Abstract
The rapid growth of published literature makes biomedical text mining increasingly invaluable for unpacking implicit knowledge hidden in unstructured text. We employed biomedical text mining and biological networks analyses to research the process of sperm egg recognition and binding (SERB). We selected from the literature the molecules expressed either on spermatozoa or on oocytes thought to be involved in SERB and, using an automated literature search software (Agilent Literature Search), we realized a network, SERBN, characterized by a hierarchical scale free and a small world topology. We used an integrated approach, either based on selection of hubs or by a cluster analysis, to discern the key molecules of SERB. We found that in most cases some of them are not directly situated on spermatozoa and oocyte, but are dispersed in oviductal fluid or embedded in exosomes present in the perivitelline space. To confirm and validate our results, we performed further analyses using STRING and Reactome FI software. Our findings underscore that the fertility is not a property of gametes in isolation, but rather depends on the functional integrity of the entire reproductive system. These observations collectively underscore the importance of integrative biology in exploring biological systems and in rethinking of fertility mechanisms in the light of this innovative approach.
Introduction
Omics is a systems science raising both promises and challenges from cell to society to bioeconomy (Akondi and Lakshmi, 2013; Bernabò et al., 2013; Birch and Tyfield, 2013; Bowler et al., 2013; Dove, 2013; Rajan, 2013; Vivar et al., 2013). A key challenge and opportunity is the rapid growth of published literature that makes biomedical text mining increasingly invaluable so as to unpack implicit knowledge hidden in unstructured text. Not surprisingly, biomedical text mining is now greatly used in medical research and integrative biology (Zhu et al., 2013).
One area where biomedical text mining offers much potential is reproductive physiology, due to its rapid growth as a field of postgenomics inquiry. It is noteworthy that more research is needed on the sperm–egg recognition and binding process (Fardilha et al., 2013), and importantly, with a view to more comprehensive understanding of the system level determinants of fertility. Mammalian spermatozoa during coitus are released in deep vagina or in uterus, depending on the species, and reach the utero-tubal junction (UTJ), where they form a reservoir. Here, the male gametes pass a window of time, ranging from hours to days, waiting for the oocyte and interaction with the oviductal epithelial cells. In this context, the spermatozoa are exposed to different gradients of either activating (i.e., bicarbonate, pH, calcium concentration, progesterone, serum proteins) or inhibiting (i.e., endocannabinoids) factors, which influence their metabolic activity and reactivity (Alasmari et al., 2013; Barboni et al., 2011; Ijiri et al., 2012; Wertheimer et al., 2013). Here, the spermatozoa undergo the process of capacitation, acquiring the full ability to fertilize. A few hours before ovulation, the intratubal concentration of progesterone markedly increases, thus activating the detachment of spermatozoa from the tubal epithelium and stimulating hyperactivated motility, necessary to reach the cumulus oophorus and to penetrate its matrix (Chang and Suarez, 2010; Olson et al., 2011; Suarez, 2008). Finally, the spermatozoa contact the external involvement of oocyte, the zona pellucida (ZP) in a process known as sperm–egg recognition and binding (SERB). The interaction between spermatozoa and cumulus oophorus and ZP causes the exocytosis of acrosome content, the so-called acrosome reaction (AR), which exposes surface antigens with release of numerous enzymes that are responsible for the ZP lysis, thus allowing spermatozoa to reach the perivitelline space where fusion between oocyte and spermatozoa membrane occurs (Gadella 2012; Okabe, 2013).
The molecular mechanism involving the interaction between spermatozoa and oocyte, the SERB process, has been extensively studied in mammals, in particular, in mouse, and some dogmas have been challenged. Indeed, until a few years ago, it was believed that acrosome-intact sperm only could pass through the cumulus oophorus and that the exocytosis of acrosome content was triggered by binding to the ZP: only acrosome-intact sperm were observed on the surface of the ZP and only acrosome reacted sperm were observed in the perivitelline space (Saling et al., 1979). This model was strengthened by evidence of vesiculated acrosomal shrouds on the zona surface (VandeVoort et al., 1997; Wakayama et al., 1996; Yanagimachi and Phillips, 1984) and by the ability of solubilized ZP or isolated mouse ZP3 glycoproteins to induce AR (Bleil and Wassarman, 1983).
In recent years, experimental evidence questioned this sequence of events. In particular, in a video microscopic in vitro fertilization experiment, it was recently found that double transgenic male mice spermatozoa, expressing enhanced green fluorescent protein (EGFP) in their acrosomes and red fluorescent protein (Ds-Red2) in their midpiece, undergo AR before reaching the ZP (Jin et al., 2011). In other words, it was shown that AR occurs during the interaction with cumulus and it is not necessary to penetrate ZP, thus confirming the role of receptors for cumulus extracellular matrix proteins observed in spermatozoa of several species (Barboni et al., 2001; Gadella, 2012; Hong et al., 2009). Furthermore, recently, the concept that the spermatozoa that have already undergone AR are unable to bind the ZP and have lost their fertilizing ability has been revamped. Indeed, Inoue and co-workers used AR spermatozoa collected from the perivitelline space of in vitro fertilized oocytes from Cd9-null mice to successfully fertilize eggs from WT mice, obtaining live offspring (Inoue et al., 2011). In addition, several molecules believed to play a key role in these SERB, once removed in KO mice, failed to exert their hypothesized role during fertilization (the KO mice were fertile).
These new data pose several intriguing question about molecules involved in triggering AR and, more broadly, about the interaction mechanism that takes place between male and female gametes (Avella and Dean, 2011). In order to explore this process, we used a computational modeling-based approach in the spirit of a systems science approach. To this aim we adopted a biological text mining and networks-based approach, which obtains hidden information from biomedical literature and brings this information in a network model whose topology could be studied, thus obtaining quantitative information. More in detail, a model that represents the molecules involved in the interaction of spermatozoa and eggs as a mathematical object called “graph” has been designed. This graph was constituted by nodes (the molecules) linked by edges (the interactions among them), thus originating a network. The topological analysis of the network allowed us to draw inferences about biological events, otherwise impossible to obtain (Bernabò et al., 2010, 2011). Finally we compared the text mining results with those obtained using Reactome Functional Interaction (FI) and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) software, to confirm and validate that approach.
Materials and methods
Realization of sperm-egg recognition and binding network (SERBN)
The molecules involved in SERB process were manually selected by examining the scientific literature available on PubMed (http://www.ncbi.nlm.nih.gov/pubmed). Particularly considered was the literature of the last 10 years that concerned the molecules involved in that process and, possibly, the data from knock-out or immunoneutralization mice experiments, when available (Table 1). For each molecule, we automatically realized a network (when it was possible) representing its interaction with the other components of the system found in literature as unweighted links by using the Cytoscape 3.0.2. App Agilent Literature Search 3.0.3 beta (LitSearch version 2.68), using the data contained in PubMed database. The checked molecules were used as “term,” and we entered sperm egg recognition and binding as “context.” Max Engine Matches was set at 1.000 (which always was higher than the number of articles found, thus in all the cases all the available information was processed), the “Use Aliases,” the “Use Context,” and the “Concept Lexicon Restrict Search” options were set. As Concept Lexicon we used “Mus musculus;” the data have been accessed until March 15, 2014. Finally, we created the Sperm-Egg Recognition and Binding Network (SERBN) by merging all the obtained networks and removing all the self-loops and the duplicated edges.
Table 1.
Proteins Involved in SERB Process and Correspondent Genes
The analysis of SERBN and of main connected component of SERBN (MC-SERBN) was carried out, as an undirected network, by the Network Analyze plug-in of Cytoscape 3.0.2 to measure the most relevant topological parameters (see Supplementary Table S1).
As a comparison, a scale free network following the Barabasì-Albert model was created (BA network, BAN) by the Cytoscape plug-in Random Networks (function: Generate Random Network) set on Barabási-Albert Model, Number of Nodes (n)=438; Minimum Edge per Node (m)=1, initial Number of Nodes (s)=3, as undirected network.
