Abstract
Cereblon, a member of the cullin 4 ring ligase complex (CRL4), is the molecular target of the immunomodulatory drugs (IMiDs) lenalidomide and pomalidomide and is required for the antiproliferative activity of these agents in multiple myeloma (MM) and immunomodulatory activity in T cells. Cereblon's central role as a target of lenalidomide and pomalidomide suggests potential utility as a predictive biomarker of response or resistance to IMiD therapy. Our studies characterized a cereblon monoclonal antibody CRBN65, with high sensitivity and specificity in Western analysis and immunohistochemistry that is superior to commercially available antibodies. We identified multiple cereblon splice variants in both MM cell lines and primary cells, highlighting challenges with conventional gene expression assays given this gene complexity. Using CRBN65 antibody and TaqMan quantitative reverse transcription polymerase chain reaction assays, we showed lack of correlation between cereblon protein and mRNA levels. Furthermore, lack of correlation between cereblon expression in MM cell lines and sensitivity to lenalidomide was shown. In cell lines made resistant to lenalidomide and pomalidomide, cereblon protein is greatly reduced. These studies show limitations to the current approaches of cereblon measurement that rely on commercial reagents and assays. Standardized reagents and validated assays are needed to accurately assess the role of cereblon as a predictive biomarker.
Keywords: immunomodulatory drugs, cereblon, myeloma, cullin 4 ring ligase complex, DNA damage binding protein 1
Immunomodulatory drugs (IMiDs®) are thalidomide analogues that have efficacy in haematological malignancies, including multiple myeloma (MM), non-Hodgkin lymphoma and chronic lymphocytic leukaemia. The protein cereblon (CRBN) was first described to be the molecular target of thalidomide in a seminal paper by Ito et al (2010), linking its role to teratogenic effects by thalidomide in zebrafish and chicks. Cereblon is a ubiquitously expressed protein and member of a Cullin 4 ring E3 ligase complex (CRL4) that consists of Cullin 4, RING finger protein (Roc1), and DNA damage binding protein 1 (DDB1; Groisman et al, 2003). Cereblon is proposed to function as a substrate receptor by recruiting substrate proteins to the CRL4 complex for ubiquitination (Lopez-Girona et al, 2012).
The CRBN gene (RefSeq NM_016302.3) is 1329 base pairs and encodes a protein of 443 amino acids that contain a nonfunctional LON protease domain and a putative leucine zipper motif. The CRBN gene consists of 11 exons, and as previously described, exons 5–7 and exons 10–11 are proposed to function in DDB1 and thalidomide binding, respectively (Ito et al, 2010). Two critical residues, tyrosine 384 and tryptophan 386, have been shown to be required for cereblon protein binding to thalidomide (Ito et al, 2010).
Cereblon plays a key role in mediating the antiproliferative and immunomodulatory activities of lenalidomide and pomalidomide in MM and T cells, respectively (Zhu et al, 2011; Lopez-Girona et al, 2012). MM cells stably transduced with CRBN shRNA, resulting in reduced CRBN mRNA and protein expression, were less sensitive than the parental cells to antiproliferative effects by lenalidomide. In addition, MM cells acquired resistance to lenalidomide and pomalidomide via long-term passaging with these drugs that resulted in reduced cereblon protein and RNA expression, indicating the importance of cereblon expression for IMiD function. In T cells, reducing cereblon protein expression with siRNA abrogated the T cell costimulatory activity of the IMiD compounds, again supporting the crucial role of cereblon on IMiD compounds function (Lopez-Girona et al, 2012).
Cereblon's central role as a target of lenalidomide and pomalidomide has brought to light its potential utility as a predictive biomarker of response or resistance to IMiD compound therapy. Currently, there are no standardized reagents or assays for measuring cereblon expression accurately, although commercial antibodies and gene expression assays are available. There is also uncertainty over whether quantification of protein or gene expression level of cereblon is therapeutically relevant. Thus, the clinical value of cereblon measurement is currently unknown.
In this study, we address the relative merits for the measurement of mRNA or protein levels of cereblon, characterize the monoclonal antibody CRBN65 (previously described as anti-CRBN 65–76 by Lopez-Girona et al, 2012), and compare the properties of CRBN65 with the currently available commercial antibodies. We show that CRBN65 is a highly sensitive and specific antibody and describe 2 separate assays – Western blot analysis and immunohistochemistry (IHC) – using this reagent. Our studies with MM cell lines reveal discordance between CRBN gene expression and cereblon protein levels measured using a commercial TaqMan gene expression assay and CRBN65, respectively. Furthermore, we did not observe any correlation between CRBN gene expression or cereblon protein level to sensitivity or to intrinsic resistance to lenalidomide treatment in a diverse panel of MM cell lines. In contrast, cell lines that acquired resistance to IMiD drugs in tissue culture demonstrated a general decline in both cereblon protein and mRNA levels compared to levels in the respective parental sensitive lines. In addition, we have described the presence of multiple alternatively spliced variants of the CRBN pre-messenger RNA transcript in MM cell lines and CD138+ cells isolated from MM patients that complicate accurate measurement of gene expression. Taken together, our data show that cereblon measurements that rely on commercial reagents and assays have limitations due to reagent quality and assay characteristics. Due consideration should be given to developing standardized reagents and validated assays prior to investigating the value of cereblon measurement in clinical samples.
Materials and methods
Cell lines and recombinant protein
Cell lines NCI-H929, U266, RPMI8226 and JJN-3 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). OPM-2, KMS-12-BM and LP-1 were obtained from DSMZ (Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). KMS-11 and KMS-34 cells were obtained from the Japanese Collection of Research Bioresources Cell Bank (Health Science Research Resources Bank, Osaka, Japan). MM.1S cells were obtained from Dr Steven Rosen (Northwestern University, Chicago, IL, USA). ANBL-6, CAG and DF15 cells were obtained from Dr John Shaughnessy (University of Arkansas, Little Rock, AR, USA). Cells were grown in RPMI-I640 medium containing 10% (V/V) heat-inactivated bovine serum (Gibco, Life Technologies, Grand Island, NY, USA) supplemented with 2 mmol/l glutamine. Generation of H929- and DF15-resistant cell lines were described previously (Lopez-Girona et al, 2012). Briefly, lenalidomide-resistant H929 cells were made by continuous treatment with lenalidomide, and DF15 cells were made resistant to pomalidomide by long-term passage in the presence of pomalidomide. Prior to experiments described here, H929 lenalidomide–resistant cells and DF15R pomalidomide–resistant cells were taken out of culture with compounds for 5–7 d before use. Select primary CD138+ MM cells were purchased from AllCells (Emeryville, CA, USA). Production of recombinant human cereblon protein was previously described (Lopez-Girona et al, 2012). Briefly, baculovirus of ZZ-His-CRBN and DDB1-StrepTag (ST) were generated and coexpressed in High Five (Tni) insect cells (Life Technologies). Cells were harvested 48 h after infection, followed by additional purification steps, including Nickel-Sepharose (Qiagen, Valencia, CA, USA) and S200 Sephacryl chromatography (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA), anion exchange chromatography on an 8 ml MonoQ column and a second pass on a S-200 gel filtration column. The CRBN-DDB1 complex was identified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the CRBN-DDB1 containing fractions were pooled and stored in gel filtration buffer (50 mmol/l Tris pH 8·0, 200 mmol/l NaCl, 5% glycerol, 2 mmol/l dithiothreitol) at −70°C.
