Abstract
Intracellular calprotectin (S100A8/A9) functions in the control of the cell cycle checkpoint at G2/M. Dysregulation of S100A8/A9 appears to cause loss of the checkpoint, which frequently characterizes head and neck squamous cell carcinoma (HNSCC). In the present study, we analyzed carcinoma cells for other S100A8/A9-directed changes in malignant phenotype. Using a S100A8/A9-negative human carcinoma cell line (KB), transfection to express S100A8 and S100A9 caused selective down-regulation of MMP-2 and inhibited in vitro invasion and migration. Conversely, silencing of endogenous S100A8 and S100A9 expression in TR146 cells, a well-differentiated HNSCC cell line, increased MMP-2 activity and in vitro invasion and migration. When MMP-2 expression was silenced, cells appeared to assume a less malignant phenotype. To more closely model the architecture of cell growth in vivo, cells were grown in a 3D collagen substrate, which was compared to 2D. Growth on 3D substrates caused greater MMP-2 expression. Whereas hypermethylation of CpG islands occurs frequently in HNSCC, S100A8/A9-dependent regulation of MMP-2 could not be explained by modification of the upstream promoters of MMP2 or TIMP2. Collectively, these results suggest that intracellular S100A8/A9 contributes to the cancer cell phenotype by modulating MMP-2 expression and activity to regulate cell migration and mobility.
Keywords: calprotectin, S100A8/A9, matrix metalloproteinase-2, carcinoma, in vitro
1. Introduction
Calprotectin is a heterodimeric complex of the calcium-binding proteins S100A8 and S100A9 (S100A8/A9). As members of the S100 protein family, S100A8/A9 had been postulated to regulate cell cycle progression, cell growth and cell survival (Heizmann et al., 2002; Emberley et al., 2004). Indeed, S100A8/A9 appears to be a control of the cell-cycle checkpoint at G2/M and malignant phenotype in carcinoma cells (Khammanivong et al., 2013). Expressed in a cell- and tissue-specific manner, S100A8/A9 is differentially regulated in different tissues and types of malignancy. For example, squamous epithelial tissues constitutively express S100A8/A9; S100A8/A9 is typically down-regulated in primary human head and neck squamous cell carcinomas (HNSCC), including oral, nasopharyngeal and oropharyngeal, and esophageal (Coleman & Stanley, 1994; Fung et al., 2000; Kong et al., 2004; Wang et al., 2004; Tugizov et al., 2005). In these squamous mucosal cancers, decreased S100A8/A9 and loss of cellular differentiation correlate and are inversely related to proliferation. Conversely, S100A8/A9 expressing SCCs appear less aggressive (Wang et al., 2004; Roesch et al., 2005; Sewell et al., 2007).
Increased cell proliferation and malignant phenotype are associated with epithelial-mesenchymal transition (EMT). In contrast to normal epithelium, EMT reflects the loss of cellular adherence and contact growth, involving dysregulation of adhesion molecules and increased breakdown of the extracellular matrix by matrix metalloproteinases (MMPs) (Guarino et al., 2007; Neth et al., 2007). MMPs are a family of zinc-dependent proteinases that degrade most extracellular matrix components (Kondo et al., 2007). Approximately 20 members of the MMP family have been identified, sharing common structural and functional elements (Zucker & Vacirca, 2004). In comparison to normal tissues, cancers often show abnormally elevated MMP-2 expression and activity, which are directly correlated with invasion, metastasis and poor prognosis (Pellikainen et al., 2004; Vihinen et al., 2005; Turpeenniemi-Hujanen, 2005). Targeting MMP-2 activity, therefore, is actively pursued as a potential anticancer therapy.