Cluster analysis
The nodes corresponding to the molecules we selected, which were also present in the MC-SERBN, were subjected to cluster analysis on the basis of their topological parameters (eccentricity, closeness centrality, clustering coefficient, neighborhood connectivity, average shortest path length, degree, radiality, stress, betweenness centrality, for explanations see Supplementary Table S2). The analysis was performed using a paired group algorithm (UPGMA) with an Euclidean similarity index (Past3).
Reactome Functional Interaction (FI) and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) experiments
With the aim of validating and completing our data, we performed two computational experiments using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) and Reactome Functional Interaction (FI) tools.
STRING is a database of known and predicted protein interactions, which includes direct (physical) and indirect (functional) associations (http://string-db.org/) (Franceschini et al., 2013). We performed a search using the same proteins used in previous experiments, and we obtained a network by converting the data as a Cytoscape file (STRING network). The network was analyzed with the Cytoscape Plugin Network Analyzer.
Reactome FI is a Cytoscape Plugin designed to find pathways and network patterns accessing the data stored in the Reactome Pathways Database (http://www.reactome.org/). (http://wiki.reactome.org/index.php/Reactome_FI_Cytoscape_Plugin_4) (Wu et al., 2010). We explored the pathways related to the Reproduction (REACT_163484.3) and we performed the Pathway Enrichment Analysis, using the gene list previously used in text mining experiments.
Results
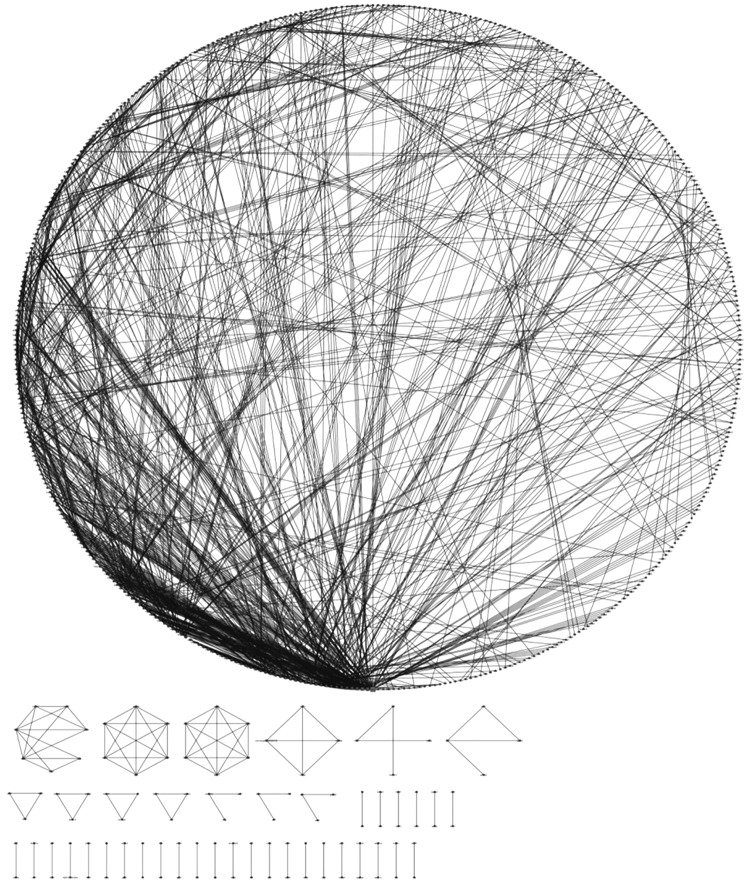
The network describing SERBN is composed by 548 nodes linked by 1356 edges, and constituted by 43 connected components: one component with 438 nodes and 1251 edges (main component of SERBN, MC-SERBN), one component with 7 nodes and 15 edges, two components with 6 nodes and 16 edges, one component with 4 nodes and 6 edges, one component with 4 nodes and 3 edges, one component with 4 nodes and 4 edges, four components with 3 nodes and 3 edges, three components with 3 nodes and 2 edges, 29 components with 2 nodes and 1 edge (Figure 1).
FIG. 1.
SERBN. It possible to distinguish MC-SERBN and other components.
The resulting analyses of SERBN and MC-SERBN topology are shown in Table 2. Interestingly, in both cases (SERBN and MC-SERBN) the node degree distribution followed an exponential law, whose exponent γ were −1.542 and −1.429 (SERBN and MC-SERBN, respectively); similarly the averaged clustering coefficient distribution had a γ exponent=−0.822 or −0.811 (SERBN and MC-SERBN, respectively) (Table 3A and 3B). The most linked nodes in MC-SERBN (the hubs) are listed in Table 4.
Table 2.
The Most Relevant Topological Parameters of SERBN and MC-SERBN
| Parameter | SERBN | MC-SERBN |
|---|---|---|
| Number of nodes | 548 | 438 |
| Number of edges | 1356 | 1251 |
| Clustering coefficient | 0.491 | 0.530 |
| Connected components | 43 | 1 |
| Network diameter | 12 | 12 |
| Network radius | 1 | 6 |
| Network centralization | 0.097 | 0.119 |
| Shortest paths | 191644 (63%) | 191406 (100%) |
| Characteristic path length | 4.405 | 4.600 |
| Avg. number of neighbors | 5.234 | 5.032 |
| Network density | 0.0008 | 0.012 |
| Network heterogeneity | 1.123 | 1.054 |
| Isolated nodes | 0 | 0 |
Table 3A.
Node Degree of SERBN MC-SERBN
| Node degree equation | SERBN | MC-SERBN |
|---|---|---|
| γ | −1.524 | −1.429 |
| r | 0.942 | 0.860 |
| R2 | 0.831 | 0.810 |
Table 3B.
Clustering Coefficient of SERBN, MC-SERBN
| Clustering coefficient vs. node degree | SERBN | MC-SERBN |
|---|---|---|
| γ | −0.822 | −0.811 |
| r | 0.644 | 0.640 |
| R2 | 0.618 | 0.610 |
Table 4.
The Most Connected Nodes in MC-SERBN
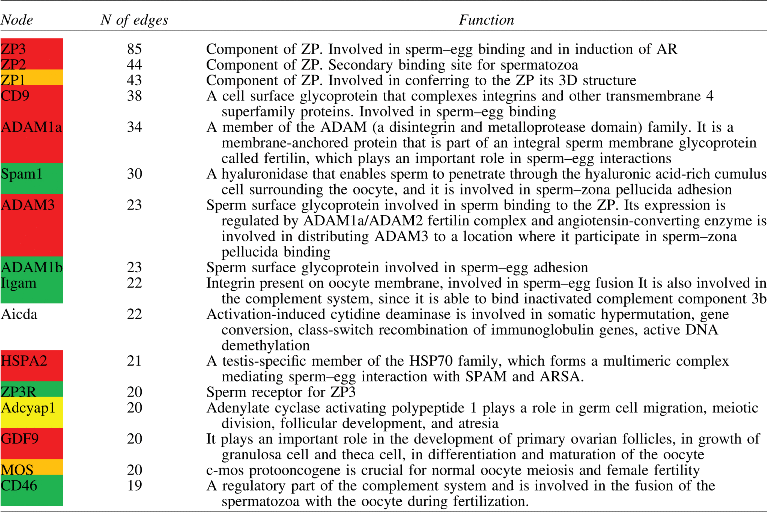 |
The color of nodes depends on the effect on fertility of the deletion of relative gene in KO mice: Red=infertile; Yellow=hypofertile; Green=fertile; White=no data.
The nodes, representing the proteins we selected, were also analyzed by cluster analysis on the basis of their topological parameters. As reported in Figure 2, it was found that three main nodes clusters exist. In particular, cluster A is only constituted by zp3, cluster B by adam2, CD46, zp1, zp2, spam1, and CD9, while the other nodes are comprised in cluster C. Interestingly, clusters A and B contain the most linked hubs of the network with the addition of CD46, which is a hub but is characterized by a relatively lower value of node degree.
FIG. 2.