Nested polymerase chain reaction
Total poly-A RNA was isolated from cell lines or primary CD138+ MM cells using Qiagen (Valencia, CA, USA) RNeasy mini kit and was treated with DNase I (New England Biolabs, Ipswich, MA, USA, Grand Island, NY, USA). RNA was reverse transcribed to cDNA using TaqMan PreAmp Master Mix (Applied Biosystems, Life Technologies), initially to enrich for CRBN cDNA using primers annealing to the 5′ and 3′ UTR regions (5′-CCTTTGCGGGTAAACAGACATGGCC-3′ and 5′-GCAATAATTTCCAAAGCAGATCTTA-3′) in a 14-cycle reaction. A second round of polymerase chain reaction (PCR) using M13-tagged primers (TGTAAAACGACGGCCAGT CATGGCCGGCGAAGGAGATCA and CAGGAAACAGCTATGAC GTTTACAAGCAAAGTATTACTTTGTCT) and Herculase II Fusion DNA polymerase (Agilent, Santa Clara, CA, USA) in a 35-cycle reaction amplified CRBN splice variants. PCR reactions were run on agarose gels, and products were gel purified, inserted into Topo vectors (Life Technologies), and their sequence determined by Sanger dideoxy method.
Generation of monoclonal CRBN antibody and Western blots
CRBN antisera production and validation was previously described by Lopez-Girona et al (2012). Briefly, CRBN antiserum against the peptide sequence of amino acids 65–76 of human CRBN was generated in a rabbit and CRBN specificity determined with peptide blocking experiments and siCRBN experiments. Subsequent hybridoma and subcloning generated the anti-CRBN monoclonal antibody CRBN65. Commercial CRBN antibodies used included Abcam ab68763 (Abcam 1) and ab98992 (Abcam 2); Aviva ARP56882_P050; Novus H00051185-B01P; ProteinTech 11435-1-AP; and Sigma HPA045910-100UL (Sigma 1) and SAB1407456-50UG (Sigma 2). These were evaluated at the manufacturers' recommended concentration and conditions. The β-actin (ACTB) antibody (Sigma, St Louis, MO, USA) was used at 1:10 000. For immunoblot analyses, cells were harvested and lysed in Chris buffer [50 mmol/l Tris pH8, 200 mmol/l NaCl, 10% glycerol, 0·1 mmol/l ethylenediaminetetraacetic acid (EDTA)] containing complete EDTA-free protease inhibitor cocktail tablets (Roche, Indianapolis, IN, USA), PhosStop phosphatase inhibitor cocktail tablets (Roche), and 20 mmol/l para-nitrophenylphosphate (PNPP; Sigma). Protein concentration was determined using a nine-point standard curve with Bio-Rad (Hercules, CA, USA) Protein Assay reagent and the tunable VERSAmax microplate reader (Molecular Devices, Sunnyvale, CA, USA). Cell extracts (50 μg protein) were loaded onto Criterion XT-Precast 4–12% Bis-Tris gels using NuPAGE 2-[N-morpholino]ethanesulfonic acid (MES) SDS running buffer (Invitrogen, Life Technologies). Protein was transferred onto Nitrocellulose Criterion Blotting Sandwiches (Bio-Rad) using NuPAGE Transfer Buffer (Invitrogen) containing 15% methanol. Standard protocols for signal detection and quantification using the LI-COR (Lincoln, NE, USA) Odyssey imager and software were applied.
Immunohistochemistry
Cereblon IHC was performed on the Bond-Max automated slide stainer (Leica Microsystems, Buffalo Grove, IL, USA) using the Bond Polymer Refine Detection system. Formalin-fixed paraffin-embedded (FFPE) tissues or cell pellets were sectioned at 4 μm thickness and deparaffinized on the Bond-Max instrument. Antigen retrieval was performed with Epitope Retrieval 2 (ER2, pH 9·0) for 20 min at 100°C. The slides were blocked for endogenous peroxidase activity with Peroxide Block for 5 min at room temperature. Sections were then incubated with the rabbit monoclonal antibody CRBN65 at a 1:4000 dilution for 15 min at room temperature. Post-primary and horseradish peroxidase-labelled polymer were incubated at the instrument's default conditions. Antigen-antibody complex was then visualized with hydrogen peroxide substrate and diaminobenzidine tetrahydrochloride (DAB) chromogen. Slides were counterstained with haematoxylin. For the peptide neutralization experiment, as part of the assay validation, the CRBN65 antibody was preincubated with a 10-fold (by mass) excess of blocking peptide at room temperature for 2 h with rotation before applying to the slides. CRBN immunoreactivity was quantified using ImagePro Plus 6.3 (Media Cybernetics, Bethesda, MD, USA) when examining assay precision. Cell pellets were fixed in 10% neutral buffered formalin overnight and were processed and embedded into paraffin blocks using standard histology procedures. Tissue microarray slides containing multiple myeloma cases and normal bone marrow were purchased from US Biomax (Catalog No. BM483a; Rockville, MD, USA), and normal colon tissue from ProteoGenex Inc. (Culver City, CA, USA).
Results
CRBN65 is a highly specific and sensitive antibody for detecting cereblon by Western analysis and immunohistochemistry
CRBN65 is a rabbit monoclonal antibody developed against cereblon amino acids 65–76 and was described previously (Lopez-Girona et al, 2012). We compared, via Western blot, the CRBN65 antibody with seven common commercially-available antibodies from five different companies using the manufacturers' recommended conditions in cell extracts from MM cell lines DF15 and DF15R. The DF15 MM cell line expressed cereblon protein but DF15R cells, which are resistant to lenalidomide and pomalidomide, have little to no cereblon protein (Lopez-Girona et al, 2012). Fig1 shows the performance of CRBN65 and the seven commercial antibodies with identical input of total protein in each case. A strong band corresponding to full-length 51 kDa cereblon protein was specifically detected by CRBN65 in DF15 cells, whereas a faint band was detected at the same location in the DF15R line. In addition to full-length CRBN protein, a number of faint lower molecular weight bands were detected in both DF15 and DF15R cell lines, which may represent degraded CRBN protein as these bands appear inconsistently among various cell preparations and are blocked by a cereblon peptide. Among the seven commercial antibodies, a putative cereblon band was only detectable in blots probed with Abcam #1, ProteinTech and Sigma #1 antibodies. However, compared with CRBN65, the commercial antibodies detected a large number of bands of varying molecular weights at varying intensities, including many higher molecular weight bands than what is expected from the full-length cereblon protein (51 kDa).
Fig 1.
