Abstract
Maize originated in the highlands of Mexico approximately 8700 years ago and is one of the most commonly grown cereal crops worldwide, followed by wheat and rice. Abiotic stresses (primarily drought, salinity, and high and low temperatures), together with biotic stresses (primarily fungi, viruses, and pests), negatively affect maize growth, development, and eventually production. To understand the response of maize to abiotic and biotic stresses and its mechanism of stress tolerance, high-throughput omics approaches have been used in maize stress studies. Integrated omics approaches are crucial for dissecting the temporal and spatial system-level changes that occur in maize under various stresses. In this comprehensive analysis, we review the primary types of stresses that threaten sustainable maize production; underscore the recent advances in maize stress omics, especially proteomics; and discuss the opportunities, challenges, and future directions of maize stress omics, with a view to sustainable food production. The knowledge gained from studying maize stress omics is instrumental for improving maize to cope with various stresses and to meet the food demands of the exponentially growing global population. Omics systems science offers actionable potential solutions for sustainable food production, and we present maize as a notable case study.
Introduction
Maize (Zea mays L.), which originated in the highlands of Mexico approximately 8700 years ago (Piperno et al., 2009), is one of the most commonly cultivated cereal crops worldwide, followed by wheat and rice. Maize is the largest crop in China in both planting area and yield (http://www.stats.gov.cn/). In 2012, global maize production was approximately 840 million tons (Barkla et al., 2013). Maize has long been a staple food of much of the world's population (particularly in South America and Africa) and a primary nutrient source for animal feed and for food industrial materials; in recent years, maize has also been used for biofuel production.
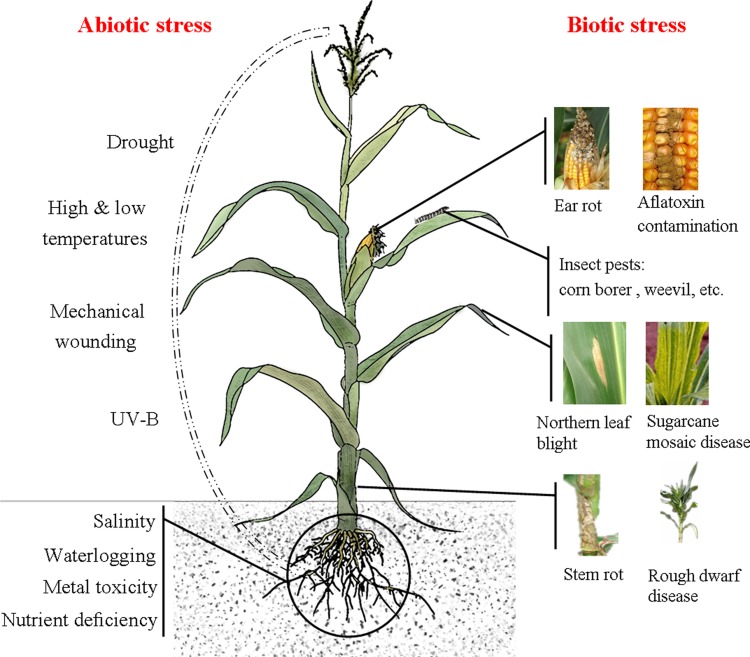
Maize belts range from the latitude 58° north to the latitude 40° south, and maize ripens every month of the year. Unsurprisingly, abiotic and biotic stresses are common in maize belts worldwide. Figure 1 illustrates the major stresses that threaten maize production. Abiotic stresses, such as drought, salinity, high and low temperatures, and nutrient deficiency, are primary environmental factors that negatively affect maize production. In particular, recent intense droughts, waterlogging, and extreme temperatures have severely affected maize growth and yield (Ahuja et al., 2010). In China, 60% of the maize planting area is prone to drought, and the resultant yield loss is 20%–30% per year; in India, 25%–30% of the maize yield is lost as a result of waterlogging each year (Zaidi et al., 2010). The biotic stresses on maize are primarily pathogens (fungal, bacterial, and viral), and the resultant syndromes, such as ear/stalk rot, rough dwarf disease, and northern leaf blight, are prevalent and cause heavy damage. Approximately 10% of the global maize yield is lost each year as a result of biotic stresses. For example, the European corn borer [ECB, Ostrinia nubilalis (Hübner)] causes yield losses of up to 2000 million dollars annually in the USA alone (Basu et al., 2010); in the northern regions of China, the maize yield loss reaches 50% during years when maize is plagued by northern leaf blight (Ji et al., 2010). In addition, abiotic and biotic stresses often are present simultaneously and severely influence maize production.
FIG. 1.
An overview of major abiotic and biotic stresses threatening maize production.
The world's food supply is inadequate compared with the demand for food. The global population is expected to reach 9000 million by 2050 (Shelden and Roessner, 2013), indicating that there will be an additional 2000 million people to feed in the coming years. Approximately 80% of human food is composed of crops, which are dominated by cereals that make up 50% of global food production (Langridge and Fleury, 2011). Moreover, by 2050, more than 50% of the world's arable land will be negatively affected by soil salinization and drought, resulting in an increased loss of arable land. Therefore, to meet the required increase in food production, crop productivity must increase by 38% annually (Shelden and Roessner, 2013).
To ensure the world's food production, crops (e.g., maize) that are well adapted to these extreme environmental conditions should be developed (Vinocur and Altman, 2005). Therefore, understanding the underlying response mechanisms of maize toward various abiotic and biotic stresses is important. Abiotic stresses were expected to be controlled through the use of commercial drought tolerant varieties of maize by 2012 or earlier in the USA and by 2017 in Sub-Saharan Africa, where maize is a staple food (Basu et al., 2010). Minimizing the maize yield loss caused by ECB was attempted through bioengineering a gene from a soil bacterium, Bacillus thuringiensis (Bt), which confers almost complete resistance to ECB. This attempt resulted in the development of high-yielding Bt-corn hybrids to fight ECB infestation (Raybould et al., 2007).
Omics approaches are important for dissecting the temporal and spatial protein expression changes that occur in maize under various stresses. Advances in maize omics have been accelerated by the availability of the genomic sequence of the B73 line of maize (Schnable et al., 2009), improved methodology and advanced apparatuses. In recent years, considerable progress has been made in maize stress research. Although several excellent reviews have summarized the responses of plants to various stresses, as determined by using proteomics (Barkla et al., 2013; Kosová et al., 2011; Ngara and Ndimba, 2013; Pechanova et al., 2013; Sobhanian et al., 2011; Zhao et al., 2013; Zagorchev et al., 2014), genomics (Shelden and Roessner, 2013), and metabolomics (Arbona et al., 2013; Balmer et al., 2013), there are no specific reviews on the advances in maize stress omics to date.
This article comprehensively analyzes the recent advances in maize stress omics, especially proteomics, describing the materials and methods used in this research, and the results obtained and the potential meanings of these research activities. New knowledge about maize stress omics will be pivotal for understanding the host-environment interactions and not to mention genetically improving the stress tolerance of maize, thereby contributing to sustainable food production.
Maize Response to Abiotic Stresses
Drought
Drought stress caused by water scarcity is one of the critical factors that limits maize yields in most maize production regions worldwide (Cooper et al., 2014). Drought tolerance is important for the success of maize hybrids grown in drought-prone regions. The maize response to drought stress is a crucial and complex process. Drought affects many processes involved in plant growth and development, including osmotic adjustment, antioxidant capabilities, photosynthetic rate reduction, and abscisic acid (ABA) accumulation (Cramer et al., 2011). These processes are controlled by many proteins, which are expressed differentially in various stress tolerant species, in various functions and biochemical pathways, and at various developmental stages.
The drought response of maize has been extensively studied in seeds, leaves, and roots, mainly using two-dimensional gel electrophoresis (2-DE)/MS/MS based proteomics (Table 1). Huang et al. (2012) analyzed the desiccation tolerance of maize seeds using 2-DE combined with MS/MS. The differentially expressed proteins (e.g., 17.4 kDa Class I HSP3, EMB564, and stress responsive protein) that they observed may be involved in drought tolerance during embryogenesis and germination. The expression levels of CAT, APX, and SOD, which are drought-protective proteins in leaves, differed between drought-tolerant and drought-sensitive maize genotypes: Higher accumulation of these proteins was observed in drought-tolerant maize (Benesova et al., 2012). The ABA pathway is one of the primary signaling pathways that mediate maize adaptation to drought stress (Bahrun et al., 2002). To further understand the mechanism by which ABA regulates the maize proteome in response to drought, we analyzed the proteomic differences between the ABA-deficient maize mutant vp5 and the wild-type Vp5 under drought stress, by using 2-DE and MS/MS (Hu et al., 2011; 2012). In maize roots, proteins associated with drought stress were primarily involved in energy and metabolism, redox homeostasis, and regulatory processes (Hu et al., 2011); in maize leaves, the identified proteins were primarily involved in processes such as ATP synthesis, protein synthesis, chlorophyll synthesis, CO2 fixation, gluconeogenesis, antioxidant defense, and signal transduction. Most of the proteins that differentially accumulated in leaves were localized to the chloroplasts and functioned in an ABA-dependent manner in response to drought and light stress (Hu et al., 2012). These results reveal an important role of ABA in regulating the synthesis of drought-induced proteins. In addition, using quantitative trait loci (QTL) (Capelle et al., 2010) and oligo-microarray (Luo et al., 2010) analyses, several QTLs or genes associated with kernel desiccation were found to be involved in ABA synthesis. Other studies also found that many drought response-related proteins or genes, such as ZmTPA (Huang et al., 2012), ZmRFP1 (Xia et al., 2012), and ZmCPK4 (Jiang et al., 2013) were induced by ABA or in an ABA-dependent manner.
Table 1.
