Abstract
The opportunistic pathogen Pseudomonas aeruginosa PAO1 has a remarkable capacity to adapt to various environments and to survive with limited nutrients. Here, we report the discovery and characterization of a novel small non-coding RNA: NrsZ (nitrogen-regulated sRNA). We show that under nitrogen limitation, NrsZ is induced by the NtrB/C two-component system, an important regulator of nitrogen assimilation and P. aeruginosa's swarming motility, in concert with the alternative sigma factor RpoN. Furthermore, we demonstrate that NrsZ modulates P. aeruginosa motility by controlling the production of rhamnolipid surfactants, virulence factors notably needed for swarming motility. This regulation takes place through the post-transcriptional control of rhlA, a gene essential for rhamnolipids synthesis. Interestingly, we also observed that NrsZ is processed in three similar short modules, and that the first short module encompassing the first 60 nucleotides is sufficient for NrsZ regulatory functions.
Introduction
The opportunistic pathogen Pseudomonas aeruginosa PAO1 has a remarkable capacity to adapt to various environments and to survive with limited nutrients. To ensure an optimal coordination of genes involved in nutrient utilization, this versatile bacterium uses two-component systems, such as NtrB/C and CbrA/B, working in concert with the alternative sigma factor RpoN. RpoN is a global regulator involved in nitrogen metabolism, carbon assimilation, nutrient transport, motility, mucoidy and quorum sensing (Potvin et al., 2008). The RpoN RNA holoenzyme binds to specific promoters of target genes with the consensus sequence TGG-24CAC-N5-TTGC-12W (bold: invariant nucleotides, W = A or T) upstream of the transcription start site (Barrios et al., 1999). The alternative sigma factor RpoN is required by two-component systems, such as NtrB/C, to activate the transcription of target genes.
The two-component system NtrB/NtrC is an important regulator of nitrogen assimilation (Merrick and Edwards, 1995; Ninfa et al., 1995; Li and Lu, 2007; Zhang and Rainey, 2008; Hervás et al., 2009; Yeung et al., 2009). Under nitrogen-limited conditions, the intracellular level of glutamine decreases and NtrB histidine kinase activity is enhanced, leading to its autophosphorylation. Activated NtrB transfers a phosphate group to its response regulator NtrC, which binds to specific upstream activating sequences, thereby activating transcription, with the alternative sigma factor RpoN, of its target genes (Kustu et al., 1989). By contrast, the intracellular glutamine level increases under nitrogen excess, promoting NtrB phosphatase activity and leading to its auto-dephosphorylation (Dixon and Kahn, 2004). Until now, little is known about the NtrB/C network controlling P. aeruginosa metabolism (Nishijyo et al., 2001; Li and Lu, 2007). In Pseudomonas putida, a transcriptome analysis in response to nitrogen availability was performed, which revealed that NtrB/C regulates various genes, such as glnA (encoding a glutamine synthetase), porin genes, amino acid transporter genes, and genes involved in urea assimilation and in carbon catabolism (Hervás et al., 2008). Furthermore, it was shown that NtrB/C is also involved in the control of P. aeruginosa swarming motility, a surface movement (Merrick and Edwards, 1995; Ninfa et al., 1995; Li and Lu, 2007; Zhang and Rainey, 2008; Hervás et al., 2009; Yeung et al., 2009).
Pseudomonas aeruginosa swarming motility is a social behaviour considered to be a virulence trait that is augmented under nitrogen limitation and in response to certain amino acids, in turn leading to an increase in P. aeruginosa resistance to multiple antibiotics (Köhler et al., 2000; Overhage et al., 2008). Furthermore, conditions for swarming are similar to those found in the lung, an organ that can be infected by P. aeruginosa (Hutchison and Govan, 1999; Breidenstein et al., 2011). To swarm, the bacterium requires flagella, type IV pili and rhamnolipids production (Köhler et al., 2000; Déziel et al., 2003; Caiazza et al., 2005; Overhage et al., 2007). Rhamnolipids are tension-active glycolipid molecules, and their biosynthesis is mediated by the rhlAB operon encoding RhlA and RhlB rhamnosyltransferases (Ochsner et al., 1994; Soberón-Chávez et al., 2005). The swarming motility of the P. aeruginosa rhlA negative strain is impaired, whereas an rhlB negative strain is able to swarm (Köhler et al., 2000; Déziel et al., 2003). Interestingly, rhamnolipids production is stimulated under unfavourable nutrient conditions, and availability of carbon, nitrogen and phosphate in particular was shown to modulate the production of these surfactants (Lang and Wullbrandt, 1999; Clarke et al., 2010).
Noteworthy, bacteria can also modulate their cellular functions at the post-transcriptional level using small non-coding RNAs (sRNAs). As regulatory molecules, sRNAs have two major advantages: they are synthesized at low energy cost for the cell and they act rapidly. sRNAs can be divided mainly into two classes: the first class includes sRNAs that capture RNA-binding proteins [e.g. RsmA/CsrA type (Lapouge et al., 2008)], while the second class of sRNAs undergo base-pairing interactions with target mRNAs, resulting in the regulation of translation and/or mRNA degradation/stabilization and often requires the RNA chaperone Hfq for function. The Hfq chaperone is an abundant, stable and hexameric protein, which was originally identified as a host factor necessary for the replication of bacteriophage Qβ in Escherichia coli (Franze de Fernandez et al., 1972). In Enterobacteriaceae, Hfq facilitates the interaction of regulatory sRNAs with their mRNAs targets and is important for the stability of the sRNAs (Storz et al., 2011). Interestingly, in P. aeruginosa, Hfq was shown to be a global regulator of virulence and stress response, and to co-immunoprecipitate, bind and stabilize sRNAs, but no evidence has been presented so far for its involvement in the sRNAs–mRNAs interaction (Sonnleitner et al., 2003; 2006; 2008). Extensive studies carried out in E. coli have shown the existence of around 100 sRNAs, representing around 2% of protein-coding genes in this bacterium (Sharma and Vogel, 2009; Waters and Storz, 2009). Until recently, only 44 sRNAs were reported in P. aeruginosa, of which only a few were experimentally validated and functionally characterized (Sonnleitner et al., 2010). These sRNAs were shown to play a crucial role in the regulation of primary and secondary metabolism (Sonnleitner and Haas, 2011; Sonnleitner et al., 2012). New studies using RNA sequencing technology have led to the identification of over 500 novel transcripts in P. aeruginosa, among which 50 were validated (Dötsch et al., 2012; Ferrara et al., 2012; Gómez-Lozano et al., 2012).
To discover novel sRNAs regulating the adaptation of P. aeruginosa to its nutritional environment, we conducted a bioinformatics-based approach and discovered a novel sRNA that we named NrsZ (nitrogen-regulated sRNA). We showed that NrsZ is highly conserved among Pseudomonads, and its induction in PAO1 under nitrogen-limited condition is regulated by the two-component system NtrB/C in concert with the alternative sigma factor RpoN. We demonstrated that NrsZ modulates P. aeruginosa motility by controlling the production of rhamnolipid surfactants, virulence factors notably needed for swarming motility. This regulation takes place through the post-transcriptional control of rhlA, a gene essential for rhamnolipids synthesis. Furthermore, we observed that NrsZ is processed to yield three similar short modules, and that the first 60 nucleotides (nt) encoding the first module are sufficient to ensure regulatory functions.
Results
Identification of NrsZ, a nitrogen-dependent, non-coding small RNA
To discover novel sRNAs involved in the regulation of nitrogen source utilization by Pseudomonas aeruginosa PAO1, we used a bioinformatics approach and searched for putative sRNAs regulated by the alternative sigma factor RpoN, which is required for the activation of target genes by the two-component system NtrB/C. Hence, we used the pattern search tool Fuzznuc (Rice et al., 2000) to query 162 intergenic regions of the PAO1 genome described by González et al. (González et al., 2008) to encode putative sRNAs using the RpoN consensus sequence (TGGCAC-N5-TTGCW) based on Barrios et al. (Barrios et al., 1999). Twelve intergenic regions containing conserved RpoN consensus sequences were identified. As expected, the RpoN consensus sequence of the described sRNA CrcZ and the RNA NalA were also found (Sonnleitner et al.,2009; Romeo et al., 2012).