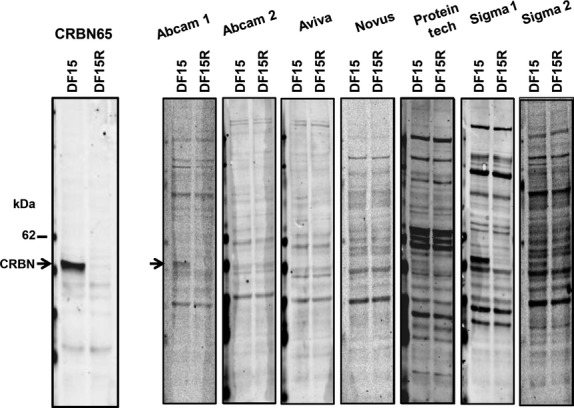
The CRBN65 antibody detects cereblon (CRBN) protein with high specificity compared with commercial antibodies. DF15 and DF15R MM cell extracts (20 μg/lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and probed with CRBN65 (1:1000) or commercial antibodies (manufacturers' recommended dilution) against CRBN. Blots were analysed on LI-COR Oddysey and arrows denote the full-length CRBN protein band [51 kiloDalton (kDa)] that appears specifically in DF15 cells but not DF15R.
In order to establish sensitivity of detection by CRBN65, we tested its ability to detect cereblon by Western blot analysis on recombinant cereblon protein in a dilution range (0·2–60 ng total protein) and in DF15 and DF15R cell lines (Fig2). As shown in Fig2A, recombinant cereblon protein was detectable at the 200 pg level by the CRBN65 antibody under the conditions employed. Linear regression analysis indicated high correlation (R2 = 0·99) between protein level and Western blot signal with a linear dynamic range of detection of 300-fold (Fig2B). This detection range is relevant and will cover a diverse sample set including the MM cell line DF15, which has 10 ng cereblon per 20 μg of total protein. These data demonstrate that the CRBN65 antibody enabled CRBN protein detection across a relevant dynamic range.
Fig 2.
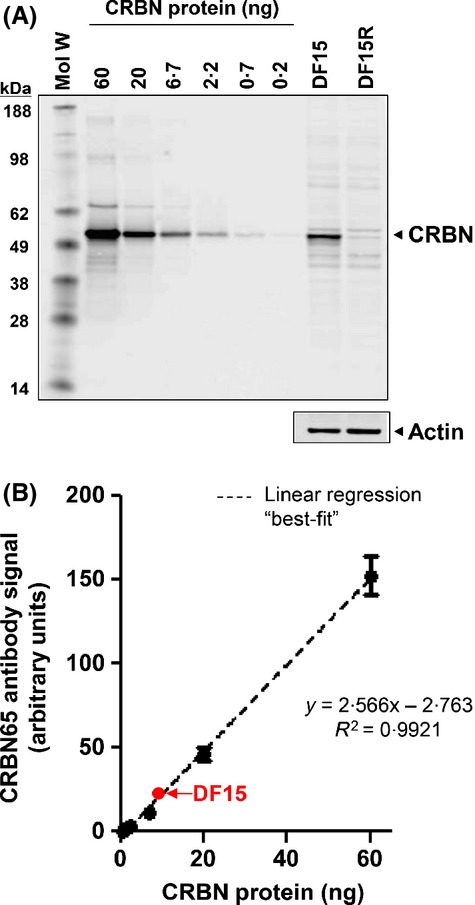
The CRBN65 antibody detects recombinant and endogenous CRBN protein with high sensitivity. (A) Recombinant human full-length CRBN (0·2–60 ng) along with DF15 and DF15R cell extracts (20 μg/lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and probed with CRBN65 antibody (1:1000). (B) Linear regression analysis using CRBN signal as measured on LI-COR Oddysey for rhCRBN is linear in the 0·2–60 ng range, and CRBN protein measured in DF15 cells falls within linear range of detection. CRBN, cereblon; kDa, kiloDalton; Mol W, molecular weight; R2, coefficient of determination.
We next tested the ability of CRBN65 to detect cereblon protein across a variety of MM cell lines that included IMiD agent sensitive (e.g., DF15, MM.1S, ANBL6), intrinsically resistant (e.g., RPMI-8226, KMS11, LP1), or acquired resistant lines (DF15R). As seen in Fig3 (upper panel), CRBN65 detected a dominant band corresponding to full-length cereblon protein in all cell lines tested except DF15R. In a number of cell lines (e.g., MM.1S, KMS34, DF15, DF15R), a few lower molecular weight faint bands were also detected. When the same samples were tested with a blocking peptide, both full length as well as the lower bands disappeared completely, suggesting that the lower bands are related to cereblon protein (Fig3 lower panel). In addition, CRBN65 recognized full-length cereblon protein in normal T cells, lymphoma cell lines, and epithelial cancer cell lines (Fig4). The observed lower molecular weight bands are likely to be proteolytic fragments of cereblon or possible isoforms of cereblon and were undetectable with blocking peptide (data not shown). These results demonstrate that most commercial antibodies are neither sensitive nor specific for detecting cereblon protein in DF15 and DF15R cell lines. Among the antibodies that could detect cereblon protein, none of them showed high specificity as compared with the CRBN65 antibody (Fig1). Thus, among the seven antibodies tested, CRBN65 was both the most sensitive and specific for detecting cereblon protein.
Fig 3.
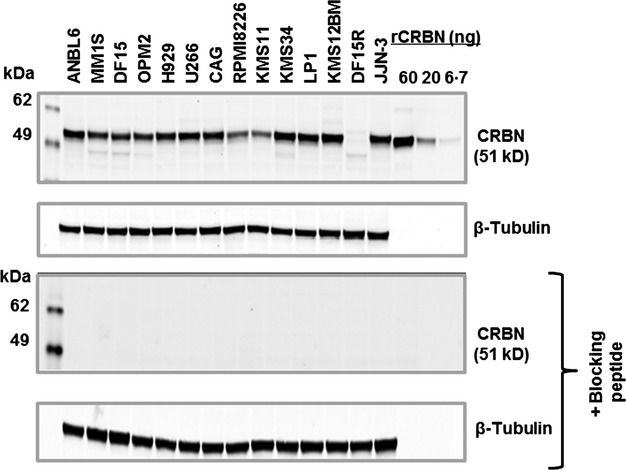
The CRBN65 antibody detects CRBN protein across a wide range of MM cell lines with high specificity. A panel of MM cell extracts (20 μg/lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and probed with CRBN65 antibody (1:1000) or β-tubulin, without blocking peptide (upper panel) or with blocking peptide (lower panel). CRBN, cereblon; kDa, kiloDalton; MM, multiple myeloma; rCRBN, recombinant cereblon.
Fig 4.
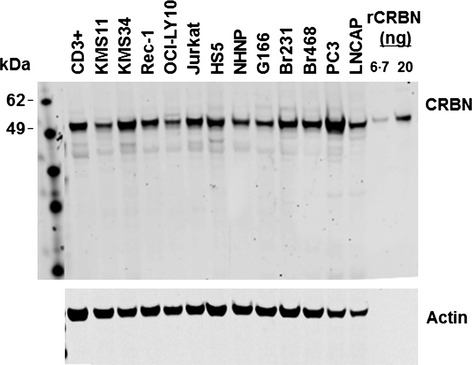
The CRBN65 antibody detects CRBN protein across a range of normal human cells and tumour cells. A panel of cell extracts (20 μg/lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and probed with CRBN65 antibody (1:1000) or actin antibody. Cell types of cell lines tested are as follows: KMS11 and KMS34, human multiple myeloma; Rec1, mantle cell lymphoma; OCI-LY10, activated B-cell–like diffuse large B-cell lymphoma; Jurkat, T-cell leukaemia; HS5, bone marrow stroma fibroblast; NHNP, normal human progenitor cells; G166, glioblastoma; Br231 and Br468, breast cancer cell lines; PC3, prostate carcinoma cell line; LNCAP, prostate adenocarcinoma cell line; CRBN, cereblon; kDa, kiloDalton.