Summary of Recent Years' Publications on Drought Response of Maize
| Maize genotype | Stage/tissue | Treatment | Technique | Protein/gene classification | Identified major proteins/genes | Reference |
|---|---|---|---|---|---|---|
| Zhengdan 958 (cultivar, China) | 14-d-old leaves | -0.7 MPa PEG for 8 h | 2-DE, MALDI-TOF | Protective proteins, metabolism | HSP17.4, HSP17.2, HSP26, guanine nucleotide-binding protein, β-subunit-like protein, granule-bound starch synthase IIa | (Hu et al., 2010) |
| Zhengdan 958 | Leaves of 3-leaf-stage seedlings | 16% (w/v) PEG for 8 h | 2-DE, MALDI-TOF | Photosynthesis, glycolysis parthway, protein metabolim, cell wall biosynthesis, disease defence, lipid biosynthesis, harpin binding protein | 50S ribosomal protein L28, chlorophyll α-β binding protein 2 precursor, ATP synthase CF1 α subunit, glucosy l transferase, ferredoxin–NADP reductase, triosephosphate isomerase | (Tai et al., 2011) |
| vp5 (ABA-deficient mutant); Vp5 (wild-type) | 14-d-old roots | -1.0 MPa PEG6000 for 6 h | 2-DE, MALDI-TOF | Regulatory proteins, energy and metabolism, redox homeostasis | Nuclear transport factor 2, glycine-rich RNA binding protein 2, pathogenesis-related protein 10, transcription factor BTF3, serine/threonine-protein kinase receptor | (Hu et al., 2011) |
| vp5, Vp5 | 14-d-old leaves | –1.0 MPa mannitol for 8 h | 2-DE, MALDI-TOF | Photosynthesis, translation elongation, proteolysis, stress response | ATP synthase, elongation factor, β-D-glucosidase, 60S ribosomal protein L32, phosphoribulokinase, glyceraldehyde-3-phosphate dehydrogenase A, putative NBS-LRR disease, glutathione S-transferase 4 | (Hu et al., 2012) |
| CE704 (drought-tolerant); 2023 (drought-sensitive) | 34-d-old leaves | Holding water for 6 d | 2-DE, iTRAQ, MALDI-TOF | Protective proteins, protein synthesis | Dehydrin RAB-17, HSP 16.9, elongation factor 1-δ, ribosomal protein S18 | (Benesova et al., 2012) |
| Nongda108 (cultivar, China) | Embryos | Desiccation to 7% moisture in a desiccator | 2-DE, MALDI-TOF-TOF | Stress response, carbohydrate and energy metabolism, protein metabolism, signal transduction, transcriptional regulation-related, etc. | Enoyl-[acyl-carrier-protein] reductase [NADH], globulin 2, EMB564, HSP, RAB17 protein, 14-3-3 like protein, transcriptional regulator, DNA binding protein | (Huang et al., 2012) |
| Zhengdan958 | 14-d-old roots | -0.7 MPa PEG for 8 h | 2-DE, MALDI-TOF | Transporters, signal transduction, metabolism, disease/defense, cell growth/division | Alcohol dehydrogenase 1, O-methyltransferase, APx1-cytosolic ascorbate peroxidase, aquaporin PIP2-5, glutathione S-transferase 4 glycine-rich RNA-binding protein 2 | (Liu et al., 2013) |
| B73 (inbred line) | 21-d-old leaves | Holding water, and re-watering for 5-60 min | Stable-isotope labeling, LC-MS/MS | Carbohydrate metabolism, cell division or cell expansion-related proteins, chromatin remodeling, phytohormone related and signaling proteins, transcription regulators, protein metabolism, energy metabolism, stress response | Calmodulin-related protein 2, auxin-repressed protein, serine/threonine-protein kinase SAPK10, SnRK2.4, putative pumilio/Mpt5 family, RNA-binding protein, proline-rich cell wall protein-like | (Bonhomme et al., 2012) |
| CN165 (Inbred line, drought-tolerant) | Roots of 6-leaf-stage seedlings | Stopped to water for 20, 40, and 60 h | Subtractive cDNA library | Signaling and regulatory, the synthesis of functional and structural metabolites, conferring stress tolerance | 14-3-3 family protein, putative protein kinase SPK-3, ATP binding kinase, chlorophyll a oxygenase, HSP, putative NADH dehydrogenase | (Li et al., 2009) |
| B73 Tex6 (inbred line) | Developing kernels | Stopped to irrigate at 18 days after pollination | Oligo-microarray | ABA, JA and phenylalanine ammonia lyase related genes | HSP101, cytochrome P450, β 1,3-glucanase, ABA stress ripening protein homolog, auxin response factor 2, ribonuclease III | (Luo et al., 2010) |
| B73 | 5th and 6th leaves | Stop watering after 21 d | qRT-PCR; microarray; 2-DE, LC-MS/MS; GC-MS | Amino acid activation, carbohydrate metabolism, protein folding, RNA regulation, glycolysis and tricarboxylic acid cycle | Class II aspartyl-tRNA synthetase, β-D-glucosidase, spermidine synthase 1, trigger factor-like protein, adenylosuccinate synthetase, 3-isopropylmalate, dehydrogenase, pyruvate dehydrogenase subunit E1b, PSII stability/assembly factor | (Virlouvet et al., 2011) |
| B73 | Fertilized ovary and leaf meristem tissue | Irrigation was withheld at the onset of silk emergence | RNA-Seq analysis | Carbon metabolism, cell cycle and division, programmed cell death, responses of antioxidant and ABA-related, sugar, phosphatidylinositol signaling-associated, phospholipase related responses | Chlorophyll α/β-binding family protein, ABA response element-binding factor, sucrose transporter 1, phosphorylase, 1,4-α-glucan-branching enzyme 2 | (Kakumanu et al., 2012) |
| LX9801, Qi319, etc. (drought-tolerant inbred lines); B73, Ye478, Ji853 (drought-sensitive inbred lines) | 14-d-old | Water stress | Whole-genome resequencing | Plant hormone regulation, carbohydrate and sugar metabolism, signaling molecules regulation and redox reaction | 524 nsSNPs | (Xu et al., 2014) |
| Six maize hybrids | Leaf blades, ears, husks, sheath, and silks | Stopping irrigation for 12 d before flowering | GC–MS | Amino acids, sugars, sugar alcohols, and intermediates of the TCA cycle, etc | – | (Witt et al., 2014) |
Recently, a robust, quantitative proteomic methodology based on stable-isotope labeling has been widely used to analyze maize proteome and phosphoproteome dynamics under drought stress. Bonhomme et al. (2012), using this stable-isotope labeling method, identified 3664 unique phosphorylation sites on 2496 proteins. These phosphopeptides included proteins that influence epigenetic control, transcriptional regulation, cell cycle-dependent processes, phytohormone-mediated responses, cell cycle control, histone modification, DNA methylation, and ABA-, ethylene-, auxin- and/or jasmonate-related responses. Using both LC-based isobaric tags for relative and absolute quantitation (iTRAQ) and gel-based 2-DE analysis, Benesova et al. (2012), studied drought-induced changes in the maize leaf proteome. They identified 326 proteins by iTRAQ and 11 proteins by 2-DE combined with MS/MS analysis. Only four proteins were identified by both methods, which indicated that the two technologies were compatible but only partially overlapping. Thus, more proteins were identified by the stable-isotope labeling method, which is therefore likely to contribute to a better understanding of the molecular basis of the maize response to drought stress.
As determined by omics studies, the proteins involved in the maize drought response primarily include protective proteins (such as HSPs) (Benesova et al., 2012; Hu et al., 2010a; Li et al., 2009; Luo et al., 2010), late embryogenesis-abundant proteins (LEAs) (Benesova et al., 2012; Huang et al., 2012), stress response-related proteins (such as NBS-LRR resistance-like protein) (Hu et al., 2012), 14-3-3-like proteins (Huang et al., 2012; Li et al., 2009), phytohormone-related proteins, and signaling proteins (such as auxin-repressed protein and serine/threonine protein kinase) (Bonhomme et al., 2012; Hu et al., 2011; Luo et al., 2010).
Salinity
Salinity, resulting mainly from NaCl, is one of the most significant abiotic stresses that affects crop growth and yield. More than 20% of agricultural land worldwide is affected by salinity (Zhu, 2001), and salinization is expected to become an increasingly severe problem. The high salt concentration of soil causes a water deficit in crops, resulting initially in osmotic stress and later in ion-specific toxicity (Munns, 2005). Subsequently, salinity can limit crop growth, development, and yield and even cause crop death under severe conditions. The effect of salt stress on plant growth depends on the plant species. Maize is sensitive to hyperosmotic stresses, showing marked production decreases in saline soils (Ngara et al., 2012), and many changes in the proteome, transcriptome, and metabolome have been observed in response to salt stress. Several comprehensive reviews describing the biological responses of plants (including maize) (Barkla et al., 2013; Sobhanian et al., 2011), roots (Zhao et al., 2013), and cell walls (Zagorchev et al., 2014) to salt stress exist.
To improve maize salt tolerance, omics tools have been applied to study the response of maize to various salt concentrations over time (Table 2). Alterations in the maize proteome in response to salt, especially in the leaves, roots, and seeds, have been studied using the powerful proteomics tool of 2-DE combined with MS/MS. The chloroplasts in the leaves are the main organs that respond to salt stress. The levels of salt-responsive chloroplast proteins (e.g., ferredoxin NADPH reductase, the 23 kDa polypeptide of photosystem II, and the FtsH-like protein) were found to increase, possibly attenuating the detrimental effects of Na+ on the photosynthetic machinery (Zörb et al., 2009). In particular, some proteins, including the ATP synthase CF1 epsilon chain and a Ca2+-sensing receptor, showed rather transient responses under salt stress (Zörb et al., 2009), as was the case in the roots, where calmodulin was found to be involved in the initial response after the adjustment to the salt conditions (Zörb et al., 2010). The Ca2+-sensing receptor and calmodulin could be members of the Ca2+/calmodulin signaling pathway, which is involved in mediating transient salt stress responses in maize.
Table 2.
Summary of Recent Years' Publications on Salt Response of Maize
| Maize genotype | Treatment | Tissue | Technique | Target | Differentially expressed proteins/genes | Primary results | Reference |
|---|---|---|---|---|---|---|---|
| R12 (salt-resistant hybrid) | 25 mM; 1, 2, 4 h | Chloroplasts | 2-DE, MALDI-TOF/TOF | Total proteins | 20 | Na+ accumulates quickly and excessively in maize chloroplasts under moderate short-term salt stress. A defined set of specific chloroplast proteins in maize change instantaneously in response to salt stress. | (Zörb et al., 2009) |
| SR12 | 25 mM; 1 h | Roots | 2-DE, MALDI-TOF/TOF | Total proteins; phosphoproteins; dephosphoproteins | 14; 10; 6 | This study provides new insights for the initial reaction of the proteome and phosphoproteome of maize roots after adjustment to saline conditions and reveals early members of sugar signalling and cell signalling pathways, e.g. calmodulin, 14-3-3 proteins. | (Zörb et al., 2010) |
| BeiDan1 (cultivar, China) | 0.1 M, 7 d; 0.2 M, 7 d | Embryos | 2-DE-MALDI-TOF, 2-DE-MALDI-TOF/TOF | Total proteins | 51; 79 | Identified proteins are mainly involved in seed storage, energy metabolism, stress response, and protein metabolism. Notably, the expression of proteins that respond to ABA signals increased in response to salt stress. | (Meng et al., 2014) |
| Nongda108 (cultivar, China) | 150 mM; 24 h | Leaves | PVA-phosphate-Ti4+ IMAC, ESI-Q-TOF | Phosphoproteins; dephosphoproteins | 47 | Cell signaling pathway members (e.g. calmodulin and 14-3-3 proteins) are regulated in response to 24-h salt stress, multiple putative salt-responsive phosphoproteins seem to be involved in the regulation of photosynthesis-related processes. | (Hu et al., 2013) |
| NC286 (salt-tolerant inbred line) Huangzao4 (salt-sensitive inbred lines) |
200 mM; 0.5, 5, 24 h | Roots | miRNA microarray, qRT-PCR | miRNA | 98 | Salt-responsive miRNAs are involved in the regulation of metabolic, morphological and physiological adaptations of maize seedlings at the post-transcriptional level. | (Ding et al., 2009) |
| YQ7-96 (inbred line) | 100 mM; 72 h | Leaves and roots | cDNA microarray hybridization, qRT-PCR | Total RNA | 296 | The salt-induced genes are related to catalytic activity, nucleic acid binding, protein binding and structural molecule activity. | (Qing et al., 2009) |
| A cultivar from Zeneca Agrochemicals | 50 or 150 mM; 2, 4 and 6 d | Roots and shoots | NMR spectroscopy | Metabolites | - | The levels of alanine, glutamate, asparagine, glycine-betaine and sucrose are increased and malic acid, trans-aconitic acid and glucose are decreased saline treated shoots. | (Gavaghan et al., 2011) |
High concentrations of salt inhibit shoot growth in salt-sensitive maize varieties (Geilfus et al., 2010). Notably, β-expansin, which acts as a softening factor on the cell wall, was found to accumulate in low abundance in the shoots under salt stress and positively correlated with reduced shoot growth. This decrease may be induced by the downregulation of ZmExpB2, ZmExpB6, and ZmExpB8 transcripts. In shoots of salt-tolerant maize, however, the aforementioned transcripts were upregulated, sustaining the stable expression of β-expansin, which may be necessary for the maintenance of shoot growth under salt stress. β-Expansin responded similarly in leaves and shoots (Geilfus et al., 2011). Moreover, the levels of sucrose synthase 1, cytosolic 3-phosphoglycerate kinase, fructokinase 2, and voltage-dependent anion channel protein were found to be increased in the root proteome and are involved in glucose metabolism, phosphorylation, fructose activation, and metabolism transport, respectively (Zörb et al., 2010). Recently, Meng et al. (2014) monitored the alterations in seed proteomes when the seeds were treated with NaCl. Most of the differentially expressed salt-responsive-proteins in seeds were primarily involved in seed storage, energy metabolism, stress response, and protein metabolism. Most of these alterations resulted in delays in storage-reversal catabolism and in the accumulation of ATP, negatively affecting germination (delay or death). However, the stress-responsive proteins were changed, likely protecting metabolic pathways from salt stress. On the whole, these proteomic studies provide important clues to how NaCl is a stressor on the physiology of leaves, roots, and seeds, and they reveal potential molecules involved in salt-responsive pathways.