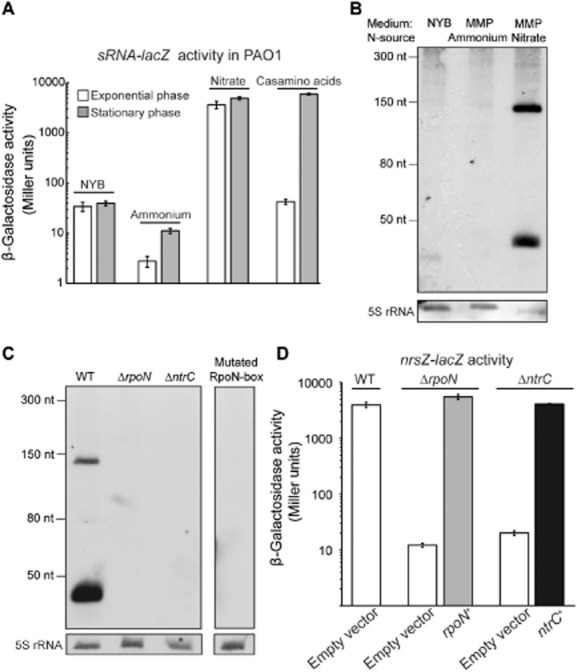
One intergenic region was chosen for further investigation due to the prediction of the well-conserved RpoN consensus sequence TGG-24CACAGCCCCTGC-12A, and its position between the tail-to-tail genes ntrC and PA5126 (Fig. S1). To investigate the transcriptional activity of the predicted RpoN promoter of a putative sRNA located in the ntrC-PA5126 intergenic region, we measured β-galactosidase activity of the chromosomal sRNA-lacZ transcriptional reporter fusion inserted into the PAO1 wild-type (WT) strain (PAO6850). The promoter cloned in this fusion encompassed the predicted −309 to +1 region relative to the start site of transcription. Then, regulation of the predicted promoter was investigated during growth with different nitrogen sources (N-sources), taking into account that RpoN is a regulator of nitrogen metabolism. We detected very low activity of the fusion in the rich medium nutrient yeast broth (NYB) and in the minimal medium P (MMP) supplemented with ammonium as the sole N-source (Fig. 1A). By contrast, the high activity of the fusion was measured when nitrate or casamino acids were used as sole N-sources. Furthermore, we observed that RpoN promoter activity was growth-phase independent in minimal medium containing nitrate as nitrogen source, whereas activity increased with cell density in minimal medium supplemented with a low concentration of casamino acids as N-source (Fig. 1A). The latter may be explained by the fact that in this condition, growth in 0.1% casamino acids medium is limited by N-source availability, which suggests that nitrogen depletion triggers activity of the promoter. To test this hypothesis, we measured the growth of the PAO1 WT strain and the activity of the sRNA-lacZ fusion in MMP medium supplemented with different concentration of casamino acids. As expected, an increase in casamino acids concentration led to an increase in PAO1 growth rate and to a decrease in the expression of the fusion (Fig. S2). These results reveal that transcriptional activity of the RpoN promoter is induced under nitrogen limitation when a poor N-source, such as nitrate, is present or when cells undergo nitrogen starvation, i.e. growing to stationary phase with a limited N-source, such as low concentration of casamino acids.
Fig 1.
The sRNA NrsZ of P. aeruginosa PAO1 is induced during nitrogen limitation by the NtrB/C-RpoN cascade.
A. β-Galactosidase activities of the chromosomal reporter fusion sRNA-lacZ (nrsZ-lacZ) under various nitrogen-limited conditions. The PAO1 WT strain carrying nrsZ-lacZ (PAO6750) was grown in NYB, MMP supplemented with glucose as carbon source and ammonia, nitrate or casamino acids (0.1%) as nitrogen source. Activity of the nrsZ-lacZ chromosomal fusion was measured in exponential phase and when stationary phase was reached. Each value represents the average of triplicate cultures ± standard deviation.
B. Northern blot detection of the sRNA: RNA was isolated from PAO1 (WT) grown to stationary phase in NYB, MMP supplemented with succinate as carbon source and ammonium or nitrate as nitrogen source. 7.5 μg of cross-linked total RNA was hybridized with the ssRNA probe NrsRNA. As loading control, the membrane was re-probed with the 5SDNA, which detects 5S rRNA.
C. Northern blot detection of the sRNA: Total RNA was extracted from strains PAO1 WT, ΔrpoN (PAO6358), ΔntrC (PAO6764), and from the strain mutated in the RpoN box of the sRNA promoter (PAO6846) grown to stationary phase in MMP supplemented with glucose and casamino acids (0.1%). 5 μg of cross-linked total RNA was hybridized with the ssRNA probe NrsRNA. As loading control, the membranes were re-probed with the 5SDNA detecting the 5S rRNA.
D.β-Galactosidase activities of the chromosomal reporter fusion nrsZ-lacZ in different strains. The WT (PAO6750), ΔrpoN (PAO6847) and ΔntrC (PAO6842) strains carrying the pME6001 empty vector, the strain ΔrpoN complemented with rpoN+ (pME6001::rpoN, pME10389) and the strain ΔntrC complemented with ntrC+ (pME6001::ntrC, pME10390) were grown in MMP supplemented with glucose and casamino acids (0.1%). nrsZ-lacZ activity was measured when stationary phase was reached. Each value represents the average of triplicate cultures ± standard deviation.
To detect the putative sRNA encoded downstream of the RpoN consensus sequence, Northern blot analysis was performed. Total RNA was extracted from PAO1 cultures grown to stationary phase in NYB or in MMP supplemented with different N-sources. Northern blot experiments were carried out using a single-stranded RNA (NrsRNA) probe of 242 nt complementary to the region encoding the putative sRNA. As expected, transcripts were only detected when PAO1 was grown in MMP supplemented with nitrate (Fig. 1B) or with a limited concentration of casamino acids as the sole N-source (Fig. 1C). Surprisingly, two major transcripts of around 40 nt and 140 nt were detected. To investigate if the RpoN promoter generated both transcripts, Northern blot analysis was performed on the PAO6846 strain (where the RpoN consensus sequence of the WT PAO1 was mutated, as indicated in bold T(GG→AA)CACAGCCCCT(GC→TT)A) grown under nitrogen-limited conditions. In this strain, no transcript was detected (Fig. 1C), demonstrating that both the 40 nt and the 140 nt RNA forms are transcribed from the same RpoN promoter.
Taken together, these results show that an sRNA is encoded in the ntrC-PA5126 intergenic region of PAO1, that its transcription is induced during nitrogen limitation, and that it is processed into two short transcripts of around 40 nt and 140 nt. Therefore, due to the nitrogen source-dependent expression of the sRNA, we named the sRNA ‘NrsZ’, an acronym for nitrogen-regulated sRNA.
NrsZ is induced during nitrogen limitation by the NtrB/C-RpoN cascade
As suggested above, transcription of NrsZ is activated by the alternative sigma factor RpoN under nitrogen-limited conditions. Previous work demonstrated that the NtrB/C two-component system is activated under nitrogen-limited conditions and that nitrate used as a nitrogen source simulates the condition of nitrogen limitation (Kustu et al., 1989; Rashedi et al., 2005). As NtrB/C is known to activate transcription in concert with the alternative sigma factor RpoN, we reasoned that expression of NrsZ may also be regulated by NtrB/C.
To confirm that the transcription of NrsZ was NtrB/C- and RpoN-dependent, Northern blot analysis was performed on total RNA extracted from cultures of PAO1 (WT), ΔrpoN (PAO6358) and ΔntrC (PAO6764) mutant strains grown to stationary phase in MMP supplemented with glucose and casamino acids. Glucose was used as the carbon source to allow growth of the ΔrpoN mutant and casamino acids as a limited nitrogen source to allow growth of the ΔntrC mutant. The abundant 40 nt and 140 nt NrsZ transcripts observed in the WT strain were absent in both mutant strains (Fig. 1C), indicating that expression of NrsZ is RpoN- and NtrB/C-dependent. Moreover, as expected, NrsZ promoter activity was dramatically reduced in the ΔntrC (PAO6842) and ΔrpoN (PAO6847) strains carrying the chromosomal nrsZ-lacZ reporter fusion (Fig. 1D). Complementation of the ΔrpoN and ΔntrC mutant strains with the plasmid pME10389 or pME10390, carrying respectively the rpoN and ntrC genes under the control of the lac promoter, restored the expression of the nrsZ-lacZ reporter fusion to the WT level (Fig. 1D).
These results demonstrate that NrsZ is induced during nitrogen limitation by the NtrB/C two-component system in concert with RpoN. It is noteworthy that NrsZ is the first regulatory sRNA discovered to be activated by the NtrB/C-RpoN cascade in P. aeruginosa PAO1 in reaction to nitrogen starvation.
NrsZ is processed in three similar small forms
The observation that two transcripts of around 40 nt and 140 nt were generated from the same promoter led us to investigate the 3′ and 5′ ends of the respective RNA transcripts. For this purpose, we analyzed RNA deep sequencing data of directional full-length RNA from two fractionated small RNA samples. Briefly, RNA sequencing was performed on total RNA separated into two fractions, ranging from 30 nt to 200 nt (small fraction) and from 150 nt to 450 nt (medium fraction). One thousand four hundred eighty-one reads were mapped onto the nrsZ encoding strand located in the intergenic region ntrC-PA5126 (455 reads in the small fraction and 1026 reads in the medium fraction) (Fig. 2A and B), and covered all potential RNA molecules transcribed and derived from the nrsZ genomic position.
Fig 2.
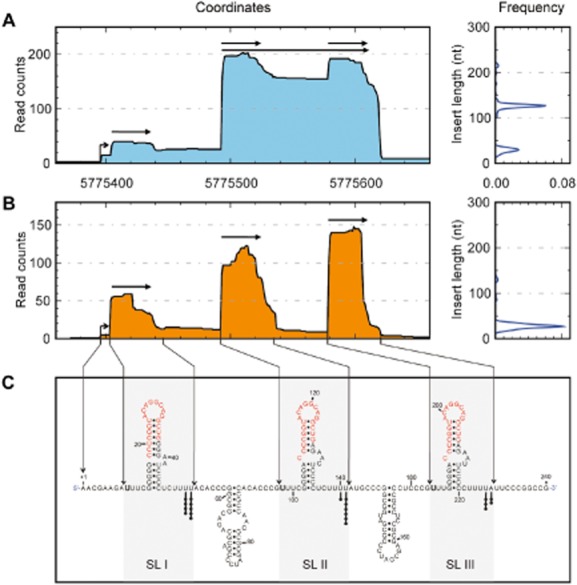
NrsZ is processed in three small forms. The RNA deep sequencing profile of NrsZ was determined from RNA extracted from PAO1 grown in minimum medium supplemented with nitrate and fractioned in two parts, (A, left panel) the medium fraction ranging from 150 nt to 450 nt, and (B, left panel) the small fraction ranging from 30 nt to 200 nt. (A and B, right panels): insert length distributions of the transcripts corresponding to NrsZ in both fractions. Base numbering starts from the +1 transcription start site represented by a bent arrow, and horizontal arrows underline the major transcripts positions and sizes. (C) Predicted secondary structure of the NrsZ primary transcript. Each black dot represents one 3′ extremity of NrsZ obtained by 3′ RACE experiment. 5′-matured terminal nucleotides determined by RNA deep sequencing are indicated in bold. Conserved motifs in the stem-loop sequences are shown in red. SL: stem-loop structure. The Mfold program was used to predict RNA secondary structures (Zuker, 2003).
