Abstract
For the establishment of an effective root nodule symbiosis, a coordinated regulation of the infection processes between the epidermis and cortex is required. However, it remains unclear whether the symbiotic genes identified so far are involved in epidermal and/or cortical infection, e.g. epidermal and cortical infection thread formation or cortical cell division. To analyze the symbiotic gene requirements of the infection process, we have developed an epidermis-specific expression system (pEpi expression system) and examined the symbiotic genes NFR1, NFR5, NUP85, NUP133, CASTOR, POLLUX, CCaMK, CYCLOPS, NSP1 and NSP2 for involvement in the infection process in the epidermis and cortex. Our study shows that expression of the upstream common symbiosis genes CASTOR, POLLUX, NUP85 and NUP133 in the epidermis is sufficient to induce formation of infection threads and cortical cell division, leading to the development of fully effective nodules. Our system also shows a requirement of CCaMK, CYCLOPS, NSP1 and NSP2 for the entire nodulation process, and the different contributions of NFR1 and NFR5 to cortical infection thread formation. Based on these analyses using the pEpi expression system, we propose a functional model of symbiotic genes for epidermal and cortical infection.
Keywords: Lotus japonicus, root nodule symbiosis, infection threads, cortical cell division, root epidermis, root cortex, epidermis-specific expression system (pEpi expression system)
Introduction
Leguminous plants have the ability to establish endosymbiosis with soil bacteria (collectively termed rhizobia), and form root nodules in which rhizobia fix atmospheric nitrogen (Kouchi et al., 2010). Recent studies of symbiotic mutants of the model legumes Lotus japonicus and Medicago truncatula have led to the identification of a number of host genes that regulate root nodule and/or arbuscular mycorrhizal symbioses (Parniske, 2008). In L. japonicus, two LysM receptor-like kinases, NFR1 and NFR5, are essential for specific recognition of Nod factors secreted from Mesorhizobium loti (Madsen et al., 2003; Radutoiu et al., 2003). Downstream of these receptors, a leucine-rich repeat receptor kinase (SYMRK; Stracke et al., 2002), three components of the nucleopore (NUP133, NUP85 and NENA; Kanamori et al., 2006; Saito et al., 2007; Groth et al., 2010), two cation channel proteins (CASTOR and POLLUX; Ané et al., 2004; Imaizumi-Anraku et al., 2005), Ca2+/calmodulin-dependent protein kinase (CCaMK; Lévy et al., 2004; Gleason et al., 2006; Tirichine et al., 2006) and a coiled-coil domain-containing protein (CYCLOPS; Yano et al., 2008) have been identified as components of the ‘common symbiosis pathway’ that is required for both root nodule and arbuscular mycorrhizal symbioses. Following the perception of Nod factors through NFR1 and NFR5, Ca2+ influx at the tip of the root hair and Ca2+ spiking at the perinuclear region of the root hair are induced. Among the eight common symbiosis pathway genes, SYMRK, NUP85, NUP133, NENA, CASTOR and POLLUX are essential for the generation of Ca2+ spiking in response to Nod factors (Miwa et al., 2006; Groth et al., 2010). Therefore, these symbiotic genes are denoted as ‘upstream genes’ of the common symbiosis pathway (Hayashi et al., 2010). In contrast, CCaMK and CYCLOPS are positioned downstream of Ca2+ spiking (Miwa et al., 2006). Two GRAS family transcription factor genes (NSP1 and NSP2; Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2006) and a putative transcription factor gene (NIN, Schauser et al., 1999) are located downstream of Ca2+ spiking (Miwa et al., 2006), and are necessary for both nodule organogenesis and rhizobial infection, accompanied by the formation of infection threads (ITs).
Mutational perturbation of the symbiotic genes mentioned above leads to severe impairment of IT formation in root hairs. Thus, the majority of symbiotic mutants have a non-nodulation phenotype, suggesting that these symbiotic genes function at least in the root epidermis, but the role of these genes in cortical IT formation and cortical cell division remains unclear. However, in the case of the symrk–14 and nena–1 mutants, although epidermal IT formation is impaired, cortical IT formation and cortical cell division occur, resulting in the formation of effective nodules (Groth et al., 2010; Kosuta et al., 2011). In contrast, although epidermal IT formation occurs, arrested development of cortical ITs was observed in ccamk–14 mutant (Liao et al., 2012), making it difficult to understand the functionality of those symbiotic genes in both the epidermis and cortex. Functional analyses of symbiotic genes, using a combination of loss-of-function mutants transformed with gain-of-function CCaMK or LHK1 (which encodes a cytokinin receptor kinase), have revealed regulation pathways for both nodule organogenesis (Gleason et al., 2006; Tirichine et al., 2006, 2007; Marsh et al., 2007; Yano et al., 2008) and rhizobial infection (Hayashi et al., 2010; Madsen et al., 2010). However, it remains uncertain whether expression of these genes in the epidermis or cortex or both is a prerequisite for the accommodation of rhizobia. Using the pLeEXT (epidermis) and pCO2 (cortex) tissue-specific promoters, Rival et al. (2012) proposed a model for the control of nodule organogenesis by NFP and DMI3, orthologs of NFR5 and CCaMK, respectively, in M. truncatula (Rival et al., 2012). However, the involvement of other symbiotic genes for nodule organogenesis remains elusive, and the roles of the symbiotic genes in epidermal and cortical IT formation require clarification. In order to dissect the symbiotic cellular responses that occur in two cell layers, we isolated Lotus expansin genes that are expressed specifically in the root epidermis, and developed a root epidermis-specific expression system called the ‘pEpi expression system’, which makes it possible to express a symbiotic gene in the epidermis. Using the pEpi expression system, we analyzed the requirements of the symbiotic genes NFR1, NFR5, NUP85, NUP133, CASTOR, POLLUX, CCaMK, CYCLOPS, NSP1 and NSP2, for epidermal and cortical rhizobial infection and nodule organogenesis.
Results
A root epidermis-specific expression system using the promoter region of Lotus expansinA7 orthologs
To develop a root epidermis-specific expression system (pEpi expression system) in L. japonicus, we searched for Lotus orthologs of expansinA7 (AtEXPA7), an expansin gene that is known to be expressed specifically in the root hair cells of Arabidopsis (Cho and Cosgrove, 2002). A blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using AtEXPA7 and AtEXPA18 as query sequences resulted in identification of two orthologs in L. japonicus. The amino acid sequence between 55 (Gly) and 181 (Ile) of AtEXPA7, and the corresponding regions of AtEXPA18, MtEXPA7, MtEXPA8 (Kim et al., 2006) and Lotus expansin protein sequences were aligned using clustalw (http://clustalw.ddbj.nig.ac.jp/) (Figure S1). Based on this phylogenetic tree, we named two Lotus expansin orthologs LjEXPA7 and LjEXPA8 [accession numbers AP010346 (region 34 276–34 741) and AP009544 (region 25 411–25 974), respectively]; these genes were closely related to MtEXPA7 and MtEXPA8, respectively. To evaluate epidermis-specific expression of LjEXPA genes, root hairs of Lotus roots were isolated. The RH101 and RH102 genes, both of which have been reported to be expressed specifically in root hairs (Maekawa et al., 2005), were highly expressed in the isolated root hairs in comparison with stripped roots that contain stele, cortex and epidermal cells but not root hairs (Figure S2). The LjEXPA7 and LjEXPA8 transcripts also showed a root hair-specific expression pattern (Figure1a).
Figure 1.