CRBN65 immunohistochemistry assay
CRBN65 was validated in an IHC assay for detecting cereblon in FFPE tissues. Multiple parameters, including assay specificity, precision, accuracy, and detection range were investigated. Specificity was demonstrated by the staining of fixed DF15 and DF15R cell pellets. The cereblon IHC results correlated well with Western blot data, such that the parental DF15 had high cereblon immunoreactivity, and the pomalidomide-resistant DF15R had very low to almost absent cereblon immunoreactivity (Fig5A). In addition, blocking peptide was able to completely eliminate cereblon immunoreactivity in the DF15 cell pellet and normal colon tissue (Fig5B). The cereblon IHC assay demonstrated an acceptable detection range in tissues and cell types tested. As shown in Fig5C, a range of staining intensity from low to high was detected on the MM tissue cores. Both cytoplasmic and nuclear immunoreactivity was observed. The normal bone marrow cores on this tissue microarray had sporadic cereblon positive cells.
Fig 5.
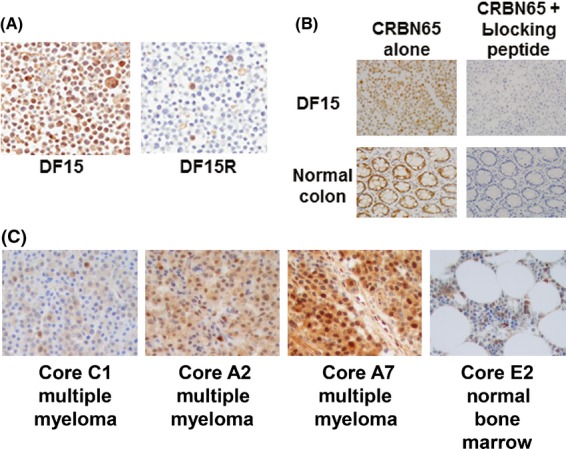
Cell model staining and peptide blocking experiments demonstrate the specificity of the CRBN65 antibody. (A) CRBN immunohistochemistry in DF15 and DF15R cell pellets. (B) DF15 cell pellet blocks and normal colon section was stained with CRBN65 with and without blocking peptide. (C) Representative photomicrographs from MM and adjacent normal bone marrow microarray. Objective = 40×. CRBN, cereblon.
Staining intensities of CRBN65 antibody were consistent across intrarun and interrun slides on colon specimen M1-Colon 1 (Fig S1). The precision of the assay was expressed as coefficient of variation (CV) that was calculated from the mean pixel intensity values from the nine stained slides across three runs. The CV of the imaging scores for the cereblon IHC assay was 9%. This exceeds US Food and Drug Administration and National Cancer Institute guidelines that suggest that CV values should be <15% for analytical assays (International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use, 1995; National Cancer Institute Division of Cancer Prevention, 1999). To further test the accuracy of the assay, DF15 and DF15R cells were mixed at different proportions, including 100%/0%, 75%/25%, 50%/50%, 25%/75% and 0%/100% DF15/DF15R to mimic a range of cereblon protein expression. IHC staining of the proportionally mixed cells correlated with Western analysis data of corresponding cell lysates (Fig S2), suggesting that the assay accurately detects cereblon protein. In summary, together with data from DF15 and DF15R cell models, the cereblon IHC assay utilizing the CRBN65 antibody specifically and reliably detected varying levels of cereblon immunoreactivity in FFPE tissue.
MM cells express multiple splice variants of CRBN messenger RNA
A number of splice variants of CRBN primary transcript have been described in the public domain, including two reported in National Center for Biotechnology Information RefSeq (NM_016302.3, NM_001173482.1) and 13 reported in Ensembl (Flicek et al, 2012; Ovsyannikova et al, 2012). It should be noted that the two RefSeq transcripts are also contained in the Ensembl set and that only a total of four transcripts are believed to be protein coding. However, no splice variant of CRBN mRNA has been experimentally demonstrated yet. In order to identify potential splice variants of CRBN mRNA, we performed a two-step nested amplification of CRBN cDNA from H929 cells. Double stranded PCR-amplified CRBN cDNA was separated on an agarose gel. Fig6A is an illustrative example showing a number of PCR fragments, including a band ≈1300 bp that is consistent with the full-length coding sequence of CRBN mRNA (1329 bp). The bands marked with an arrow indicate CRBN mRNA splice variants that were successfully subcloned and subsequently sequenced. A number of other PCR fragments that were not sequenced could represent additional alternative splice variants of CRBN mRNA. Nested PCR amplification of CRBN mRNA in primary CD138+ MM tumour cells also generated multiple PCR fragments (Fig6B). Based on Sanger sequencing of the seven gel-isolated bands from H929 MM cells, we identified full length CRBN mRNA and six additional unique and novel splice variants of CRBN mRNA, designated crbncc-001 to crbncc-007, respectively. Nested PCR in additional MM cell lines expanded the CRBN mRNA splice variant profile to an additional 15 distinct isoforms in MM cell lines, making a total of 22 CRBN splice variants thus far. CRBN splice variants apparently utilize canonical, as well as noncanonical, splice donor/acceptor sited within the exon sequences. The organizational structure of these isoforms and their nomenclature are shown in Fig7. A number of isoforms are notable for their exon composition and potential function. First, crbncc-002 has exon 10 deleted while retaining an open reading frame of CRBN mRNA, potentially encoding a protein of 399 aa. Ito et al (2010) previously showed that deletion of exons 10 and 11 results in loss of thalidomide binding. Thus exon 10-deleted cereblon is not expected to bind IMiDs. Similarly, crbncc-003 is generated by a noncanonical splicing event that joins a partial segment of exon 2 with a portion of exon 5, retaining an open reading frame that encodes a truncated CRBN protein. If translated, crbncc-003 could produce a protein of 274 aa lacking a portion of the putative DDB1-binding domain, while retaining the drug-binding region at the C-terminus. Finally, variant crbncc-004 encodes an open-reading frame that completely lacks the putative DDB1-binding domain. All other isoforms contain multiple stop codons in their primary sequences and are not likely to produce translated versions of cereblon protein.
Fig 6.
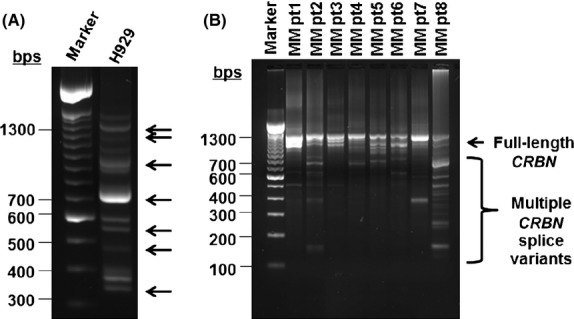
A complex profile of CRBN splice variants exists in both MM cell lines and primary CD138+ MM tumour cells. (A) Nested polymerase chain reaction (PCR) reaction for CRBN splice variants in H929 MM cells were separated on an agarose gel and arrows indicate those bands that were gel purified and sequenced. (B) Nested PCR reaction for CRBN splice variants in 8 MM patient (pt) cells were separated on an agarose gel and multiple bands indicate complexity of CRBN mRNA profile. CRBN, cereblon; MM, multiple myeloma.