Protein phosphorylation plays an important role in plant responses to salt stress (Kersten et al., 2006). Phosphoproteomic analysis of maize roots exposed to NaCl stress has revealed several phosphorylated or dephosphorylated proteins, as detected by 2-DE and fluorescence staining with PhosTag (Zörb et al., 2010). The levels of phosphorylated maize proteins, including fructokinase, UDP-glucosyl transferase BX9, and 2-Cys-peroxyredoxine, were increased, whereas isocitrate-dehydrogenase, calmodulin, maturase, and 40-S-ribosomal protein were dephosphorylated after adjusting to salt conditions. Xyloglucan endotransglycosylase (XET), a cell wall growth regulator, was phosphorylated immediately after the adjustment to salt stress. XET is responsible for cleaving and rejoining intermicrofibrillar xyloglucan chains, enabling the cell wall loosening that is required for cell expansion. This enzyme is thought to be responsible for growth inhibition under salt stress.
14-3-3 proteins play important roles in stress responses, including signaling, transcriptional activation, and defense. 14-3-3 protein levels were found to decrease in roots (Zörb et al., 2010) and to be phosphorylated in leaves (Hu et al., 2013) under salt stress. The changes in 14-3-3 proteins may affect the plasma membrane ATPase activity and lead to a change in the apoplastic pH in response to phosphorylation. Another salt-responsive phosphoproteomic analysis has been performed in maize leaves using Ti4+-IMAC enrichment and ESI-Q-TOF MS (Hu et al., 2013). Seven novel phosphoproteins were identified: phototropin-1 (a translationally controlled tumor protein homolog), rad23 (a DNA repair protein), protein phosphatase inhibitor 2-containing protein, pyruvate orthophosphate dikinase, p-pyruvate carboxylase, and dehydrin, which seem to be involved in the regulation of photosynthesis-related processes. Phosphoproteomic analysis provides new insight into how maize proteins contribute to resistance to salt stress, possibly through post-translational modifications that allow them to receive and conduct signals.
The transcriptional profiles of maize leaves and roots under salt stress have been analyzed (Qing et al., 2009). In total, 296 genes were found to be regulated specifically by salt stress in leaves and roots. The primary effects of salt stress on cellular function included catalytic activity, nucleic acid binding, protein binding, and structural molecule activity (Qing et al., 2009). In addition, the genes whose levels clearly changed under salt stress or that were specific were selected for future analysis. Several genes, such as ZmCOI6.1 (Guerra-Peraza et al., 2009), mitogen-activated protein kinase 3 (Wang et al., 2010), FK506-binding protein genes (Yu et al., 2012), mitogen-activated protein kinase kinase 1 (Cai et al., 2014) and 4 (Kong et al., 2011), and protein phosphatase 2C (Tan, 2010), were shown to be involved in maize salt tolerance.
To determine the role of miRNAs in maize responses to salt stress, an inbred salt-tolerant maize line and a salt-sensitive maize line were used to identify salt stress-responsive miRNAs by using microarray hybridization (Ding et al., 2009). The results revealed that 98 miRNAs from 27 plant miRNA families had significantly altered expression after salt treatment. Of these miRNAs, 18 were expressed only in the salt-tolerant maize line, and 25 showed a delayed regulation pattern in the salt-sensitive line.
NMR-based metabolomics have also been applied to investigate the maize response to salt stress (Gavaghan et al., 2011). The metabolic effect of high salinity was found to be consistent with that of osmotic stress and was stronger in the shoots than the roots.
Mechanical wounding
Mechanical wounding is an abiotic stress caused by processes such as wind, rain, and hail and is the first step in both pathogen infection and herbivore attack. A localized injury can stimulate the metabolism and activate signal transduction pathways throughout the plant for defense and for recovery, not only in the damaged tissues but also in non-wounded areas (Schilmiller and Howe, 2005). Phosphoproteomic methods and molecular techniques have been used to determine the response of maize at wounding sites, as well as the signal transduction process from the wounding site to other regions.
The changes in the phosphoproteome of maize leaves wounded by abrading were analyzed using multiplex staining of high-resolution 2-DE gels for protein (SYPRO Ruby) and phosphorylation (Pro-Q Diamond) for quantifying changes in phosphorylation stoichiometry (Lewandowska-Gnatowska et al., 2011). Most of the identified phosphorylated proteins, such as PEP carboxykinase, pyruvate Pi dikinase, and 14-3-3 scaffold protein, are involved in mechanical wounding. Ribosome inactivating protein (RIP) and allene oxide synthase (AOS) were locally induced by mechanical wounding (Engelberth et al., 2012), and RIP2 accumulated in the leaves of an insect-resistant inbred maize line upon caterpillar attack (Chuang et al., 2014). When the fall armyworm (Spodoptera frugiperda) attacked maize leaves, RIP, AOS, and other genes were differentially expressed in the roots (Ankala et al., 2013). Therefore, the signaling response to foliar herbivory can be conveyed to the roots from leaves. When maize leaves were touched, the activity of maize calcium-dependent protein kinases increased, and the expression of ZmCPK11 was induced (Szczegielniak et al., 2012).
Waterlogging
Waterlogging, caused by flooding, long periods of rain, and poor drainage, is a severe abiotic stress that affects crop yields worldwide (Visser et al., 2003). In India, 25%–30% of maize production is lost annually as a result of waterlogging (Zaidi et al., 2010). A primary feature of waterlogging is oxygen depletion, which mainly affects the plant roots (which are underground). The slow diffusion of oxygen and its rapid consumption result in insufficient oxygen supply to plant roots, and this supply is a priority for plant survival.
Although many studies of the molecular mechanism of plant (e.g., Arabidopsis) tolerance to waterlogging (e.g., in Arabidopsis) have been reported, relatively few studies on proteome and metabolome changes in maize have been conducted in recent years. There are only a few studies on maize root waterlogging at the transcriptomic and miRNA levels (Table 3). During the early stages of waterlogging, maize senses the lack of oxygen around the root system, which triggers initial changes in gene expression. The characterization of miRNAs in response to short-term waterlogging conditions in the roots of tolerant, semi-tolerant, and sensitive inbred maize lines (Liu et al., 2012) revealed that miRNAs exhibited differential expression as a result of signal transduction pathways. miRNAs such as miR159, miR164, miR167, miR393, miR408, and miR528 were found to be key regulators in post-transcriptional regulatory mechanisms and to be primarily involved in root development and stress responses.
Table 3.