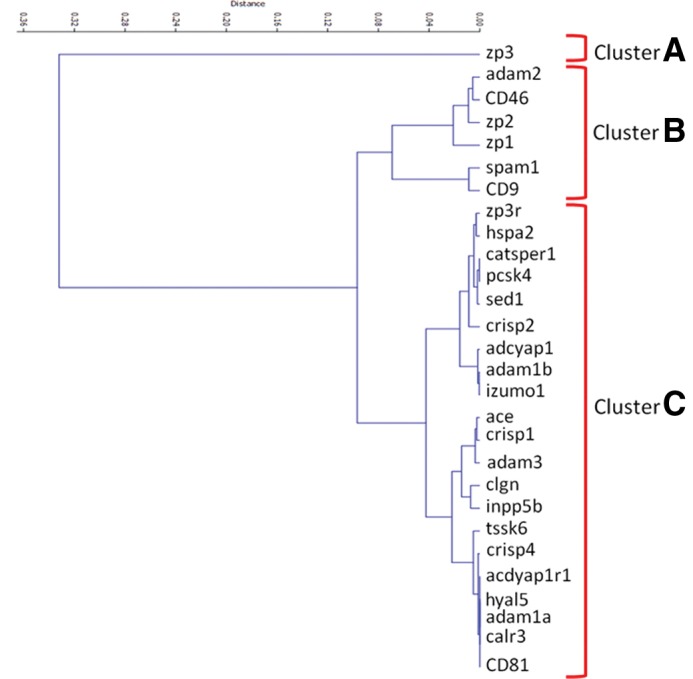
Results of cluster analysis (see text for explanation).
To compare the SERB network and MC-SERBN network with a scale-free network, following the Barabási–Albert model, we used a computer generated network, whose main topological parameters are shown in Table 5. It has one connected component and 438 nodes and 438 links, and it is characterized by a clustering coefficient value of 0. The correlation between node degree and clustering coefficient is 0.
Table 5.
The Most Relevant Topological Parameters of BA Computer Generated Network
| Parameter | BA computer generated network |
|---|---|
| Number of nodes | 438 |
| Number of edges | 438 |
| Clustering coefficient | 0.0 |
| Connected components | 1 |
| Network diameter | 16 |
| Network radius | 8 |
| Network centralization | 0.055 |
| Shortest paths | 191406 (100%) |
| Characteristic path length | 6.518 |
| Avg. number of neighbours | 2 |
| Network density | 0.005 |
| Network heterogeneity | 1.249 |
| Isolated nodes | 0 |
From the Reactome FI analysis we obtained a network of 22 nodes and 61 links, constituted of three connected components, with 10 nodes and 45 links, 7 nodes and 10 links, and 5 nodes and 4 links. Interestingly, the Pathway Enrichment Analysis shows that the proteins used in our text mining experiment belong to several pathways, involved in very different biological functions of relevant importance in mammalian biology (Table 6).
Table 6.
Results of Reactome FI Pathway Enrichment Analysis
| Pathway | FDR | % of Hit genes present in the pathway | N° of molecules of the pathway | N° of Hit Genes | P | Hit Genes |
|---|---|---|---|---|---|---|
| Fibronectin matrix formation | 0 | 100,000 | 3 | 3 | <1.111e-04 | ITGB1,FN1,ITGA5 |
| Interaction with zona pellucida | 0 | 50,000 | 10 | 5 | <2.500e-04 | ADAM2,ZP1,ZP2,ZP3,B4GALT1 |
| Fertilization | 0 | 33,333 | 24 | 8 | <3.333e-04 | ADAM2,ZP1,ZP2,ZP3,CATSPER1,ACR,CD9,B4GALT1 |
| Reproduction | 0 | 33,333 | 24 | 8 | <3.333e-04 | ADAM2,ZP1,ZP2,ZP3,CATSPER1,ACR,CD9,B4GALT1 |
| Syndecan interactions | 0 | 20,000 | 20 | 4 | <1.000e-04 | ITGB5,ITGB3,ITGB1,ITGA6 |
| Signal transduction by L1 | 0 | 19,048 | 21 | 4 | <9.091e-05 | ITGB3,ITGB1,ITGA9,ITGA5 |
| Molecules associated with elastic fibres | 0 | 16,667 | 30 | 5 | <1.250e-04 | ITGB5,ITGB3,ITGB1,ITGB6,ITGA8 |
| Integrin cell surface interactions | 0 | 15,152 | 66 | 10 | <1.000e-03 | ITGB5,ITGB3,ITGB1,ITGB6,FN1,ITGA4,ITGA9,ITGA6,ITGA5,ITGA8 |
| Elastic fibre formation | 0 | 14,634 | 41 | 6 | <2.000e-04 | ITGB5,ITGB3,ITGB1,ITGB6,ITGA5,ITGA8 |
| ECM proteoglycans | 0 | 10,909 | 55 | 6 | <1.429e-04 | ITGB5,ITGB3,ITGB1,ITGB6,ITGA9,ITGA8 |
| Non-integrin membrane-ECM interactions | 0 | 9,524 | 42 | 4 | 5,39E-01 | ITGB5,ITGB3,ITGB1,ITGA6 |
| Cell surface interactions at the vascular wall | 0 | 6,316 | 95 | 6 | <8.333e-05 | ITGB3,ITGB1,FN1,ITGA4,ITGA6,ITGA5 |
| Extracellular matrix organization | 0 | 3,802 | 263 | 10 | <1.667e-04 | ITGB5,ITGB3,ITGB1,ITGB6,FN1,ITGA4,ITGA9,ITGA6,ITGA5,ITGA8 |
| L1CAM interactions | 0,0004 | 4,255 | 94 | 4 | 1,19E+01 | ITGB3,ITGB1,ITGA9,ITGA5 |
| Hemostasis | 0,0013 | 1,522 | 460 | 7 | 3,46E+01 | ITGB3,ITGB1,CD9,FN1,ITGA4,ITGA6,ITGA5 |
| GRB2:SOS provides linkage to MAPK signaling for Intergrins | 0,0015 | 13,333 | 15 | 2 | 3,62E+01 | ITGB3,FN1 |
| p130Cas linkage to MAPK signaling for integrins | 0,0015 | 13,333 | 15 | 2 | 3,62E+01 | ITGB3,FN1 |
| Platelet degranulation | 0,0031 | 3,896 | 77 | 3 | 6,84E+01 | ITGB3,CD9,FN1 |
| Laminin interactions | 0,0035 | 8,696 | 23 | 2 | 7,36E+01 | ITGB1,ITGA6 |
| Response to elevated platelet cytosolic Ca2+ | 0,0037 | 3,659 | 82 | 3 | 7,39E+01 | ITGB3,CD9,FN1 |
| Basigin interactions | 0,0041 | 8,000 | 25 | 2 | 7,77E+01 | ITGB1,ITGA6 |
| Integrin alphaIIb beta3 signaling | 0,0048 | 7,407 | 27 | 2 | 8,56E+01 | ITGB3,FN1 |
| Platelet aggregation (plug formation) | 0,0088 | 5,405 | 37 | 2 | 1,45E+02 | ITGB3,FN1 |
| Localization of the PINCH-ILK-PARVIN complex to focal adhesions | 0,0152 | 25,000 | 4 | 1 | 2,58E+02 | ITGB1 |
| Axon guidance | 0,0161 | 1,544 | 259 | 4 | 2,65E+02 | ITGB3,ITGB1,ITGA9,ITGA5 |
| Sperm:Oocyte membrane binding | 0,019 | 20,000 | 5 | 1 | 2,91E+02 | CD9 |
| NGF-independant TRKA activation | 0,019 | 20,000 | 5 | 1 | 2,91E+02 | ADCYAP1 |
| Activation of TRKA receptors | 0,0227 | 16,667 | 6 | 1 | 3,47E+02 | ADCYAP1 |
| Attachment of GPI anchor to uPAR | 0,0265 | 14,286 | 7 | 1 | 4,03E+02 | PGAP1 |
| Post-translational protein modification | 0,0294 | 1,695 | 177 | 3 | 4,32E+02 | PGAP1,B4GALT1,ARSA |
| Sperm motility and taxes | 0,0302 | 12,500 | 8 | 1 | 4,21E+02 | CATSPER1 |
| Cross-presentation of particulate exogenous antigens (phagosomes) | 0,0302 | 12,500 | 8 | 1 | 4,21E+02 | ITGB5 |
| CHL1 interactions | 0,0339 | 11,111 | 9 | 1 | 4,35E+02 | ITGB1 |
| N-Glycan antennae elongation | 0,0339 | 11,111 | 9 | 1 | 4,35E+02 | B4GALT1 |
| Type I hemidesmosome assembly | 0,0339 | 11,111 | 9 | 1 | 4,35E+02 | ITGA6 |
| Cell junction organization | 0,0391 | 2,439 | 82 | 2 | 4,94E+02 | ITGB1,ITGA6 |
| Platelet activation, signaling and aggregation | 0,04 | 1,500 | 200 | 3 | 4,95E+02 | ITGB3,CD9,FN1 |
| N-glycan antennae elongation in the medial/trans-Golgi | 0,045 | 8,333 | 12 | 1 | 5,51E+02 | B4GALT1 |
| PECAM1 interactions | 0,045 | 8,333 | 12 | 1 | 5,51E+02 | ITGB3 |
| The activation of arylsulfatases | 0,0486 | 7,692 | 13 | 1 | 5,82E+02 | ARSA |
| Platelet adhesion to exposed collagen | 0,0486 | 7,692 | 13 | 1 | 5,82E+02 | ITGB1 |