Fig 7.
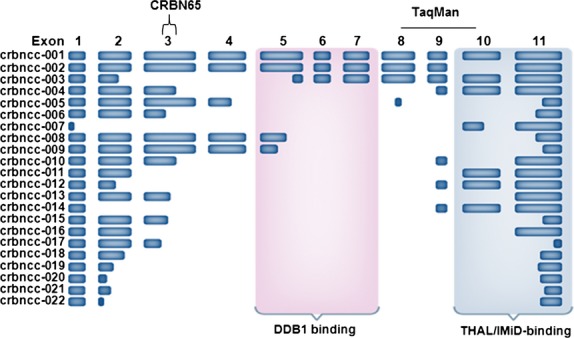
A schematic of CRBN splice variants crbncc-001 to crbncc-022 sequenced from MM cell lines with regions recognized by CRBN65 and TaqMan denoted. CRBN, cereblon; DDB1, DNA damage binding protein 1; IMiD, IMiD immunomodulatory drug; THAL, thalidomide.
Neither CRBN messenger RNA nor protein levels correlate with intrinsic sensitivity or resistance to the lenalidomide and pomalidomide in MM cell lines
We utilized CRBN65 antibody to measure cereblon protein levels in a panel of 12 MM cell lines by Western blot analysis. Cereblon protein levels were first normalized to β-tubulin (TUBB), and then further normalized to cereblon protein levels in DF15 cells (Fig8). In parallel, CRBN gene expression in these lines was measured using the ‘best coverage’ TaqMan assay and normalized to HPRT1 mRNA expression, then further normalized to CRBN mRNA levels in DF15 cells. A number of important observations emerged as CRBN gene and cereblon protein expression were compared across these cell lines. First, as seen in Fig8, based on protein expression level, the cell lines can be grouped into three categories relative to the expression in DF15 cells (>1·25, 0·75–1·25 and <0·75): cell lines (ANBL6, U266, KMS34, LP1, KMS12BM and JJN-3) that express high levels of cereblon protein (>1·25-fold higher relative to DF15), cell lines (OPM2, H929, CAG) that express cereblon protein at a similar level as in DF15 (0·75–1·25-fold of DF15), and finally cell lines (RPMI8266, KMS11) that express a low level of cereblon protein (<0·75-fold of DF15 cells). However, in both the high and medium groups, the relative level of CRBN mRNA was either comparable (i.e., DF15, U266, JJN, CAG and H929) or lower (ANBL6, KMS34, LP1 and KMS12BM) than the relative protein levels. Specifically, there was no clear correlation between gene expression levels to protein across the 13 cell lines tested (r2 = 0·32, data not shown).
Fig 8.
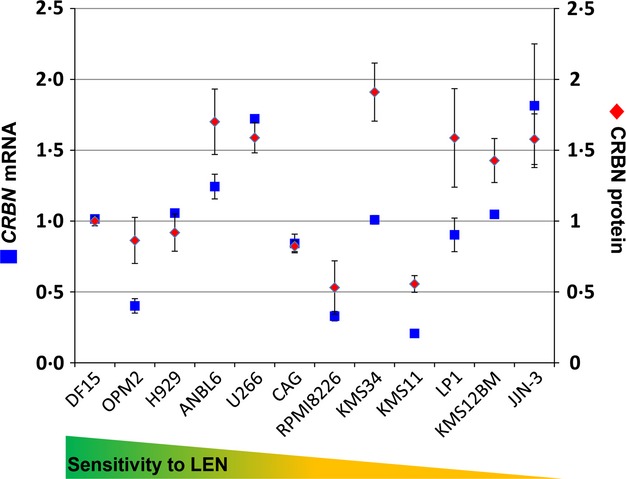
CRBN mRNA and CRBN protein levels do not correlate in MM cell lines with intrinsic sensitivity or resistance to the IMiD agent lenalidomide. CRBN mRNA and CRBN protein levels were measured in 12 human MM cell lines by TaqMan polymerase chain reaction and Western blot, respectively. mRNA and protein were normalized to HRPT1 and β-tubulin, respectively. Graph represents the average of 3–4 experiments. CRBN, cereblon; LEN, lenalidomide; MM, multiple myeloma; mRNA, messenger RNA.
Our second observation was that there was no clear correlation between CRBN mRNA or cereblon protein levels with the antiproliferative sensitivity of cell lines to lenalidomide or pomalidomide For example, only two (RPMI-8226 and KMS11) of the six intrinsically resistant cell lines had low cereblon protein expression; the other four intrinsically resistant cell lines (KMS34, LP-1, KMS12BM and JJN-3) had high levels of the protein, suggesting the underlying cause of IMiD resistance in these lines is probably not due to limited availability of cereblon protein. In contrast, among the sensitive cell lines, cereblon protein level was higher or comparable to DF15 level. Similarly, there was a discrepancy among sensitive and resistant lines with respect to the level of CRBN transcripts. The levels of CRBN mRNA relative to DF15 CRBN mRNA levels were not predictive of the sensitivity or resistance in these cell lines (Fig8).
Acquired resistance to lenalidomide and pomalidomide in MM cell lines is accompanied by a reduction in CRBN mRNA and protein levels
DF15 and H929 cells were cultured long-term in pomalidomide and lenalidomide respectively, to generate pomalidomide-resistant cells DF15R and lenalidomide-resistant lines H929-1051, -1052, -1053 and -1054. The methods for generation of these resistant cell lines were described by Lopez-Girona et al (2012). We tested CRBN gene expression and cereblon protein levels in each cell line and compared their normalized values with their respective parental lines (Fig9). In DF15R cells, cereblon protein is reduced nearly 100%, and the CRBN mRNA level is reduced by 77% compared with parental DF15 cells (Fig9A). Following a similar trend, lenalidomide-resistant H929 clones 1051, 1052, 1053 and 1054 had 42%, 23%, 47% and 68% less CRBN mRNA and 50%, 70%, 54% and 34% less protein respectively, when compared with the parental H929 line. While the general trend of lower CRBN mRNA and protein in resistant lines versus parental cells is maintained in all resistant lines, the relative levels of CRBN gene expression and cereblon protein levels in 1052 and 1054 were opposite of each other. Even though both cell lines have lower mRNA and protein levels compared with the parental cell line, 1052 has higher level of RNA than protein, while the opposite is true in resistant 1054 line (Fig9B). A smaller percentage of cells expressed cereblon protein in the H929 1054 cell line compared with the parental H929 cells, as shown by cereblon IHC (Fig9C). Thus, the discrepancy of mRNA and protein level observed in sensitive and intrinsically resistant lines is also seen in cell lines with acquired resistance upon IMiD exposure in vitro. Taken together, these data demonstrate that CRBN gene expression and cereblon protein level per se do not predict whether a cell line is sensitive to IMiD treatment. However when cell lines acquire resistance by long-term passaging with lenalidomide or pomalidomide, our data is consistent with previous clinical observations that CRBN gene expression levels decrease from diagnosis to relapse in MM patients treated with lenalidomide (Zhu et al, 2011).