Summary of Recent Years' Publications on Mechanical Wounding, Waterlogging, High and Low Temperatures, UV-B, Nutrient Deficiency, and Metal Toxicity Stresses Response of Maize
| Maize genotype | Treatment | Tissue | Technique | Related proteins or genes | Reference |
|---|---|---|---|---|---|
| Mechanical wounding | |||||
| B73 (inbred line) | Abrade lamina with sand paper | Leaves from 2-week-old seedlings | 2-DE, Pro-Q Diamond staining | HSP81, 14-3-3-like protein, PEP carboxylase, pyruvate Pi dikinase, Chlorophyll a–b binding protein, Dual specificity protein phosphatase, ADP-glucose pyrophosphorylase, etc. | (Lewandowsk a-Gnatowska et al., 2011) |
| A cultivar from USA | Scratch leaves and then add the elicitor to the wound | Leaves form 2- or 3-week-old seedlings | Semi-quantitative PCR | Allene oxide synthase (AOS), transcription factor MYC7, ribosome inactivating protein (RIP), etc. | (Engelberth et al., 2012) |
| Mp708 (inbred line) | The whorls of 4- or 5-week-old seedlings infested by fall armyworm | Roots and whorls | cDNA synthesis, qRT-PCR | LOX1, LOX2, AOS, AOC, 12-OPR, ASK2, etc. | (Ankala et al., 2013) |
| Waterlogging | |||||
| HZ32 (tolerant inbred line) | Submerged 3-leaf-stage seedlings in water with all leaves in air | Roots treated for (12 h, 16 h, 20 h and 24 h) | Suppression subtractive hybridization | Genes involved in complex pathways, such as signal transduction, protein degradation, ion transport, carbon and amino acid metabolism, and transcriptional and translational regulation | (Zou et al., 2010) |
| HZ32 (tolerant inbred line); B73 (mid-tolerant); Mo17 (sensitive inbred line) | Submerge 2-week-old maize to a container that filled with water 2–3 cm above the sand surface | Roots | qRT-PCR | miR159, miR164, miR167, miR393, miR408, miR528, etc. | (Liu et al., 2012) |
| HKI1105 (tolerant inbred line); V372 (sensitive inbred line) | Maintain 5 cm of standing water | Roots on the 42th day after sowing | RNA hybridization, microarray | Transcription factors ERFs, MYB, HSPs, MAPK, LOB-domain protein, etc. | (Thirunavukkarasu et al., 2013) |
| High and low temperatures | |||||
| Zhengdan958 (cultivar, China) | Raise temperature from 28 to 42°C at 2°C/h interval and then kept at 42°C for 1 h, for a total of 8 h | Expanding 2nd leaves from the top | 2-DE, MS | sHSP17.4, sHSP17.2, sHSP26, guanine nucleotide-binding protein b-subunit-like protein, putative uncharacterized protein, etc. | (Hu et al., 2010) |
| UV-B | |||||
| W23 (inbred line) | UV-B-irradiate maize plants for 10 min, 30 min, 1, 2 or 6 h | Treated leaves and immature ears | Microarray experiments; GC-MS | - | (Casati et al., 2011a) |
| W23 | The topmost leaves received 1, 2, 4, 6 h UV-B exposure | Leaves and ears | Microarray experiments; 2-DE, MS; GC-MS | - | (Casati et al., 2011b) |
| W23 | Two adult leaves per plant irradiated with UV-B during 4 h | Irradiated and shielded leaves from field or greenhouse maize plants | Microarray experiments; 2-DE, MS; GC-MS | - | (Casati et al., 2011c) |
| Nutrient deficiency | |||||
| T250 (inbred line) | N starvation for 17 d | Roots and leaves | 2-DE, LC-ESI-MS/MS | Enzymes involved in nitrate assimilation and in metabolic pathways (in roots); proteins related to regulation of photosynthesis (in leaves). | (Prinsi et al., 2009) |
| Ye478 (inbred line) | 0.04 mM nitrate | Leaves and roots from15-d-old seedlings | Microarray hybridization, RT-PCR | miR164, miR169, miR172, miR397, miR398, miR399, miR408, miR528, and miR827 (in leaves); miR160, miR167, miR168, miR169, miR319, miR395, miR399, miR408, and miR528 (in roots), etc. | (Xu et al., 2011) |
| B73 | 2-week-old seedlings were supplied with N-free nutrient solutions for 2 d | Shoots and roots | RT-PCR | miR169, miR171, miR398, miR395, miR827, miRC1, miR528, miRC19, miRC37, etc. | (Zhao et al., 2012) |
| A cultivar from Hannover, Germany | 0, 100 or 500 μM Fe-EDTA | Roots of 18-d-old seedlings | LC-MS/MS | Proteins related to signaling proteins, PM-bound redox proteins, redox signaling and redox homeostasis, etc. | (Hopff et al., 2013) |
| Zheng58 (inbred line) | Seedlings cultured in Hoagland's solution without 20 μM Fe-EDTA for 1, 2, 4 and 7 d | Roots | qRT-PCR | Genes involved in 2′-deoxy-mugineic acid (DMA) synthesis, secretion, Fe(III)–DMA uptake, plant hormones, protein kinases, protein phosphatases | (Li et al., 2014) |
| Metal toxicity | |||||
| Zheng58 | 300 mg/L potassium dichromate for 24 h | The second and third leaves after 1, 6 and 24 h | 2-DE, MS/MS | Proteins involved in ROS detoxification and defense Responses, photosynthesis and chloroplast organization, post-transcriptional processing of mRNA and rRNA, protein synthesis and folding, DNA damage response, cytoskeleton, etc. | (Wang et al., 2013) |
| 178 (inbred line) | 3 mM Pb(NO3)2 | Roots | Library construction and Solexa sequencing | Gene related to related to cellular processes and signaling, information storage and processing or metabolism functions, etc. | (Shen et al., 2013) |
However, during the later stages of waterlogging, signal transduction pathways that provide morphological and metabolic adaptations have been activated. Zou et al. (2010) characterized maize roots transcriptomes at a late stage of waterlogging. Numerous genes were found to be upregulated by waterlogging, and these genes were mainly involved in protein degradation, carbon metabolism, amino acid metabolism, oxidation-reduction, secondary metabolism, energy, signal transduction, stress induction, transcriptional regulation, translation regulation, and transporter facilitation for adaptation to waterlogging. The crosstalk between carbon and amino acid metabolism reveals that amino acid metabolism performs two main roles at this late stage: regulation of cytoplasmic pH, and supply of energy through breakdown of the carbon skeleton. Another study analyzed the genome-wide expression of maize root transcriptomes (Thirunavukkarasu et al., 2013). This study found that related genes involved in ethylene and auxin synthesis, cell wall metabolism, G-protein activation, and aerenchyma and adventitious root formation were upregulated in the tolerant maize genotype HKI 1105 under waterlogging stress.
The aforementioned recent experiments indicate that there is still a shortage of proteomic and metabolomic studies of maize, and of cross-talk through omics, in response to waterlogging stress. In addition, roots are the first organ to respond to waterlogging. Do other tissues (e.g., leaves) receive the signal at the same time? When and how does the signal move from roots to other tissues? What alterations can be induced? These scientific problems need to be further studied to fully understand the mechanism of the maize response to waterlogging stress.
High and low temperatures
Global warming will cause frequent extreme temperatures, resulting in more extreme hot and cold days (Nguyen et al., 2009). High or low temperatures could affect the germination, seedling growth, and productivity of maize. High temperature stress may be exacerbated in northern China (Piao et al., 2010), and in the United States, maize yields decrease sharply when the plants are exposed to temperatures greater than approximately 29°–30°C (Schlenker and Roberts, 2009). Perhaps different crop growing strategies or selective crop breeding can effectively guide adaptation to future changes (Hawkins et al., 2013). Currently, there are only a few omics reports on the response of maize to extreme temperatures (Table 3). Maize tolerance to heat stress has been studied in leaves by using a proteomics approach: 2-DE combined with MS (Hu et al., 2010a). In response to heat stress, maize was found to synthesize sHSPs, such as sHSP17.4, sHSP17.2, and sHSP26, throughout the plant. Clearly, sHSPs play an important role in thermotolerance. In addition, other heat-resistant proteins, including LEA D34, HSP1, ubiquitin and alcohol dehydrogenase, were found to be induced earlier in heat-tolerant maize lines (Andrade et al., 2012). Conversely, ZmCOI6.1, ZmACA1, ZmDREB2A, ZmERF3 (Nguyen et al., 2009), ZmPP2C2 (Hu et al., 2010b), ZmMKK1 (Cai et al., 2014), and ZmMKK4 (Kong et al., 2011) were induced by low temperatures. ZmFKBP (Yu et al., 2012) responded to both heat and cold stresses.
UV-B
Excess ultraviolet (UV-B) affects crop yields (Casati, 2011b). Under normal solar fluence, UV-B damage to macromolecules is balanced by subsequent repair or replacement within plants. However, under conditions of ozone depletion, terrestrial plants are exposed to periodic but unpredictable spikes in UV-B. The protective ability of the ozone shield is gradually declining; thus, sessile plants (e.g., maize), particularly high altitude landraces, must develop strong tolerance to high UV-B fluences, similar to their acclimation to normal fluence (Casati et al., 2011a).
Omics methods have recently been used to analyze UV-B-mediated signaling in maize (Casati et al., 2011a, b, c, Table 3). UV-B exposure was found to induce changes in transcripts, proteins, and metabolites in irradiated leaves and in shielded tissues of maize. During the first few minutes of exposure, the number of UV-B-regulated transcripts rapidly increased with exposure length, and the transcript diversity dramatically decreased, which indicated the susceptibility of maize to short-term UV-B exposure. These early events in all tissues may be elicited by common signaling pathways. After the first few minutes of exposure, overlapping transcriptome changes occur in the irradiated and shielded organs, and these become significantly different with exposure length. However, after 6 h of UV-B exposure, most transcripts are specific to each tissue. For example, some phenylpropanoid pathway genes were expressed only in irradiated leaves. Therefore, the responses became more organ-specific at longer exposure times (Casati et al., 2011a, b).
The proteomes of shielded and irradiated maize organs were also analyzed, and most of the detected protein changes occurred quickly under UV-B exposure. The proteins with altered levels primarily included photosynthetic proteins, transcriptional regulators and proteins involved in signal transduction, protein synthesis, and secondary metabolism (Casati et al., 2011b). Metabolic profiling identified several metabolites that rapidly increased in irradiated leaves and shielded organs after UV-B exposure, and the pathways associated with the synthesis, sequestration, or degradation of some of these potential signal molecules were UV-B-responsive. Importantly, myoinositol is a candidate UV-B responsive metabolite because of the rapid modulation of its levels in all organs (Casati et al., 2011a, b).
Most of the experiments were conducted using maize plants grown in greenhouses. However, these conditions do not occur in the natural environment, where plants are constantly exposed to UV-B from solar radiation and consequently become acclimated to UV-B. The responses to UV-B of various organs of maize in the greenhouse and in the field have been compared. Greenhouse and field plants have substantially different responses to UV-B exposure, as shown by differences in the transcriptomes, proteomes, and metabolomes. The majority of transcripts, proteins, and metabolites affected by UV-B exposure were distinct to either field or greenhouse conditions. Prior acclimation to UV-B of maize plants in the field resulted in less transcript and protein downregulation and fewer metabolite changes (Casati et al., 2011c).
Nutrient deficiency
Plants require at least 14 essential mineral nutrients to complete their life cycle. In natural soils, the availability of most essential mineral nutrients is extremely low and does not meet the demands of plants. Nitrogen (N) is a macronutrient for plant growth and is the essential constituent of many important primary and secondary organic compounds in plants, such as proteins, nucleic acids, and chlorophyll (Xu et al., 2012). N deficiency can severely inhibit maize growth, subsequently reducing maize yield (Liang et al., 2013).
A proteomic analysis of maize roots and leaves in response to N deficiency has been performed using 2-DE and LC-ESI-MS/MS (Prinsi et al., 2009). In leaves, TaWIN2, methionine synthase protein, oxygen-evolving enhancer protein 2, ATP synthase subunit alpha, and 23 kDa polypeptide of photosystem II were found to contribute to maize acclimation to N deficiency. The levels of many proteins altered in maize roots are enzymes involved in nitrate assimilation and are components of metabolic pathways implicated in the balance of the energy and redox status of the cell. By contrast, most altered proteins in abundance in maize leaves were involved in photosynthesis regulation. The nutritional status of the plant may affect two post-translational modifications of phosphoenolpyruvate carboxylase, monoubiquitination and phosphorylation in roots and leaves, respectively.
A genome-wide analysis of miRNAs that respond to chronic, transient low N availability in maize has been performed (Xu et al., 2011). miR164, miR169, miR172, miR397-399, miR408, miR528, and miR827 in leaves, and miR160, miR167-169, miR319, miR395, miR399, miR408, and miR528 in roots were detected in response to N-limiting conditions. These miRNAs are mainly involved in the gene expression regulation, signal transduction, energy metabolism, oxidative species scavenging, and encoding the RNA slicer enzyme. Some of the miRNAs, such as miR169, miR398, miR408, miR528, miR827, and miR395, detected above, such as miR169 and miR395, were subsequently demonstrated to respond to N-deficient conditions in maize roots (Zhao et al., 2012).