As observed in Fig. 2A and B, RNA size selection allowed the distinction of multiple processed forms of NrsZ. Although NrsZ is produced as a transcript of at least a 226 nt, only few molecules covered this entire sequence, indicating efficient maturation processes. In addition, abundant transcripts of around 40 nt and 130 nt long were detected. To better understand NrsZ processing, NrsZ secondary structure analysis was performed using the Mfold program (Zuker, 2003) (Fig. 2C). NrsZ was predicted to form five stem-loop structures (SL), of which three present conserved residues in the loop (SL I–III). Analysis of the RNA sequencing in the medium fraction showed that two major forms of NrsZ were prevalent (Fig. 2A): short forms of around 44 nt composed by the SL I, II and III (starting at residue 9 and ending at residue 52, 98 to 142 and 184 to 226), and a long form of 127 nt composed of the SL II and SL III RNA (starting from residue 98 and ending at residue 226; Fig. 2A and 2C). These two major transcripts matched with the two major bands detected by Northern blot experiments (Fig. 1B and C). RNA sequencing results of the small fraction demonstrated that NrsZ was also detected as three abundant small forms of around 44, 43 and 41 nt, corresponding to the sequences of the three individual SL I–II–III respectively. Interestingly, these three forms were present in a relatively equal amount. Surprisingly, the RNA sequencing mapping indicated that only few transcripts started at the predicted transcription start site located 12 nt downstream of the RpoN consensus sequence, and that the 5′ and 3′ extremities of the transcripts enclosed three and four U residues respectively (Fig. 2C). To confirm the RNA deep sequencing results, 3′ and 5′ RACE (rapid amplification of cDNA ends) experiments were performed. Twenty-one independent sequences were obtained from the 3′RACE experiments, and were mapped to the U51-U52, U142-U141 and U225-A226 residues, as presented in Fig. 2C. The 3′RACE results were in agreement with those obtained from RNA sequencing that the terminal 3′ extremities were located after each SL I–III (Fig. 2C). Furthermore, it is noteworthy that the 127 nt long transcript detected in the medium fraction, encompassing SL II and SL III, is also processed into two shorter fragments of 43 nt and 41 nt that are detected in the small fraction and encompass SL II and SL III respectively. 5′ RACE analysis was also attempted, but technical problems did not permit the identification of the 5′ extremities of NrsZ (data not shown).
NrsZ regulates P. aeruginosa swarming motility
To identify the biological function(s) of NrsZ in P. aeruginosa PAO1, we used a phenotypic approach. We demonstrated that NrsZ expression is NtrB/C-dependent, and is therefore induced under nitrogen-limited conditions. It was previously demonstrated that nitrogen-limited conditions are necessary to sustain P. aeruginosa swarming motility (Köhler et al., 2000). Consequently, we tested the swarming motility of P. aeruginosa PAO1 (WT) and PAO1 NrsZ negative strain (PAO6846, nrsZ−) carrying the empty plasmid pME4510. The mutant strain showed a complete swarming defect that could be restored by introducing the WT nsrZ gene under control of its native promoter (pME9995, Fig. 3A). Interestingly, the 60 first nucleotides of the nrsZ gene encoding the SLI (pME10138) were also able to revert the swarming-defective phenotype of the PAO1 nrsZ− mutant, indicating that the first 60 nt of NrsZ is sufficient to regulate swarming motility in P. aeruginosa PAO1 (Fig. 3A). Twitching and swimming motilities of the nsrZ mutant strain were also tested, but no modulation of these phenotypes was observed (data not shown). These results show that NrsZ is a regulator of P. aeruginosa swarming motility and that the first 60 nt of NrsZ containing the SLI acts as a functional unit sufficient for swarming regulation.
Fig 3.
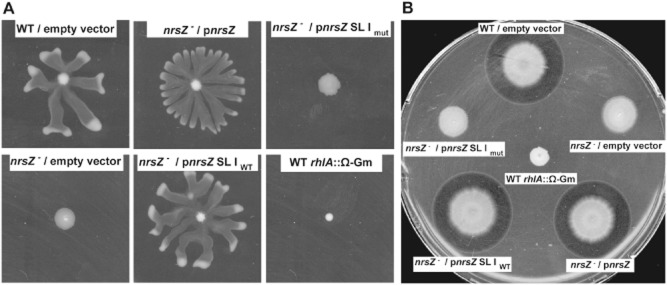
The P. aeruginosa nrsZ– mutant is defective for swarming motility and rhamnolipids production. Swarming motility (A) and rhamnolipids production (B) were assayed as described in the Experimental procedures for strains PAO1 WT and PAO1 nrsZ− (PAO6846) containing the empty vector pME4510 or its derivative plasmids pME9995, pME10138 and pME10191, encoding full length nrsZ (pnrsZ), truncated nrsZ (pnrsZ SL IWT) or the mutated truncated nrsZ (pnrsZ SL Imut) respectively. As control, motility- and rhamnolipid-defective mutant PT712 (PAO1rhlA::Ω-Gm) was tested in the same condition. All experiments were performed in four replicates. The pictures depict a representative plate for each experiment.
NrsZ regulates rhamnolipids production
In order to swarm, P. aeruginosa secretes rhamnolipid biosurfactants (Caiazza et al., 2005). Furthermore, rhamnolipids production was observed to be enhanced in the presence of nitrate in comparison to ammonium as nitrogen source (Guerra-Santos et al., 1984; Lang and Wullbrandt, 1999; Maier and Soberón-Chávez, 2000). As is the case in rhamnolipids production, NrsZ is induced during nitrogen limitation, and as we showed positively regulates swarming motility. These considerations led us to investigate the involvement of NrsZ in rhamnolipids production by P. aeruginosa PAO1. We tested the production of rhamnolipids on plates containing the same nutrient composition used for swarming assays. In comparison to PAO1 WT, PAO1 nrsZ− carrying the empty plasmid pME4510 was clearly deficient for rhamnolipids production (Fig. 3B). The complementing plasmids pME9995 (carrying nrsZ) or pME10138 (carrying the first 60 nucleotides of nrsZ) fully restored rhamnolipids production of the PAO1 nrsZ− mutant. These results indicate that NrsZ, and in particular its first 60 nt, positively regulates rhamnolipids production.
NrsZ activates rhlA expression at the post-transcriptional level
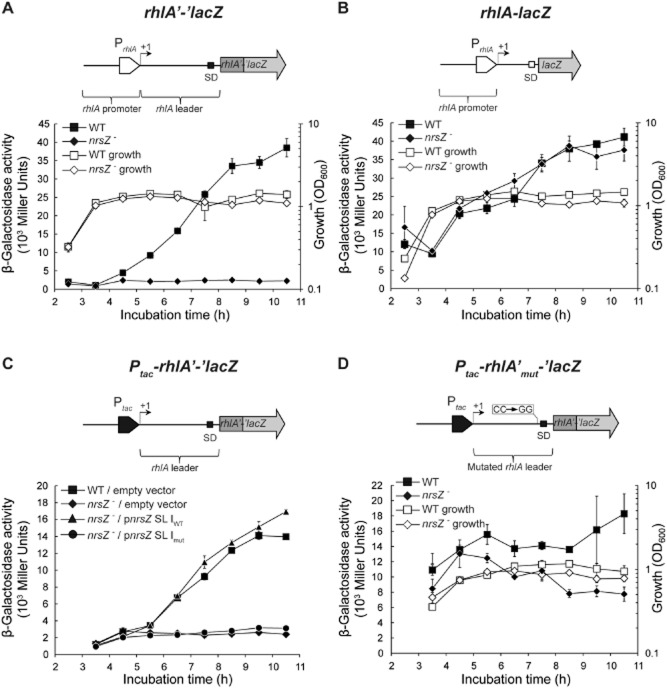
Synthesis of the rhamnolipid biosurfactants in P. aeruginosa is mediated by a biosynthetic pathway involving the rhlAB gene cluster (Maier and Soberón-Chávez, 2000; Soberón-Chávez et al., 2005). Importantly, an rhlA mutant is defective in rhamnolipids production and swarming motility (Köhler et al., 2000; Déziel et al., 2003). As NrsZ regulates rhamnolipids production, we investigated whether NrsZ regulates rhlA expression. Expression of the translational rhlA’-‘lacZ reporter fusion (pECP60) was first measured in the PAO1 WT strain grown in medium containing the same nutrient composition used for swarming motility and rhamnolipids analyses. In these conditions, rhlA expression increased with increasing cell density to reach a very high expression level (40·103 Miller units). We then measured β-galactosidase activity of the rhlA’-‘lacZ fusion in the PAO1 nrsZ− mutant strain and observed that the expression of rhlA was completely repressed in the mutant strain in comparison with the WT strain, indicating that NrsZ positively regulates rhlA expression (Fig. 4A). By contrast, no difference of the transcriptional rhlA-lacZ fusion (pME3838) activity was detected between both strains (Fig. 4B). The expression of the Ptac-rhlA'-‘lacZ fusion (pME10173), carrying the constitutive strong tac promoter, was increasing following growth in the WT strain (Fig. 4C), similarly to the nrsZ expression profile observed in the same condition (Fig. S2). However, the Ptac-rhlA'-‘lacZ fusion activity was subject to a strong repression in PAO1 nrsZ− and stayed at a basal level during all cell growth (Fig. 4C). In addition, the complementing plasmid pME10138, carrying the first 60 nt of NrsZ fully restored rhlA expression in PAO1 nrsZ− (Fig. 4C).