We now report that S100A8/A9 contributes to control of MMP-2 expression and malignant phenotype in carcinoma cells. In S100A8/A9-negative human carcinoma cells (KB cell line), transfection to express S100A8 and S100A9 selectively down-regulated MMP-2 and inhibited in vitro invasion and migration. Conversely, silencing endogenous S100A8/A9 expression in TR146 buccal carcinoma cells increased MMP-2 activity and in vitro invasion and migration. In contrast, silencing MMP-2 expression appears to drive cells to a less malignant phenotype. S100A8/A9-dependent expression of MMP-2 was not apparently related to epigenetic changes in the upstream promoters of either MMP2 or TIMP2. MMP-2 levels were greater in 3D than 2D cell cultures on collagen substrate, more closely modeling the architecture of cell growth in vivo.
2. Materials and Methods
2.1. Cell culture
S100A8/A9 was studied in S100A8/A9-negative KB carcinoma cells as we previously described (Nisapakultorn et al., 2001; Khammanivong et al., 2013). In brief, KB cells (ATCC CCL-17), a HeLa-like S100A8/A9-negative human carcinoma cell line, had been co-transfected to express S100A8 (MRP8) and S100A9 (MRP14) in the same cell to generate KB-S100A8/A9 cells (same as KB-MRP8/14 cells reported previously) (Nisapakultorn et al., 2001). A sham-control transfectant, KB-EGFP, was also generated. The transfected cells were compared experimentally to the parent KB carcinoma cells. As we described previously (Sorenson et al., 2012), TR146 cells, a buccal carcinoma cell line, which constitutively expresses S100A8/A9, was transfected with short hairpin RNA (shRNA) to silence S100A8 and S100A9 (termed TR146-S100A8/A9-shRNA). TR146-shRNA-control cells were produced as a negative control cell line for S100A8/A9 gene silencing by transfecting with non-specific shRNA for any mammalian gene.
KB cells were maintained in Minimum Essential Medium (MEM), whereas TR146 cells were cultured in Dulbecco’s Modified Eagle’s Medium/Ham’s F-12 (DMEM/F-12; 1:1 volume ratio) (Mediatech Inc., Manassas, VA); both media were supplemented with 10% fetal bovine serum. MCF-7 cells were maintained in DMEM supplemented with 5% fetal bovine serum. KB-EGFP and KB-S100A8/A9 were maintained in 700 μg/ml Geneticin® (G418) sulfate (Mediatech), whereas TR146-shRNA-control and TR146-S100A8/A9-shRNA were maintained in 250 μg/ml G418 sulfate. The wild-type KB and TR146 cells were grown in complete medium without G418 sulfate (Sorenson et al., 2012).
MMP-2 expression in KB cells was knocked-down using small interfering RNA (siRNA) for MMP-2 (sc-29398; Santa Cruz Biotech) as described in the manufacturer’s instructions. Briefly, KB cells were washed with siRNA transfection medium (sc-36868, Santa Cruz Biotech) and treated with MMP-2 siRNA, resuspended to 10 μM in RNAse-free water, or with scrambled siRNA (control) in transfection reagent (sc-29528, Santa Cruz Biotech). After 72 h, cells were collected and lysed and the efficiency of MMP-2 knockdown was determined by Western Blotting (Ke et al., 2006).
2.2. 2D collagen substrate cultures
For two-dimensional collagen cultures, CytoOne 6-well plates (USA Scientific, Ocala, FL) were coated by incubating with 1 mg/mL collagen type I (BD Biosciences, San Jose, CA) for 1 h at 37°C. Each well was rinsed with PBS. Cells were then plated at a density of approximately 3 × 105 cells/mL.
2.3. 3D collagen matrix cell cultures
Collagen type 1 stock solution (BD Biosciences, San Jose, CA) was diluted to 1 mg/mL at 4°C as recommended by the manufacturer. The diluted collagen solution (1 mL) was mixed with 3 × 105 cells, pipetted into the wells of 6-well plates as above and incubated (37°C, 5% CO2) for 1 h to allow complete polymerization. After polymerization, culture media (1 mL) was added on top of the collagen gel (Chen et al., 2012).
2.4. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Cellular expression of S100A8, S100A9 and MMPs was verified using polymerase chain reaction (PCR) analysis (Schröpfer et al., 2010). For RT-PCR, total cellular RNA was isolated using TRIzol reagent (Invitrogen) and concentration and purity were determined by analyzing absorption at 260/280 nm. Genomic contamination was eliminated using the RNeasy® Mini Kit (Qiagen), and 1 μg of total RNA per sample was used to generate cDNA using the iScript™ enzyme (Bio-Rad Laboratories, Hercules, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control.