Iron (Fe) is a micronutrient required for maize growth. However, owing to its low solubility in aerobic and neutral pH environments, Fe availability in soils is extremely low. Proteomic analysis revealed that under low and high Fe conditions, many proteins change in abundance in the plasma membranes of maize roots. These proteins are mainly transport proteins, signaling proteins, membrane trafficking proteins, stress-related proteins, redox proteins, metabolism proteins, cell wall-related proteins, cytoskeleton, and protein folding proteins (Hopff et al., 2013). Changes in the transcriptomic profiles of maize roots in response to Fe-deficiency stress were also monitored (Li et al., 2014). Genes involved in 2-deoxy-mugineic acid (DMA) synthesis, secretion and Fe (III)-DMA uptake were significantly induced. Many genes genetically related to plant hormones, protein kinases, and protein phosphatases responded to Fe deficiency stress. As a result, maize responded to Fe deficiency stress in different ways at the proteomic and transcriptomic levels.
Metal toxicity
Heavy metal contamination, primarily induced by industrial pollution, urban activities, and agricultural practices (Pilon-Smits, 2005), negatively affects the growth and development of maize. Excess heavy metal concentrations, such as Cr, Pb, and Al, can impair plant growth and productivity.
A proteomic study of the effect of short-term hexavalent Cr exposure on maize leaves (Wang et al., 2013) has shown that the most differentially expressed proteins are primarily involved in ROS detoxification and defense responses, photosynthesis and chloroplast organization, post-transcriptional processing of mRNA and rRNA, protein synthesis and folding, DNA damage response, and the cytoskeleton. Of these Cr stress-responsive proteins, some were shown to correlate with abiotic stress responses, including ATP sulfurylase (Gao et al., 2009), aspartate aminotransferase (Rocha et al., 2010) and chloroplastic thiamine thiazole synthase 1 and 2 (Rapala-Kozik et al., 2012).
A genome expression profile analysis of maize roots has also revealed important transcripts in the response to Pb stress (Shen et al., 2013). Under Pb stress, more than 4000 genes were found to be differentially regulated. These gene products were associated with cellular processes and signaling, information storage, and processing or metabolism. Several pathways, including ribosome, photosynthesis, and carbon fixation were affected, with the ribosome pathway being significantly upregulated, although the meaning of this result is unclear. Association and linkage analyses (Krill et al., 2010) showed that four genes may contributed to Al tolerance in maize: Zea mays AltSB-like (ZmASL), Zea mays aluminum-activated malate transporter2 (ALMT2), S-adenosyl-L-homocysteinase (SAHH), and malic enzyme (ME). These genes are promising candidates for future biochemical and physiological studies on the mechanisms of Al tolerance in maize.
Maize Response to Biotic Stresses
Biotic stress, commonly induced by diseases or by insect pests, is a primary factor in maize yield losses (Lodha et al., 2013). The most prevalent maize diseases are northern leaf blight, ear rot, maize rough dwarf disease, sugarcane mosaic disease, and aflatoxin contamination. Maize is also plagued by pests, including European, Mediterranean, and tropical corn borers and the storage pest maize weevil. The hemibiotrophic fungus Colletotrichum graminicola, which induces maize anthracnose, is responsible for annual losses of up to 1000 million dollars in the USA (Balmer et al., 2013; Frey et al., 2011). Lepidopteran stalk boring larvae cause significant economic losses in maize production worldwide. One of the primary corn borer pests is ECB. This pest reduces the maize yield not only by damaging kernels through direct feeding but also by tunneling into stalks, causing plant lodging at harvest. The maize weevil (Sitophilus zeamais Motsch.) is a destructive insect that feeds on stored maize worldwide. Subsistence farmers in tropical and subtropical agroecosystems often experience grain damage exceeding 30% during on-farm storage (Tigar et al., 1994). Recent omics studies on the maize response to pathogens are summarized in Table 4.
Table 4.
Summary of Recent Years' Publications on Maize Response to Pathogens
| Maize genotype | Disease (pathogen) | Inoculated time and tissue | Sample | Technique | Differentially expressed proteins/genes | Primary results | Reference |
|---|---|---|---|---|---|---|---|
| Northern leaf blight | |||||||
| A619Ht2 (resistant line) | Setosphaeria turcica (fungus) | 4-leaf-stage; seedlings | 4th Leaf at 72 hpia | 2-DE, MALDI-TOF | 137 | A complex regulatory network functions in interaction between maize and S. turcica. The resistance process mainly resides on directly releasing defense proteins, modulation of primary metabolism, affecting photosynthesis and carbohydrate metabolism. | (Zhang et al., 2014) |
| OH43 (inbred line) | Exserohilum turcicum (fungus) | 4- to 6-leaf stage; whorls | Whorls at 0, 1, 3, 5, 7, and 9 dpib | miRNA microarray | 5 | Five miRNAs are identified as novel miRNAs, four miRNAs (miR811, miR829, miR845 and miR408) are differentially regulated in response to E. turcicum. | (Wu et al., 2014) |
| Ear rot | |||||||
| CO441 (resistant line) B73 (susceptible line) |
Fusarium graminearum (fungus) | 11-12 DASc; ears | Kernels at 48 hpi | 2-DE, iTRAQ, MS/MS | 96 | Many proteins associated with the defense response are more abundant after infection, especially pathogenesis-related protein, chitinases, xylanase inhibitors, protease inhibitors and a class III peroxidase. | (Mohammadi et al., 2011) |
| L4637 (resistant line) L4674 (susceptible line) |
Fusarium verticillioides (fungus) | 4 DAS; silks | Kernels of 18-20% moisture | Microarray hybridization | Inoculation with F. verticillioides caused no important changes in transcriptional and metabolomic profiles in the resistant line compared with susceptible inbred, suggesting that a preformed or constitutive defense mechanism may confer resistant line an advantage against F. verticillioides infection. | (Campos-Bermudez et al., 2013) | |
| CO441 (resistant line) CO354 (susceptible line) |
Fusarium verticillioides (fungus) | 15 DAP; ears | Seeds at 48 and 96 hpi | Microarray hybridization | - | In the resistant kernels, the defense-related genes are transcribed at high levels before infection and provided basic defense against the fungus. In the susceptible kernels, the defense-related genes are induced from a basal level, responding specifically to pathogen infection. | (Alessandra et al., 2010) |
| Sugarcane mosaic disease | |||||||
| Siyi (resistant line) Mo17 (susceptible line) |
Sugarcane mosaic virus | 3-leaf-stage; the lowest two leaves | Apical leaves at 12 dpi | 2-D DIGE | 93 | SCMV-responsive proteins are mainly involved in energy and metabolism, stress and defense responses, photosynthesis, and carbon fixation. | (Wu et al.,2013) |
| Siyi, Mo17 | The lowest two leaves at 6 dpi | 2-DE, MALDI-TOF | 96 | There are overlapping and specific phytohormone responses to SCMV infection between resistant and susceptible maize genotypes. | (Wu et al., 2013) | ||
| Maize rough dwarf disease | |||||||
| Ye478 (susceptible line) | Rice black streaked dwarf virus | 2-leaf-stage; seedlings | 15th leaf with typical symptoms | 2-DE | 91 | MRDD, a complicated disease controlled by multigene participating in different pathways, results in dramatic changes in the fundamental metabolism and eventually the significant differences in morphology and development between virus-infected and normal plants. | (Li et al., 2011) |
| Aflatoxin contamination | |||||||
| Mp313E, Mp420 (Resistant line) B73 and SC212m |
Aspergillusflavs (fungus) | 15 DAS; ears | Rachis at 6, 10 and 35 dpi | 2-D DIGE, MALDI-TOF/TOF | 91 | Resistant rachis relies on constitutive defenses, while susceptible rachis is more dependent on inducible defenses. | (Pechanova et al., 2011) |
hpi, hours post inoculation; bdpi, days post inoculation; cDAS: days after silking.
Northern leaf blight
Northern leaf blight, caused by Exserohilum turcicum (E. turcicum), is a primary fungal foliar disease that occurs in almost all maize-growing areas worldwide, particularly in cool climate regions with temperatures ranging from 20°–25°C, relative humidity from 90%–100%, and low luminosity (Wu et al., 2014). In China, yield losses have approached approximately 50% in the northern regions, where crops have suffered overwhelming E. turcicum infections (Ji et al., 2010).
A comparative proteomic study using 2-DE and MS has been conducted to explore the molecular mechanisms underlying the defense responses of the maize line A619 Ht2, which is resistant to Setosphaeria turcica race 13 (Zhang et al., 2014). Many proteins, involved in energy metabolism, protein destination and storage, and disease defense, were found to have altered levels in response to northern leaf blight. On inoculation with S. turcica, some defense-related proteins, such as β-glucosidase, SOD, polyamines oxidase, and PPIases, were upregulated, whereas photosynthesis- and metabolism-related proteins were downregulated. These results indicate that maize plants resistant to S. turcica may rely on directly releasing defense proteins, modulating primary metabolism, and affecting photosynthesis and carbohydrate metabolism.
Accumulating evidence supports the idea that miRNAs are hypersensitive to various processes. The miRNA expression patterns in maize in response to E. turcicum stress have been investigated using a plant miRNA microarray platform (Wu et al., 2014). In total, 118 miRNAs were detected, including miR530, miR811, miR829, and miR845, which had not previously been identified in maize. In addition, miR811, miR829, miR845, and miR408 were differentially regulated in response to E. turcicum infection. Stress-responsive miRNAs regulated metabolic, morphological, and physiological adaptations in maize seedlings at the post-transcriptional level. Furthermore, miR811 and miR829 conferred high degrees of resistance to E. turcicum and can therefore be used in maize breeding programs.
Ear rot and stalk rot
Ear rot and stalk rot can usually be induced by more than 20 types of mold, including Fusarium graminearum, Fusarium verticillioides, Fusarium proliferatum, Penicillium spp, Cladosporium spp, and Trichothecium spp. These diseases are present in many arable regions of the world, particularly in low-rainfall, high-humidity environments, such as the southern USA and some lowland tropics (van Egmond et al., 2007). Moldy and mycotoxin-contaminated grain not only results in yield losses but also in toxicity problems for livestock and humans. Mohammadi et al. (2011) presented a global proteomics approach to document early infection with F. graminearum of the tolerant inbred maize line B73 and the susceptible line CO441. In infected developing kernels, many proteins associated with the defense response to ear rot accumulated after infection, including pathogenesis-related-10, xylanase inhibitors, chitinases, proteinase inhibitors, and a class III peroxidase. Defense-related proteins accumulated to higher levels in the kernels of the susceptible line than in the tolerant line, which suggested that these proteins may provide a basal defense against F. graminearum infection in the susceptible line.