Fig 4.
RhlA expression is activated by NrsZ at the post-transcriptional level. Cell density dependent β-galactosidase activities of (A) the translational rhlA'-'lacZ reporter fusion (pECP60) and of (B) the transcriptional rhlA-lacZ fusion (pME3838) in PAO1 WT and PAO1 nrsZ− (PAO6846). (C) Expression of the translational Ptac-rhlA'-'lacZ fusion (pME10173) in PAO1 WT and PAO1 nrsZ−, containing the empty vector pME4510 or its derivative plasmids pME10138 and pME10191, which encode for truncated nrsZ (pnrsZ SL IWT) and mutated, truncated nrsZ (pnrsZ SL Imut) respectively. (D) β-galactosidase activities of the translational Ptac-rhlAmut'-'lacZ (pME10182) fusion in PAO1 WT and PAO1 nrsZ−. Cultures were grown in MMP supplemented with glucose and casamino acids (0.1 %); growth curves are represented with the indicated symbols. Each value represents the average of three cultures ± standard deviation. Schematics for each fusion are represented: the promoters, the transcription start sites (+1), the Shine-Dalgarno sequences (SD) and the mutations (in a box) are indicated.
Taken together, these results showed that NrsZ regulates positively rhlA at the post-transcriptional level and confirmed that NrsZ short transcript is a sufficient functional unit for the activation of rhlA expression.
The conserved motif ACAGGCAG in the loop of NrsZ is essential for rhlA activation
The predicted secondary structure of NrsZ suggests the presence of three repeated SL with a conserved ACAGGCAG motif in their loop (Fig. 2C). Therefore, we hypothesize that this sequence is involved in the activation of rhlA expression. To test this hypothesis, we constructed the plasmid pME10191, carrying the first 60 nt of NrsZ mutated in the ACA(GG→CC)CAG motif and transformed the plasmid into PAO6846. The plasmid did not restore rhlA expression in PAO1 nrsZ− (Fig. 4C), and neither swarming motility nor rhamnolipids production of this strain was recovered (Figs 3A and B, and 4C). These results indicate that the ACAGGCAG stem-loop motif of NrsZ is crucial for NrsZ regulatory function.
Next, to identify the binding site of NrsZ on rhlA mRNA, we hypothesized that the ACAGGCAG motif of NrsZ is exposed and involved in NrsZ-rhlA RNA duplex formation. We, therefore, screened the leader sequence of rhlA mRNA for a potential complementary sequence to this motif. Our analysis predicted that the NrsZ stem-loop motif interacts with a region present upstream of the rhlA ribosome binding site (Fig. 5). We then constructed a Ptac-rhlA'-‘lacZ fusion mutated in the NrsZ binding site GUUUG(CC→GG)UGUUCGA (Ptac-rhlAmut'-‘lacZ; pME10182), and tested its expression in PAO1 WT and PAO1 nrsZ−. In exponential growth phase, the expression of the Ptac-rhlAmut’-‘lacZ fusion was clearly de-repressed in the WT strain compared with the Ptac-rhlA'-‘lacZ fusion in the same condition (Fig. 4C and D). In addition, the expression of the Ptac-rhlAmut’-‘lacZ fusion was similar in both the WT and nrsZ− mutant. However, a slight increase of the Ptac-rhlAmut’-‘lacZ fusion activity was observed in stationary growth phase in the WT strain compared with the mutant strain (Fig. 4D), suggesting that this mutation is not sufficient to impair NrsZ regulation. These results suggest a role for this region in the regulation of rhlA expression and in NrsZ-mediated regulation.
Fig 5.
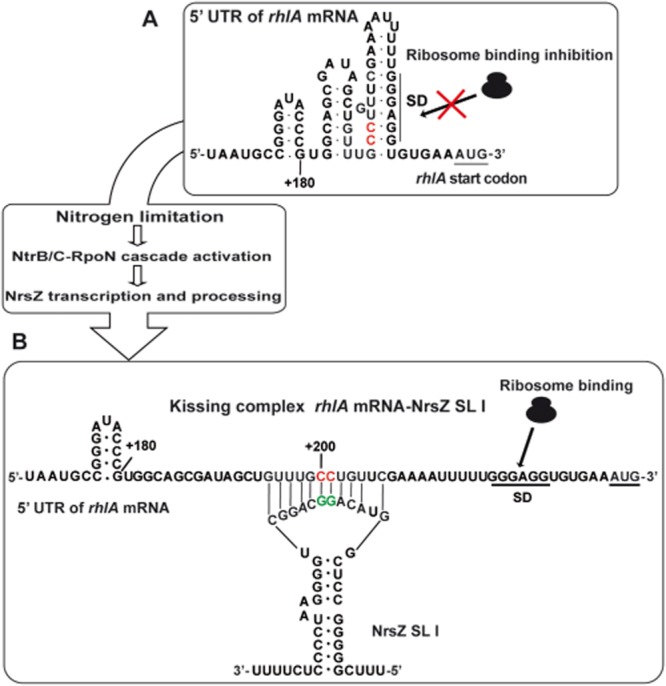
Model of the rhlA-NrsZ kissing complex formation that leads to the activation of rhlA mRNA translation.
A. In the absence of NrsZ, rhlA translation is repressed by a self-inhibitory secondary structure on the 5' untranslated region (5' UTR) of its mRNA. This structure sequesters the rhlA Shine-Dalgarno sequence (SD), and therefore ribosome binding, leading to the inhibition of rhlA mRNA translation.
B. Under nitrogen limitation, NrsZ is induced by the NtrB/C-RpoN cascade and is processed in short, functional stem-loop molecules (NrsZ SL I, II, III). These molecules base-pair with their identical loop sequence (GUACAGGCAGGC) with the rhlA 5' UTR forming a kissing complex. This base-pairing allows melting of the self-inhibitory structure of rhlA, liberating the SD for ribosome binding. Therefore, rhlA mRNA translation is activated, triggering rhamnolipids production and swarming motility. For simplification, only the NrsZ SL I molecule is presented. Nucleotides mutated (CC→GG) in the fusion Ptac-rhlAmut'-'lacZ (pME10182) are indicated in red. Nucleotides mutated (GG→CC) in the plasmid expressing NrsZ SL Imut (pME10191) are indicated in green. The Mfold program was used to predict RNA secondary structures (Zuker, 2003).
Discussion
NrsZ is, until now, the only described sRNA regulated by the NtrB/C system and shown to positively modulate P. aeruginosa swarming motility. Our study shows that during nitrogen limitation, NrsZ modulates swarming motility of P. aeruginosa by activating rhlA expression, leading to an increase in the production of rhamnolipids, important virulence factors of P. aeruginosa (Figs A and B, and 4). Previous studies already demonstrated that rhlA expression and rhamnolipids production are dependent on nitrogen availability in P. aeruginosa (Mulligan and Gibbs, 1989; Ochsner et al., 1994) and on the alternative sigma factor RpoN (Pearson et al., 1997). However, the mechanisms leading to these regulations were not fully elucidated. Nevertheless, it has been suggested, using translational reporter fusions, that RpoN activates rhlA expression indirectly through the activation of the transcription of rhlR (encoding an rhlAB transcriptional activator) (Medina et al b 2003a). Based on our results, however, we propose that activation of rhlA expression by nitrogen limitation and RpoN is indirect and post-transcriptional due to the regulation of rhlA expression at the post-transcriptional level by NrsZ, an sRNA induced by the NtrB/C-RpoN cascade.
To obtain more insight into NrsZ regulatory function, Mfold analysis of NrsZ secondary structure was performed and predicted the formation of three highly conserved stem-loop structures separated by two stem-loop spacers (Fig. 2C). Furthermore, our study demonstrated that NrsZ is processed into two transcripts of around 40 nt and 140 nt, and that NrsZ regulatory activity can be mediated solely by the short abundant processed fragment encompassing the first predicted stem-loop structure (SL I), which exposes the conserved RNA sequence motif ACAGGCAG in the loop. These results would support the theory that NrsZ belongs to a category of sRNAs, such as ArcZ and MicX, that require processing after transcription to produce active regulatory molecules (Davis and Waldor, 2007; Papenfort et al., 2009). Our results suggest that specific and efficient endoribonucleolytic cleavage of the 5′ end and 3′ exonucleolytic trimming of NrsZ occur, and we hypothesize that these mechanisms are triggered by RNAseE or RNAse G, and PNPase, RNAseR or RNase II respectively. It is noteworthy that the two spacers, positioned between SL I–SL II and SL II–SL III, and having the same size and structure but different sequences, seem unstable and are quickly degraded by ribonucleases. This leads to the production of short and efficient matured regulatory NrsZ transcripts. Intriguingly, the predicted NrsZ structure resembles that of the CRISPR system that is composed of multiple structured short direct repeats separated by spacers. However, as revealed by a recent study, P. aeruginosa strain PAO1 lacks a CRISPR/Cas system (Ferrara et al., 2012). To better characterize NrsZ function, the roles played by all three highly conserved stem-loop structures are currently being investigated.