| Immunoregulatory interactions between lymphoid and non-lymphoid cell | 0,0577 | 1,293 | 232 | 3 | 7,11E+02 | CD81,ITGB1,ITGA4 |
| Cell–extracellular matrix interactions | 0,0595 | 6,250 | 16 | 1 | 7,05E+02 | ITGB1 |
| Metabolism of angiotensinogen to angiotensins | 0,0595 | 6,250 | 16 | 1 | 7,05E+02 | ACE |
| Pre-NOTCH processing in Golgi | 0,0667 | 5,556 | 18 | 1 | 7,93E+02 | B4GALT1 |
| Other semaphorin interactions | 0,0703 | 5,263 | 19 | 1 | 8,14E+02 | ITGB1 |
| Developmental biology | 0,0704 | 0,966 | 414 | 4 | 8,05E+02 | ITGB3,ITGB1,ITGA9,ITGA5 |
| Post-translational modification: synthesis of GPI-anchored proteins | 0,0774 | 4,762 | 21 | 1 | 8,73E+02 | PGAP1 |
| Smooth muscle contraction | 0,0809 | 4,545 | 22 | 1 | 8,98E+02 | ITGB5 |
| Regulation of complement cascade | 0,0845 | 4,348 | 23 | 1 | 9,17E+02 | CD46 |
| Cell–Cell communication | 0,0846 | 1,575 | 127 | 2 | 9,02E+02 | ITGB1,ITGA6 |
| Transport to the Golgi and subsequent modification | 0,0915 | 4,000 | 25 | 1 | 9,53E+02 | B4GALT1 |
| Keratan sulfate biosynthesis | 0,0984 | 3,704 | 27 | 1 | 9,82E+02 | B4GALT1 |
| PTM: gamma carboxylation, hypusine formation and arylsulfatase activation | 0,0984 | 3,704 | 27 | 1 | 9,82E+02 | ARSA |
| Glucagon-type ligand receptors | 0,1088 | 3,333 | 30 | 1 | 1,00E+03 | ADCYAP1 |
| Glycosphingolipid metabolism | 0,1088 | 3,333 | 30 | 1 | 1,00E+03 | ARSA |
| Keratan sulfate/keratin metabolism | 0,1122 | 3,226 | 31 | 1 | 1,00E+03 | B4GALT1 |
| Pre-NOTCH expression and processing | 0,1588 | 2,222 | 45 | 1 | 1,00E+03 | B4GALT1 |
| Muscle contraction | 0,1717 | 2,041 | 49 | 1 | 1,00E+03 | ITGB5 |
| Metabolism of proteins | 0,1796 | 0,686 | 583 | 4 | 1,00E+03 | PGAP1,ACE,B4GALT1,ARSA |
| Sphingolipid metabolism | 0,1812 | 1,923 | 52 | 1 | 1,00E+03 | ARSA |
| Golgi associated vesicle biogenesis | 0,1875 | 1,852 | 54 | 1 | 1,00E+03 | NAPA |
| Assembly of collagen fibrils and other multimeric structures | 0,1875 | 1,852 | 54 | 1 | 1,00E+03 | ITGA6 |
| Clathrin derived vesicle budding | 0,2092 | 1,639 | 61 | 1 | 1,00E+03 | NAPA |
| trans-Golgi network vesicle budding | 0,2092 | 1,639 | 61 | 1 | 1,00E+03 | NAPA |
| Asparagine N-linked glycosylation | 0,2153 | 1,587 | 63 | 1 | 1,00E+03 | B4GALT1 |
| Semaphorin interactions | 0,2243 | 1,515 | 66 | 1 | 1,00E+03 | ITGB1 |
| Antigen processing-Cross presentation | 0,2624 | 1,266 | 79 | 1 | 1,00E+03 | ITGB5 |
| Peptide hormone metabolism | 0,271 | 1,220 | 82 | 1 | 1,00E+03 | ACE |
| Class B/2 (Secretin family receptors) | 0,2766 | 1,190 | 84 | 1 | 1,00E+03 | ADCYAP1 |
| Collagen formation | 0,2822 | 1,163 | 86 | 1 | 1,00E+03 | ITGA6 |
| Adaptive Immune System | 0,3403 | 0,519 | 770 | 4 | 1,00E+03 | CD81,ITGB5,ITGB1,ITGA4 |
| Signaling by NOTCH | 0,3461 | 0,909 | 110 | 1 | 1,00E+03 | B4GALT1 |
| MPS IIID - Sanfilippo syndrome D | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS VII - Sly syndrome | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS IX - Natowicz syndrome | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| Glycosaminoglycan metabolism | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS IIIB - Sanfilippo syndrome B | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS II - Hunter syndrome | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| Mucopolysaccharidoses | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS IIIA - Sanfilippo syndrome A | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS I - Hurler syndrome | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS IV - Morquio syndrome B | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS IV - Morquio syndrome A | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS VI - Maroteaux-Lamy syndrome | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| MPS IIIC - Sanfilippo syndrome C | 0,3512 | 0,893 | 112 | 1 | 1,00E+03 | B4GALT1 |
| G alpha (s) signaling events | 0,3711 | 0,833 | 120 | 1 | 1,00E+03 | ADCYAP1 |
| Membrane trafficking | 0,501 | 0,559 | 179 | 1 | 1,00E+03 | NAPA |
| Complement cascade | 0,5296 | 0,515 | 194 | 1 | 1,00E+03 | CD46 |
| NGF signalling via TRKA from the plasma membrane | 0,537 | 0,505 | 198 | 1 | 1,00E+03 | ADCYAP1 |
| Immune system | 0,5453 | 0,397 | 1261 | 5 | 1,00E+03 | CD46,CD81,ITGB5,ITGB1,ITGA4 |
| Metabolism of carbohydrates | 0,6001 | 0,426 | 235 | 1 | 1,00E+03 | B4GALT1 |
| Class I MHC mediated antigen processing & presentation | 0,6277 | 0,395 | 253 | 1 | 1,00E+03 | ITGB5 |
| Signaling by NGF | 0,6644 | 0,358 | 279 | 1 | 1,00E+03 | ADCYAP1 |
| GPCR ligand binding | 0,8202 | 0,231 | 433 | 1 | 1,00E+03 | ADCYAP1 |
| Metabolism of lipids and lipoproteins | 0,8397 | 0,217 | 461 | 1 | 1,00E+03 | ARSA |
| Innate immune system | 0,9382 | 0,145 | 688 | 1 | 1,00E+03 | CD46 |
| Signal transduction | 0,9442 | 0,221 | 1806 | 4 | 1,00E+03 | ITGB3,ADCYAP1,FN1,B4GALT1 |
| Metabolism | 0,9716 | 0,154 | 1296 | 2 | 1,00E+03 | B4GALT1,ARSA |
| GPCR downstream signaling | 0,9794 | 0,106 | 939 | 1 | 1,00E+03 | ADCYAP1 |
| Disease | 0,9802 | 0,105 | 948 | 1 | 1,00E+03 | B4GALT1 |
| Signaling by GPCR | 0,9869 | 0,096 | 1039 | 1 | 1,00E+03 | ADCYAP1 |
FRD, False Discovery Rate; Hit gene, genes used in text mining analysis present in the pathway; N° of Hit Genes, number of the genes used in text mining analysis present in the pathway; N° of molecules of the pathway, number of molecules constituting the pathway; P, probability level; Pathway, name of the pathway, as reported in Reactome Database; % of Hit genes present in the pathway, percentage of the genes used in text mining analysis present in the pathway.