2.5. Real-Time quantitative PCR
To analyze expression of S100A8, S100A9 and MMP2 mRNA, total RNA was isolated as above and cDNA was synthesized using the SuperScript™ III First-Strand Synthesis System (Invitrogen). MMP2 mRNA was quantified using real-time quantitative PCR (TaqMan® Reverse Transcription Kit, Invitrogen). For human S100A8 and S100A9, primers were obtained from Integrated DNA Technologies (Coralville, IA) and for MMP2, PrimeTime pre-designed qRT-PCR assays were used (Integrated DNA Technologies, Coralville, Iowa) and GAPDH (Integrated DNA Technologies) was used as an internal control.
2.6. MMP activity assay
MMP activity was assayed by zymography as previously described (Gerlach et al., 2007). Conditioned serum-free medium was collected, equal amounts of protein were loaded onto 10% polyacrylamide gels containing 1 g/L gelatin, and proteins were separated electrophoretically. The gels were re-natured in 2.5% Triton-X-100 with gentle agitation for 30 min at room temperature, placed into developing buffer (5 mM CaCl2, 50 mM Tris, 0.2 mM NaCl, and 0.02% Brij35, pH 7.5) for 30 min at room temperature, then incubated overnight at 37°C, stained with Coomassie Brilliant Blue R-250 for 30 min, destained, and digestion of gelatin was visualized as clear, unstained bands.
2.7. Western blot analysis
Cells were washed twice with 1 to 2 ml ice-cold (4°C) Dulbecco’s-PBS and lysed in standard radioimmunoprecipitation assay (RIPA) buffer (Thermo Scientific, Rockford, IL, USA). After centrifugation, soluble protein concentrations were measured using Bicinchoninic Acid (BCA) assay. Total protein (50 μg) was resolved using SDS-PAGE and transferred onto nitrocellulose membranes. Anti-MMP-1 (ab2461), anti-MMP-2 (ab2462), anti-MMP-9 (ab3159) and anti-MMP-15 (ab53770) were purchased from ABcam (Cambridge, MA, USA). Rabbit anti-β-actin (DB070, Delta Biolabs, Gilory, CA) was used as control. Membranes were visualized using PIERCE® ECL Western Blotting Substrate (Thermo Scientific, Rockford, IL, USA) and exposed to Amersham Hyperfilm ECL film (GE Healthcare Biosciences, Piscataway, NJ).
2.8. Cell migration assay
Cells were grown at 37°C with 5% CO2 to approximately 80-90% confluence in 6-well plates and were incubated for 2 to 3 hours with 10 μg/ml Mitomycin C from Streptomyces caespitosus (M4287, Sigma-Aldrich, Saint Louis, MO), an inhibitor of DNA synthesis. After incubation, the medium containing Mitomycin C was removed and cells were washed once with 2 mL of Dulbecco’s-PBS per well. A center swathe of cells of reproducible dimensions was scraped from the monolayer using a sterile 200 μl pipette tip. Dislodged cells and debris were removed by rinsing twice with MEM. For KB, KB-EGFP and KB-S100A8/A9 cell lines, incubation continued for up to 48 h in fresh MEM supplemented with 10% fetal bovine serum, and images were recorded immediately (time zero), 24 and 48 h later. TR146, TR146-S100A8/A9-shRNA, TR146-shRNA-control, and the KB-MMP-2 knocked-down cells were each incubated for up to 24 h and images were recorded at time zero and 24 h later. To assure optimal silencing of MMP-2 during cell migration assays, growing KB cells were plated at 50-60% confluency in 6-well plates for 12 to 18 h before transfection. A swathe was scraped 72 hrs after transfection and MMP-2 knockdown efficiency was confirmed by Western blotting. Cell migration into the denuded area was visualized at a magnification of 200X using an upright Nikon AZ100 Motorized Microscope (Tokyo, Japan) and documented with a Nikon DS-Ri1 color 12-bit camera. Captured images were processed using Nikon Elements AR software for multi-dimensional data collection. Migration of cells was quantified using a planimetric approach. Denuded swathes of images at the identical magnification were measured as the weight of paper corresponding to the area of at least four randomly selected fields. The total weight of paper was subsequently converted into the area of remaining denuded surface. Assays were performed in triplicate for each cell line and repeated at least three times.