Fig 9.
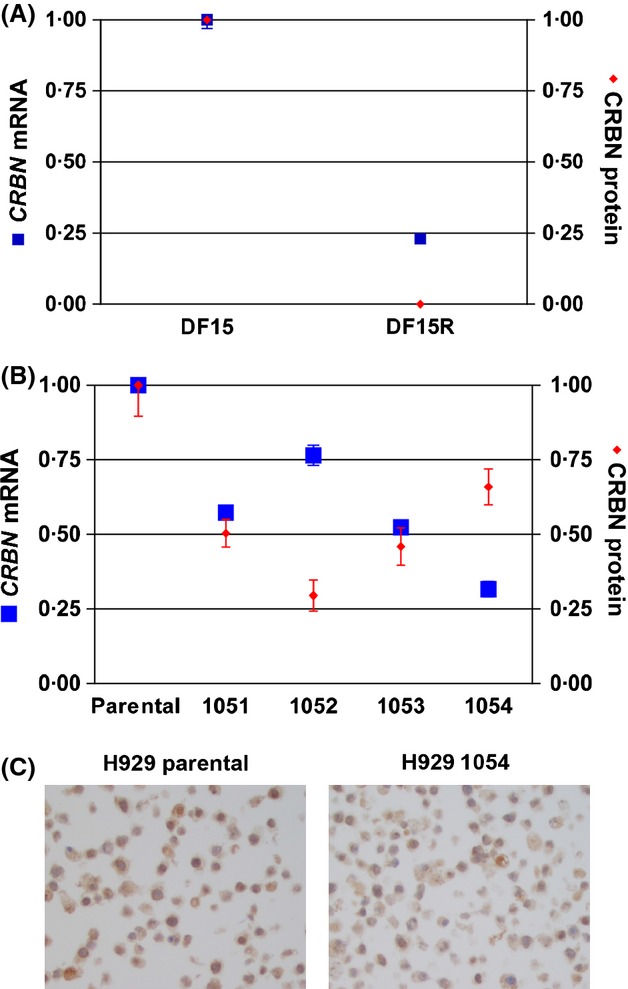
Acquired resistance to immunomodulatory drugs in MM cell lines leads to less CRBN mRNA; however, a disconnect exists between CRBN mRNA and CRBN protein levels. Quantification of mRNA and protein, and immunohistochemistry (IHC) were performed as described in the text. (A) Comparison of CRBN protein and CRBN mRNA in DF15 and DF15R resistant cells. (B) Comparison of CRBN protein and CRBN mRNA in parental H929 versus lenalidomide-resistant H929 lines 1051, 1052, 1053 and 1054. (C) CRBN IHC in H929-parental cell versus H929 lenaliomide-resistant line 1054. CRBN, cereblon; MM, multiple myeloma; mRNA, messenger RNA.
Discussion
The discovery of cereblon as a target of thalidomide and the IMiDs, lenalidomide and pomalidomide, has enabled a pursuit of better understanding of the molecular mechanism of action by these agents. In parallel, there is significant clinical interest to investigate the possible role of cereblon as a biomarker for the IMiD compounds response or resistance. Currently, there are no validated assays for measuring cereblon protein levels or gene expression in clinical samples. It is also unclear whether measurement of cereblon expression at the mRNA or protein level or both will be optimal for clinical studies.
Measurement of cereblon protein is associated with a number of assay limitations. Currently available commercial antibodies are neither sensitive nor specific for reliable detection of cereblon protein. We have characterized a monoclonal antibody CRBN65 and compared its properties with the commonly used, currently available commercial antibodies against cereblon. We showed that CRBN65 is highly sensitive and specific in Western blot and IHC analysis. Furthermore, it has a broad dynamic range and can detect as little as 200 pg of cereblon protein via Western blot. Against this, results using commercial antibodies of cereblon must be viewed with caution. For example, a recent study of 42 newly diagnosed MM patients, which measured CRBN levels by IHC using a commercial antibody, found an association between CRBN protein and response (overall survival and progression-free survival) to lenalidomide. However, 16·1% of patients in the CRBN-high group failed to respond to lenalidomide, and 54·5% of patients in the cereblon-low group also failed to respond, suggesting a significant portion of responders were CRBN-low (Klimowicz et al, 2012). Without an assessment of the sensitivity and specificity of the antibody used in this study, it is difficult to interpret the significance of the observed correlation presented in this study.
Current methods of gene expression measurement predominately employ Affymetrix array or quantitative reverse-transcription PCR (qRT-PCR)–based methods to detect and quantify transcripts. The measurement of CRBN by qRT-PCR typically uses commercial assays, such as predesigned TaqMan assays, where primer and probe sets are optimized with respect to known gene splicing information. For CRBN mRNA, the current best coverage assay detects the exon 8-exon 9 junction and exon 10 as a measure of gene expression (data not shown; https://bioinfo.invitrogen.com/genome-database/details/gene-expression/Hs00372271_m1). However, this assay needs to be fully validated given the multiple splice variants. The presence of multiple CRBN splice variants complicates the transcript measurement by Affymetrix, which leads to inaccurate overestimation of full-length CRBN transcripts. A number of CRBN splice variants, such as crbncc-002 and crbncc-003, contain the target region of the probes and have deletions in other exons. Even though these transcripts will not give rise to full-length cereblon protein, they will be measured by the Affymetrix probesets 222533_at and 218142_s_at. The Affymetrix chip, such as U133 plus 2, contains 2 probe sets 218142_s_at and 222533_at for the detection of CRBN transcripts (Fig S3). The 222533_at probe set hybridizes entirely within the CRBN 3′UTR. The second probe set 218142_s_at is designed to hybridize with sequences that span exon 10 and 11. This probe set will not be able to measure the exon 10–deleted splice variant crbncc-002. CRBN measurement has been reported based on a combined value from the two probe sets and described correlation of CRBN level measured in diagnostic samples to clinical outcomes of pomalidomide/dexamethasone or thalidomide-maintenance (Schuster et al, 2012; Broyl et al, 2013).
In a study of 53 relapsed MM patients treated with pomalidomide and dexamethasone, CRBN mRNA levels, measured by Affymetrix chip prior to therapy initiation, were used to correlate with response (Schuster et al, 2012). However, the cut-off values used to distinguish low-, mid- and high-CRBN expression gave a narrow range (0·89 vs. 1·03 vs. 1·21). This highlights the potential problem associated with poor dynamic range in microarray-based gene expression measurement that may not reflect the true range in patient samples. Validation of clinical microarray-based assays that segregate patients with such narrow differences poses technical challenges. In contrast to microarray-based assays, qRT-PCR, in principle, provides a broader dynamic assay range and could provide a more accurate representation of CRBN mRNA transcript levels in patients. In another study of 49 MM patients treated with lenalidomide plus dexamethasone, median CRBN gene expression was measured by qRT-PCR and ranged from 0·31 to 28·74; median CRBN mRNA expression indicated a correlation with clinical response (Heintel et al, 2013). However, there were broad confidence intervals and exceptions, such as a complete responder with a low CRBN mRNA expression level of 0·81.
