, Qiu Q, Watson SS, Schweitzer R, Johnson RL. Uncoupling skeletal and connective tissue patterning: conditional deletion in cartilage progenitors reveals cell-autonomous requirements for Lmx1b in dorsal-ventral limb patterning. Development. 2010;137:1181–1188. doi: 10.1242/dev.045237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- Mackie EJ, Tatarczuch L, Mirams M. The skeleton: a multi-functional complex organ. The growth plate chondrocyte and endochondral ossification. J. Endocrinol. 2011;211:109–121. doi: 10.1530/JOE-11-0048. [DOI] [PubMed] [Google Scholar]
- Marcelino J, Carpten JD, Suwairi WM, Gutierrez OM, Schwartz S, Robbins C, Sood R, Makalowska I, Baxevanis A, Johnstone B. CACP, encoding a secreted proteoglycan, is mutated in camptodactyly-arthropathy-coxa vara-pericarditis syndrome. Nat. Genet. 1999;23:319–322. doi: 10.1038/15496. [DOI] [PubMed] [Google Scholar]
- Marini JC, Forlino A. Replenishing cartilage from endogenous stem cells. N. Engl. J. Med. 2012;366:2522–2524. doi: 10.1056/NEJMcibr1204283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maye P, Fu Y, Butler DL, Chokalingam K, Liu Y, Floret J, Stover ML, Wenstrup R, Jiang X, Gooch C, Rowe D. Generation and characterization of Col10a1-mcherry reporter mice. Genesis. 2011;49:410–418. doi: 10.1002/dvg.20733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic D. Development of the diarthrodial joints in the rat embryo. Am. J. Anat. 1978;151:475–485. doi: 10.1002/aja.1001510403. [DOI] [PubMed] [Google Scholar]
- Mundy C, Yasuda T, Kinumatsu T, Yamaguchi Y, Iwamoto M, Enomoto-Iwamoto M, Koyama E, Pacifici M. Synovial joint formation requires local Ext1 expression and heparan sulfate production in developing mouse embryo limbs and spine. Dev. Biol. 2011;351:70–81. doi: 10.1016/j.ydbio.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13:225–237. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Neubert D, Merker HJ, Tapken S. Comparative studies on the prenatal development of mouse extremities in vivo and in organ culture. Naunyn Schmiedebergs Arch. Pharmacol. 1974;286:251–270. doi: 10.1007/BF00498309. [DOI] [PubMed] [Google Scholar]
- Onyekwelu I, Goldring MB, Hidaka C. Chondrogenesis, joint formation, and articular cartilage regeneration. J. Cell. Biochem. 2009;107:383–392. doi: 10.1002/jcb.22149. [DOI] [PubMed] [Google Scholar]
- Pacifici M, Koyama E, Iwamoto M. Mechanisms of synovial joint and articular cartilage formation: recent advances, but many lingering mysteries. Birth Defects Res. C Embryo Today. 2005;75:237–248. doi: 10.1002/bdrc.20050. [DOI] [PubMed] [Google Scholar]
- Pazin DE, Gamer LW, Cox KA, Rosen V. Molecular profiling of synovial joints: Use of microarray analysis to identify factors that direct the development of the knee and elbow. Devel. Dynam. 2012;241:1816–1826. doi: 10.1002/dvdy.23861. [DOI] [PubMed] [Google Scholar]
- Pitsillides A, Ashhurst DE. A critical evaluation of specific aspects of joint development. Devel. Dynam. 2008;237:2284–2294. doi: 10.1002/dvdy.21654. [DOI] [PubMed] [Google Scholar]
- Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Devel. Dynam. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- Ray K. Therapy: Tissue engineering: harnessing stem cells in cartilage repair. Nat. Rev.s Rheum. 2012;8:308–308. doi: 10.1038/nrrheum.2012.62. [DOI] [PubMed] [Google Scholar]
- Rountree RB, Schoor M, Chen H, Marks ME, Harley V, Mishina Y, Kingsley DM. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2004;2:e355. doi: 10.1371/journal.pbio.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LJ. Etiology of osteoarthritis: genetics and synovial joint development. Nat. Rev. Rheum. 2012;8:77–89. doi: 10.1038/nrrheum.2011.199. [DOI] [PubMed] [Google Scholar]
- Sanz-Ezquerro JJ, Tickle C. Fgf signaling controls the number of phalanges and tip formation in developing digits. Curr. Biol. 2003;13:1830–1836. doi: 10.1016/j.cub.2003.09.040. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- Seo H-S, Serra R. Deletion of Tgfbr2 in Prx1-cre expressing mesenchyme results in defects in development of the long bones and joints. Dev. Biol. 2007;310:304–316. doi: 10.1016/j.ydbio.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra R, Karaplis A, Sohn P. Parathyroid Hormone–related Peptide (PTHrP)-dependent and -independent Effects of Transforming Growth Factor β (TGF-β) on Endochondral Bone Formation. Journal Cell Biol. 1999;145:783–794. doi: 10.1083/jcb.145.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EL, Kanczler JM, Oreffo RO. A new take on an old story: chick limb organ culture for skeletal niche development and regenerative medicine evaluation. Eur. Cell Mater. 2013;26:91–106. doi: 10.22203/ecm.v026a07. discussion 106. [DOI] [PubMed] [Google Scholar]