2.9. Matrigel invasion assay
Transwell inserts of 8 μm pore size polyethylene terephthalate (PET) membranes covered with MATRIGEL Basement Membrane Matrix (#354480, BD BioCoat, BD Biosciences, Bedford, MA) were placed into transwell chambers and used for this study of cell invasion. Cells (1.25 × 105 in 500 μL of serum free medium) were added to the upper chamber inserts, and 750 μL DMEM with 20% serum was added to the lower chamber. After 22 h at 37°C in 5% CO2, cells remaining on the upper side of the membrane (non-invading) were removed using cotton swabs. Cells that invaded to the underside were fixed in ice-cold methanol at room temperature for 30 min at room temperature and then stained with crystal violet solution. KB cells were transfected with the MMP-2 siRNA 72 hrs before the matrigel invasion assays. KB-MMP-2-siRNA cells were grown to 70 to 80% confluency in 10 cm plates (about 72 hrs of culture), the efficiency of MMP-2 knockdown was confirmed by Western blot analysis, and harvested cells were seeded on the inserts. The number of invaded cells in 10 high power fields (hpfs) was counted by visualizing at 100× magnification under an inverted Leica MZ FL III Stereomicroscope using a Leica DFC420 C camera. Assays were performed in four replicates (inserts) for each cell line and repeated at least 3 times.
2.10. DNA methylation analysis
Genomic DNAs from MCF-7, KB, KB-EGFP and KB-S100A8/A9 cells were isolated using the PureLink™ genomic DNA mini kit (Invitrogen). The purified genomic DNA for each cell line was bisulfite converted using the CpGenome Fast DNA Modification Kit (Millipore, MA) according to the manufacturer's recommendations. Bisulfite-treated DNA was then analyzed by methylation-sensitive PCR (MSP) analysis and bisulfite DNA sequencing. All PCR amplifications were carried out using Platinum Taq polymerase (Life Technologies, Grand Island, NY). MSP analysis for human MMP-2 was performed as described previously (Chernov et al. 2009; Farias et al. 2012) (see Fig. S1A). Bisulfite-treated genomic DNA from human blood (Promega) and MCF-7 cells were used as positive controls for unmethylated and methylated specific amplification, respectively. Amplified products were separated by gel electrophoresis. Bisulfite DNA sequencing of BIS1 and BIS2 region of MMP-2 (see Fig. S1B) or TIMP-2 (see Fig. S2) in KB-EGFP and KB-S100A8/A9 cells was performed as described previously (Chernov et al., 2009).
2.11. Statistical analysis
Differences among multiple independent groups were evaluated for significance using one-way ANOVA and subsequent comparisons were made with the Tukey test. Differences between two independent groups were analyzed using Student’s t-test. For all statistical analyses, the criterion for significance was p < 0.05.