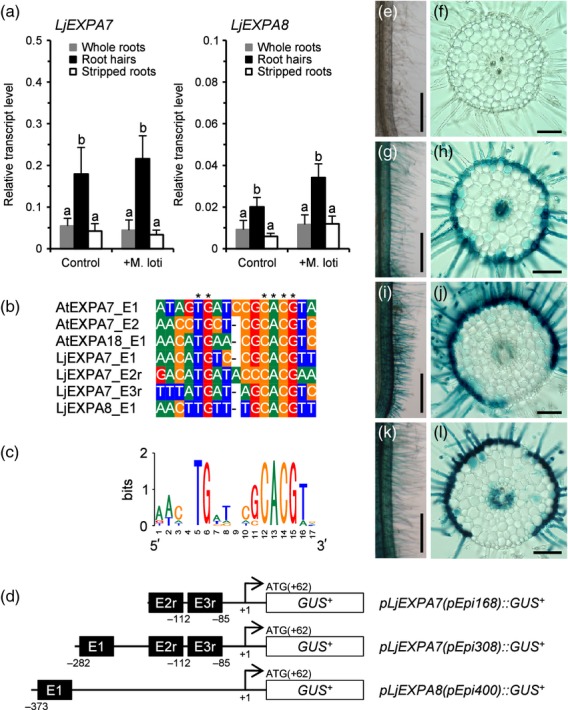
Root epidermis-specific expression system using promoter regions of AtEXPA7 orthologs in Lotus. (a) Real-time RT–PCR analysis of LjEXPA7 and LjEXPA8 transcripts in root hairs, stripped roots or whole roots 2 days after mock inoculation (Control) or inoculation with Mesorhizobium loti (+M. loti). Values are means and SD of three or four biological replications. Bars with different letters are significantly different (Tukey–Kramer multiple comparison test, P < 0.05). (b) Alignment of root hair-specific cis element (RHE) core sequences of promoter regions of EXPA orthologs. Asterisks indicate highly conserved nucleotides within the RHE core. (c) Sequence logo representing the conservation of nucleotides at each position in the RHE cores. The sequence logo was generated using WebLogo (http://weblogo.berkeley.edu/). (d) Illustrations of three types of RHE-containing promoter fused to the β–glucuronidase reporter gene (GUSplus; GUS+). The numbers below the sequence are the starting nucleotide positions of the RHE core (black boxes). Nucleotide positions are numbered relative to the attR1 site of a Gateway destination vector cassette (the ATG start codon is at nucleotide positions 62–64). An ‘r’ after the RHE number indicates a reverse orientation of the RHE. (e–l) Promoter GUS analysis. Lotus roots were transformed with an empty vector (e, f), pEpi168::GUS+ (g, h), pEpi308::GUS+ (i, j) or pEpi400::GUS+ (k, l) by Agrobacterium rhizogenes-mediated hairy root transformation. (f, h, j, l) Transverse sections of transformed roots. Scale bars = 500 μm (e, g, i, k) and 100 μm (f, h, j, l).
The promoter regions of AtEXPA7 and AtEXPA18 contain conserved root hair-specific cis elements (RHEs) that confer root hair-specific expression of expansins in angiosperms (Kim et al., 2006). The RHE core consists of 16 or 17 nucleotides, and includes two conserved motifs, TG and CACG. As well as the AtEXPA genes, multiple RHEs were found in the promoter regions of both LjEXPA7 and LjEXPA8 (Figures1b,c and S3). We selected three types of RHE-containing promoters, and tested their potential to drive root epidermis-specific expression of the β–glucuronidase gene (GUSplus; GUS+) in Lotus roots (Figure1d). pEpi::GUS+ constructs were introduced by Agrobacterium rhizogenes-mediated hairy root transformation. GUS expression was observed mainly in epidermal cells and partially in the stele of transformed roots (Figure1 g–l). In particular, pEpi308, which includes three RHEs, exhibited the most specific expression in epidermis (Figure1i,j), and this epidermal expression was not affected by inoculation of M. loti (Figure S4). This is consistent with the fact that the abundance of the LjEXPA7 transcript in root hairs was not changed in response to M. loti (Figure1a). Thus we selected pEpi308 for establishment of the Epi expression system. To avoid the possibility that gene expression under the control of the Epi promoter interferes with rhizobial infection processes, we examined the rhizobial infection phenotypes on wild-type Gifu B–129 (wt) roots expressing GUS+ under the control of the pEpi (wt/pEpi::GUS+) or CaMV 35S promoter (wt/p35S::GUS+). Upon inoculation of DsRed-labeled M. loti (Maekawa et al., 2009), infection events, including IT formation and nodule organogenesis, occurred on the roots of wt/pEpi::GUS+ to an equivalent extent as the roots of wt/p35S::GUS+. These observations indicate that heterologous gene expression via pEpi does not affect rhizobial infection processes (Table1 and Figure S5).
Table 1.
Complementation analysis of symbiosis-defective phenotypes using the pEpi expression system
| Lotus line | Construct | Root hair phenotype (%) | Nodulation phenotype | Total number of plants | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No colonization | Micro-colony | Short ITs | ITs | (%) | Nodules per nodulated plant (mean ± SE) | |||||
| Nod− | Bump | Nod | ||||||||
| Wild-type | Empty | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 17.5 ± 1.1 | 31 |
| Wild-type | pEpi::GUS+ | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 22.7 ± 1.8 | 35 |
| Wild-type | p35S::GUS+ | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 20.9 ± 1.5 | 30 |
| nfr1–4 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 20 |
| nfr1–4 | pEpi::NFR1 | 0.9 | 34.9 | 44.9 | 19.3 | 7.3 | 67.9 | 24.8 | 1.1 ± 0.1 | 109 |
| nfr1–4 | p35S::NFR1 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 18.3 ± 0.8 | 65 |
| nfr5–2 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 51 |
| nfr5–2 | pEpi::NFR5 | 0 | 12.2 | 50 | 37.8 | 4.9 | 18.3 | 76.8 | 6.0 ± 0.6 | 82 |
| nfr5–2 | p35S::NFR5 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 13.5 ± 1.0 | 44 |
| nup85–2 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 59 |
| nup85–2 | pEpi::NUP85 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 15.3 ± 0.6 | 102 |
| nup85–2 | p35S::NUP85 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 18.0 ± 0.8 | 93 |
| nup133–3 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 67 |
| nup133–3 | pEpi::NUP133 | 0 | 0 | 0 | 100 | 2.3 | 0 | 97.7 | 9.8 ± 0.6 | 87 |
| nup133–3 | p35S::NUP133 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 11.2 ± 0.7 | 60 |
| castor–4 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 21 |
| castor–4 | pEpi::CASTOR | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 23.0 ± 1.8 | 54 |
| castor–4 | p35S::CASTOR | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 19.4 ± 1.2 | 48 |
| pollux–3 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 50 |
| pollux–3 | pEpi::POLLUX | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 20.5 ± 1.3 | 55 |
| pollux–3 | p35S::POLLUX | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 18.7 ± 1.3 | 64 |
| ccamk–3 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 88 |
| ccamk–3 | pEpi::CCaMK | 0 | 3.7 | 0.9 | 95.4 | 58.3 | 13.9 | 27.8 | 2.2 ± 0.4 | 108 |
| ccamk–3 | p35S::CCaMK | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 11.5 ± 0.9 | 55 |
| cyclops–3 | Empty | 0 | 86.1 | 13.9 | 0 | 0 | 100 | 0 | – | 36 |
| cyclops–3 | pEpi::CYCLOPS | 0 | 0 | 0 | 100 | 0 | 100 | 0 | – | 90 |
| cyclops–3 | p35S::CYCLOPS | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 12.5 ± 1.0 | 78 |
| nsp1–1 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 61 |
| nsp1–1 | pEpi::NSP1 | 0 | 0 | 0 | 100 | 58.1 | 17.7 | 24.2 | 1.7 ± 0.4 | 62 |
| nsp1–1 | p35S::NSP1 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 16.1 ± 1.0 | 67 |
| nsp2–1 | Empty | 100 | 0 | 0 | 0 | 100 | 0 | 0 | – | 61 |
| nsp2–1 | pEpi::NSP2 | 5.3 | 11.6 | 8.4 | 74.7 | 96.8 | 2.1 | 1.1 | 1.0 | 95 |
| nsp2–1 | p35S::NSP2 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 16.4 ± 1.9 | 39 |
The numbers indicate the percentage of plants that showed infection events, i.e. formation of micro-colonies or infection threads (ITs) in the epidermis, and nodulation events, i.e. formation of empty nodules (bump) or infected nodules (Nod) in the cortex, 4 weeks after inoculation of DsRed-labeled Mesorhizobium loti. Data were compiled from more than two independent experiments. Wild-type, Gifu B–129; Empty, empty vector; pEpi, Epi308 promoter; p35S, CaMV 35S promoter.