- Soeda T, Deng JM, de Crombrugghe B, Behringer RR, Nakamura T, Akiyama H. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis. 2010;48:635–644. doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Estrada KD, Lyons KM. Smad signaling in skeletal development and regeneration. Cytokine Growth Factor Rev. 2009;20:379–388. doi: 10.1016/j.cytogfr.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnoli A, O'Rear L, Chandler RL, Granero-Molto F, Mortlock DP, Gorska AE, Weis JA, Longobardi L, Chytil A, Shimer K. TGF- signaling is essential for joint morphogenesis. J. Cell Biol. 2007;177:1105. doi: 10.1083/jcb.200611031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee S-J. Limb alterations in brachypodism mice due to mutations in a new member of the TGF[beta]-superfamily. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development. 1996;122:3969–3979. doi: 10.1242/dev.122.12.3969. [DOI] [PubMed] [Google Scholar]
- Towers M, Tickle C. Growing models of vertebrate limb development. Development. 2009;136:179–190. doi: 10.1242/dev.024158. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Frank S, Pap T, Bertrand J. Cartilage biology, pathology, and repair. Cell. Mol. Life Sci. 2010;67:4197–4211. doi: 10.1007/s00018-010-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Kraan P, Blaney Davidson E, Blom A, Van den Berg W. TGF-beta signaling in chondrocyte terminal differentiation and osteoarthritis: modulation and integration of signaling pathways through receptor-Smads. Osteoarthr.and Cartil. 2009;17:1539–1545. doi: 10.1016/j.joca.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Wang Y, Belflower RM, Dong Y-F, Schwarz EM, O'Keefe RJ, Drissi H. Runx1/AML1/Cbfa2 Mediates Onset of Mesenchymal Cell Differentiation Toward Chondrogenesis. J. Bone Miner. Res. 2005;20:1624–1636. doi: 10.1359/JBMR.050516. [DOI] [PubMed] [Google Scholar]
- Xu T, Zhang M, Laurent T, Xie M, Ding S. Concise Review: Chemical Approaches for Modulating Lineage-Specific Stem Cells and Progenitors. Stem Cells Trans. Med. 2013;2:355. doi: 10.5966/sctm.2012-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara R, Ohta Y, Yuasa T, Kondo N, Hoang T, Addya S, Fortina P, Pacifici M, Iwamoto M, Enomoto-Iwamoto M. Roles of β-catenin signaling in phenotypic expression and proliferation of articular cartilage superficial zone cells. Lab. Invest. 2011;91:1739–1752. doi: 10.1038/labinvest.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Pacifici M. Vascular regression is required for mesenchymal condensation and chondrogenesis in the developing limb. Dev. Dyn. 2001;222:522–533. doi: 10.1002/dvdy.1212. [DOI] [PubMed] [Google Scholar]
- Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Core-binding factor β interacts with Runx2 and is required for skeletal development. Nat. Genet. 2002;32:633–638. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- Zeller R, Lopez-Rios J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat. Rev. Gene.t. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Zhen G, Wen C, Jia X, Li Y, Crane JL, Mears SC, Askin FB, Frassica FJ, Chang W, Yao J, Carrino JA, Cosgarea A, Artemov D, Chen Q, Zhao Z, Zhou X, Riley L, Sponseller P, Wan M, Lu WW, Cao X. Inhibition of TGF-[beta] signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat. Med. 2013;19:704–712. doi: 10.1038/nm.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig.1. Alcian blue staining of E12.5 forelimbs at time of harvest and after 96h in ex vivo organ culture with DMSO control (10ul/mL) or KGN (1μM).
Supplemental Fig.2. Development of forelimb digit tips in response to KGN. (A-E) Safranin-O staining of E12.5 forelimb digits after 96h in DMSO control (10ul/mL) or KGN (1μM). The middle phalange (mp) (A, C) was well developed in control samples, while cells condensed to form a distal phalange (dp) in response to KGN (B, D). (E-F) Whole mount in situ hybridization for Fgf10 mRNA
Supplemental Fig. 3. Expression Prg4/lubricin in control early postnatal Prg4-mCherry × Col2a1-ECFP Reporter mice. (A-C) mCherry fluorescence was strong in superficial cells of the knee and (D-E) wrist at postnatal day 2.
Supplemental Fig. 4. RT-PCR analysis of Gli1 expression in E12.5 limb bud mesenchymal cells in micromass that were treated (+) or co-treated with 1 μM KGN, 2.5 ng/ml TGFβ1 and/or 5μM cyclopamine (CPN). Note that Gli1 was induced by treatment with KGN or TGFβ1 and was counteracted in either case by CPN co-treatment.
Supplemental Fig. 5. Kartogenin modulates multiple components of the TGFβ signaling pathway. hMSC were treated for 24h with 100nM of KGN in serum free DMEM. Ingenuity Pathway Analysis software was used to evaluate regulation of TGFβ signaling. Network components surrounded by a purple border are significantly disregulated by KGN (p<0.01).
Supplemental Table 1. Sequences of primer sets used for RT-PCR analyses.
Supplemental Table 2. Fold change in genes related to TGFβ/BMP Signaling after KGN treatment. RNA isolated from putative joint regions of E12.5 forelimbs after 96h of culture in DMSO or 1μM KGN was used to analyze gene expression on an RT-Profiler TGFβ/BMP Signaling quantitative RT-PCR array. Fold regulation is calculated as change in expression of KGN treated limbs relative to controls.