3. Results
3.1. Stable expression of S100A8/A9 in KB cells inhibits MMP-2 expression
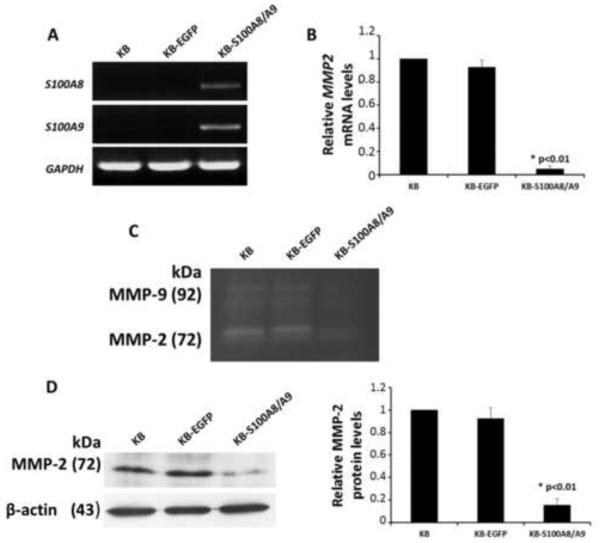
KB-S100A8/A9 cells stably expressed S100A8 and S100A9 mRNAs as analyzed by PCR (Fig. 1A). Neither mRNA was detected in the parent KB cells or in KB-EGFP, which received the EGFP vector. We analyzed the three KB cell lines for expression of MMP1, MMP2, MMP3, MMP7, MMP9, MMP13, MMP15, MMP17, MMP23, MMP24, and MMP28 mRNA using PCR (Fig. S1A). Expression of MMP1, MMP2, MMP9 and MMP15 mRNAs differed between the cell lines, but only MMP-2 differed at the protein level using Western blots (Fig. S1B).
Figure 1. Endogenous expression of MMP2 in KB cells.
(A) Semi-quantitative RT-PCR analysis confirming expression of S100A8 and S100A9 mRNA in KB cells. (B) mRNA levels of MMP2 in KB, KB-EGFP and KB-S100A8/A9 cell lines using quantitative RT-PCR. All mRNA values were normalized to GAPDH, a housekeeping gene, and expressed relative to KB control cells. (C) Zymography analysis for gelatinase activity. (D) Levels of MMP-2 protein in KB, KB-EGFP and KB-S100A8/A9 cell lines using Western blot analysis. Protein levels were normalized to β-actin and expressed relative to KB cell controls. MMP-2 protein and mRNA levels are analyzed by one-way ANOVA and the statistical analysis reveal differences between the cell lines (P < .05).
Therefore, KB, KB-EGFP, and KB-S100A8/A9 cells were compared for MMP2 expression using real-time PCR (Fig. 1B), and MMP-2 activity using zymographic (Fig. 1C), and immunoreactive protein (Fig. 1D) analyses. Stable expression of S100A8/A9 inhibited MMP2 mRNA expression, MMP-2 enzyme activity and protein production.
3.2. S100A8/A9 expression decreased KB cell motility and matrigel invasion
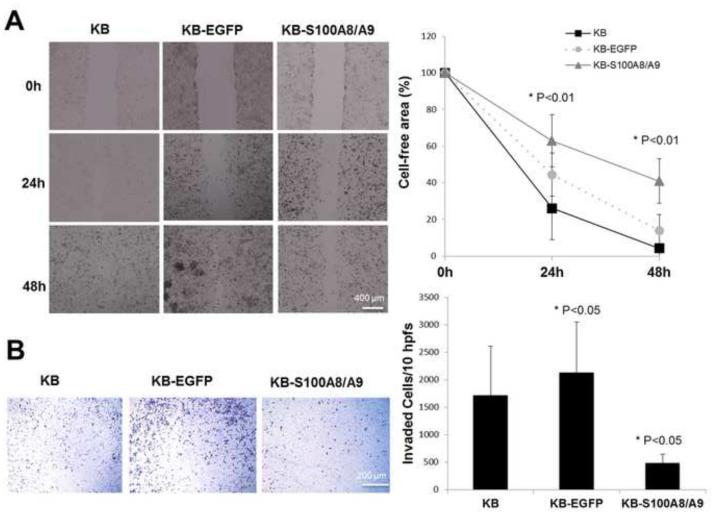
To determine whether S100A8/A9 expression affects the potential for KB cells to migrate and invade, KB, KB-EGFP and KB-S100A8/A9 cells were analyzed in vitro. KB-S100A8/A9 cells showed significantly reduced motility (reduced cell-free area) for up to 48 h when compared to KB and KB-EGFP cells (Fig. 2A). Likewise, S100A8/A9 expression in KB cells significantly reduced the ability of the cells to invade the matrigel when compared to KB or KB-EGFP cells (Fig. 2B). Unexpectedly, invasiveness increased as a result of traansfection to create the KB-EGFP cells.