The experiment performed with STRING allowed us to obtain a network by merging all the networks representing the interactions of all the proteins used in text mining experiment. After removal of self loops and duplicate edges, we obtained a network of 341 nods and 1311 edges (STRING network) composed of eight connected components, the main of which (MC-STRING network) had 255 and 988 edges (Table 7 and Table 8A and B).
Table 7.
The Most Relevant Topological Parameters of STRING and MC-STRING Networks
| Parameter | STRING | MC-STRING |
|---|---|---|
| Number of nodes | 358 | 273 |
| Number of edges | 1366 | 1043 |
| Clustering coefficient | 0.698 | 0.657 |
| Connected components | 8 | 1 |
| Network diameter | 10 | 10 |
| Network radius | 1 | 5 |
| Network centralization | 0.108 | 0.142 |
| Shortest paths | 74878 (58%) | 73712 (99%) |
| Characteristic path length | 4.642 | 4.691 |
| Avg. number of neighbours | 7.631 | 7.641 |
| Network density | 0.021 | 0.028 |
| Network heterogeneity | 0.790 | 0.848 |
| Isolated nodes | 0 | 0 |
Table 8A.
Node Degree of STRING and MC-STRING
| Node degree equation | STRING | MC-STRING |
|---|---|---|
| γ | −1.251 | −0.353 |
| r | 0.647 | 0.755 |
| R2 | 0.764 | 0.481 |
Table 8B.
Clustering Coefficient of STRING and MC-STRING
| Clustering coefficient vs. node degree | STRING | MC-STRING |
|---|---|---|
| γ | −0.357 | −0.353 |
| r | 0.761 | 0.755 |
| R2 | 0.502 | 0.481 |
Discussion
The analysis carried out on SERBN and MC-SERBN allowed us to classify them on the basis of their topology. The simplest network topology is that of random networks, following the Erdös–Rényi (ER) model (Erdös and Rényi, 1959), where N nodes are connected with probability p, thus creating a network with approximately pN(N–1)/2 randomly placed links. The node degree (i.e., the expected number of links per node) follows a Poisson distribution, consequently most of the nodes have approximately the same number of links (close to the average degree), which will define the network scale. The tail (high k region) of the degree distribution P(k) decreases exponentially, thus indicating that the nodes that significantly deviate from the average are extremely rare. The clustering coefficient is independent of the node degree, and the mean path length is proportional to the logarithm of the network size: l ∼ log N (Erdös and Rényi, 1959). These networks are used to model physical phenomena such as phase transition or percolation, but often fail to describe biological events such as control of gene expression, epidemiology of infectious diseases, and social behavior. Indeed, in these contexts, a strong heterogeneity in the number of links per node is evident, due to the coexistence of a small number of highly linked nodes and a large number of poorly linked nodes in the same network. Often the node degree follows a power-law distribution in which the probability that a node has k links follows the equation P(k)∼k –γ, where γ is the degree exponent. These networks are characterized by a strong dependence on a small number of highly connected nodes (the hubs), and it is impossible to define a scale of system (scale-free topology). It is the case of networks following the Barabási–Albert (BA) model (Barabási and Albert, 2002), which are characterized by a power-law degree distribution and in which the clustering coefficient C(k) is independent of k and the average path length follows the equation l ∼ log log N, significantly shorter than the log N typical of random networks (Albert and Barabási, 2002).
It has been found that the networks representing important biological functions (such as gene interaction networks), as well as nonbiological entities (WW, actor network, internet at the domain level), are characterized by the coexistence of a scale-free topology with an highly clustered structure (Ravasz and Barabási, 2003), thus originating a hierarchical pattern. The most relevant feature of hierarchical networks is the scaling law of the clustering coefficient, which follows a power law. This implies that sparsely connected nodes are part of highly clustered areas, with a few hubs maintaining communication between different, highly clustered neighbours.
The analysis of SERBN allows classification as a scale-free hierarchical network. In our opinion, this is an interesting finding because it is possible to correlate the network topology with the biological features of the process. First, it is possible to identify the super-connected nodes (i.e., those nodes that play a key role in determining the network topological (and biological) characteristics). It is immediately evident that the three most connected nodes, which are also included in cluster A and B, are components of ZP. In mammals it is composed of either three or four proteins. In particular, mouse ZP is composed of ZP1 (623 amino acids), ZP2 (713 aa), and ZP3 (424aa). Pig, cow, and dog ZP are composed of Z2, ZP3, and ZP4 proteins. Differently, the ZP of rats, hamsters, bonnet monkeys, and humans are composed of four glycoproteins: ZP1, ZP2, ZP3, and ZP4. In humans, ZP1 has a 638-aa polypeptide backbone; ZP2 has 745 aa; ZP3 has 424 aa, and ZP4 has 540 aa. In all cases, the ZP proteins are heavily glycosylated, having N- and O-linked glycans (Gupta and Bhandari, 2011).
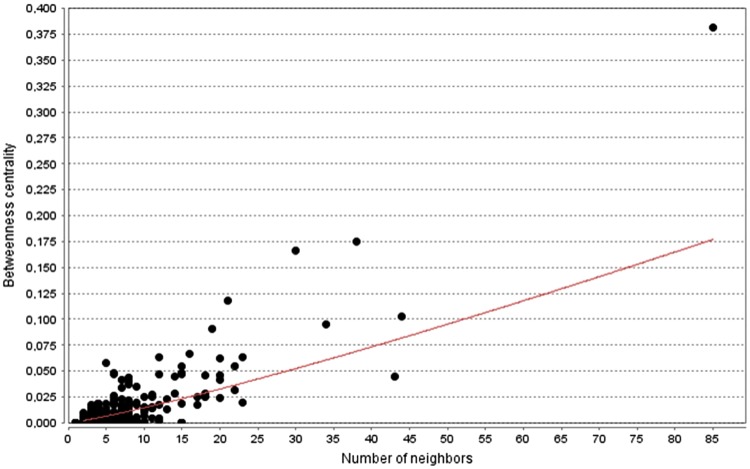
ZP3, in the mouse model, represents the higher affinity ligand for spermatozoa, and it is the primary site for the two gametes interaction. It has been reported that the initial adhesion of spermatozoon to the oocyte involves about 30,000 binding sites (300 molecules/μm2) (Thaler and Cardullo, 1996). After the sperm binding, ZP3 is able to induce the AR by interacting at least with two different receptors: the pertussis toxin-sensitive Gi protein-coupled receptor linked to PLCβ1 and a putative tyrosine kinase receptor coupled to PLCγ. In our model, the biological relevance of ZP3 in SERBN is strengthened by its topology: it has the higher node degree value (85) within the network, the higher value of betweenness centrality (0.17), and it is the only node belonging to cluster A. The number of neighbors and the betweenness centrality in SERBN are correlated (r=0.876) and ZP3 is clearly the most central node in the network (Fig. 3). This role of ZP3 is confirmed by the experimental evidence that its removal causes the loss of function of SERB in mouse model: the ZP3-/- mice are infertile.
FIG. 3.
Results of plotting of betweenness centrality vs. number of neighbors, MC-SERBN.
ZP2, in mice, are unable to interfere with sperm–egg binding or with induction of the AR, and monoclonal and polyclonal antibodies anti-ZP2 do not affect the initial binding of the sperm to the oocyte, whereas they are significantly able to inhibit the binding of acrosome-reacted sperm to the ZP (Gupta and Bhandari, 2011). This suggests that ZP2 could act as a secondary receptor for sperm during fertilization (Bleil et al., 1988).
In mouse it has been established that ZP1 is not able to induce AR and that ZP1 purified proteins are unable to interfere with the sperm-egg binding (Bleil and Wassarman, 1980), thus suggesting that ZP1 is not active in sperm egg interaction or signaling, but it was supposed to exert a mechanical function. In particular, it has been suggested that it cross-links the filaments formed by ZP2–ZP3 heterodimers, thus providing stability and structural and topological integrity to the ZP structure (Greve, and Wassarman, 1985). Importantly, the removal of ZP1 and ZP2 nodes have very important consequences on SERB process, in fact Zp1-/- and Zp2-/- mice display a severely reduced or an absent fertility, respectively.