Another group of resistant and susceptible inbred maize lines was used to identify genes and metabolites that may be involved in the response to Fusarium (Campos-Bermudez et al., 2013). Upon inoculating maize leaves with Fusarium, gene expression data were obtained from microarray hybridizations of maize kernels. Fungal inoculation did not produce considerable changes in gene expression or metabolites in the resistant line. However, the defense-related gene levels changed in the kernels of the susceptible lines, specifically in response to the pathogen infection. These results indicate that resistance may be primarily due to constitutive defense mechanisms that prevent fungal infection (Campos-Bermudez et al., 2013). A similar result was obtained by studying resistant and susceptible maize genotypes (Alessandra et al., 2010). The assayed defense-related genes were found to be transcribed at high levels in the resistant line before infection, whereas these genes were expressed at basal levels in the susceptible line.
From comparisons of the responses of the resistant and susceptible maize lines to Fusarium, it can be concluded that in the resistant lines, defense-related proteins/genes are present at higher levels than in the susceptible lines, not only during or after infection but also before infection; upon infection, the susceptible lines need to enhance their resistance through increasing the levels of defense-related proteins/genes from a basal level.
Maize rough dwarf disease
Maize rough dwarf disease (MRDD) is a destructive disease that causes great maize yield loss. MRDD is primarily caused by three pathogens: maize rough dwarf virus (MRDV), Mal de Río Cuarto virus (MRCV), and rice black-streaked dwarf virus (RBSDV) (Dovas et al., 2004). MRDV and MRCV are the primary MRDD pathogens in Europe and in South America, respectively (Dovas et al., 2004). RBSDV is considered the causal agent of MRDD in China and is transmitted by Laodelphax striatellus (Wang et al., 2003).
In recent years, only a few omics studies have been conducted on MRDD (Table 4). A comparative proteomic analysis of leaves from virus-infected and normal plants has been performed using 2-DE and MS/MS (Li et al., 2011). The proteins that were found to be differentially expressed between RBSDV-inoculated maize and control maize mostly belonged to metabolic/biochemical pathways: glycolysis, starch metabolism, and morphology. In addition, MRDD was found to increase the demands for G-proteins, antioxidant enzymes, UDP-glucosyltransferase BX9, and lipoxygenases, which may play important roles in plant responses to virus infection. MRDD is a complex disease that is controlled by multiple genes participating in different pathways.
Sugarcane mosaic disease
Plant diseases caused by viruses are a severe limiting factor for food production. Sugarcane mosaic virus (SCMV) is an important viral pathogen that has caused severe losses in grain and forage yield. A high incidence of SCMV has occurred in maize in China (Xu et al., 2008) and Argentina (Perera et al., 2008).
Changes in the protein profiles of SCMV-resistant and -susceptible maize during SCMV infection have been analyzed using a DIGE-based proteomics approach (Wu et al., 2013). Ninety-three proteins were found to be differentially expressed after virus inoculation, and these were primarily involved in energy, metabolism, stress and defense responses, photosynthesis, and carbon fixation. SCMV-responsive proteins were also identified and analyzed in the maize lines Siyi and Mo17 in response to SCMV infection by using 2-DE and MALDI-TOF-MS/MS. Most of the identified proteins were present in chloroplasts, chloroplast membranes, and the cytoplasm (Wu et al., 2013). Further study of the roles of these proteins in maize-virus interactions will be valuable.
Aflatoxin contamination
The fungal metabolite aflatoxin, which is primarily produced by Aspergillus flavus (A. flavus), is carcinogenic (Klich, 2007). Warm, humid conditions favor the growth of the A. flavus fungus, resulting in severe ear rot, whereas hot, dry weather favors high aflatoxin production. Infecting maize with A. flavus and the consequent contamination with aflatoxin are persistent and serious agricultural problems, causing disease and significant maize losses worldwide. Moreover, abiotic stresses (e.g., heat, drought) exacerbate aflatoxin contamination of maize. Enhancing host plant resistance may eliminate A. flavus infection and aflatoxin contamination.
A comparative proteomic analysis of rachises from a resistant and a susceptible maize genotype has been performed (Pechanova et al., 2011). The resistant line was found to primarily contain higher levels of abiotic stress-related proteins and phenylpropanoid metabolism related-proteins, whereas the susceptible line contained pathogenesis-related proteins. At 10 to 35 days after A. flavus infection, many stress/defense proteins were differentially expressed in rachises, revealing that the resistant rachises rely on constitutive defenses, whereas susceptible rachises are more dependent on inducible defenses.
To examine the roles of stress-related genes in aflatoxin contamination, the expression levels of 94 genes selected from previous studies were analyzed using RT-PCR in six resistant and susceptible maize lines (Jiang et al., 2011). The inbred maize lines B73 and Mo17 were found to have higher levels of aflatoxin contamination and lower levels of overall gene expression than the inbred maize lines Tex6 and A638. The creation of the Corn Fungal Resistance Associated Sequences Database (CFRAS-DB) (http://agbase.msstate.edu/) has facilitated the identification of genes that are important for aflatoxin resistance in maize and has supported queries across different datasets (Kelley et al., 2010).
Outlook and Executive Topline Points
We live in exciting times, when omics research is increasingly affecting facets of the life sciences such as agriculture, food production, and traditional medicine (Manaa et al., 2013; Mishra et al., 2013; Misra et al., 2013; Wang and Chen, 2013). Looking into the near future, abiotic and biotic stresses will, however, continue to be major challenges to sustainable maize and food production. Fully dissecting the mechanisms underlying the maize stress response, and breeding maize lines adapted to various stresses, remain challenges.
Maize stress omics is often used and focuses on identifying each stress-related molecule (e.g., proteins, genes, miRNAs). Stress proteomics focuses on the qualitative analysis and identification of differentially expressed stress-responsive proteins. Thus, the functions and mechanisms of these bio-molecules in the maize stress response must be verified using molecular techniques (e.g., loss- and gain-of function). Major genes that control the response to stresses can be candidates for transforming maize plants to enhance maize stress tolerance (Rabara et al., 2014). Moreover, the interaction network among RNA, DNA, and proteins should be constructed to elucidate the maize stress response.
The PTMs of stress-responsive proteins should be studied in maize. PTMs are important for regulating protein function, subcellular localization, and protein stability. Thus far, the only PTM that has been studied is protein phosphorylation during maize salt stress (Hu et al., 2013; Zörb et al., 2010) and mechanical wounding (Lewandowska-Gnatowska et al., 2011).
More attention should be given to the temporal and spatial system-level changes in the maize stress response. The dynamic response phases to various stresses (i.e., an initial shock phase, an acclimation phase, a maintenance phase, an exhaustion phase, and/or a recovery phase after the cessation of stress treatment) and the influence of stress at different developmental stages should be defined in terms of maize growth and final yields. Mature maize plants have seldom been used in stress omics: seedlings are commonly used, primarily because of their long growth period and large size. Exposure to stress during the mature stage will directly reduce maize yield to a considerably extent. Maize stress omics studies using mature plants (particularly at flowering time) will allow a better exploration of the maize stress response and of effect of stress on maize production.
The maize stress response should be analyzed on a cellular or subcellular level, integrated with studies on whole plants, organs, or tissues. Currently, sampling methods, biochemical procedures, and MS instruments allow scientists to perform “in depth” analysis of the bio-molecules of any tissue of interest (Longuespée et al., 2014). For example, laser microdissection can isolate specific cell types of interest from sectioned specimens of heterogeneous tissues under direct microscopic visualization with the assistance of a laser beam (Suwabe et al., 2008). Cellular omics, especially proteomics, plays an essential role in determining the functions of cellular compartments and the mechanisms underlying protein/gene targeting and trafficking.
Moreover, the methodology of maize stress omics studies has room for improvement. For example, most maize stress proteomics studies are gel-based, and the protein detection methods use CBB. However, one disadvantage of 2-DE is its low resolution, particularly for membrane proteins. Therefore, gel-free methods with increased sensitivity, such as iTRAQ, should be introduced into maize stress proteomics studies.
In addition, in current maize stress proteomics studies, many local inbred lines and native cultivars are used. These maize genotypes differ greatly in their genetic backgrounds; thus, explaining the results is complex. Determining the mechanisms underlying the maize stress response will be facilitated by using a mutant line (resistant or sensitive) and the corresponding wild type.
Finally, rapid advances in high-throughput omics technologies, such as proteomics, transcriptomics, genomics, and metabolomics, make it possible to use a systems biology approach to understand plant responses to abiotic stress. Recent emerging studies of nitrogen metabolism in crop plants, published in October 2014, further attest to the role that will be played by systems sciences and integrated omics approaches in understanding the maize biology and its diverse responses to a complex environment (Fukushima and Kusano, 2014; Simons et al., 2014). The availability of these novel methodologies will likely accelerate our understanding of the maize stress response(s) at the molecular, metabolic, and physiological levels.
Acknowledgments
Work in our laboratory was supported by the National Natural Science Foundation of China (Grant no. 31371543, 31100200 and 31171470) and by Plan for Scientific Innovation Talent of Henan Province (Grant No. 144200510012).