Figure 2. S100A8/A9 up-regulation inhibited motility and matrigel invasion.
(A) KB, KB-EFGP and KB-S100A8/A9 cells migrate to fill a uniform gap in the monolayer. Cells were treated with Mitomycin-C before the motility assay to inhibit cell proliferation. Monolayer closure expressed as a function of time after removal of uniform swath of cells was observed, photographed, and measured at times up to 48 h. The experiments were performed in triplicate for each cell line and repeated at least three times with similar results. (B) Matrigel invasion capacity of KB, KB-EFGP and KB-S100A8/A9 cells using transwell analysis. Cells invading through to the underside of the Matrigel-coated transwell membrane were fixed with ice-cold methanol for 30 minutes, stained with crystal violet and observed using an inverted microscope at 100x magnification. For each cell line, 10 hpfs were randomly chosen, the number of invaded cells per field was counted, and the total number of cells in the 10 hpfs was reported. Assays were performed in four replicates (inserts) for each cell line and repeated 3 times. Values were analyzed by one-way ANOVA and significant differences reported (P < .05). Data are expressed as mean ± SD.
3.3. Knockdown of S100A8/A9 in TR146 cells promotes MMP-2 expression
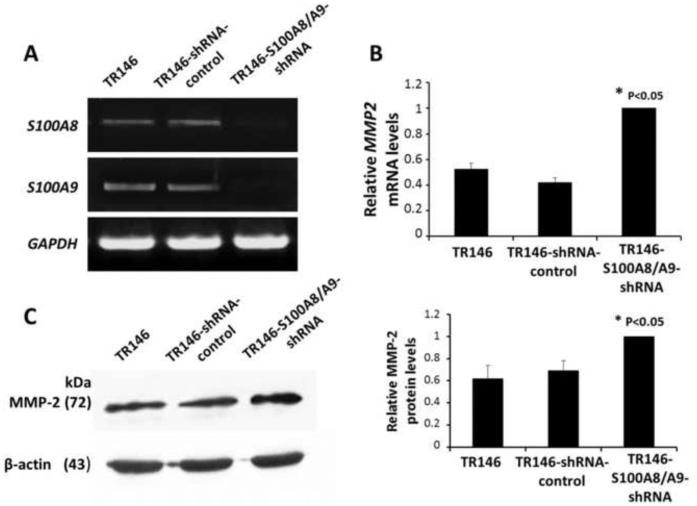
To confirm that expression of S100A8/A9 regulates expression of MMP2, TR146 cells were transfected with a mammalian shRNA expression vector, silencing both S100A8 and S100A9 genes. When compared to TR146 or TR146 cells with control shRNA, TR146-S100A8/A9-shRNA cells show no detectable expression of S100A8 or S100A9 (Fig. 3A) and significant increases in MMP2 mRNA (Fig. 3B) and MMP-2 protein (Fig. 3C). S100A8/A9-dependent control of MMP2 expression was not idiosyncratic to the KB cell line.
Figure 3. Endogenous expression of MMP2 in TR146 cells.
(A) Confirmation of knockdown of endogenous S100A8 and S100A9 in TR146 cells using PCR analysis. (B) MMP2 mRNA levels in TR146, TR146-shRNA-control, TR146-S100A8/A9-shRNA cell lines determined using quantitative RT-PCR. All mRNA values were normalized to GAPDH, a housekeeping gene, and expressed relative to TR146 control cells. (C) Levels of MMP-2 protein in TR146, TR146-shRNA-control, and TR146-S100A8/A9-shRNA cells as determined using Western blots. Protein levels were normalized to β-actin and expressed relative to TR146 cells. Assays were performed in triplicate and repeated three times. The protein and mRNA levels were analyzed by one-way ANOVA and statistical differences between the cell lines were reported (P < 0.05).