One challenge in studying novel sRNAs is to decipher their mode of action. The sRNAs characterized so far and involved in post-transcriptional regulation exert their regulatory functions using two main mechanisms: capture of small RNA-binding proteins of the RsmA/CsrA type or base-pairing with an mRNA target. To test the hypothesis that NrsZ titrates an RNA-binding protein, we carried out a transposon mutagenesis in the P. aeruginosa PAO1 nrsZ− strain containing the translational rhlA’-‘gfp fusion. In this condition, rhlA expression is impaired due to repression by the RNA-binding protein. If the gene coding for this RNA-binding protein repressor is mutated, rhlA expression will be restored. Unfortunately, we did not identify any putative protein targets despite having carried out an extensive screening (data not shown), suggesting that either the putative protein is essential or that NrsZ regulation does not involve a protein partner.
To activate translation initiation, sRNAs have been generally shown to bind to the 5′ region of their mRNA targets (Waters and Storz, 2009). In line with this theory, we predicted that the NrsZ stem-loop motif could interact with a region upstream the rhlA Shine-Dalgarno (SD) by forming a kissing complex. Using the secondary structure prediction Mfold program, we observed that the rhlA leader mRNA seems to be well structured and that a stem-loop structure might engulf the SD, conceivably leading to an inhibition of translation. Upon binding of NrsZ, this stem-loop structure would dissociate and liberate the rhlA SD for initiation of translation (Fig. 5). To validate the predicted interaction, we performed electrophoretic mobility shift assays, as previously described (Møller et al., 2002; Lease and Woodson, 2004), using the short transcript form of NrsZ and the rhlA leader mRNA. However, no binding was detected (data not shown). We also carried out mutational analyses. Expression of the rhlA reporter fusion mutated in the putative NrsZ binding site was dramatically less subjected to NrsZ regulation, indicating that the two mutated residues are probably part of the rhlA mRNA-NrsZ recognition and binding site (Fig. 4D). However, we observed that the level of expression of the Ptac-rhlAmut’-‘lacZ fusion was similar to that of the Ptac-rhlA'-‘lacZ fusion in late stationary phase (Fig. 4D). This result could be explained by the fact that the mutated residues are involved in stem formation of the stem-loop structure engulfing the SD, and therefore breakage of this stem (a result of the sequence mutation) would lead to liberation of the rhlA SD and increase of its expression. To counteract this undesirable regulation, we envisaged construction of a new Ptac-rhlAmut’-‘lacZ fusion carrying compensatory mutations to preserve integrity of the stem-loop structure. However, compensatory mutations in this region would alter the SD sequence, leading to dramatic effects on the expression of the reporter fusions. Therefore, the NrsZ mechanism of action is still unknown and is currently under investigation.
To emphasize the global role of NrsZ in Pseudomonas, we predict that the NrsZ stem-loop structures with an ACAGGCAG motif in their loop are highly conserved and widely distributed among Pseudomonads, independently of their ecological features (Fig. S3A and B). In addition, a conserved RpoN promoter is predicted to control transcription of the different nrsZ species. Interestingly, the copy number of the NrsZ conserved stem-loop sequence and structure is variable among different Pseudomonas strains. For example, the secondary structure of P. protegens Pf-5, P. putida KT2442, P. entomophila L48 and P. syringae pv. tomato DC3000 NrsZ is predicted to form six, three, two and only one conserved stem-loop structures separated by stem-loop spacer respectively (Fig. S3B). Interestingly, the length of the spacers (from 3′-U triplet to 5′-U triplet) in each Pseudomonas specie is well conserved, and only small variations in length are observed for the last spacer sequence, while the length of the spacers in between species is variable (Fig. S3B). This observation raises numerous questions on the acquisition and divergence of NrsZ sequence in Pseudomonads.
It is noteworthy that complementation experiments showed that when the PAO1 nrsZ− mutant strain, deficient for rhamnolipids production, was complemented with plasmids carrying either P. protegens Pf5, P. putida KT2442, P. entomophila L48 or P. syringae pv. tomato DC3000 NrsZ under the control of their native promoter, rhamnolipids production was restored (Fig. S4). These results demonstrate that NrsZ can be exchanged and functional in the different Pseudomonads strains. Similarly, swarming motility impaired in the mutant strain was restored by complementation experiments (data not shown).
It is well known that in addition to its capacity to colonize eukaryotic hosts like other Pseudomonas species, P. aeruginosa is widely distributed in soil as well as in fresh or sea water (Green et al., 1974; Hardalo and Edberg, 1997; Kimata et al., 2004). In most of these niches, nitrogen is probably limited. Therefore, we hypothesize that the regulatory sRNA NrsZ is an important regulator of P. aeruginosa virulence and is crucial for the adaptation of Pseudomonads in these oligotrophic conditions, ensuring an optimal physiological response to thwart nitrogen starvation.
Experimental procedures
Bacterial strains and growth conditions
Strains and plasmids used in this study are listed in Table 1 and the oligonucleotide sequences in Table S1. Cells were grown at 37°C in the NYB medium, in Luria-Bertani broth medium (LB) or in minimal medium P (MMP) (Haas et al., 1977) supplemented with 20 mM succinate, mannitol or glucose as carbon source, and 20 mM ammonium (NH4Cl), 20 mM nitrate (KNO3) or 0.1% (w/v) casamino acids as nitrogen source. When required, antibiotics for P. aeruginosa were used at the following concentrations: 2000 μg ml−1 carbenicillin, 25 μg ml−1 gentamicin and 125 μg ml−1 tetracycline, and 100 μg ml−1 ampicillin, 10 μg ml−1 chloramphenicol, 25 μg ml−1 kanamycin, 10 μg ml−1 gentamicin and 25 μg ml−1 tetracycline for E. coli.
Table 1.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Descriptiona | References |
|---|---|---|
| Strains | ||
| Pseudomonas | ||
| P. aeruginosa | ||
| PAO1 | Wild-type | Holloway and colleagues (1979) |
| PAO6358 | PAO1 ΔrpoN | Heurlier and colleagues (2003) |
| PAO6750 | PAO1, nrsZ-lacZ; Tcr | This study |
| PAO6764 | PAO1 ΔntrC carrying a 1.41 Kb in-frame deletion in ntrC | This study |
| PAO6842 | PAO6764, nrsZ-lacZ; Tcr | This study |
| PAO6846 | PAO1 nrsZ−, mutation in the RpoN-box of the nrsZ promoter | This study |
| PAO6847 | PAO6358, nrsZ-lacZ; Tcr | This study |
| P. protegens | ||
| Pf-5 | Wild-type | Paulsen and colleagues (2005) |
| P. putida | ||
| KT2442 | Spontaneous rifampicin-resistant mutant of the KT2440 wild-type strain | Franklin and colleagues (1981), and Nelson and colleagues (2002) |
| P. entomophila | ||
| L48 | Wild-type | Vodovar and colleagues (2006) |
| P. syringae | ||
| pv. tomato DC3000 | Wild-type | Buell and colleagues (2003) |
| E. coli | ||
| DH5α | recA1, endA1, hsdR17, deoR, thi-1, supE44, gyrA96, relA1, Δ(lacZYA-argF), U169(φ80dlacZΔM15) | Sambrook and colleagues (2001) |
| HB101 | hsdS, recA, proA2, leu-6, ara-14, galK2, lacY1, xyl-5, mtl-1, rpsL20, thi-1, supE44 | Sambrook and colleagues (2001) |
| Plasmids | ||
| pECP60 | rhlA'-'lacZ fusion; Apr, Cbr | Pesci and colleagues (1997) |
| pGEM-T easy | Cloning vector; Apr | Promega |
| pUX-BF13 | mini-Tn7 transposition helper plasmid; Apr | Bao and colleagues (1991) |
| pME497 | Mobilizing plasmid, IncP-1, Tra RepA (Ts); Apr | Voisard and colleagues (1994) |
| pME3087 | Suicide plasmid, Co1E1 replicon; Tcr | Voisard and colleagues (1994) |
| pME3838 | rhlA-lacZ fusion; Tcr | Heurlier and colleagues (2004) |
| pME4510 | Multicopy broad host range plasmid; Gmr | Rist and Kertesz (1998) |
| pME6001 | Cloning vector; Gmr | Blumer and colleagues (1999) |
| pME6015 | Translational lacZ fusion cloning vector; Tcr | Schnider-Keel and colleagues (2000) |
| pME6016 | Transcriptional lacZ fusion cloning vector; Tcr | Schnider-Keel and colleagues (2000) |
| pME6552 | pUK21::Ptac; Kmr | Blumer and colleagues (1999) |
| pME7549 | Mini-Tn7-Tc delivery plasmid; Tcr, Apr | C. Reimmann (unpublished) |
| pME9989 | pME6016 carrying a 0.314 kb insert of the nrsZ promoter (nrsZ-lacZ); Tcr | This study |
| pME9991 | Suicide plasmid for deletion of ntrC; Tcr | This study |
| pME9995 | pME4510 derivative carrying a 1.015 kb insert encompassing nrsZ; Gmr | This study |
| pME10129 | pME7549 derivative carrying the nrsZ-lacZ fusion; Tcr, Apr | This study |
| pME10134 | Suicide plasmid for mutation of the RpoN-box of the nrsZ promoter; Tcr | This study |
| pME10138 | pME4510 derivative carrying a 0.4 kb insert containing the first 60 nucleotides of nrsZ; Gmr | This study |
| pME10142 | pME4510 derivative carrying a 0.797 kb insert encompassing nrsZ of P. protegens Pf-5; Gmr | This study |
| pME10143 | pME4510 derivative carrying a 0.563 kb insert encompassing nrsZ of P. putida KT2242; Gmr | This study |
| pME10144 | pME4510 derivative carrying a 0.425 kb insert encompassing nrsZ of P. entomophila L48; Gmr | This study |
| pME10145 | pME4510 derivative carrying a 0.408 kb insert encompassing nrsZ of P. syringae pv. tomato DC3000; Gmr | This study |
| pME10171 | pME6015 derivative carrying the Ptac promoter; Tcr | This study |
| pME10173 | pME10171 derivative carrying the Ptac –rhlA'-'lacZ fusion; Tcr | This study |
| pME10182 | pME10173 derivative with CC(200–201) → GG(200–201)b mutations, Ptac –rhlAmut'-'lacZ fusion; Tcr | This study |
| pME10191 | pME10138 derivative with GG(28–29) → CC(28–29)b mutations; Gmr | This study |
| pME10389 | pME6001 derivative carrying rpoN in a 1.579 kb insert; Gmr | This study |
| pME10390 | pME6001 derivative carrying ntrC in a 1.552 kb insert; Gmr | This study |
Antibiotic resistance phenotypes are indicated by r: Ap, ampicillin, Gm, gentamicin; Tc, tetracycline; Km, kanamycin; Cb, carbenicillin.