Our findings in MM cell lines reveal a lack of correlation (r2 = 0·32) between CRBN gene level and cereblon protein level measured using a commercial TaqMan gene-expression assay and CRBN65 antibody in MM cell lines (data not shown). Furthermore, no correlation was observed between CRBN gene or cereblon protein expression levels to sensitivity or intrinsic resistance to lenalidomide. In contrast, cell lines that acquire resistance to IMiD agents through long-term passage showed a decline in both CRBN protein and CRBN mRNA level compared with the parental sensitive lines. This decline in protein and mRNA levels indicates that loss of cereblon protein and CRBN mRNA may play a role in acquired resistance; however, proper measurement is still required.
We identified multiple splice variants of CRBN mRNA with both canonical and noncanonical splice patterns. The noncanonical splicing may be due to dysregulation of splicing factors rather than splice site mutations since mutations in CRBN exons are a rare event (Gandhi et al, 2013). Notably, defects in the spliceosomal machinery have been demonstrated across multiple haematological malignancies, such as myelodysplastic syndromes, chronic lymphocytic leukaemia, and acute lymphoblastic leukaemia (Papaemmanuil et al, 2011; Rossi et al, 2011; Wang et al, 2011; Yoshida et al, 2011; Lasho et al, 2012). Spliceosome dysregulation in MM is not widely described, although genome analyses from patients have identified mutations in RNA processing genes (Chapman et al, 2011).
As noted earlier, several detected splice variants are missing crucial regions for the IMiD compounds function, including THAL/IMiD- and DDB1-binding domains, suggesting that these RNA species may confer IMiD compound resistance. We are currently investigating whether any of the isoforms are expressed as proteins in MM cells. Notwithstanding the lack of this information, careful consideration should be given to the presence of splice forms when designing quantitative gene expression assays.
The presence of alternatively spliced variants of CRBN in MM cells raise questions relating to their potential biological function. Full-length and other isoforms were identified from cell lines and primary samples. Whether IMiD compounds treatment changes alternative splicing of CRBN is an interesting question under investigation. However, alternative splicing of genes is common in MM. The occurrence of alternative splicing in MM patients was studied in 170 newly diagnosed MM patients using Affymetrix Exon 1·0 ST array on purified CD138+ cells (Munshi et al, 2009). Over 100 genes (exons) were identified as candidates that showed alternative splicing patterns in subsets of patients, and this observation suggests the potential to identify subgroups of patients that display similar levels of splice isoform expression alongside similar prognostic outcomes. More importantly, the study demonstrated that alternative splicing occurs in a significant proportion of MM patients and thus the process may be relevant from both disease-biology and drug-development perspectives (Moalli et al, 1993; Bakkour et al, 2007; Lee et al, 2008).
In this study, we explored CRBN protein and CRBN gene expression measurement. We characterized CRBN65, a highly sensitive and specific monoclonal antibody. In addition, we analysed CRBN mRNA and detected a large number of splice variants that reveal a complex post-transcriptional processing of the transcribed gene. While the clinical value of CRBN expression as a predictive or prognostic biomarker is an important question, current approaches to measuring CRBN levels that rely on commercial reagents and assays have limitations due to reagent quality and assay characteristics. Appropriate antibodies and validated assays for cereblon protein detection and CRBN gene expression that account for gene complexity are needed for cereblon measurement in the clinic.
Beyond the technical limitations mentioned above, however, the diagnostic assessment of CRBN is further complicated by a number of clinical considerations. First, MM treatment regimens are becoming more combination-based, with an increasing use of novel agents in addition to an IMiD. Thus, CRBN measurement would have to be examined in the combination setting in randomized controlled trials to ascertain its true utility. Second, the disease stage (newly diagnosed versus relapsed and refractory setting) represents another complexity, as the natural history of the disease plays a role in response to therapy. Third, there is currently no clinical data to describe how CRBN level may affect response/resistance to individual IMiD drugs, such as lenalidomide or pomalidomide. This is particularly critical as ≈30% of relapsed/refractory MM patients who are resistant/refractory to lenalidomide-dexamethasone combination therapy respond to pomalidomide-dexamethasone combination therapy (San Miguel et al, 2013). And finally, other members of the CRL4CRBN, DDB1, Cullin4, and the substrates for IMiD activity in addition to CRBN may have significant role(s) in IMiD response/resistance. Proper understanding of the role of CRBN measurement in the clinic will emerge from addressing the technical as well as these clinical considerations in the future.
Acknowledgments
We thank Sharon L Aukerman (Celgene Corp, Summit, NJ, USA) for helpful discussions, Michael Gonzales (Celgene Corp, San Francisco, CA, USA) for reviewing splice variant data and John Shaughnessy, PhD (University of Arkansas, Little Rock, AR, USA) for the DF15 cell line. The authors received editorial support in the preparation of this manuscript from Graham Clinthorne, PhD and Stacey Garrett, PhD (MediTech Media), funded by Celgene Corporation. The authors are fully responsible for the content and editorial decisions for this manuscript.
Author contributions
DM, GC, ER, KM, SG, AM, PJ and MA designed and performed experiments and/or analysed data related to CRBN65 antibody testing. MW and GC performed experiments and/or analysed data related to splice variants. YR, MW and MB performed immunohistochemistry studies. AKG, AT and RC wrote the manuscript. AKG, BC, PHS, TOD, ALG, AT and RC provided intellectual input and experimental design and analysis.
Conflict of interest
AKG, DM, MW, GC, ER, KM, SG, YR, MW, MB, GC, AM, PJ, MA, BC, PHS, TOD, ALG, AT, and RC are employees of Celgene Corporation. There are no other competing financial interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Intrarun and interrun IHC results demonstrate the precision of the assay.
Precision of the IHC assay with the CRBN65 antibody.
Alignment of CRBN Affymetrix probe sets 218142_S_AT and 222533_AT with exon 10, 11, and the UTR of the CRBN gene.