CD9 antigen is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. It is a cell-surface protein characterized by the presence of four hydrophobic domains, which is involved in the regulation of cell development, activation, growth, and motility (Le Naour et al., 2000). CD9 takes part in the SERB process by forming complexes with integrins, and the gametes of CD9 knocked-out mice are unable to undergo fusion (Le Naour et al., 2000). On oocytes, it is located in the microvillar membrane where it is involved in maintaining the normal shape of microvilli. At present, two different models of interaction with integrins during SERB process have been proposed: in one, fertilin β (ADAM 2) binds to α6β1 that is physically associated with CD9 and tethered to the actin cytoskeleton, whereas in the other model, fertilin β binds to α6β1 that is tethered to the actin cytoskeleton and CD9 indirectly influences the association of α6β1 with the actin cytoskeleton and, therefore, its ability to bind fertilin β (Chen et al., 1999). It has been recently proposed that CD9, together with CD81, could “play a role as ‘extracellular components’ in sperm-oocyte membrane fusion” (Ohnami et al., 2012). Indeed, CD9 and CD81 are present in exosomes released from oocytes: CD9 localizes in the perivitelline space, while CD81 is in zona pellucida (Ohnami et al., 2012). When the reacted spermatozoa reach the perivitelline space, possibly they acquire CD9 from a transfer of membrane fragments from the oocyte plasma membrane. This process seems to be crucial for the spermatozoa membrane reorganization, required for fusion with the oocyte (Barraud-Lange et al., 2012; Jégou et al., 2011). The key role of CD9, supposed by its topological parameters, is confirmed by the finding that its removal in CD9-/- mice caused infertility.
Sperm Adhesion Molecule 1 (Spam1), also known as PH-20 hyaluronidase, is an enzyme that degrades hyaluronic acid, a major structural proteoglycan present in extracellular matrices and in basement membranes. In spermatozoa it is present on the plasma membrane and on the inner acrosome membrane, where, during fertilization, it has been proposed to exert enzymatic and nonenzymatic roles: cumulus penetration, zona binding, and receptor for hyaluronic acid (Martin-Deleon, 2011). Despite Spam1 being a key player in the fertilization process, Spam1-deficient mice are fertile. This finding is not surprising, when considering the complex trafficking of that protein. Indeed, Spam1 is independently synthesized in the testis and epididymis. In particular, in epididymis Spam1 is present in the luminal fluid either in soluble and insoluble form (in epididymosomes) and is acquired by sperm membrane. Once the spermatozoa are released in the female genital tract, they are exposed to Spam1 present in uterine fluid. Here, the lipid exchange on the sperm surface has been described. Clusterin, and other lipid carriers such as APOA1, serve as GPI-linked protein monomer stabilizers, donors to the sperm plasma membrane (PM), and as acceptors of cholesterol. This interesting datum was confirmed by the finding that when Spam1-/- were exposed to accessible Spam1 there was a significant difference in cumulus dispersal efficiency (Griffiths et al., 2008), thus suggesting that the sperm surface remodeling occurring prior to and during capacitation could involve the functional expression of Spam1, in an integrated cross-talk between male and female structures. This specific peculiarity, could explain the fertility of Spam1-/- mice.
ADAM2, also known as fertilin beta, Ftnb, or Ph30-beta, is a member of the ADAM (a disintegrin and metalloprotease domain) family and is able to di/trimerize with other ADAM family members to form complexes that play a key role in sperm migration through the female genital tract and in sperm-egg interaction. In particular, it is involved in the formation of ADAM1B–ADAM2, ADAM2–ADAM3–ADAM4, ADAM2–ADAM3–ADAM5, and ADAM2–ADAM3–ADAM6 complexes. Specifically the eterodimer ADAM1B–ADAM2, also known as fertilin, has a key role in the sperm migration trough the utero-tube junction (UTJ) and in the SERB process.
Two aspects of ADAM2 biology could have a great relevance. First, its binding partner on oocyte membrane is α6β1 integrin (see CD9). Second, recent evidence has shown that ADAM2 is part of a complex chain of protein trafficking events. Indeed, in several KO mice experiments from many laboratories it was found that at least two testis-specific molecular chaperones, calmegin (CLGN) and calsperin (CALR3), are involved in control of ADAMs expression. In particular, CLGN folds ADAM1A, ADAM1B, and ADAM2, and in the absence of Clgn gene the formation of ADAM1A/2 heterodimer is impaired. The failure of ADAM1A/2 formation leads to the loss of expression of ADAM3 on sperm surface, which also seems to involve the tyrosylprotein sulfotransferase-2, TPST2, and the angiotensin-converting enzyme, ACE. Importantly, Adam2-/- mice are infertile.
CD46, is one of the most connected nodes of SERBN and belongs to the cluster B, and may play an important role in the SERB process. It is also known as Human Membrane Cofactor protein (MCP), and it is an ubiquitously expressed protein involved in cell protection from complement attack. In spermatozoa, it has been found on the inner acrosomal membrane (Clift et al., 2009; Inoue et al., 2003). Surprisingly, mice carrying a null mutation in the CD46 gene were healthy, females were fully fertile, and male fertility appeared to be facilitated by disruption of the CD46 gene. Indeed, the average number of pups born from CD46−/− males was significantly greater than that of wild-type males and the incidence of the spontaneous acrosome reaction doubled in CD46−/− sperm, when compared to wild-type sperm (Inoue et al., 2003). These data suggest that CD46 may be involved in regulation of sperm AR (Inoue et al., 2003; Johnson et al., 2007).
The other hubs are listed in Table 6 and participate in the SERB process in various ways: ZP3R is involved in ZP binding process, ADAM3, ADAM1b, CD46, Itgam are involved in sperm-oocyte membrane recognition and fusion, HSPA2 is a chaperonin involved in regulation of ASPAM and ARSA, CD46 is involved also in regulation of complement, c-MOS, GDF9, Aicda, Adcyap1 are involved in oocyte maturation process. Importantly, the results of analysis carried out on network topology either in the case of node degree (identification of hubs) or on the basis of cluster analysis (definition of cluster A, B, and C), seems to be in keeping with experimental findings: the most connected nodes (those belonging to cluster A and B) represent molecules whose knock-out mice are infertile or severely subfertile.
In other words, the computational approach we used seems to be able to identify the most important molecules involved in SERB process. Only in a few cases the inference from nodes topology and the data from knock-out mice are incongruent. For instance, the Izumo1-/- mice are infertile but izumo1 is not a hub and it belongs to the cluster C. This, in our opinion, could be due to different reasons. First, in many cases our knowledge of molecular interaction of elements involved in SERB is incomplete or in rapid development (see the recent article from Bianchi et al., 2014 describing the interaction of izumo1 with Juno). In addition, our investigations have an intrinsic limit due to the impossibility to analyze the SERBN as a directed network (the networks produced by Agilent Literature Search 3.0.3 beta are undirected). Finally, the topology of the SERBN network is characterized by a high level of robustness and redundancy. The robustness against random failure is due to the marked heterogeneity in the number of links per node. Indeed the large majority of nodes will be scarcely connected, thus the random damage of nodes will affect mainly the less linked ones. In this case, the functional stability of SERB process will be unaffected by the damage. Vice versa, the hubs are the less frequent nodes, thus the probability that their failure will happen is low. The redundancy is due to the relatively high value of clustering coefficient, when compared with other networks representing molecular events involved in signal transduction. For instance, in a previous independent work, we found that the networks representing the sperm activation in sea urchin, Caenorhabditis elegans, and human, and other biologically relevant processes (i.e., smooth and striated muscular contraction, release cycle of neurotransmitters, visual phototransduction, insulin signaling pathway, the p53 pathway, regulation of retinoblastoma protein, mitochondrial ATP metabolism, glucose metabolism, signalling events mediated by stem cell factor receptor c-Kit; and the circadian clock) the clustering coefficient ranged between 0 and 0.074 (Bernabò et al., 2014); in other words, it was about 10% of that found in SERBN.