Nucleotide numbers correspond to the +1 transcription starts nucleotides.
Construction of plasmids and strains
The intergenic region ntrC-PA5126 containing nrsZ and the sequence containing the promoter to the 60th nucleotide after the predicted start of transcription of nrsZ were amplified using chromosomal DNA of P. aeruginosa PAO1, and primer pairs 2913FW/2913REV and NRS-HP1-REV/sRNA2913FW2 respectively. The obtained Polymerase Chain Reaction (PCR) products of 1.015 and 0.4 kb were digested with BamHI and HindIII, and ligated into the corresponding sites in pME4510, resulting in pME9995 and pME10138 respectively. pME10191 was constructed by ligating a PCR fragment of 0.4 kb, amplified using primers sRNA2913FW2 and NrsZRevmodifIV, and digested with BamHI and HindIII, into pME4510 digested with the same restriction enzymes.
The intergenic region encoding for the homologs of nrsZ in P. protegens Pf-5, P. putida KT2442, P. entomophila L48 and P. syringae pv. tomato DC3000 was amplified using chromosomal DNA of each strains, and primer pairs Pf5-2913FW/Pf5-2913REV, KT2442-2913FW/KT2442-2913REV, L48-2913FW/L48-2913REV and DC3000-2913FW/DC3000-2913REV respectively. For the three first strains (Pf-5, KT2442, L48), the corresponding 797 bp, 563 bp and 425 bp PCR products were digested with EcoRI and BamHI, and ligated in the corresponding site of pME4510, resulting in plasmid pME10142, pME10143 and pME10144 respectively. For the fourth strain (DC3000), the 408 bp PCR product was digested with BamHI and HindIII, and ligated into the corresponding site of pME4510, resulting in plasmid pME10145.
To construct the nrsZ transcriptional reporter fusions, the promoter region of nrsZ was amplified by PCR using primers sRNA2913FW2 and sRNA2913REV2 with pME9995 as DNA template. The PCR fragment of 314 bp was digested with EcoRI and BamHI, and ligated into pME6016 digested with the same enzymes, resulting in pME9989 (nrsZ-lacZ). The nrsZ promoter fused to lacZ was excised from pME9989 by digestion with BamHI and XhoI, and the 3.5 kb fragment was blunted and ligated into pME7549 digested with StuI, resulting in pME10129.
To construct pME10173 carrying the Ptac-rhlA'-'lacZ reporter fusion, a fragment of 300 bp containing the sequence from the start of transcription [previously determined (Pearson et al., 1997)] to the 24th codon of the rhlA gene was amplified by PCR using the primer pair RhlA-FW-K/RhlA-REV-P. The fragment was digested with KpnI and PstI, and cloned into the corresponding sites in pME10171.
Plasmid pME10171 was constructed from ligation of the Ptac promoter (0.9 kb) excised from pME6552 with StuI and EcoRI, and a 2 kb fragment of pME6015 digested with EcoRI and SacI into pME6015 digested with HindIII blunted and SacI. The plasmid pME10182 carrying the Ptac-rhlAmut'-'lacZ mutant reporter fusion was constructed as follows: a 217 bp and 129 bp fragments were amplified by PCR from pME10173 using primer pairs RhlA-FW-K/RhlA-MutIIIA and RhlA-FW-P/RhlA-MutIIIB respectively. The two fragments were fused and amplified by overlap extension PCR (Heckman and Pease, 2007) using primers RhlA-FW-K and RhlA-FW-P, resulting in a 320 bp fragment that was digested with KpnI and PstI, and ligated into the corresponding sites in pME10171.
The nrsZ-lacZ transcriptional fusion carried by the mini-Tn7 delivery plasmid pME10129 was inserted into the Tn7 attachment site of PAO1, PAO6764 and PAO6358, yielding strains PAO6750, PAO6842 and PAO6847, respectively, using the pUXBF-13 transposition helper plasmid (Bao et al., 1991).
Chromosomal inactivation of the ntrC gene in P. aeruginosa PAO1 was performed as follows: PCR fragments of 500–600 bp corresponding to the flanking regions of ntrC were amplified from P. aeruginosa genomic DNA with primer pairs ntrC1/ntrC2 and ntrC3/P33REV1. Fragments were fused together by overlap extension PCR, digested with BamHI and HindIII, and ligated into the corresponding sites of the suicide vector pME3087, resulting in pME9991. Triparental mating was performed to mobilize plasmid pME9991 from E. coli DH5α into P. aeruginosa PAO1 using the mobilizing plasmid pME497 carried by E. coli HB10, and merodiploids were resolved as described before (Ye et al., 1995). Strain PAO6764 carrying an in-frame ΔntrC mutation was generated.
For inactivation of the nrsZ gene, a mutation in the RpoN-box (TAACACAGCCCCTTTA in place of TGG-24CACAGCCCCTGC-12A) of the nrsZ promoter region was created. Two 670 bp fragments were amplified by PCR using the primer pairs 2913FW/sRNA2913rpoN1 and sRNA2913rpoN2/sRNA2913del5. Fragments were fused by overlap extension PCR, digested with BamHI and HindIII, and ligated into the corresponding sites of the suicide vector pME3087. The resulting plasmid pME10134 was used as described above to construct PAO6846.
The plasmids pME10389 or pME10390 used for complementation experiments were obtained by cloning, respectively, the rpoN and ntrC genes in the pME6001 multi-copy plasmid under the lac promoter. rpoN and ntrC were amplified by PCR with primer pairs rpoN-FWcomp/rpoN-REVcomp or ntrC-FWcomp/ntrC-REVcomp, respectively, and cloned into pME6001 after digestion with EcoRI/BamHI.
Northern blot analysis
The total RNA was isolated from cells grown in different media (as described in figure legends) using the hot acid phenol method, as previously described (Leoni et al., 1996). RNA fractions were treated with DNase I recombinant (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's recommendations. The 5S rRNA DIG-labelled, double-stranded DNA (dsDNA) probe (5SDNA) was synthesized, as described before (Sonnleitner et al., 2009), with the primer pair 5S-rRNA-1/5S-rRNA-2.
The anti-NrsZ DIG-labelled, single-stranded RNA (ssRNA) probe of 242 nt (NrsRNA), covering a region encompassing the predicted +1 nucleotide of NrsZ (12 nt after the G nt of the −12 box of the RpoN-box sequence) to the last 17 nt of the PA5126 gene, was synthesized with T7 RNA polymerase (Promega, Madison, WI, USA) from a PCR DNA template obtained using pME9995, and the primer pair Nrs_FW_P and T7-2913 probe, according to the manufacturer's recommendations (DIG RNA Labeling Mix, Roche Diagnostics). Prior to hybridization, dsDNA and ssRNA DIG-labelled probes were heat-denatured for 10 min at 100°C or 68°C respectively. Of the total RNA, 5′10 μg were loaded onto the denaturing urea-polyacrylamide gels and analyzed by Northern blotting, as previously described (González et al., 2008). Primer sequences are listed in Table S1.
Rapid amplification of cDNA 3′ ends (3′ RACE)
The 3′ extremities of NrsZ transcripts were determined by 3′-RACE according to the modified protocol of Argaman et al. (Argaman et al., 2001). Briefly, 40 μg of total RNA, extracted from the PAO1 WT strain grown in MMP supplemented with 20 mM glucose and 20 mM nitrate to an OD600 of ∼ 2, was 5′-dephosphorylated with rAPID alkaline phosphatase (Roche Diagnostics). Dephosphorylated RNA was ligated with 500 pmol of the 3′ RNA adapter P-RIBOLI (Table S1) using 50 U of T4 RNA ligase (New England Biolabs, Hertfordshire, England) in a buffer containing 50 mM HEPES/NaOH (pH 8.0), 20 mM MgCl2, 3 mM DTT, 10 % (v/v) dimethyl sulfoxide (DMSO), 10 μg ml−1 BSA, 2 U RNasin (Promega) and 0.2 mM ATP in a final volume of 50 μl. Reverse transcription was carried out using Maxima Reverse Transcriptase (Fermentas, Pittsburgh, PA, USA) and the DEOXYLI primer complementary to the P-RIBOLI RNA adapter (Table S1) according to the manufacturer's recommendations but in the presence of 5% of DMSO. cDNAs complementary to NrsZ were PCR amplified (40 cycles with an annealing temperature of 57°C) using GoTaq polymerase (Promega) and primers DEOXYLI and NrsZ-3′RACE (Table S1). After agarose gel analyses of the resulting PCR products, the DNA fragments of interest were directly ligated into pGEM-T easy vector (Promega). Bacterial colonies obtained by transformation were screened for the presence of the expected fragments by PCR using the DEOXYLI and NrsZ-3′RACE primers, and plasmids carrying the inserts of interest were isolated and sequenced by the Sanger method using primer M13__40_long (Table S1).