Expression analysis of symbiotic genes in root hairs
In addition to the LjEXPA genes, we examined relative expression levels of several symbiotic genes in root hairs and stripped roots with or without M. loti inoculation. Two days after inoculation of M. loti or mock inoculation, all genes were found to be expressed in root hairs as well as in stripped roots. Among the genes examined, NUP85 and NSP2 showed a significant root hair expression pattern (Figure S6). Inoculation of M. loti caused increases in both NSP1 and NIN expression in both root hairs and stripped roots (Figure S6). In contrast, the expression of other symbiotic genes remained unchanged after inoculation of M. loti (Figure S6).
pEpi::CCaMK induces the formation of ITs in epidermis, but infection events in the cortex do not occur
CCaMK is involved not only in rhizobial infection through ITs (Hayashi et al., 2010) but also in nodule organogenesis in the cortex (Tirichine et al., 2006). Spatio-temporal expression of CCaMK, analyzed by introduction of the GUS gene driven by the CCaMK promoter (pCCaMK::GUS), showed both epidermal and cortical expression in Lotus roots (Figure2a–d). Based on the expression pattern of CCaMK, we examined whether the Epi promoter confers epidermis-specific expression of target genes in L. japonicus by complementation of the symbiotic phenotypes of the ccamk–3 mutant using the pEpi expression system. Neither IT formation nor nodule formation was induced on ccamk roots transformed with the empty vector control upon M. loti inoculation (Figure S7), or on any symbiotic mutant examined in this study (Figures S7 and S8). In contrast, the roots of ccamk/p35S::CCaMK plants formed nodules associated with the development of ITs from curled root hairs to the cortex (Figure2h–j). Introduction of pEpi::CCaMK resulted in rescue of epidermal infection events, including root hair curling and well-developed ITs within root hairs, while penetration of ITs into the cortical cell layer was rarely observed (Figure2e–g).
Figure 2.
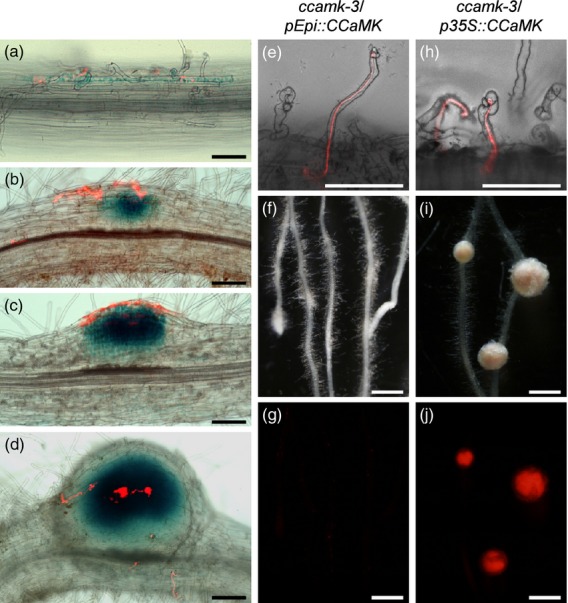
Histochemical localization of GUS activity in wild-type roots transformed with pCCaMK::GUS vector, and complementation tests of rhizobial infection and nodule organogenesis phenotypes of the ccamk–3 mutant transformed with CCaMK driven by pEpi or p35S. (a–d) GUS expression patterns for wild-type roots transformed with pCCaMK::GUS vector after inoculation with DsRed-labeled Mesorhizobium loti. Bright-field and red fluorescence images were merged into single images. (a) Root hairs of wt/pCCaMK::GUS roots observed 7 days after inoculation with M. loti. (b–d) Development of nodules on the wt/pCCaMK::GUS roots 10 days after inoculation with M. loti. Blue staining was observed in curled root hairs (a), in cortical cells before cortical infection thread (IT) formation (b), and during cortical cell division (c) and cortical infection (d). (e–j) Symbiotic phenotypes of transformed roots were observed 4 weeks after inoculation with DsRed-labeled M. loti. (e, h) Root hairs of ccamk–3/pEpi::CCaMK and ccamk–3/p35S::CCaMK, shown as merged bright-field and red fluorescence images. ITs are visible inside the curled root hairs. Bright-field images (f, i) and corresponding red fluorescence images (g, j) for nodulation phenotypes are shown. (i) Nodules formed on the roots of ccamk–3/p35S::CCaMK plants. (j) Infection of DsRed-labeled M. loti was observed as red fluorescence in the central zone of nodules. (f, g) Nodule organogenesis was not induced on the roots of ccamk–3/pEpi::CCaMK. Scale bars = 100 μm (a–e, h) and 1 mm (f, g, i, j).
To evaluate the effectiveness of the pEpi expression system more thoroughly, the number of nodules per nodulated plant and the frequency distribution of cortical cell division were investigated 4 weeks after inoculation (Table1 and Figure S9). Mature nodules and nodule primordia in which infected cells are visualized with red fluorescence (derived from DsRed-labeled M. loti) are defined as ‘nodules’. In contrast, empty nodule-like structures, in which red fluorescence was not observed, are defined as ‘bumps’. In the case of ccamk/p35S::CCaMK, all plants formed nodules (indicated by red bars in Figure S9) and the mean number of nodules per nodulated plant was 11.5 ± 0.9 (Table1). On the roots of ccamk/pEpi::CCaMK, over 50% of the plants showed a non-nodulation phenotype (Nod−, indicated by black bars in Figure S9). Although cortical cell division was induced, the number of nodules per nodulated plant was lower (2.2 ± 0.4; Table1) than that of ccamk/p35S::CCaMK. These data demonstrate that expression of CCaMK via the pEpi expression system is sufficient for epidermal IT formation, but is not sufficient to induce cortical IT formation or cortical cell division. These phenotypes are in agreement with the report by Rival et al. (2012), in which epidermal expression of DMI3 genes via the LeEXT promoter rescues only the epidermal infection phenotype of the dmi3 mutant of M. truncatula.
As previously reported (Hayashi et al., 2010), introduction of the gain-of-function gene CCaMKT265D under the control of the CaMV 35S promoter (p35S) induced spontaneous nodulation in the absence of rhizobia (Figure S10). To examine whether epidermal expression of CCaMKT265D induces spontaneous nodulation, pEpi::CCaMKT265D was introduced into wild-type roots. Spontaneous nodulation was not observed on the roots of wt/pEpi::CCaMKT265D or those of wt/pEpi::CCaMKT265T even 6 weeks after transplantation (Figure S10 and Table S1). These results suggest that expression of CCaMKT265D in the cortex is indispensable for spontaneous nodule formation.
Taking these results together, the pEpi expression system can be regarded as an epidermis-specific expression system in Lotus roots, based on the phenotypes observed on the roots of both ccamk/pEpi::CCaMK and wt/pEpi::CCaMKT265D. Furthermore, the pEpi expression system shows that there is a requirement for cortical CCaMK expression in cortical IT formation and nodule organogenesis.
Expression of upstream genes in the epidermis is sufficient for both epidermal and cortical infection events
The nup85, nup133, castor and pollux mutants transformed with their corresponding genes driven by p35S formed nodules upon M. loti inoculation (Table1 and Figure S11), while symrk/p35S::SYMRK did not form effective nodules. Accordingly, we examined the function of NUP85, NUP133, CASTOR and POLLUX by the pEpi expression system. Expression of these genes driven by pEpi rescued the symbiotic defects in their corresponding mutants, resulting in the formation of nodules (Table 1 and Figure3). A sequence of infection events, including root hair curling, IT development from epidermis to cortex and the formation of nodules, were observed, comparable to those in mutants transformed with p35S to drive the corresponding genes (Table 1, Figures 3 and S11). These results indicate that expression of upstream genes in the epidermis is sufficient for both rhizobial infection and nodule organogenesis.
Figure 3.
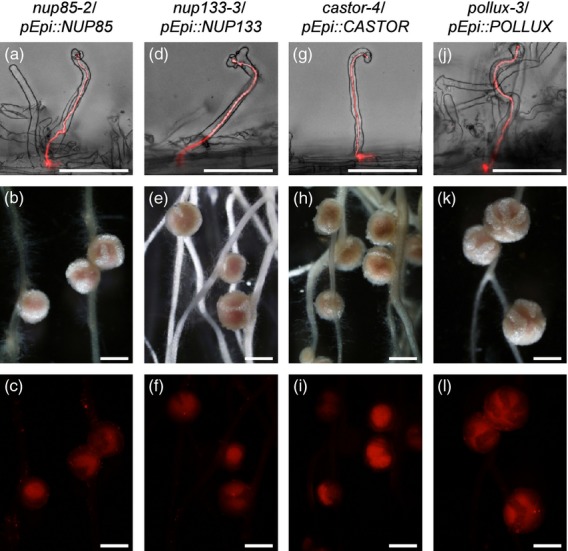
Complementation tests of rhizobial infection and nodule organogenesis phenotypes of nup85–2, nup133–3, castor–4 and pollux–3 mutants transformed with the corresponding genes driven by pEpi. Symbiotic phenotypes of transformed roots were observed 4 weeks after inoculation with DsRed-labeled Mesorhizobium loti. Root phenotypes of nup85–2/pEpi::NUP85 (a–c), nup133–3/pEpi::NUP133 (d–f), castor–4/pEpi::CASTOR (g–i) and pollux–3/pEpi::POLLUX (j–l). (a, d, g, j) ITs are visible inside the curled root hairs, shown as merged bright-field and red fluorescence images. Bright-field images (b, e, h, k) and corresponding red fluorescence images (c, f, i, l) for nodulation phenotypes are shown. (c, f, i, l) Infection of DsRed-labeled M. loti was observed as red fluorescence in the central zone of nodules. Scale bars = 100 μm (a, d, g, j) and 1 mm (b, c, e, f, h, i, k, l).