References
- Bakkour N, Lin Y, Maire S, Ayadi L, Mahuteau-Betzer F, Nguyen CH, Mettling C, Portales P, Grierson D, Chabot B, Jeanteur P, Branlant C, Corbeau P. Tazi J. Small-molecule inhibition of HIV pre-mRNA splicing as a novel antiretroviral therapy to overcome drug resistance. PLoS Pathogens. 2007;3:e159. doi: 10.1371/journal.ppat.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyl A, Kuiper R, van Duin M, van der Holt B, el Jarari L, Bertsch U, Zweegman S, Buijs A, Hose D, Lokhorst HM, Goldschmidt H. Sonneveld P. High cereblon expression is associated with better survival in patients with newly diagnosed multiple myeloma treated with thalidomide maintenance. Blood. 2013;121:624–627. doi: 10.1182/blood-2012-06-438101. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G. Golub TR. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kähäri AK, Keefe D, Keenan S, Kinsella R, Komorowska M, Koscielny G, Kulesha E, Larsson P, Longden I, McLaren W, Muffato M, Overduin B, Pignatelli M, Pritchard B, Riat HS, Ritchie GRS, Ruffier M, Schuster M, Sobral D, Tang YA, Taylor K, Trevanion S, Vandrovcova J, White S, Wilson M, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernández-Suarez XM, Harrow J, Herrero J, Hubbard TJP, Parker A, Proctor G, Spudich G, Vogel J, Yates A, Zadissa A. Searle SMJ. Ensembl 2012. Nucleic Acids Research. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi AK, Lentzsch S, Schey SA, Calle Y, Chelliah R, Orlowski RZ, Bjorklund C, Madan A, Waldman M, Ning Y, Mendy D, Lopez-Girona A, Schafer P, Aukerman L, Thakurta A. Chopra R. Exon mutations in cereblon (CRBN) and DNA damage binding protein 1 (DDB1) genes are rare in myeloma cells and patients. Clinical Lymphoma Myeloma and Leukemia. 2013;13:S182–S183. doi: 10.1038/leu.2013.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K. Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Heintel D, Rocci A, Ludwig H, Bolomsky A, Caltagirone S, Schreder M, Pfeifer S, Gisslinger H, Zojer N, Jäger U. Palumbo A. High expression of cereblon (CRBN) is associated with improved clinical response in patients with multiple myeloma treated with lenalidomide and dexamethasone. British Journal of Haematology. 2013;161:695–700. doi: 10.1111/bjh.12338. [DOI] [PubMed] [Google Scholar]
- International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. 1995. Guidelines for industry: text on validation of analytical procedures http://www.labcompliance.de/documents/international/ich/h-307-ich-fda-methods-terminology-ichq2a.pdf. Accessed September 9, 2013. [DOI] [PMC free article] [PubMed]
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y. Handa H. Identification of a primary target of thalidomide teratogenicity. Science (New York, N.Y.) 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Klimowicz A, Neri P, Belch A, Belch A, Dean M, Ren L, Gratton K, Slaby J, Johnson J, Duggan P, Steward DA. Bahlis NJ. High cereblon protein expression correlates with improved response and survival in myeloma patients treated with lenalidomide. Blood. 2012;120:931. [Google Scholar]
- Lasho TL, Finke CM, Hanson CA, Jimma T, Knudson RA, Ketterling RP, Pardanani A. Tefferi A. SF3B1 mutations in primary myelofibrosis: clinical, histopathology and genetic correlates among 155 patients. Leukemia. 2012;26:1135–1137. doi: 10.1038/leu.2011.320. [DOI] [PubMed] [Google Scholar]
- Lee T, Ma W, Zhang X, Giles F, Cortes J, Kantarjian H. Albitar M. BCR-ABL alternative splicing as a common mechanism for imatinib resistance: evidence from molecular dynamics simulations. Molecular Cancer Therapeutics. 2008;7:3834–3841. doi: 10.1158/1535-7163.MCT-08-0482. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J, Karasawa S, Carmel G, Jackson P, Abbasian M, Mahmoudi A, Cathers B, Rychak E, Gaidarova S, Chen R, Schafer PH, Handa H, Daniel TO, Evans JF. Chopra R. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moalli PA, Pillay S, Krett NL. Rosen ST. Alternatively spliced glucocorticoid recep-tor messenger RNAs in glucocorticoid-resistant human multiple myeloma cells. Cancer Research. 1993;53:3877–3879. [PubMed] [Google Scholar]
- Munshi NC, Li C, Minvielle S, Moreau P, Amin S, Magrangeas M, Anderson K. Avetloiseau H. Use of alternate splicing to identify subgroups in uniformly treated newly-diagnosed myeloma. Journal of Clinical Oncology. 2009;27:8602. [Google Scholar]
- National Cancer Institute Division of Cancer Prevention. 1999. Requirements for pharmacokinetic and biomarker methods development http://prevention.cancer.gov/files/clinical-trials/biomarkers.pdf. Accessed September 9, 2013.
- Ovsyannikova IG, Kennedy RB, O'Byrne M, Jacobson RM, Pankratz VS. Poland GA. Genome-wide association study of antibody response to smallpox vaccine. Vaccine. 2012;30:4182–4189. doi: 10.1016/j.vaccine.2012.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, Mudie LJ, Stephens PJ, McLaren S, Massie CE, Tarpey PS, Varela I, Nik-Zainal S, Davies HR, Shlien A, Jones D, Raine K, Hinton J, Butler AP, Teague JW, Baxter EJ, Score J, Galli A, Della Porta MG, Travaglino E, Groves M, Tauro S, Munshi NC, Anderson KC, El-Naggar A, Fischer A, Mustonen V, Warren AJ, Cross NC, Green AR, Futreal PA, Stratton MR, Campbell PJ Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. The New England Journal of Medicine. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, Fangazio M, Vaisitti T, Monti S, Chiaretti S, Guarini A, Del Giudice I, Cerri M, Cresta S, Deambrogi C, Gargiulo E, Gattei V, Forconi F, Bertoni F, Deaglio S, Rabadan R, Pasqualucci L, Foa R, Dalla-Favera R. Gaidano G. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Miguel JS, Weisel K, Moreau P, Lacy M, Song K, Delforge M, Karlin L, Goldschmidt H, Banos A, Oriol A, Alegre A, Chen C, Cavo M, Garderet L, Ivanova V, Martinez-Lopez J, Belch A, Palumbo A, Schey S, Sonneveld P, Yu X, Sternas L, Jacques C, Zaki M. Dimopoulos M. Pomalidomide plus low-dose dexamethasone versus high-dose dexamethasone alone for patients with relapsed and refractory multiple myeloma (MM-003): a randomised, open-label, phase 3 trial. The Lancet Oncology. 2013;14:1055–1066. doi: 10.1016/S1470-2045(13)70380-2. [DOI] [PubMed] [Google Scholar]
- Schuster SR, Kortum KM, Zhu YX, Braggio E, Shi C-X, Bruins LA, Schmidt JE, Ahmann GJ, Kumar S, Rajikumar V, Mikhael J, Roy V, LaPlant B, Laumann K, Barlogie B, Shaughnessy J, Fonseca R, Bergsagel PL, Lacy M. Stewart AK. Cereblon expression predicts response, progression free and overall survival after pomalidomide and dexamethasone therapy in multiple myeloma (oral presentation) Blood (ASH Annual Meeting Abstracts) 2012;120:194. [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, Zhang W, Vartanov AR, Fernandes SM, Goldstein NR, Folco EG, Cibulskis K, Tesar B, Sievers QL, Shefler E, Gabriel S, Hacohen N, Reed R, Meyerson M, Golub TR, Lander ES, Neuberg D, Brown JR, Getz G. Wu CJ. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. The New England Journal of Medicine. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S. Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Zhu YX, Braggio E, Shi C, Bruins LA, Schmidt JE, Van Wier S, Chang X, Bjorklund CC, Fonseca R, Bergsagel PL, Orlowski RZ. Stewart AK. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118:4771–4779. doi: 10.1182/blood-2011-05-356063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intrarun and interrun IHC results demonstrate the precision of the assay.
Precision of the IHC assay with the CRBN65 antibody.
Alignment of CRBN Affymetrix probe sets 218142_S_AT and 222533_AT with exon 10, 11, and the UTR of the CRBN gene.