RNA deep sequencing and qualitative analysis
Pseudomonas aeruginosa PAO1 was grown in MMP medium supplemented with 20 mM mannitol and 20 mM nitrate. Total RNA was isolated as previously mentioned. RNA size selection was performed following the modified protocol of Gómez-Lozano et al. (Gómez-Lozano et al., 2012). After separation on denaturing polyacrylamide gel (urea 10 M), two gel fractions corresponding to RNA molecules ranging from 30 nt to 200 nt and from 150 nt to 450 nt were excised, eluted in 0.4 M NaCl and treated with tobacco alkaline pyrophosphatase for 1 h at 37°C. Treated RNA samples were used to perform two multiplexed libraries according to the TruSeq Small RNA Sample Preparation kit (Illumina, San Diego, CA, USA) without fragmentation to preserve molecule integrity. High-GC Accuprime Polymerase (Invitrogen, New York, NY, USA) was used in PCR reactions to overcome elongation difficulties due to the high GC-content of the PAO1 transcriptome. 2 × 100 paired-end sequencing was performed on a TruSeq Illumina HiSeq2000 machine. After de-multiplexing, paired-end reads were mapped using bowtie (v0.2.19) (Langmead et al., 2009) and samtools (v0.1.18) (Li et al., 2009) software on the PAO1 genome (NC_002516.2). 2 × 100 paired-end reads were merged to produce artificial orientated single-end reads ranging from 30 nt up to 450 nt.
β-Galactosidase assays
β-Galactosidase activities were determined by the Miller method (Miller, 1972) using cells grown at 37°C and permeabilized with 5% (v/v) toluene. All experiments were performed in triplicate.
Swarming motility assays
Swarming motility was assayed on MMP medium supplemented with 20 mM glucose and 0.1% (w/v) casamino acids plates solidified with 0.5% (w/v) bacteriological Agar (Oxoid) (modified from Yeung et al., 2011). Bacteria were grown in LB medium to mid-logarithmic-growth-phase (OD600 0.5–1) and 1 μl of culture was plated in four independent replicates. Swarming was observed after 24 h of incubation at 37°C.
Rhamnolipids production assays
Rhamnolipids production was tested on the MMP medium supplemented with 20 mM glucose, 0.1% (w/v) casamino acids, 0.02% (w/v) cetyltrimethylammonium bromide and 0.0005% (w/v) methylene blue solidified with 1.2% (w/v) bacteriological agar (Oxoid) (modified from Pinzon and Ju, 2009). The plates were inoculated with single colonies grown overnight at 37°C on agar plates. Rhamnolipids production was estimated by measuring violet halos formed on plates around the colonies after 72 h incubation at 37°C, followed by an incubation at room temperature for the same duration.
Acknowledgments
We thank Cornelia Reimmann for providing the pME7549 plasmid, Sébastien Ritzman for helping in the construction of the plasmids pME10142-10145, and Christoph Keel, Onya Opota and Inés Canosa for providing the P. protegens Pf-5, P. entomophila L48 and P. putida KT2442 strains respectively. Karine Lapouge was supported by the Sandoz Family Foundation (Programme for Academic Promotion), the Swiss National Foundation for Scientific Research (Project 31003A-127587) and the Fondation Herbette.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. Schematic of the nrsZ locus. The sequence of nrsZ from its putative +1 transcription start site to the PA5126 gene stop codon is indicated in bold. The RpoN promoter of nrsZ is boxed, and the invariant −24 and −12 nucleotides are underlined. The RpoN promoter sequence degenerated in the strain PAO6846 (nrsZ−) is indicated in a grey box. The promoter region cloned in the nrsZ-lacZ is indicated and encompasses the sequence from the T nucleotide labelled by a star to the vertical arrow.
Fig. S2. nrsZ-lacZ expression in PAO1 WT strain grown with different casamino acids concentration. The PAO1 WT strain carrying nrsZ-lacZ (PAO6750) was inoculated at an initial optical density (OD600) of 0.05, and was grown in MMP supplemented with glucose as carbon source and casamino acids (CA) at different concentrations [0.05%, 0.1%, 0.2% or 0.5% (w/v)]. The growth (OD600) (A) and the activity of the nrsZ-lacZ chromosomal fusion (B) were followed in the different conditions during 10.5 h of incubation. Each value represents the average of triplicate cultures ± standard deviation.
Fig. S3. NrsZ promoter and structure are conserved among Pseudomonads.
A. Alignment of the promoters of nrsZ in different Pseudomonads. The RpoN promoter sequence identified in P. aeruginosa PAO1 is in bold. Nucleotides at −12 and −24 are in red box, and conserved nucleotides are indicated with a star. The predicted transcriptional start site of NrsZ in PAO1 and other Pseudomonads is in bold. The matured 5′ extremity of the first stem loop is indicated by an arrow.
B. Predicted secondary structures of NrsZ in different Pseudomonads (using the Mfold software). The conserved motifs of NrsZ are in red, and the unconserved spacer sequences represented by a square.
Fig. S4. Rhamnolipids production in the nrsZ− mutant strain is restored by nrsZ of other Pseudomonads. Rhamnolipids production was assayed as described in the Experimental procedures for strains PAO1 WT and PAO1 nrsZ− (PAO6846) containing the empty vector pME4510, or its derivative plasmids pME9995 (pnrsZPAO1), pME10142 (pnrsZPf-5), pME10143 (pnrsZKT2442), pME1044 (pnrsZL48) and pME10145 (pnrsZDC3000), carrying respectively the nrsZ genes of P. aeruginosa PAO1, P. protegens Pf-5, P. putida KT2442, P. entomophila L48 and of P. syringae pv. tomato DC3000, under the control of their native promoter.
Table S1. List of primers used in this study.
References
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H. Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Bao Y, Lies DP, Fu H. Roberts GP. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene. 1991;109:167–168. doi: 10.1016/0378-1119(91)90604-a. [DOI] [PubMed] [Google Scholar]
- Barrios H, Valderrama B. Morett E. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer C, Heeb S, Pessi G. Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci USA. 1999;96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenstein EBM, de la Fuente-Núñez C. Hancock REW. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, Shanks RM. O'Toole GA. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol. 2005;187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KG, Ballot F. Reid SJ. Enhanced rhamnolipid production by Pseudomonas aeruginosa under phosphate limitation. World J Microbiol Biotechnol. 2010;26:2179–2184. [Google Scholar]
- Davis BM. Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol Microbiol. 2007;65:373–385. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E, Lépine F, Milot S. Villemur R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology. 2003;149:2005–2013. doi: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- Dixon R. Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- Dötsch A, Eckweiler D, Schniederjans M, Zimmermann A, Jensen V, Scharfe M, et al. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS ONE. 2012;7:e31092. doi: 10.1371/journal.pone.0031092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara S, Brugnoli M, De Bonis A, Righetti F, Delvillani F, Dehò G, et al. Comparative profiling of Pseudomonas aeruginosa strains reveals differential expression of novel unique and conserved small RNAs. PLoS ONE. 2012;7:e36553. doi: 10.1371/journal.pone.0036553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin FCH, Bagdasarian M, Bagdasarian MM. Timmis KN. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Hayward WS. August JT. Bacterial proteins required for replication of phage Qβ ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J Biol Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- Gómez-Lozano M, Marvig RL, Molin S. Long KS. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol. 2012;14:2006–2016. doi: 10.1111/j.1462-2920.2012.02759.x. [DOI] [PubMed] [Google Scholar]
- González N, Heeb S, Valverde C, Kay E, Reimmann C, Junier T. Haas D. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics. 2008;9:167. doi: 10.1186/1471-2164-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SK, Schroth MN, Cho JJ, Kominos SD. Vitanzajack VB. Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl Environ Microbiol. 1974;28:987–991. doi: 10.1128/am.28.6.987-991.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Santos L, Käppeli O. Fiechter A. Pseudomonas aeruginosa biosurfactant production in continuous culture with glucose as carbon source. Appl Environ Microbiol. 1984;48:301–305. doi: 10.1128/aem.48.2.301-305.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D, Holloway BW, Schambock A. Leisinger T. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol Gen Genet. 1977;154:7–22. doi: 10.1007/BF00265571. [DOI] [PubMed] [Google Scholar]
- Hardalo C. Edberg SC. Pseudomonas aeruginosa: assessment of risk from drinking water. Crit Rev Microbiol. 1997;23:47–75. doi: 10.3109/10408419709115130. [DOI] [PubMed] [Google Scholar]
- Heckman KL. Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Hervás AB, Canosa I. Santero E. Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J Bacteriol. 2008;190:416–420. doi: 10.1128/JB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás AB, Canosa I, Little R, Dixon R. Santero E. NtrC-dependent regulatory network for nitrogen assimilation in Pseudomonas putida. J Bacteriol. 2009;191:6123–6135. doi: 10.1128/JB.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurlier K, Denervaud V, Pessi G, Reimmann C. Haas D. Negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185:2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, et al. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J Bacteriol. 2004;186:2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BW, Krishnapillai V. Morgan AF. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison ML. Govan JRW. Pathogenicity of microbes associated with cystic fibrosis. Microbes Infect. 1999;1:1005–1014. doi: 10.1016/s1286-4579(99)80518-8. [DOI] [PubMed] [Google Scholar]
- Kimata N, Nishino T, Suzuki S. Kogure K. Pseudomonas aeruginosa isolated from marine environments in Tokyo Bay. Microb Ecol. 2004;47:41–47. doi: 10.1007/s00248-003-1032-9. [DOI] [PubMed] [Google Scholar]