Dispensability of Ca2+ signal-responsive domains of CCaMK for cortical infection processes
Expression of upstream genes by the pEpi expression system is sufficient to induce the complete nodulation process. In contrast, the complementation results for both ccamk/pEpi::CCaMK and wt/pEpi::CCaMKT265D suggest that there is a requirement for cortical CCaMK expression for cortical IT formation and nodule organogenesis. Upstream genes encode the components of machinery for Ca2+ spiking, which is believed to be an activator for CCaMK (Singh and Parniske, 2012). Therefore, our results raise questions about whether CCaMK activation by cortical Ca2+ spiking via upstream components is essential for cortical infection processes. CCaMK contains a calmodulin-binding domain (CaMBD) and Ca2+-binding EF hands at its C–terminus. The CaMBD and EF hands respond to Ca2+ signals and act as regulatory domains for CCaMK. Among the C–terminus-deleted CCaMKs examined in our previous work, CCaMK1–314 (lacking CaMBD and EF hands) and CCaMK1–340 (lacking EF hands) lost the ability to bind calmodulin and Ca2+ in vitro (Shimoda et al., 2012). Complementation analysis of the ccamk mutant by the truncated CCaMKs showed a loss of function of both CCaMKs for rhizobial infection, demonstrating that CaMBD and EF hands play an essential role in IT formation. In contrast to CCaMK1–340, which also showed a loss of function for nodule organogenesis, CCaMK1–314 showed a gain of function for nodule organogenesis, in the sense that spontaneous nodulation was induced on the roots of ccamk/p35S::CCaMK1–314 (Shimoda et al., 2012).
To evaluate the requirement of the C–terminus regulatory domains of CCaMK for cortical infection processes, we developed a simultaneous expression system comprising two target genes expressed under the control of two distinct promoters, p35S and pEpi (Figure4). Under the control of p35S, the first target gene is expressed in whole roots including the epidermis and cortex, while pEpi allows expression of the second target gene only in the epidermis. Using the pEpi+p35S double expression system, we introduced pEpi::CCaMK+p35S::CCaMK1–314 into ccamk roots. Following the formation of epidermal ITs, cortical ITs associated with cortical cell division occurred, resulting in the formation of nodules (Table 2, Figures 4 and S12). Similar phenotypes were also observed on the roots of ccamk/pEpi::CCaMK+p35S::CCaMK1–340. In the case of pEpi::CCaMK+p35S::CCaMK1–314, spontaneous nodule-like structures were occasionally formed (Table2 and Figure S13). This phenotype is considered to be an indication of a gain of function of CCaMK1-314 for nodule organogenesis. These results show not only an absolute requirement of full-length CCaMK for epidermal IT formation, but dispensability of the Ca2+-responsive domains of CCaMK for cortical infection processes. Taking these results into account, our results suggest that activation of CCaMK via upstream genes in the epidermis is essential for the formation of epidermal ITs, but the symbiotic function of CCaMK in the cortex does not necessarily depend on Ca2+ and calmodulin.
Figure 4.
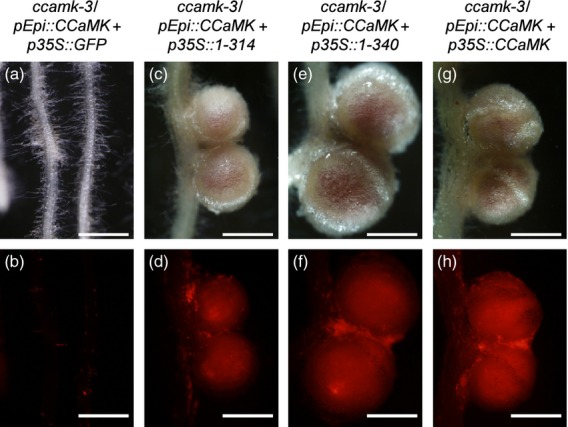
Complementation tests of nodulation phenotypes of the ccamk–3 mutant by co-transformation with CCaMK under the control of pEpi and truncated CCaMK1–314 or CCaMK1–340 under the control of p35S. Nodulation phenotypes of transformed roots were observed 4 weeks after inoculation with DsRed-labeled Mesorhizobium loti. Bright-field images (a, c, e, g) and their corresponding red fluorescence images (b, d, f, h) for nodulation phenotypes are shown. Nodule organogenesis was not induced on the roots of ccamk–3/pEpi::CCaMK+p35S::GFP (a, b). Nodules formed on the roots of ccamk–3/pEpi::CCaMK+p35S::CCaMK1–314 (c, d), ccamk–3/pEpi::CCaMK+p35S::CCaMK1–340 (e, f) and ccamk–3/pEpi::CCaMK+p35S::CCaMK (g, h). Infection of DsRed-labeled M. loti was observed as red fluorescence in the central zone of nodules (d, f, h). Scale bars = 1 mm.
Table 2.
Complementation analysis of symbiosis-defective phenotypes using the pEpi+p35S double expression system
| Lotus line | Construct | Nodulation phenotype | Total number of plants | ||||
|---|---|---|---|---|---|---|---|
| (%) | Nodules per nodulated plant (mean ± SE) | Spontaneous nodule-like structures per nodulated plant (mean ± SE) | |||||
| Nod− | Bump | Nod | |||||
| ccamk–3 | pEpi::CCaMK+p35S::GFP | 46.3 | 11.9 | 41.8 | 2.5 ± 0.3 | – | 67 |
| ccamk–3 | pEpi::CCaMK+p35S::CCaMK1–314 | 9.8 | 9.8 | 80.4 | 5.2 ± 0.4 | 0.7 ± 0.1 | 92 |
| ccamk–3 | pEpi::CCaMK+p35S::CCaMK1–340 | 15.7 | 1.2 | 83.1 | 7.6 ± 0.6 | – | 83 |
| ccamk–3 | pEpi::CCaMK+p35S::CCaMK | 3.3 | 0 | 96.7 | 9.6 ± 0.7 | – | 91 |
The numbers indicate the percentage of plants that showed nodulation events, i.e. formation of empty nodules (bump) or infected nodules (Nod) in the cortex, 4 weeks after inoculation of DsRed-labeled Mesorhizobium loti. Data were compiled from more than two independent experiments. pEpi, Epi308 promoter; p35S, CaMV 35S promoter.
CYCLOPS appears to be indispensable for IT formation in both the epidermis and cortex
On the roots of cyclops mutants, IT development accompanied by rhizobial infection is arrested in the epidermis, resulting in the formation of small bumps without infected cells (Figure S14). pEpi::CYCLOPS restored epidermal infection defects in the cyclops mutant, as did p35S::CYCLOPS (Table1 and Figure S14), but cortical IT development was not rescued, resulting in the formation of bumps (Table1, Figures S14 and S15). These results show that CYCLOPS expression under the control of pEpi is insufficient for cortical IT development, suggesting that cortical expression of CYCLOPS is indispensable for bacterial entry into the cortex.
Epidermal expression of NSP1 and NSP2 only restores epidermal IT formation on the roots of the corresponding mutants
On the roots of both nsp1/pEpi::NSP1 and nsp2/pEpi::NSP2, micro-colony and IT formation in the epidermis were restored (Table1 and Figure S16). However, neither cortical cell division nor IT development into the cortex occurred on these roots. The majority of the plants showed a non-nodulation phenotype (Figure S15), and the number of nodules per nodulated plant was lower than those of nsp1/p35S::NSP1 and nsp2/p35S::NSP2 plants (Table1 and Figure S15). Taken together, NSP1 and NSP2 appear to be indispensable not only for IT formation in the epidermis, but also for IT development and nodule organogenesis in the root cortex.
Different requirements of NFR1 and NFR5 expression for IT development
NFR1 and NFR5, LysM-like receptor kinases that bind to Nod factors derived from M. loti, are considered to be the starting point of root nodule symbiosis in Lotus (Broghammer et al., 2012). Their corresponding mutants are deficient in any symbiotic responses, including deformation of root hairs and the formation of ITs (Radutoiu et al., 2003). Introduction of both genes into the corresponding mutants via the pEpi expression system led to the deformation of root hairs, the colonization of bacteria in curled root hairs, and the formation of ITs within root hairs (Table 1 and Figure5a,d). However, the frequency of epidermal ITs (19.3% on the roots of nfr1/pEpi::NFR1; 37.8% on the roots of nfr5/pEpi::NFR5) were lower than those on nfr1/p35S::NFR1 or nfr5/p35S::NFR5 (100%; Table1 and Figure5g,j).