To confirm this supposition, we carried out the comparison between SERBN and a network of the same size (438 nodes) generated by computer following the Barabasi–Albert model (Table 5). In this case, the clustering coefficient value was 0, thus strengthening that SERBN and MC-SERBN did not follow the BA model. This finding could suggest that SERB process has evolved redundancy as safety strategy. An elegant example of that are acrosin and prss21 serine proteases. Indeed, spermatozoa express functional proteases in acrosomal matrix (Acrosin) and on plasma membrane (PRSS21), which are thought to be involved in enzymatic digestion of ZP and in sperm-egg binding. When the fertility of acrosin-null mice was tested, it was found that they are fertile: they only display in vitro a delayed penetration on ZP, probably due to the reduced ability to disperse the acrosomal components. Thus, per se the acrosin seems to be not necessary for fertilization.
At the same time, PRSS21 seems also not to be essential for fertilization, because the Prss21-deficient male mice exhibit normal fertility. Interestingly, when both serine proteases have been removed in a double knock-out experiment, it was found that acrosin/Prss21-deficient mice were subfertile: in vivo they shown a reduced litter sizes and the successful fertilization of about only 20%–35% of eggs; in vitro epididymal spermatozoa from Acrosin/Prss21-/- fail to fertilize metaphase II-arrested eggs. Easily, it is possible to explain this finding by the coexistence of a redundant mechanism as a safety strategy: prss21 and acrosin exert similar (but likely not identical) function during the SERB process, thus the failure of a only one of them did not affected the efficiency of whole process. Only the contemporaneous removal of both of them caused a detectable negative effect (Kawano et al., 2010).
Another important topological feature of SERBN is its easily navigability. Indeed, it is characterized by low values of characteristic path length and of the averaged number of neighbours, which confers it a “small world” structure (Barabási and Albert, 1999). As demonstrated by Latora and Marchirori (2001), the analysis of real data indicates that small world networks are both globally and locally highly efficient. This implies that the information flux through the SERBN is efficient and specific, that it is in agreement with the importance of SERB process and its safety and specificity. In addition, this finding must be interpreted on the light of high spermatozoa differentiation and of the fact that they are “disposable” cells. The signal transduction cascade is designed to reach a specific target (the ability to recognize and bind oocytes) and not for assuring to sperm cells the ability to survive for long times, as it could happen, for instance, for neurons or oocytes.
Ultimately, male and female gametes must be able to recognize each other, avoiding, at the same time, to be activated by other cells. In particular, spermatozoa pass several hours or some day in the female genital tract, where they interact with a myriad of different kind of cells, and gain the ability to fertilize the oocyte, then avoiding to undergo AR in response to the interaction with others cells. They must be able to specifically recognize and interact with one single cell in an ocean of others cells. From our data (high clustering and redundancy), the picture emerges of a complex series of interactions involving several molecular mechanisms acting simultaneously. Moreover, not only molecules directly located on spermatozoa and oocyte are involved, but also molecules present in oviductal fluid (complement and SPAM1) or in exosomes (CD9).
To complete and validate the data obtained with the text mining approach, we performed two further computational experiments. First we compared the networks obtained by searching in STRING database of protein interaction (STRING network and MC-STRING network), and we saw that they display topological features very similar to those of corresponding SERB and MC-SERB networks. In our opinion, this datum seems to confirm the reliability of the approach we used, indeed this independent analysis, event taking into account the differences intrinsic to the two systems, arrives at a similar result. Interestingly, the further Pathway Enrichment Analysis, carried out with Reactome FI, to explore the pathways in which the protein of our interest were involved, shows that they are involved in several pathways of great biological relevance and not also in reproduction, thus strengthening the concept that the fertilization is a complex process interconnected with all other organic functions.
Conclusions
The approach we used to explore the SERB process, the adoption of a biomedical text mining strategy, is a useful way forward to discover novel information hidden in biomedical literature and to manage it in computational models. On the other hand, the data we obtained with this strategy could be affected by potential bias due to the literature source. But it is a hitherto underutilized approach that warrants further consideration in future omics systems science research and development. Indeed, the difference in the amount of molecular data concerning the different proteins and the inhomogeneity of data availability on the different biological processes under investigation could potentially influence the results of text mining research and, as a consequence, the inference from the model.
From our analysis, it emerges that the SERB process is a multistep multi-molecular event, involving not only the gametes but virtually all the female genital tract, which, in turn, is functionally and anatomically connected with the systemic compartment. We can imagine a multi keys–multi locks systems, in which the keys and the locks are on spermatozoa, oocytes, and in the oviductal environment.
In addition, the key role of zona pellucida emerges, in particular ZP3, in the SERB process. Indeed, from our data it is possible to speculate that ZP could be considered the conductor of that process. Indeed, this structure has a central role in reproduction since it is the main agent in many events of pivotal importance. It plays several important roles in reproduction such as the participation to the species-specificity of sperm–egg recognition and binding, the physiological induction of sperm acrosomal exocytosis, the avoidance of polyspermy, and the protection of embryo during the early phases of its development. In addition, the findings strengthen the concept that the in vitro approach used to model the SERB process is markedly different from what occurs in vivo, because some of the environmental elements are omitted. This requires use of nonphysiological parameters such as higher sperm/egg ratio, longer length of gametes co-incubation, to use of methylxantines and other drugs, to increase the calcium concentration, among others. Our findings suggest that it will be hypothetically possible to improve the performance of in vitro systems, in parallel obtaining condition closer to the physiological ones, adding to the systems new players, such as epithelial cells cultures, oviductal fluid, conditioned media. Finally, it is conceivable that fertility depends on the functional integrity of the whole reproductive system, which includes not only gametes, but also the female genital tract, the female endocrine axis, and ultimately, the male and female systemic environment, thus strengthening the importance of an integrated approach on the study of reproductive medicine.