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Ahuja I, de Vos RC, and Bones AM. (2010). Plant molecular stress responses face climate change. Trends Plant Sci 15, 664–674 [DOI] [PubMed] [Google Scholar]
- Alessandra L, Luca P, and Adriano M. (2010). Differential gene expression in kernels and silks of maize lines with contrasting levels of ear rot resistance after Fusarium verticillioides infection. J Plant Physiol 167, 1398–1406 [DOI] [PubMed] [Google Scholar]
- Andrade T, Von Pinho EV, Von Pinho RG, et al. (2012). Physiological quality and gene expression related to heat-resistant proteins at different stages of development of maize seeds. Genet Mol Res 12, 3630–3642 [DOI] [PubMed] [Google Scholar]
- Ankala A, Kelley RY, Rowe DE, et al. (2013). Foliar herbivory triggers local and long distance defense responses in maize. Plant Sci 199, 103–112 [DOI] [PubMed] [Google Scholar]
- Arbona V, Manzi MD, and Gómez-Cadenas A. (2013). Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int J Mol Sci 14, 4885–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrun A, Jensen CR, Asch F, and Mogensen VO. (2002). Drought-induced changes in xylem pH, ionic composition and ABA concentration act as early signals in field-grown maize (Zea mays L.). J Exp Bot 53, 251–263 [DOI] [PubMed] [Google Scholar]
- Balmer D, Flors V, Glauser G, and Mauch-Mani B. (2013). Metabolomics of cereals under biotic stress: Current knowledge and techniques. Front Plant Sci 4, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, and Pantoja O. (2013). Progress and challenges for abiotic stress proteomics of crop plants. Proteomics 13, 1801–1815 [DOI] [PubMed] [Google Scholar]
- Basu SK, Dutta M, and Goyal A. (2010). Is genetically modified crop the answer for the next green revolution? GM Crops 1, 68–79 [DOI] [PubMed] [Google Scholar]
- Benesova M, Hola D, Fischer L, et al. (2012). The physiology and proteomics of drought tolerance in maize: Early stomatal closure as a cause of lower tolerance to short-term dehydration? PLoS ONE 7, e38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme L, Benoît V, Tardieu F, and Zivy M. (2012). Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves. Mol Cell Proteomics 11, 957–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Wang G, Wang L, et al. (2014). ZmMKK1, a novel group A mitogen-activated protein kinase kinase gene in maize, conferred chilling stress tolerance and was involved in pathogen defense in transgenic tobacco. Plant Sci 214, 57–73 [DOI] [PubMed] [Google Scholar]
- Cai G, Wang GD, Wang L, Liu Y, Pan JW, and Li DQ. (2014). A maize mitogen-activated protein kinase kinase, ZmMKK1, positively regulated the salt and drought tolerance in transgenic Arabidopsis. J Plant Physiol 171, 1003–1016 [DOI] [PubMed] [Google Scholar]
- Campos-Bermudez VA, Fauguel CM, Tronconi MA, et al. (2013). Transcriptional and metabolic changes associated to the infection by Fusarium verticillioides in maize inbreds with contrasting ear rot resistance. PLoS ONE 8, e61580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelle V, Remoué C, Moreau L, et al. (2010). QTLs and candidate genes for desiccation and abscisic acid content in maize kernels. BMC Plant Biol 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Campi M, Morrow DJ, et al. (2011b). Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics 12, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Campi M, Morrow DJ, et al. (2011c). Transcriptomic, proteomic and metabolomic analysis of maize responses to UV-B. Plant Signal Behav 6, 1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Morrow DJ, Fernandes JF, and Walbot V. (2011a). UV-B signaling in maize: Transcriptomic and metabolomic studies at different irradiation times. Plant Signal Behav 6, 1926–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang WP, Herde M, Ray S, et al. (2014). Caterpillar attack triggers accumulation of the toxic maize protein RIP2. New Phytol 201, 928–939 [DOI] [PubMed] [Google Scholar]
- Cooper M, Gho C, Leafgren R, Tang T, and Messina C. (2014). Breeding drought-tolerant maize hybrids for the US corn-belt: Discovery to product. J Exp Bot, doi: 10.1093/jxb/eru064 [DOI] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, et al. (2011). Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol 11, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Zhang L, Wang H, et al. (2009). Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot 103, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovas CI, Eythymiou K, and Katis NI. (2004). First report of maize rough dwarf virus (MRDV) on maize crops in Greece. Plant Pathol 53, 238–238 [Google Scholar]
- Engelberth J, Contreras CF, and Viswanathan S. (2012). Transcriptional analysis of distant signaling induced by insect elicitors and mechanical wounding in Zea mays. PLoS ONE 7, e34855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey TJ, Weldekidan T, Colbert T, Wolters PJCC, and Hawk JA. (2011). Fitness evaluation of Rcg1, a locus that confers resistance to Colletotrichum graminicola (Ces.) G.W. Wils. using near-isogenic maize hybrids. Crop Sci 51, 1551–1563 [Google Scholar]
- Fukushima A, and Kusano M. (2014) A network perspective on nitrogen metabolism from model to crop plants using integrated 'omics' approaches. J Exp Bot 65, 5619–5630 [DOI] [PubMed] [Google Scholar]
- Gao F, Zhou Y, Zhu W, et al. (2009). Proteomic analysis of cold stress-responsive proteins in Thellungiella rosette leaves. Planta 230, 1033–1046 [DOI] [PubMed] [Google Scholar]
- Gavaghan CL, Li JV, Hadfield ST, et al. (2011). Application of NMR-based metabolomics to the investigation of salt stress in maize (Zea mays). Phytochem Anal 22, 214–224 [DOI] [PubMed] [Google Scholar]
- Geilfus CM, Neuhaus C, Mühling KH, and Zörb C. (2011). β-expansins are divergently abundant in maize cultivars that contrast in their degree of salt resistance. Plant Sign Behav 6, 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geilfus CM, Zörb C, and Mühling KH. (2010). Salt stress differentially affects growth-mediating β-expansins in resistant and sensitive maize (Zea mays L.). Plant Physiol Bioch 48, 993–998 [DOI] [PubMed] [Google Scholar]
- Guerra-Peraza O, Nguyen HT, Stamp P, and Leipner J. (2009). ZmCOI6. 1, a novel, alternatively spliced maize gene, whose transcript level changes under abiotic stress. Plant Sci 176, 783–791 [Google Scholar]
- Hawkins E, Fricker TE, Challinor AJ, et al. (2013). Increasing influence of heat stress on French maize yields from the 1960s to the 2030s. Global Change Biol 19, 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopff D, Wienkoop S, and Lüthje S. (2013). The plasma membrane proteome of maize roots grown under low and high iron conditions. J Proteomics 91, 605–618 [DOI] [PubMed] [Google Scholar]
- Hu X, Liu L, Xiao B, et al. (2010b). Enhanced tolerance to low temperature in tobacco by over-expression of a new maize protein phosphatase 2C, ZmPP2C2. J Plant Physiol 167, 1307–1315 [DOI] [PubMed] [Google Scholar]
- Hu X, Wu X, Li C, et al. (2012). Abscisic acid refines the synthesis of chloroplast proteins in maize (Zea mays) in response to drought and light. PLoS ONE 7, e49500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XL, Li YH, Li CH, et al. (2010a). Characterization of small heat shock proteins associated with maize tolerance to combined drought and heat stress. J Plant Growth Regul 29, 455–464 [Google Scholar]
- Hu XL, Lu MH, Li CH, et al. (2011). Differential expression of proteins in maize roots in response to abscisic acid and drought. Acta Physiol Plant 33, 2437–2446 [Google Scholar]
- Hu Y, Guo S, Li X, and Ren X. (2013). Comparative analysis of salt-responsive phosphoproteins in maize leaves using Ti4+–IMAC enrichment and ESI-Q-TOF MS. Electrophoresis 34, 485–492 [DOI] [PubMed] [Google Scholar]
- Huang H, Mølle IM, and Song SQ. (2012). Proteomics of desiccation tolerance during development and germination of maize embryos. J Proteomics 75, 1247–1262 [DOI] [PubMed] [Google Scholar]
- Ji W, Hen H, and Zhao S. (2010). Identification of physiological races of Setosphaeria turcica in northeast corn region of Heilongjiang. J Maize Sci 18, 128–130 [Google Scholar]
- Jiang S, Zhang D, Wang L, et al. (2013). A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol Bioch 71, 112–120 [DOI] [PubMed] [Google Scholar]
- Jiang T, Zhou B, Luo M, et al. (2011). Expression analysis of stress-related genes in kernels of different maize (Zea mays L.) inbred lines with different resistance to aflatoxin contamination. Toxins 3, 538–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumanu A, Ambavaram MM, Klumas C, et al. (2012). Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 160, 846–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RY, Gresham C, Harper J, et al. (2010). Integrated database for identifying candidate genes for Aspergillus flavus resistance in maize. BMC Bioinformatics 11, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten B, Agrawal GK, Iwahashi H, and Rakwal R. (2006). Plant phosphoproteomics: A long road ahead. Proteomics 6, 5517–5528 [DOI] [PubMed] [Google Scholar]
- Klich MA. (2007). Aspergillus flavus: The major producer of aflatoxin. Mol Plant Pathol 8, 713–722 [DOI] [PubMed] [Google Scholar]
- Kong X, Pan J, Zhang M, et al. (2011). ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant Cell Environ 34, 1291–1303 [DOI] [PubMed] [Google Scholar]
- Kosová K, Vítámvás P, and Prášil IT. (2011). Plant proteome changes under abiotic stress-contribution of proteomics studies to understanding plant stress response. J Proteomics 74, 1301–1322 [DOI] [PubMed] [Google Scholar]
- Krill AM, Kirst M, Kochian LV, et al. (2010). Association and linkage analysis of aluminum tolerance genes in maize. PLoS ONE 5, e9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge P, and Fleury D. (2011). Making the most of ‘omics’ for crop breeding. Trends Biotechnol 29, 33–40 [DOI] [PubMed] [Google Scholar]
- Lewandowska-Gnatowska E, Johnston ML, Antoine W, et al. (2011). Using multiplex-staining to study changes in the maize leaf phosphoproteome in response to mechanical wounding. Phytochemistry 72, 1285–1292 [DOI] [PubMed] [Google Scholar]
- Li HY, Huang SH, Shi YS, et al. (2009). Isolation and analysis of drought-induced genes in maize roots. Agr Sci China 8, 129–136 [Google Scholar]
- Li K, Xu C, and Zhang J. (2011). Proteome profile of maize (Zea may L.) leaf tissue at the flowering stage after long-term adjustment to rice black-streaked dwarf virus infection. Gene 485, 106–113 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang N, Zhao F, et al. (2014). Changes in the transcriptomic profiles of maize roots in response to iron-deficiency stress. Plant Mol Biol 85, 349–363 [DOI] [PubMed] [Google Scholar]
- Liang CY, Tian J, and Liao H. (2013). Proteomics dissection of plant responses to mineral nutrient deficiency. Proteomics 13, 624–636 [DOI] [PubMed] [Google Scholar]
- Liu TX, Zhang L, Yuan ZL, et al. (2013). Identification of proteins regulated by ABA in response to combined drought and heat stress in maize roots. Acta Physiol Plant 35, 501–513 [Google Scholar]
- Liu Z, Kumari S, Zhang L, Zheng Y, and Ware D. (2012). Characterization of miRNAs in response to short-term waterlogging in three inbred lines of Zea mays. PLoS ONE 7, e39786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha T, Hembram P, and Basak N. (2013). Proteomics: A successful approach to understand the molecular mechanism of plant-pathogen interaction. Am J Plant Sci 4, 1212–1226 [Google Scholar]
- Longuespée R, Fléron M, Pottier C, et al. (2014). Tissue proteomics for the next decade? Towards a molecular dimension in histology. OMICS 18, 539–552 [DOI] [PubMed] [Google Scholar]
- Luo M, Liu J, Lee RD T. S.cully B, and Guo BZ. (2010). Monitoring the expression of maize genes in developing kernels under drought stress using oligo-microarray. J Integr Plant Biol 52, 1059–1074 [DOI] [PubMed] [Google Scholar]