Figure 5.
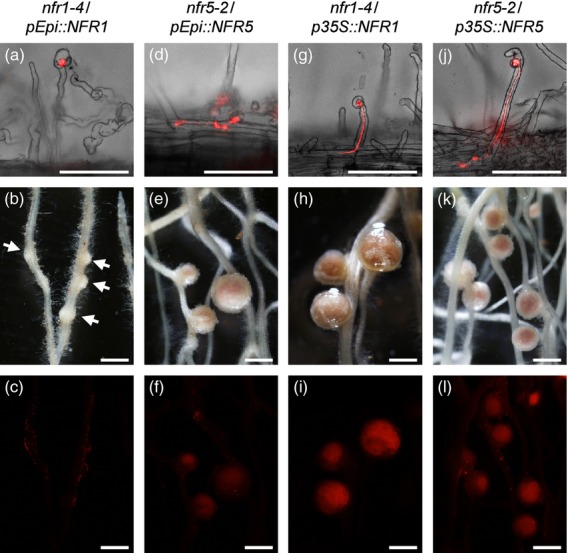
Complementation tests of rhizobial infection and nodule organogenesis phenotypes of nfr1–4 and nfr5–2 mutants transformed with the corresponding genes driven by pEpi or p35S. Symbiotic phenotypes of transformed roots were observed 4 weeks after inoculation with DsRed-labeled Mesorhizobium loti. Root hairs of nfr1–4/pEpi::NFR1 (a), nfr5–2/pEpi::NFR5 (d), nfr1–4/p35S::NFR1 (g) and nfr5–2/p35S::NFR5 (j) are shown as merged images of bright-field and red fluorescence images. (a) Although deformation of root hairs and colonization of bacteria in curled root hairs occurred, development of ITs within root hairs rarely occurred on the roots of nfr1–4/pEpi::NFR1. (d, g, j) ITs are visible inside the curled root hairs of nfr5–2/pEpi::NFR5 (d), nfr1–4/p35S::NFR1 (g) and nfr5–2/p35S::NFR5 (j). Bright-field images (b, e, h, k) and their corresponding red fluorescence images (c, f, i, l) of nodulation phenotypes are shown. (b, c) Almost 70% of cortical cell division was arrested, resulting in bump structures (arrows), and effective nodules rarely formed on the roots of nfr1–4/pEpi::NFR1. Nodules formed on the roots of nfr5–2/pEpi::NFR5 (e), nfr1–4/p35S::NFR1 (h) and nfr5–2/p35S::NFR5 (k). (f, i, l) Infection of DsRed-labeled Mesorhizobium loti was observed as red fluorescence in the central zone of nodules. Scale bars = 100 μm (a, d, g, j) and 1 mm (b, c, e, f, h, i, k, l).
In the cortex of nfr1/pEpi::NFR1 roots, although cortical cell division was induced, almost all of nodule primordia were arrested at bumps, and nodules were rarely formed (Table 1, Figures5b,c and S15). In contrast, the majority of cortical cell division resulted in formation of nodules on the roots of nfr5/pEpi::NFR5, while the number of nodules per nodulated plant was lower than that on nfr5/p35S::NFR5 (Table 1, Figures5e,f,k,l and S15). These results suggest that expression of NFR1 and NFR5 in root epidermis is sufficient for the induction of cortical cell division, but the involvement of NFR1 and NFR5 for cortical IT formation appears to be different from each other.
Discussion
Construction and evaluation of the pEpi expression system in Lotus japonicus
We have previously reported epistatic relationships among symbiotic genes in both IT formation and nodule organogenesis (Hayashi et al., 2010). However, the individual requirements of symbiotic genes for cell-layer events including epidermal IT formation, cortical IT formation and cortical cell division remain unclear. In this study, we developed a ‘pEpi expression system’ using the promoter regions of two Lotus expansin genes, LjEXPA7 and LjEXPA8. As reported for AtEXPA7 and orthologous genes of angiosperm species (Kim et al., 2006), the promoter regions of both LjEXPA7 and LjEXPA8 contain conserved RHEs. Detailed evaluation of our pEpi expression system was performed based on: (i) gene expression analysis of root hairs and stripped roots of Lotus by quantitative RT–PCR, and (ii) spatio-expression analysis of pEpi::GUS+ constructs. Among the candidates examined, we selected pEpi308, a promoter region of LjEXPA7 containing three RHE motifs (Figure1), and developed the pEpi expression system. Subsequently, we examined the symbiotic phenotypes of ccamk mutants transformed with CCaMK and CCaMKT265D using the pEpi expression system. The roots of ccamk/pEpi::CCaMK showed IT formation in the epidermis, but IT development into the cortex and cortical cell division were not recovered (Figure2e–g). Moreover, no spontaneous nodules were formed on the roots of ccamk/pEpi::CCaMKT265D (Figure S10). Collectively, these data show the utility of the pEpi expression system to analyze the effects of epidermal expression of other symbiotic genes on epidermal and cortical infection processes.
Cell layer-specific differences in requirements for upstream gene expression during nodulation
Epidermal expression of upstream genes, including NUP85, NUP133, CASTOR and POLLUX, by the pEpi expression system, perfectly rescued all of the symbiotic defects in the corresponding gene mutants, leading to formation of nodules (Table 1, Figures 3 and S9). There is the possibility of leakage of gene products from epidermis to cortex, but, in the case of upstream genes, almost all cortical cell division resulted in the formation of nodules, as shown in Figure S9. These clear-cut complementation results were restricted to the upstream genes examined (Figures3 and S9), and were not observed for CCaMK, CYCLOPS, NSP1 and NSP2 (Figures2 and S9, S14, S15 and S16). On the basis of these results, we suggest that, once the epidermal infection process is completed, cortical infection processes no longer require expression of the upstream components in the cortex (Figure6).
Figure 6.
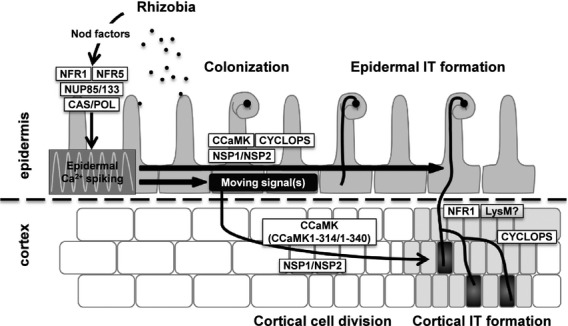
Model for coordinated infection processes regulated by symbiotic genes examined in this study. Ca2+ spiking, induced through NFR1/NFR5 and upstream genes, is indispensable for the development of epidermal ITs within root hairs. Downstream of the epidermal Ca2+ spiking, CCaMK, CYCLOPS and NSP1/NSP2 function to form epidermal ITs. The epidermal Ca2+ spiking is also involved in generation of unidentified ‘moving signal(s)’ that trigger cortical cell division. Once epidermal ITs form, Ca2+ spiking in the cortex is not necessarily required for development of cortical ITs. NFR1 and other LysM receptors may act together to perceive Nod factors, leading to invasion of rhizobia through cortical ITs. CAS, CASTOR; POL, POLLUX.
The upstream genes are responsible for the induction of Ca2+ spiking, which acts as a second messenger activating the downstream symbiotic pathway. Actually, almost all symbiotic mutants of upstream genes showed a non-nodulation phenotype. In the case of L. japonicus, two of the upstream gene mutants, nena–1 and symrk–14, also did not induce Ca2+ spiking and showed impaired ability to develop epidermal ITs. However, cortical IT formation and nodule organogenesis occurred normally, leading to the formation of nodules (Groth et al., 2010; Kosuta et al., 2011). In both cases, rhizobia invaded the host plants via intercellular 'crack entry' rather than through intracellular epidermal ITs. These phenotypes are in line with our proposed model in which the induction of Ca2+ spiking is required for epidermal IT formation but is not required for cortical IT formation in L. japonicus.