- Köhler T, Kocjancic Curty L, Barja F, van Delden C. Pechère JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S, Santero E, Keener J, Popham D. Weiss D. Expression of σ54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S. Wullbrandt D. Rhamnose lipids – biosynthesis, microbial production and application potential. Appl Microbiol Biotechnol. 1999;51:22–32. doi: 10.1007/s002530051358. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M. Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FHT. Haas D. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Lease RA. Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Leoni L, Ciervo A, Orsi N. Visca P. Iron-regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of fur and PvdS on promoter activity. J Bacteriol. 1996;178:2299–2313. doi: 10.1128/jb.178.8.2299-2313.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Lu CD. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J Bacteriol. 2007;189:5413–5420. doi: 10.1128/JB.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM. Soberón-Chávez G. Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl Microbiol Biotechnol. 2000;54:625–633. doi: 10.1007/s002530000443. [DOI] [PubMed] [Google Scholar]
- Medina G, Juárez K, Diaz R. Soberón-Chávez G. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology. 2003a;149:3073–3081. doi: 10.1099/mic.0.26282-0. [DOI] [PubMed] [Google Scholar]
- Medina G, Juárez K, Valderrama B. Soberón-Chávez G. Mechanism of Pseudomonas aeruginosa RhlR transcriptional regulation of the rhlAB promoter. J Bacteriol. 2003b;185:5976–5983. doi: 10.1128/JB.185.20.5976-5983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick MJ. Edwards RA. Nitrogen control in bacteria. Microbiol Mol Biol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG. Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Mulligan CN. Gibbs BF. Correlation of nitrogen metabolism with biosurfactant production by Pseudomonas aeruginosa. Appl Environ Microbiol. 1989;55:3016–3019. doi: 10.1128/aem.55.11.3016-3019.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Dos Santos VAP, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Ninfa AJ, Atkinson MR, Kamberov ES, Feng JL. Control of nitrogen assimilation by the NRI-NRII two-component system of enteric bacteria. In: Silhavy TJ, Ninfa EG, Hoch JA, editors. Two-Component Signal Transduction. Washington, DC, USA: American Society for Microbiology Press; 1995. pp. 67–88. [Google Scholar]
- Nishijyo T, Haas D. Itoh Y. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol Microbiol. 2001;40:917–931. doi: 10.1046/j.1365-2958.2001.02435.x. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Fiechter A. Reiser J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem. 1994;269:19787–19795. [PubMed] [Google Scholar]
- Overhage J, Lewenza S, Marr AK. Hancock REW. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5lux mutant library. J Bacteriol. 2007;189:2164–2169. doi: 10.1128/JB.01623-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J, Bains M, Brazas MD. Hancock REW. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol. 2008;190:2671–2679. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC. Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GSA, Mavrodi DV, et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Pesci EC. Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci EC, Pearson JP, Seed PC. Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzon NM. Ju LK. Improved detection of rhamnolipid production using agar plates containing methylene blue and cetyl trimethylammonium bromide. Biotechnol Lett. 2009;31:1583–1588. doi: 10.1007/s10529-009-0049-7. [DOI] [PubMed] [Google Scholar]
- Potvin E, Sanschagrin F. Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2008;32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- Rashedi H, Jamshidi F, Mazaheri Assadi M. Bonakdarpour B. Isolation and production of biosurfactant from Pseudomonas aeruginosa isolated from Iranian southern wells oil. Int J Environ Sci Technol. 2005;2:121–127. [Google Scholar]
- Rice P, Longden I. Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rist M. Kertesz MA. Construction of improved plasmid vectors for promoter characterization in Pseudomonas aeruginosa and other gram-negative bacteria. FEMS Microbiol Lett. 1998;169:179–183. doi: 10.1111/j.1574-6968.1998.tb13315.x. [DOI] [PubMed] [Google Scholar]
- Romeo A, Sonnleitner E, Sorger-Domenigg T, Nakano M, Eisenhaber B. Bläsi U. Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD-PA1779-cobA operon. Microbiology. 2012;158:1543–1552. doi: 10.1099/mic.0.053850-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF. Maniatis T. Molecular Cloning: A Laboratory Manual (Third Edition) Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schnider-Keel U, Seematter A, Maurhofer M, Blumer C, Duffy B, Gigot-Bonnefoy C, et al. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J Bacteriol. 2000;182:1215–1225. doi: 10.1128/jb.182.5.1215-1225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM. Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Soberón-Chávez G, Lépine F. Déziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E. Haas D. Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl Microbiol Biotechnol. 2011;91:63–79. doi: 10.1007/s00253-011-3332-1. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Abdou L. Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 106:21866–21871. doi: 10.1073/pnas.pnas.0910308106. and (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, González N. Small RNAs of Pseudomonas spp. In: Filloux A, Ramos J-L, Haas D, editors. Pseudomonas—Molecular Microbiology, Infection and Biodiversity. Vol. 6. Dordrecht, The Netherlands: Springer; pp. 3–28. and (2010) [Google Scholar]
- Sonnleitner E, Romeo A. Bläsi U. Small regulatory RNAs in Pseudomonas aeruginosa. RNA Biol. 9:364–371. doi: 10.4161/rna.19231. and (2012) [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP. Blasi U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol Microbiol. 59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. and (2006) [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jager KE. Bläsi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Sorger-Domenigg T, Madej MJ, Findeiss S, Hackermuller J, Huttenhofer A, et al. Detection of small RNAs in Pseudomonas aeruginosa by RNomics and structure-based bioinformatic tools. Microbiology. 2008;154:3175–3187. doi: 10.1099/mic.0.2008/019703-0. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J. Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovar N, Vallenet D, Cruveiller S, Rouy Z, Barbe V, Acosta C, et al. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol. 2006;24:673–679. doi: 10.1038/nbt1212. [DOI] [PubMed] [Google Scholar]
- Voisard C, Bull C, Keel C, Laville J, Maurhofer M, Schnider M. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches. In: O'Gara F, Dowling DN, Boesten B, et al., editors. Molecular Ecology of Rhizosphere Microorganisms. Weinheim, Germany: VCH Publishers; 1994. pp. 67–89. [Google Scholar]
- Waters LS. Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye RW, Haas D, Ka JO, Krishnapillai V, Zimmermann A, Baird C. Tiedje JM. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J Bacteriol. 1995;177:3606–3609. doi: 10.1128/jb.177.12.3606-3609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung AT, Bains M. Hancock RE. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol. 193:918–931. doi: 10.1128/JB.00911-10. and (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung AT, Torfs EC, Jamshidi F, Bains M, Wiegand I, Hancock RE. Overhage J. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol. 2009;191:5592–5602. doi: 10.1128/JB.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XX. Rainey PB. Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics. 2008;178:185–195. doi: 10.1534/genetics.107.081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Schematic of the nrsZ locus. The sequence of nrsZ from its putative +1 transcription start site to the PA5126 gene stop codon is indicated in bold. The RpoN promoter of nrsZ is boxed, and the invariant −24 and −12 nucleotides are underlined. The RpoN promoter sequence degenerated in the strain PAO6846 (nrsZ−) is indicated in a grey box. The promoter region cloned in the nrsZ-lacZ is indicated and encompasses the sequence from the T nucleotide labelled by a star to the vertical arrow.
Fig. S2. nrsZ-lacZ expression in PAO1 WT strain grown with different casamino acids concentration. The PAO1 WT strain carrying nrsZ-lacZ (PAO6750) was inoculated at an initial optical density (OD600) of 0.05, and was grown in MMP supplemented with glucose as carbon source and casamino acids (CA) at different concentrations [0.05%, 0.1%, 0.2% or 0.5% (w/v)]. The growth (OD600) (A) and the activity of the nrsZ-lacZ chromosomal fusion (B) were followed in the different conditions during 10.5 h of incubation. Each value represents the average of triplicate cultures ± standard deviation.
Fig. S3. NrsZ promoter and structure are conserved among Pseudomonads.
A. Alignment of the promoters of nrsZ in different Pseudomonads. The RpoN promoter sequence identified in P. aeruginosa PAO1 is in bold. Nucleotides at −12 and −24 are in red box, and conserved nucleotides are indicated with a star. The predicted transcriptional start site of NrsZ in PAO1 and other Pseudomonads is in bold. The matured 5′ extremity of the first stem loop is indicated by an arrow.
B. Predicted secondary structures of NrsZ in different Pseudomonads (using the Mfold software). The conserved motifs of NrsZ are in red, and the unconserved spacer sequences represented by a square.
Fig. S4. Rhamnolipids production in the nrsZ− mutant strain is restored by nrsZ of other Pseudomonads. Rhamnolipids production was assayed as described in the Experimental procedures for strains PAO1 WT and PAO1 nrsZ− (PAO6846) containing the empty vector pME4510, or its derivative plasmids pME9995 (pnrsZPAO1), pME10142 (pnrsZPf-5), pME10143 (pnrsZKT2442), pME1044 (pnrsZL48) and pME10145 (pnrsZDC3000), carrying respectively the nrsZ genes of P. aeruginosa PAO1, P. protegens Pf-5, P. putida KT2442, P. entomophila L48 and of P. syringae pv. tomato DC3000, under the control of their native promoter.
Table S1. List of primers used in this study.