- Manaa A, Faurobert M, Valot B, Bouchet JP, Grasselly D, Causse M, and Ahmed HB. (2013). Effect of salinity and calcium on tomato fruit proteome. OMICS 17, 338–352 [DOI] [PubMed] [Google Scholar]
- Meng LB, Chen YB, Lu TC, et al. (2014). A systematic proteomic analysis of NaCl-stressed germinating maize seeds. Mol Biol Rep 41, 3431–3443 [DOI] [PubMed] [Google Scholar]
- Mishra M, Kanwar P, Singh A, Pandey A, Kapoor S, and Pandey GK. (2013). Plant omics: Genome-wide analysis of ABA repressor1 (ABR1) related genes in rice during abiotic stress and development. OMICS 17, 439–450 [DOI] [PubMed] [Google Scholar]
- Misra N, Panda PK, and Parida BK. (2013). Agrigenomics for microalgal biofuel production: An overview of various bioinformatics resources and recent studies to link OMICS to bioenergy and bioeconomy. OMICS 17, 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Anoop V, Gleddie S, and Harris LJ. (2011). Proteomic profiling of two maize inbreds during early gibberella ear rot infection. Proteomics 11, 3675–3684 [DOI] [PubMed] [Google Scholar]
- Munns R. (2005). Genes and salt tolerance: Bringing them together. New Phytol 167, 645–663 [DOI] [PubMed] [Google Scholar]
- Ngara R, and Ndimba BK. (2013). Understanding the complex nature of salinity and drought stress response in cereals using proteomics technologies. Proteomics 14, 611–621 [DOI] [PubMed] [Google Scholar]
- Ngara R, Ndimba R, Jensen JB, et al. (2012). Identification and profiling of salinity stress-responsive proteins in Sorghum bicolor seedlings. J Proteomics 75, 4139–4150 [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Leipner J, Stamp P, and Guerra-Peraza O. (2009). Low temperature stress in maize (Zea mays L.) induces genes involved in photosynthesis and signal transduction as studied by suppression subtractive hybridization. Plant Physiol Biochem 47, 116–122 [DOI] [PubMed] [Google Scholar]
- Pechanova O, Pechan T, Williams WP, and Luthe DS. (2011). Proteomic analysis of the maize rachis: Potential roles of constitutive and induced proteins in resistance to Aspergillus flavus infection and aflatoxin accumulation. Proteomics 11, 114–127 [DOI] [PubMed] [Google Scholar]
- Pechanova O, Takáč T, and Šamaj J. (2013). Maize proteomics: An insight into the biology of an important cereal crop. Proteomics 13, 637–662 [DOI] [PubMed] [Google Scholar]
- Perera MF, Filippone MP, Ramallo CJ, et al. (2008). Genetic diversity among viruses associated with sugarcane mosaic disease in Tucumán, Argentina. Phytopathology 99, 38–49 [DOI] [PubMed] [Google Scholar]
- Piao SL, Ciais P, Huang Y, et al. (2010). The impacts of climate change on water resources and agriculture in China. Nature 467, 43–51 [DOI] [PubMed] [Google Scholar]
- Pilon-Smits E. (2005). Phytoremediation. Annu Rev Plant Biol 56, 15–39 [DOI] [PubMed] [Google Scholar]
- Piperno DR, Ranereb AJ, Holstb I, et al. (2009). Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc Natl Acad Sci USA 106, 5019–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinsi B, Negri AS, Pesaresi P, et al. (2009). Evaluation of protein pattern changes in roots and leaves of Zea mays plants in response to nitrate availability by two-dimensional gel electrophoresis analysis. BMC Plant Biol 9, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing DJ, Lu HF, Li N, et al. (2009). Comparative profiles of gene expression in leaves and roots of maize seedlings under conditions of salt stress and the removal of salt stress. Plant Cell Physiol 50, 889–903 [DOI] [PubMed] [Google Scholar]
- Rabara RC, Tripathi P, and Rushton PJ. (2014). The potential of transcription factor-based genetic engineering in improving crop tolerance to drought. OMICS 18, 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapala-Kozik M, Wolak N, Kujda M, and Banas AK. (2012). The upregulation of thiamine (vitamin B1) biosynthesis in Arabidopsis thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response. BMC Plant Biol 12, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould A, Stacey D, and Vlachos D. (2007). Non-target organism risk assessment of MIR604 maize expressing mCry3A for control of corn root worm. J Appl Entomol 131, 391–399 [Google Scholar]
- Rocha M, Sodek L, Licausi F, et al. (2010). Analysis of alanine aminotransferase in various organs of soybean (Glycine max) and in dependence of different nitrogen fertilisers during hypoxic stress. Amino Acids 39, 1043–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, and Howe GA. (2005). Systemic signaling in the wound response. Curr Opin Plant Biol 8, 369–377 [DOI] [PubMed] [Google Scholar]
- Schlenker W, and Roberts MJ. (2009). Nonlinear temperature effects indicate severe damages to U.S. crop yields under climate change. Proc Natl Acad Sci USA 106, 15594–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, and Fulton RS. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326, 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shelden MC, and Roessner U. (2013). Advances in functional genomics for investigating salinity stress tolerance mechanisms in cereals. Front Plant Sci 4, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhang Y, Chen J, et al. (2013). Genome expression profile analysis reveals important transcripts in maize roots responding to the stress of heavy metal Pb. Physiol Plant 147, 270–282 [DOI] [PubMed] [Google Scholar]
- Simons M, Saha R, Guillard L, et al. (2014). Nitrogen-use efficiency in maize (Zea mays L.): from 'omics' studies to metabolic modelling. J Exp Bot 65, 5657–5671 [DOI] [PubMed] [Google Scholar]
- Sobhanian H, Aghaei K, and Komatsu S. (2011). Changes in the plant proteome resulting from salt stress: Toward the creation of salt-tolerant crops? J Proteomics 74, 1323–1337 [DOI] [PubMed] [Google Scholar]
- Suwabe K, Suzuki G, Takahashi H, et al. (2008). Separated transcriptomes of male gametophyte and tapetum in rice: Validity of a laser microdissection (LM) microarray. Plant Cell Physiol 49, 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczegielniak J, Borkiewicz L, Szurmak B, et al. (2012). Maize calcium-dependent protein kinase (ZmCPK11): local and systemic response to wounding, regulation by touch and components of jasmonate signaling. Physiol Plant 146, 1–14 [DOI] [PubMed] [Google Scholar]
- Tai FJ, Yuan ZL, Wu XL, et al. (2011). Identification of membrane proteins in maize leaves, altered in stress through polyethylene glycol treatment. Plant Omics 4, 250–256 [Google Scholar]
- Tan MP. (2010). Analysis of DNA methylation of maize in response to osmotic and salt stress based on methylation-sensitive amplified polymorphism. Plant Physiol Biochem 48, 21–26 [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu N, Hossain F, Mohan S, et al. (2013). Genome-wide expression of transcriptomes and their co-expression pattern in subtropical maize (Zea mays L.) under waterlogging stress. PLoS ONE 8, e70433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigar BJ, Osborne PE, Key GE, Flores-S ME, and Vazquez AM. (1994). Insect pest associated with rural maize stores in Mexico with particular reference to Prostephanus truncates (Coleoptera: bostrichidae). J Stored Prod Res 30, 267–281 [Google Scholar]
- van Egmond HP, Schothorst RC, and Jonker MA. (2007). Regulations relating to mycotoxins in food; Perspectives in a global and European context. Anal Bioanal Chem 389, 147–157 [DOI] [PubMed] [Google Scholar]
- Vinocur B, and Altman A. (2005). Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr Opin Biotech 16, 123–132 [DOI] [PubMed] [Google Scholar]
- Virlouvet L, Jacquemot M, Gerentes D, et al. (2011). The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant Physiol 157, 917–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser EJW, Voesenek LACJ, Vartapetian BB, and Jackson MB. (2003). Flooding and plant growth. Ann Bot 91, 107–109 [Google Scholar]
- Wang J, Ding H, Zhang A, et al. (2010). A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. J Integr Plant Biol 52, 442–452 [DOI] [PubMed] [Google Scholar]
- Wang P, and Chen Z. (2013). Traditional Chinese medicine ZHENG and Omics convergence: A systems approach to post-genomics medicine in a global world. OMICS 17, 451–459 [DOI] [PubMed] [Google Scholar]
- Wang R, Gao F, Guo BQ, et al. (2013). Short-term chromium-stress-induced alterations in the maize leaf proteome. Int J Mol Sci 14, 11125–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Fang SG, Xu JL, et al. (2003). Sequence analysis of the complete genome of rice black-streaked dwarf virus isolated from maize with rough dwarf disease. Virus Genes 27, 163–168 [DOI] [PubMed] [Google Scholar]
- Witt S, Galicia L, Lisec J, et al. (2012). Metabolic and phenotypic responses of greenhouse-grown maize hybrids to experimentally controlled drought stress. Mol Plant 5, 401–417 [DOI] [PubMed] [Google Scholar]
- Wu F, Shu J, and Jin W. (2014). Identification and validation of miRNAs associated with the resistance of maize (Zea mays L.) to Exserohilum turcicum. PLoS ONE 9, e87251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Han Z, Wang S, et al. (2013). Comparative proteomic analysis of the plant-virus interaction in resistant and susceptible ecotypes of maize infected with sugarcane mosaic virus. J Proteomics 89, 124–140 [DOI] [PubMed] [Google Scholar]
- Wu L, Wang S, Chen X, et al. (2013). Proteomic and phytohormone analysis of the response of maize (Zea mays L.) seedlings to sugarcane mosaic virus. PLoS ONE 8, e70295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Liu Q, Wu J, and Ding J. (2012). ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize. Gene 495, 146–153 [DOI] [PubMed] [Google Scholar]
- Xu DL, Park JW, Mirkov TE, and Zhou GH. (2008). Viruses causing mosaic disease in sugarcane and their genetic diversity in southern China. Arch Virol 153, 1031–1039 [DOI] [PubMed] [Google Scholar]
- Xu G, Fan X, and Miller AJ. (2012). Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63, 153–182 [DOI] [PubMed] [Google Scholar]
- Xu J, Yuan Y, Xu Y, et al. (2014). Identification of candidate genes for drought tolerance by whole-genome resequencing in maize. BMC Plant Boil 14, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zhong S, Li X, et al. (2011). Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 6, e28009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zhang H, Li W, et al. (2012). Genome-wide analysis and environmental response profiling of the FK506-binding protein gene family in maize (Zea mays L.). Gene 498, 212–222 [DOI] [PubMed] [Google Scholar]
- Zagorchev L, Kamenova P, and Odjakova M. (2014). The role of plant cell wall proteins in response to salt stress. Sci World J 2014, 764089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi PH, Maniselvan P, and Srivastava A. (2010). Genetic analysis of waterlogging tolerance in tropical maize (Zea mays L.). Maydica 55, 17–26 [Google Scholar]
- Zhang XL, Si BW, Fan CM, Li HJ, and Wang XM. (2014). Proteomics identification of differentially expressed leaf proteins in response to Setosphaeria turcica infection in resistant maize. J Integr Agric 13, 789–803 [Google Scholar]
- Zhao M, Tai H, Sun S, et al. (2012). Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS ONE 7, e29669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Zhang H, Wang T, Chen S, and Dai S. (2013). Proteomics-based investigation of salt-responsive mechanisms in plant roots. J Proteomics 82, 230–253 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2001). Plant salt tolerance. Trends Plant Sci 6, 66–71 [DOI] [PubMed] [Google Scholar]
- Zörb C, Herbst R, Forreiter C, and Schubert S. (2009). Short-term effects of salt exposure on the maize chloroplast protein pattern. Proteomics 9, 4209–4220 [DOI] [PubMed] [Google Scholar]
- Zörb C, Schmitt S, and Mühling KH. (2010). Proteomic changes in maize roots after short-term adjustment to saline growth conditions. Proteomics 10, 4441–4449 [DOI] [PubMed] [Google Scholar]
- Zou X, Jiang Y, Liu L, Zhang Z, and Zheng Y. (2010). Identification of transcriptome induced in roots of maize seedlings at the late stage of waterlogging. BMC Plant Biol 10, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]