Different requirements of Ca2+-responsive regulatory domains of CCaMK for infection processes in L. japonicus
It is generally accepted that Ca2+-dependent activation of CCaMK is essential for intracellular accommodation of rhizobia. Furthermore, we revealed the importance of CaMBD and EF hands of CCaMK for IT formation (Shimoda et al., 2012). To evaluate the hypothesis that cortical infection processes do not require CCaMK activation by cortical Ca2+ spiking, we expressed Ca2+-responsive domain-deleted CCaMKs under the control of p35S, concurrently with expression of full-length CCaMK under the control of pEpi, in a ccamk mutant. When accompanied by epidermal expression of full-length CCaMK, these domain-deleted CCaMKs regulated cortical infection processes (Table 2 and Figure 4), strengthening the hypothesis that CCaMK activation by cortical Ca2+ spiking is not essential for cortical infection processes in L. japonicus (Figure6).
In contrast to the reports in L. japonicus, an important role for cortical Ca2+ spiking for intracellular cortical infection has been reported in M. truncatula. Cortical Ca2+ spiking was triggered during initial stages of IT formation in the outer roots cortex, and was proposed to play an important role for the navigation of cortical IT development (Sieberer et al., 2012). In this view, full-length DMI3 was thought to be required for both epidermal and cortical nodulation processes in M. truncatula. However, combined epidermal and cortical expression of full-length DMI3 via two promoters, pLeEXT (epidermis) and pCO2 (cortex), only complemented nodule organogenesis of the dmi3 mutant, resulting in the formation of empty nodules (Rival et al., 2012). The result for dmi3/pLeEXT+pCO2::DMI3 raises the question of whether pCO2 confers a sufficient level of gene expression in the cortex.
Medicago truncatula forms indeterminate nodules that retain a persistent nodule meristem. ITs grow and penetrate through the outer cortex towards the dividing inner cortex, which constitutes the nodule meristem, and the host plant cells continue to be infected by rhizobia during nodule development (Hirsch, 1992). In contrast, L. japonicus forms determinate nodules that lack a persistent nodule meristem. Following cortical cell division, which ceases early during nodule development, cell expansion accompanied by bacterial invasion through ITs occurs once. These differences between indeterminate and determinate nodulation have led to speculation that the importance of cortical Ca2+ spiking for IT formation in M. truncatula is different from that of L. japonicus. Further analyses of the requirement for cortical Ca2+ spiking and the functionality of CCaMK/DMI3 in cortex are necessary to provide missing pieces in our understanding of the importance of Ca2+-mediated signal transduction for accommodation of rhizobial bacteria, and also provide information about the various mechanisms by which indeterminate and determinate nodulation are governed.
In L. japonicus, cell type-specific regulation of rhizobial infection by CCaMK was reported by Liao et al. (2012), based on the phenotypes of a ccamk–14 mutant that showed excess epidermal IT formation in parallel with abnormal cortical IT formation. The ccamk–14 mutation is caused by a S337N substitution in the CaMBD. The S337 residue is a target of autophosphorylation, and has been reported to be responsible for negative regulation of CCaMK via Ca2+/calmodulin binding (Liao et al., 2012). The S337N mutation prevents CCaMK from the negative feedback regulation that appears to be essential for cortical IT formation (Liao et al., 2012). These phenotypes appear to be contradictory with our data; however, both CCaMK1–314 and CCaMK1–340 are considered to be kept in an inactive state and retain the ability to regulate cortical infection. Thus, the truncated CCaMKs may be regarded as potentially functional CCaMKs for the regulation of cortical IT formation.
Symbiotic genes responsible for cortical infection processes
In the case of CYCLOPS, although epidermal IT formation recovered, symbiosis-defective phenotypes, e.g. arrested development of cortical ITs and formation of bumps, were retained on the roots of cyclops/pEpi::CYCLOPS. As previously reported, CYCLOPS is not necessarily required for the induction of cortical cell division, as spontaneous nodulation occurs on the roots of cyclops/p35S::CCaMKT265D (Yano et al., 2008). Taken together, the results suggest that the loss of complementation of cortical infection defects on the roots of cyclops/pEpi::CYCLOPS is due to incomplete development of cortical ITs (Figure6), indicating a requirement of CYCLOPS for both epidermal and cortical IT formation.
Similarly to CCaMK, expression of NSP1 and NSP2 by pEpi did not complement cortical infection events of nsp1 and nsp2 mutants, respectively. Collectively, these results indicate that NSP1 and NSP2 are indispensable for cortical IT formation and cortical cell division (Figure6).
Different involvement of NFR1 and NFR5 in rhizobial infection processes
Each of the symbiotic genes expressed via the pEpi expression system rescued the epidermal responses in the corresponding gene mutants, except for NFR1 and NFR5 (Table 1, Figures 5 and S15). Root hair curling occurred on the roots of both nfr1/pEpi::NFR1 and nfr5/pEpi::NFR5, but development of ITs within root hairs occurred at a very low frequency compared with the other mutants examined (Table1). These results suggest that the transcript level of both NFR1 and NFR5 under the control of pEpi may be insufficient for the entire process of IT development in the epidermis.
In this study, the pEpi expression system was used to elucidated the distinct requirements of NFR1 and NFR5 for cortical infection processes in L. japonicus. On the roots of nfr5/pEpi::NFR5, the majority of nodule primordia resulted in the formation of nodules, although the number of nodules per nodulated plant was lower than that of nfr5/p35S::NFR5 plants (Table 1, Figures 5 and S15). In contrast, almost 70% of cortical cell division was arrested at bumps, and nodules were rarely formed on the roots of nfr1/pEpi::NFR1 (Table 1, Figures 5b,c and S15).
In general, rhizobia are thought to produce Nod factors continuously within developing ITs (Timmers et al., 1998). Taking this into account, our results suggest that Nod factor signaling through NFR1/NFR5 is crucial for the development of ITs in the epidermis. Complementation defects of nfr1/pEpi::NFR1 in cortical IT formation also imply the requirement of NFR1 for perception and signaling of Nod factors throughout IT development. Compared to NFR1, the involvement of NFR5 for cortical IT development is somewhat attenuated, suggesting the possibility that NFR1 may form a receptor complex with LysM receptor homologs other than NFR5 (Figure6; Arrighi et al., 2006; Lohmann et al., 2010), with which NFR1 perceives Nod factors and regulates cortical IT formation. In contrast to NFR5 in Lotus, NFP, an ortholog of NFR5 in M. truncatula, did not complement cortical IT formation on the roots of nfp/pLeEXT::NFP (Rival et al., 2012). In the case of M. truncatula, LYK3, an ortholog of NFR1, is not involved in the induction of Ca2+ spiking, and an NFR1 homolog, that may be responsible for the induction of Ca2+ spiking, has not been identified. Further analyses of LysM receptor kinases are required to broaden our understanding how epidermal and cortical infection events are coordinated by multiple LysM receptors in L. japonicus and M. truncatula.
Generation of ‘moving signal(s)’ responsible for the induction of cortical cell division
Upon inoculation with M. loti, both nfr1/pEpi::NFR1 and nfr5/pEpi::NFR5 roots showed the induction of nodule organogenesis (Table 1 and Figure 5), suggesting that the perception of Nod factors through NFR1/NFR5 in the epidermis stimulates signal(s) that are transmitted from the epidermis to cortex, leading to the induction of nodule organogenesis (Figure6).
Cytokinins are possible candidates for a ‘moving signal’ from epidermis to cortex (Oldroyd and Downie, 2008; Heckmann et al., 2011). The essential role of cytokinins in nodulation has been demonstrated by genetic studies in L. japonicus and M. truncatula (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007). Activation of a putative cytokinin receptor, Lotus histidine kinase (LHK1), has been suggested to be necessary for nodule organogenesis, as a gain-of-function mutation in LHK1 (snf2) induces spontaneous nodulation in the absence of rhizobia (Tirichine et al., 2007). Using the cytokinin-responsive ARR5 gene promoter fused to the GUS reporter gene, an increase of cytokinin levels was observed in curled and deformed root hairs and in dividing cortical cells after inoculation with M. loti (Lohar et al., 2004). These results suggest that LHK1 in cortical cells acts as a receptor for cytokinins that may be transported from the epidermis, and that the activation of LHK1 induces subsequent steps of signaling pathways for nodule organogenesis.
GRAS domain proteins have been suggested to have the ability to move between cells, and their intercellular movement plays an important role for plant development (Cui et al., 2007). Thus, NSP1 and NSP2 are also candidates for ‘moving signals’ from the epidermis to the cortex. We showed that neither nsp1/pEpi::NSP1 nor nsp2/pEpi::NSP2 roots induced nodule organogenesis upon inoculation with M. loti. Thus, movement of NSP1 or NSP2 from the epidermis to the cortex is considered not to be involved in the induction of nodule organogenesis, although we cannot rule out the possibility that the transcript levels of NSP1 and NSP2 driven by pEpi are quantitatively insufficient to allow intercellular movement and/or functioning of NSP1 and NSP2 in the cortex.