Supplementary Material
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Akondi KB, and Lakshmi VV. (2013). Emerging trends in genomic approaches for microbial bioprospecting. OMICS 17, 61–70 [DOI] [PubMed] [Google Scholar]
- Alasmari W, Costello S, Correia J, et al. (2013). Ca2+ signals generated by CatSper and Ca2+ stores regulate different behaviors in human sperm. J Biol Chem 288, 6248–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R, and Barabási AL. (2002). Statistical mechanics of complex networks. Rev Mod Phys 74, 48–97 [Google Scholar]
- Avella MA, and Dean J. (2011). Fertilization with acrosome-reacted mouse sperm: Implications for the site of exocytosis. Proc Natl Acad Sci USA 108, 19843–19844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási AL, and Albert R. (1999). Emergence of scaling in random networks. Science 286, 509–512 [DOI] [PubMed] [Google Scholar]
- Barboni B, Bernabò N, Palestini P, et al. (2011). Type-1 cannabinoid receptors reduce membrane fluidity of capacitated boar sperm by impairing their activation by bicarbonate. PLoS One 6, e23038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboni B, Lucidi P, Mattioli M, and Berardinelli P. (2001). VLA-6 integrin distribution and calcium signalling in capacitated boar sperm. Mol Reprod Dev 59, 322–329 [DOI] [PubMed] [Google Scholar]
- Barraud-Lange V, Chalas Boissonnas C, Serres C, et al. (2012). Membrane transfer from oocyte to sperm occurs in two CD9-independent ways that do not supply the fertilising ability of Cd9-deleted oocytes. Reproduction 144, 53–66 [DOI] [PubMed] [Google Scholar]
- Bernabò N, Berardinelli P, Mauro A, et al. (2011). The role of actin in capacitation-related signaling: An in silico and in vitro study. BMC Syst Biol 5, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabò N, Mattioli M, and Barboni B. (2010). The spermatozoa caught in the net: The biological networks to study the male gametes post-ejaculatory life. BMC Syst Biol 4, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabò N, Barboni B, and Maccarrone M. (2013). Systems biology analysis of the endocannabinoid system reveals a scale-free network with distinct roles for anandamide and 2-arachidonoylglycerol. OMICS 17, 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabò N, Mattioli M, and Barboni B. (2014). Signal transduction in the activation of spermatozoa compared to other signaling pathways: A biological networks study. Int J Data Min Bioinform (in press) [DOI] [PubMed] [Google Scholar]
- Bianchi E, Doe B, Goulding D, and Wright GJ. (2014). Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch K, and Tyfield D. (2013). Theorizing the bioeconomy: Biovalue, biocapital, bioeconomics or…what? Sci Technol Human Values 38, 299–327 [Google Scholar]
- Bleil JD, Greve JM, and Wassarman PM. (1988). Identification of a secondary sperm receptor in the mouse egg zona pellucida: Role in maintenance of binding of acrosome-reacted sperm to eggs. Dev Biol 128, 376–385 [DOI] [PubMed] [Google Scholar]
- Bleil JD, and Wassarman PM. (1983). Sperm–egg interactions in the mouse: Sequence of events and induction of the acrosome reaction by a zona pellucida glycoprotein. Dev Biol 95, 317–324 [DOI] [PubMed] [Google Scholar]
- Bowler RP, Bahr TM, Hughes G, et al. (2013). Integrative omics approach identifies interleukin-16 as a biomarker of emphysema. OMICS 17, 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, and Suarez SS. (2010). Rethinking the relationship between hyperactivation and chemotaxis in mammalian sperm. Biol Reprod 83, 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Tung KS, Coonrod SA, et al. (1999). Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin alpha6beta1: Implications for murine fertilization. Proc Natl Acad Sci USA 96, 11830–11835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift LE, Dvorakova-Hortova K, Frolikova M, et al. (2009). CD55 and CD59 protein expression by Apodemus (field mice) sperm in the absence of CD46. J Reprod Immunol 81, 62–73 [DOI] [PubMed] [Google Scholar]
- Dove ES. (2013). Questioning the governance of pharmacogenomics and personalized medicine for global health. Curr Pharmacogenomics Person Med 11, 253–256 [Google Scholar]
- Erdős P, and Rényi A. (1959). On random graphs I. Pub Math 6, 290–297 [Google Scholar]
- Fardilha M, Ferreira M, Pelech S, et al. (2013). “Omics” of human sperm: Profiling protein phosphatases. OMICS 17, 460–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, et al. (2013). STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41, 808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadella BM. (2012). Dynamic regulation of sperm interactions with the zona pellucida prior to and after fertilisation. Reprod Fertil Dev 25, 26–37 [DOI] [PubMed] [Google Scholar]
- Greve JM, and Wassarman PM. (1985). Mouse egg extracellular coat is a matrix of interconnected filaments possessing a structural repeat. J Mol Biol 181, 253–264 [DOI] [PubMed] [Google Scholar]
- Griffiths GS, Miller KA, Galileo DS, and Martin-DeLeon PA. (2008). Murine SPAM1 is secreted by the estrous uterus and oviduct in a form that can bind to sperm during capacitation: Acquisition enhances hyaluronic acid-binding ability and cumulus dispersal efficiency. Reproduction 135, 293–301 [DOI] [PubMed] [Google Scholar]
- Gupta SK, and Bhandari B. (2011). Acrosome reaction: Relevance of zona pellucida glycoproteins, Asian J Androl 13, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Chiu PC, Lee KF, Tse JY, Ho PC, and Yeung WS. (2009). Cumulus cells and their extracellular matrix affect the quality of the spermatozoa penetrating the cumulus mass. Fertil Steril 92, 971–978 [DOI] [PubMed] [Google Scholar]
- Ijiri TW, Mahbub Hasan AK, and Sato K. (2012). Protein-tyrosine kinase signaling in the biological functions associated with sperm. J Signal Transduct 2012, 181560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Nakanishi T, et al. (2003). Disruption of mouse CD46 causes an accelerated spontaneous acrosome reaction in sperm. Mol Cell Biol 23, 2614–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, Satouh Y, Ikawa M, Okabe M, and Yanagimachi R. (2011). Acrosome-reacted mouse spermatozoa recovered from the perivitelline space can fertilize other eggs. Proc Natl Acad Sci USA 108, 20008–20011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégou A, Ziyyat A, Barraud-Lange V, et al. (2011). CD9 tetraspanin generates fusion competent sites on the egg membrane for mammalian fertilization. Proc Natl Acad Sci USA 108, 10946–10951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Fujiwara E, Kakiuchi Y, et al. (2011). Most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the zona pellucida during in vitro fertilization. Proc Natl Acad Sci USA 108, 4892–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PM, Clift LE, Andrlikova P, et al. (2007). Rapid sperm acrosome reaction in the absence of acrosomal CD46 expression in promiscuous field mice (Apodemus). Reproduction 134: 739–747 [DOI] [PubMed] [Google Scholar]
- Kawano N, Kang W, Yamashita M, et al. (2010). Mice lacking two sperm serine proteases, ACR and PRSS21, are subfertile, but the mutant sperm are infertile in vitro. Biol Reprod 83, 359–369 [DOI] [PubMed] [Google Scholar]
- Latora V, and Marchiori M. (2001). Efficient behavior of small-world networks. Phys Rev Lett 287, 198701. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein R, Jasmin C, Prenant M, and Boucheix C. (2000). Severely reduced female fertility in CD9-deficient mice. Science 287, 319–321 [DOI] [PubMed] [Google Scholar]
- Martin-Deleon PA. (2011). Germ-cell hyaluronidases: Their roles in sperm function. Int J Androl 34, e306–318 [DOI] [PubMed] [Google Scholar]
- Ohnami N, Nakamura A, Miyado M, et al. (2012). CD81 and CD9 work independently as extracellular components upon fusion of sperm and oocyte. Biol Open 1, 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M. (2013). The cell biology of mammalian fertilization. Development 140, 4471–4479 [DOI] [PubMed] [Google Scholar]
- Olson SD, Suarez SS, and Fauci LJ. (2011). Coupling biochemistry and hydrodynamics captures hyperactivated sperm motility in a simple flagellar model. J Theor Biol 283, 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan KS. (2013). Questions of critique for big biology: Conjuncture, agency and the global postcolonial. BioSocieties 8, 486–490 [Google Scholar]
- Ravasz E, and Barabási AL. (2003). Hierarchical organization in complex networks. Phys Rev E 67, 026112. [DOI] [PubMed] [Google Scholar]
- Saling PM, Sowinski J, and Storey BT. (1979). An ultrastructural study of epididymal mouse spermatozoa binding to zonae pellucidae in vitro: Sequential relationship to the acrosome reaction. J Exp Zool 209, 229–238 [DOI] [PubMed] [Google Scholar]
- Suarez SS. (2008). Control of hyperactivation in sperm. Hum Reprod Update 14, 647–657 [DOI] [PubMed] [Google Scholar]
- Thaler CD, and Cardullo RA. (1996). The initial molecular interaction between mouse sperm and the zona pellucida is a complex binding event. J Biol Chem 271, 23289–23297 [DOI] [PubMed] [Google Scholar]
- VandeVoort CA, Yudin AI, and Overstreet JW. (1997). Interaction of acrosome-reacted macaque sperm with the macaque zona pellucida. Biol Reprod 56, 1307–1316 [DOI] [PubMed] [Google Scholar]
- Vivar JC, Pemu P, McPherson R, and Ghosh S. (2013). Redundancy control in pathway databases (ReCiPa): An application for improving gene-set enrichment analysis in Omics studies and “Big data” biology. OMICS, 414–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Ogura A, Suto J, et al. (1996). Penetration by field vole spermatozoa of mouse and hamster zonae pellucidae without acrosome reaction. J Reprod Fertil 107, 97–102 [DOI] [PubMed] [Google Scholar]
- Wertheimer E, Krapf D, de la Vega-Beltran JL, et al. (2013). Compartmentalization of distinct cAMP signaling pathways in mammalian sperm. J Biol Chem 6, 35307–35320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Feng X, and Stein L. (2010). A human functional protein interaction network and its application to cancer data analysis. Genome Biol 11, R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi R, and Phillips DM. (1984). The status of acrosomal caps of hamster sperm immediately before fertilization in vivo. Gamete Res 9, 1–20 [Google Scholar]
- Zhu F, Patumcharoenpol P, Zhang C, et al. (2013). Biomedical text mining and its applications in cancer research. J Biomed Inform 46, 200–211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.