Expression analysis of symbiotic genes in root hairs
To isolate root hairs with high viability, samples of root hairs, stripped roots and whole roots were collected 2 days after inoculation of M. loti or mock inoculation. LjEXPA7, LjEXPA8, LjRH101 and LjRH102 transcripts were expressed at very high levels in isolated root hairs in comparison with stripped roots (Figures1 and S2), indicating that enrichment of root hairs was successful. Expression analysis of symbiotic genes in root hairs and stripped roots showed that almost all symbiotic genes examined in this study are expressed in both regions (Figure S6). The root hair is a tubular outgrowth of the epidermis, thus stripped roots also contain some epidermal cells. Hence, it appears that comparison of root hairs with stripped roots is not sufficient to dissect epidermis-specific from cortex-specific expression of the symbiotic genes.
Importance of epidermal Ca2+ spiking for intracellular infection in L. japonicus
In this study, we did not directly analyze epidermal and cortical Ca2+ spiking during infection processes. However, the pEpi expression system provides evidence for cell layer-specific differences in the importance of Ca2+ spiking for IT formation. Intercellular infection via crack-entry overcame the requirement for Ca2+ spiking; however, the efficiency of intercellular infection is reduced in L. japonicus (Groth et al., 2010; Madsen et al., 2010; Kosuta et al., 2011). These reports indicated the importance of Ca2+ spiking for establishment of intracellular infection via epidermis ITs as an effective infection route. Based on the strict recognition through Nod factor perception by LysM receptor kinases, Ca2+ spiking elicited in the epidermis appears to ‘open the gate’ for rhizobia by formation of ITs from the epidermis to the cortex.
Experimental Procedures
Plant materials
Lotus japonicus B–129 accession Gifu (wild-type) and the mutants nfr1–4 (Sandal et al., 2006), nfr5–2 (Madsen et al., 2003), castor–4 and pollux–3 (Imaizumi-Anraku et al., 2005), nup85–2 (Saito et al., 2007), nup133–3 (Kanamori et al., 2006), ccamk–3 (Tirichine et al., 2006), cyclops–3 (Yano et al., 2008), nsp1–1 (Heckmann et al., 2006) and nsp2–1 (Murakami et al., 2006) were used in this study. To visualize the infection processes of rhizobia, M. loti MAFF303099 constitutively expressing DsRed (Maekawa et al., 2009) was used.
Plasmid construction
Detailed information on plasmid construction is provided in Methods S1 and Table S2.
Hairy root transformation and inoculation tests with rhizobial strains
Induction and transformation of L. japonicus hairy roots using A. rhizogenes LBA1334 were performed as described by Díaz et al. (2005) with minor modifications. Plants with GFP-positive transformed hairy roots were selected by GFP fluorescence using a Leica MZFLIII stereomicroscope (Leica, http://www.leica-microsystems.com/) or an Olympus SZX12 stereomicroscope (Olympus, http://www.olympus-global.com/en/). The transformants were transplanted into vermiculite-containing pots supplied with half-strength B&D medium (Broughton and Dilworth, 1971) supplemented with 0.5 μm ammonium nitrate. Three days after transplantation, DsRed-labeled M. loti was inoculated onto the hairy roots of L. japonicus. The plants were grown in a growth cabinet under a 16 h day per 8 h night cycle at 24°C. Four weeks after inoculation, symbiotic phenotypes of plants with GFP-positive hairy roots were analyzed using an Olympus SZX12 stereomicroscope. In the case of the pEpi::CCaMK+p35S double expression system, hairy roots with epidermal ITs were selected as transformants and their phenotypes were analyzed, because the pEpi::CCaMK+p35S double expression system did not encode the GFP gene as a fluorescence marker.
GUS staining
The transformed roots were stained using 0.5 mg ml−1 5–bromo-4–chloro-3–indolyl β–d–glucuronic acid cyclohexylammonium salt, 2.5 mm potassium ferricyanide, 2.5 mm potassium ferrocyanide and 10 mm EDTA in 100 mm sodium phosphate (pH 7.0), and incubated at 37°C in the dark. Samples were observed by using an Olympus SZX12 stereomicroscope or a light microscope (BZ–9000; Keyence, http://www.keyence.com/). For cross-sections, the stained roots were embedded in 5% agar and sectioned (50 μm thick) using a microslicer before observation.
Histological examination of rhizobial infection phenotypes
To examine the extent of rhizobial infection and nodule organogenesis, DsRed-expressing M. loti was observed using an Olympus SZX12 stereomicroscope equipped with a DP70 digital camera (Olympus). For observation of ITs, samples were analyzed under a BZ–9000 epifluorescence microscope (Keyence) using a filter set (excitation 560–600 nm band pass, emission 630–690 nm band pass). A Z–stack image was obtained using the BZ–9000 epifluorescence microscope with a step size of 2.5 μm (20 × objective) or 5 μm (10 × objective), and ten images of each stack were projected (‘full-focused’) onto a single plane to give an overall view of the samples using image processing software programs supplied with the BZ–9000 microscope.
RNA isolation from root hairs and real-time RT–PCR
The isolation of root hairs was performed as described by Maekawa et al. (2005) with some modifications. More detailed information, including the RNA extraction procedure, is provided in Methods S2. Reverse transcription was performed using a QuantiTect reverse transcription kit (Qiagen, http://www.qiagen.com/), followed by real-time quantitative PCR using LightCycler FastStart DNA MasterPLUS SYBR Green I (Roche, http://www.roche.com) in a Light Cycler system (Roche). Lotus japonicus ubiquitin (Flemetakis et al., 2000) was used as the standard. LjRH101 and LjRH102, which are expressed abundantly in the root hairs of L. japonicus (Maekawa et al., 2005), were used as markers for biological identification of root hairs. All primers used for PCR are listed in Table S3.
Acknowledgments
We are grateful to Robert Ridge (Department of Life Science, International Christian University, Mitaka, Tokyo, Japan) for English editing of this manuscript. We also thank Keisuke Yokota and Rie Iida (National Institute of Agrobiological Sciences, Japan) for technical assistance. This work was supported by a grant from the Japanese Program for the Promotion of Basic Research Activities for Innovative Biosciences (to H.I.–A.).
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Multiple alignment of expansin protein sequences by clustalw.
Figure S2. Root hair-specific expression analysis of RH101 and RH102 in response to infection by Mesorhizobium loti.
Figure S3. Promoter sequences of LjEXP7 and LjEXPA8.
Figure S4. Histochemical localization of GUS activity in wild-type roots transformed with the pEpi::GUS+ vector.
Figure S5. Rhizobial infection and nodule organogenesis phenotypes of wild-type roots transformed with the pEpi::GUS+ vector.
Figure S6. Expression analysis of symbiosis genes in root hairs, stripped roots and whole roots of Lotus japonicus.
Figure S7. Rhizobial infection and nodule organogenesis phenotypes of nup85–2, nup133–3, castor–4, pollux–3 and ccamk–3 mutant roots transformed with the empty vector.
Figure S8. Rhizobial infection and nodule organogenesis phenotypes of nfr1–4, nfr5–2, nsp1–1 and nsp2–1 mutant roots transformed with the empty vector.
Figure S9. Stacked histograms comparing frequency counts in terms of the number of nodules or bumps per mutant plants transformed with CCaMK, CASTOR, POLLUX, NUP85 and NUP133 driven by pEpi or p35S.
Figure S10. Spontaneous nodulation phenotypes of wild-type roots transformed with CCaMKT265D driven by pEpi or p35S.
Figure S11. Complementation tests of rhizobial infection and nodule organogenesis phenotypes of nup85–2, nup133–3, castor–4 and pollux–3 mutants transformed with the corresponding genes driven by p35S.
Figure S12. Stacked histograms comparing frequency counts in terms of the number of nodules or bumps per mutant plant by co-transformation with CCaMK under the control of pEpi and truncated CCaMK1–314 or CCaMK1–340 under the control of p35S.
Figure S13. Spontaneous nodule-like structure of the ccamk–3 mutant by co-transformation with CCaMK under the control of pEpi and truncated CCaMK1–314 under the control of p35S.
Figure S14. Complementation tests of rhizobial infection and nodule organogenesis phenotypes of the cyclops–3 mutant transformed with CYCLOPS driven by pEpi or p35S.
Figure S15. Stacked histograms comparing frequency counts in terms of the number of nodules or bumps per mutant plant transformed with CYCLOPS, NSP1, NSP2, NFR1 or NFR5 driven by pEpi or p35S.
Figure S16. Complementation tests of rhizobial infection and nodule organogenesis phenotypes of nsp1–1 and nsp2–1 mutants transformed with the corresponding genes driven by pEpi or p35S.
Plasmid construction.Methods S2. RNA isolation from root hairs.
Spontaneous nodulation phenotypes.
Table S2. Primer sequences used for construction.
Table S3. Primer sequences used in the expression analysis.
References
- Ané JM, Kiss GB, Riely BK, et al. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science. 2004;303:1364–1367. doi: 10.1126/science.1092986. [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, et al. The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broghammer A, Krusell L, Blaise M, et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl Acad. Sci. USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971;125:1075–1080. doi: 10.1042/bj1251075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell. 2002;14:3237–3253. doi: 10.1105/tpc.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Díaz CL, Grønlund M, Schlaman HRM, Spaink HP. Induction of hairy roots for symbiotic gene expression studies. In: Márguez AJ, editor. Lotus japonicus Handbook. Dordrecht, The Netherlands: Springer; 2005. pp. 261–277. [Google Scholar]
- Flemetakis E, Kavroulakis N, Quaedvlieg NE, Spaink HP, Dimou M, Roussis A, Katinakis P. Lotus japonicus contains two distinct ENOD40 genes that are expressed in symbiotic, nonsymbiotic, and embryonic tissues. Mol. Plant Microbe Interact. 2000;13:987–994. doi: 10.1094/MPMI.2000.13.9.987. [DOI] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Muñoz A, Poovaiah BW, Oldroyd GE. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature. 2006;441:1149–1152. doi: 10.1038/nature04812. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M, Takeda N, Perry J, et al. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell. 2010;22:2509–2526. doi: 10.1105/tpc.109.069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Banba M, Shimoda Y, Kouchi H, Hayashi M, Imaizumi-Anraku H. A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J. 2010;63:141–154. doi: 10.1111/j.1365-313X.2010.04228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA. Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 2006;142:1739–1750. doi: 10.1104/pp.106.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann AB, Sandal N, Bek AS, Madsen LH, Jurkiewicz A, Nielsen MW, Tirichine L, Stougaard J. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant Microbe Interact. 2011;24:1385–1395. doi: 10.1094/MPMI-05-11-0142. [DOI] [PubMed] [Google Scholar]
- Hirsch AM. Developmental biology of legume nodulation. New Phytol. 1992;122:211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, et al. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature. 2005;433:527–531. doi: 10.1038/nature03237. [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, et al. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl Acad. Sci. USA. 2006;103:359–364. doi: 10.1073/pnas.0508883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell. 2006;18:2958–2970. doi: 10.1105/tpc.106.045229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Held M, Hossain MS, et al. Lotus japonicus symRK–14 uncouples the cortical and epidermal symbiotic program. Plant J. 2011;67:929–940. doi: 10.1111/j.1365-313X.2011.04645.x. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010;51:1381–1397. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science. 2004;303:1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- Liao J, Singh S, Hossain MS, et al. Negative regulation of CCaMK is essential for symbiotic infection. Plant J. 2012;72:572–584. doi: 10.1111/j.1365-313X.2012.05098.x. [DOI] [PubMed] [Google Scholar]
- Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM. Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 2004;38:203–214. doi: 10.1111/j.1365-313X.2004.02038.x. [DOI] [PubMed] [Google Scholar]
- Lohmann GV, Shimoda Y, Nielsen MW, et al. Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol. Plant Microbe Interact. 2010;23:510–521. doi: 10.1094/MPMI-23-4-0510. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Hayashi M, Murooka Y. Root hair abundant genes LjRH101 and LjRH102 encode peroxidase and xyloglucan endotransglycosylase in Lotus japonicus. J. Biosci. Bioeng. 2005;99:84–86. doi: 10.1263/jbb.99.84. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Maekawa-Yoshikawa M, Takeda N, Imaizumi-Anraku H, Murooka Y, Hayashi M. Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 2009;58:183–194. doi: 10.1111/j.1365-313X.2008.03774.x. [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GE, Downie JA. Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 2006;19:914–923. doi: 10.1094/MPMI-19-0914. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S. Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 2006;13:255–265. doi: 10.1093/dnares/dsl017. [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie A. Coordinating nodule morphogenesis with rhizobial infection in legume. Annu. Rev. Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Rival P, de Billy F, Bono JJ, Gough C, Rosenberg C, Bensmihen S. Epidermal and cortical roles of NFP and DMI3 in coordinating early steps of nodulation in Medicago truncatula. Development. 2012;139:3383–3391. doi: 10.1242/dev.081620. [DOI] [PubMed] [Google Scholar]
- Saito K, Yoshikawa M, Yano K, et al. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell. 2007;19:610–624. doi: 10.1105/tpc.106.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandal N, Petersen TR, Murray J, et al. Genetics of symbiosis in Lotus japonicus: recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol. Plant Microbe Interact. 2006;19:80–91. doi: 10.1094/MPMI-19-0080. [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Han L, Yamazaki T, Suzuki R, Hayashi M, Imaizumi-Anraku H. Rhizobial and fungal symbioses show different requirements for calmodulin binding to calcium calmodulin-dependent protein kinase in Lotus japonicus. Plant Cell. 2012;24:304–321. doi: 10.1105/tpc.111.092197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Fournier J, Timmers AC, Barker DG. A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J. 2012;69:822–830. doi: 10.1111/j.1365-313X.2011.04834.x. [DOI] [PubMed] [Google Scholar]
- Singh S, Parniske M. Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr. Opin. Plant Biol. 2012;15:444–453. doi: 10.1016/j.pbi.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Timmers AC, Auriac MC, de Billy F, Truchet G. Nod factor internalization and microtubular cytoskeleton changes occur concomitantly during nodule differentiation in alfalfa. Development. 1998;125:339–349. doi: 10.1242/dev.125.3.339. [DOI] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl Acad. Sci. USA. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignment of expansin protein sequences by clustalw.
Figure S2. Root hair-specific expression analysis of RH101 and RH102 in response to infection by Mesorhizobium loti.
Figure S3. Promoter sequences of LjEXP7 and LjEXPA8.
Figure S4. Histochemical localization of GUS activity in wild-type roots transformed with the pEpi::GUS+ vector.
Figure S5. Rhizobial infection and nodule organogenesis phenotypes of wild-type roots transformed with the pEpi::GUS+ vector.
Figure S6. Expression analysis of symbiosis genes in root hairs, stripped roots and whole roots of Lotus japonicus.
Figure S7. Rhizobial infection and nodule organogenesis phenotypes of nup85–2, nup133–3, castor–4, pollux–3 and ccamk–3 mutant roots transformed with the empty vector.
Figure S8. Rhizobial infection and nodule organogenesis phenotypes of nfr1–4, nfr5–2, nsp1–1 and nsp2–1 mutant roots transformed with the empty vector.
Figure S9. Stacked histograms comparing frequency counts in terms of the number of nodules or bumps per mutant plants transformed with CCaMK, CASTOR, POLLUX, NUP85 and NUP133 driven by pEpi or p35S.
Figure S10. Spontaneous nodulation phenotypes of wild-type roots transformed with CCaMKT265D driven by pEpi or p35S.
Figure S11. Complementation tests of rhizobial infection and nodule organogenesis phenotypes of nup85–2, nup133–3, castor–4 and pollux–3 mutants transformed with the corresponding genes driven by p35S.
Figure S12. Stacked histograms comparing frequency counts in terms of the number of nodules or bumps per mutant plant by co-transformation with CCaMK under the control of pEpi and truncated CCaMK1–314 or CCaMK1–340 under the control of p35S.
Figure S13. Spontaneous nodule-like structure of the ccamk–3 mutant by co-transformation with CCaMK under the control of pEpi and truncated CCaMK1–314 under the control of p35S.
Figure S14. Complementation tests of rhizobial infection and nodule organogenesis phenotypes of the cyclops–3 mutant transformed with CYCLOPS driven by pEpi or p35S.
Figure S15. Stacked histograms comparing frequency counts in terms of the number of nodules or bumps per mutant plant transformed with CYCLOPS, NSP1, NSP2, NFR1 or NFR5 driven by pEpi or p35S.
Figure S16. Complementation tests of rhizobial infection and nodule organogenesis phenotypes of nsp1–1 and nsp2–1 mutants transformed with the corresponding genes driven by pEpi or p35S.
Plasmid construction.Methods S2. RNA isolation from root hairs.
Spontaneous nodulation phenotypes.
Table S2. Primer sequences used for construction.
Table S3. Primer sequences used in the expression analysis.