3.4. S100A8/A9 knockdown enhanced motility and matrigel invasion of TR146 cells
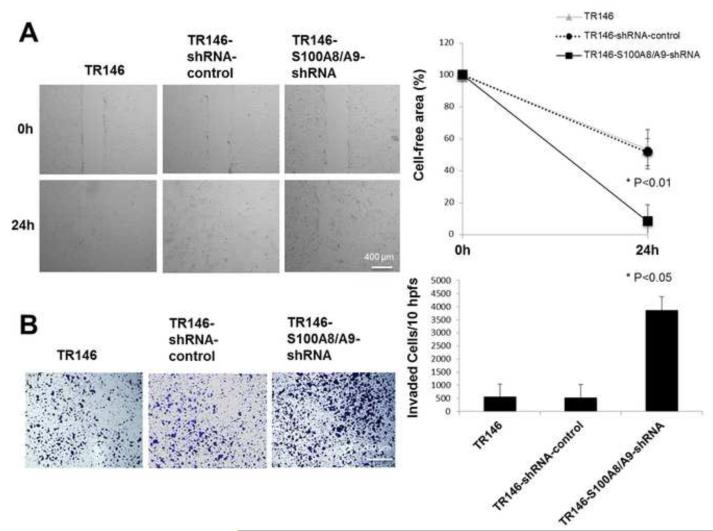
Knock-down of S100A8/A9 in TR146 cells significantly increased motility (reduced cell-free area) (Fig. 4A) and promoted invasion (Fig. 4B) in vitro in comparison to TR146 and TR146-shRNA-control cell lines, which express S100A8/A9.
Figure 4. S100A8/A9 silencing promoted motility and matrigel invasion by TR146 cells.
(A) TR146, TR146-control-shRNA and TR146-S100A8/A9-shRNA cells treated with Mitomycin-C migrate to fill a uniform gap in the monolayer as described in the Materials and Methods. The monolayer gap was photographed and measured after 24 h. The experiment was performed in triplicate for each cell line and repeated four times. (B) TR146, TR146-control-shRNA and TR146-S100A8/A9-shRNA cells invading matrigel precoated transwell membranes. Cells invading through to the underside of the membrane were fixed with ice-cold methanol for 30 minutes, stained with crystal violet and observed using an inverted microscope at 100x magnification. For each cell line, 10 hpfs were randomly chosen, the number of invaded cells per field was counted, and the total number of cells in the 10 hpfs was reported. Assays were performed in four replicates (inserts) for each cell line and repeated 3 times. These values were analyzed by one-way ANOVA and statistical differences between the cell lines were reported (P < 0.05). Data are expressed as mean ± SD.
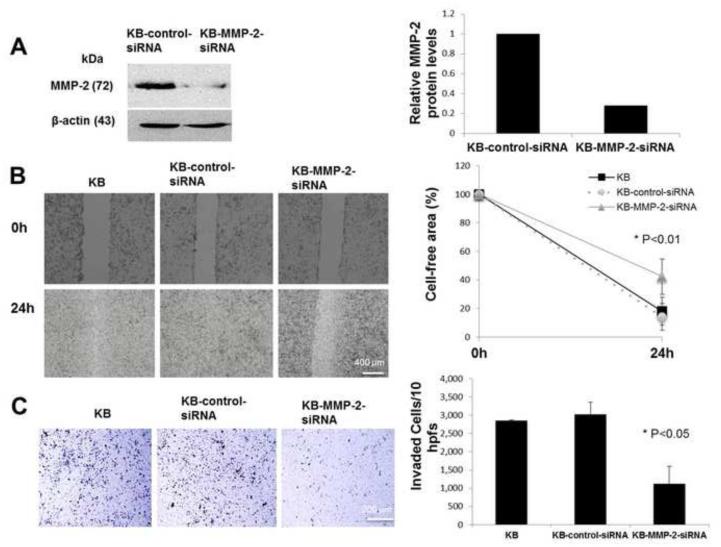
3.5. MMP-2 silencing inhibited motility and matrigel invasion of KB cells
To confirm that modulation of MMP-2 by S100A8/A9 affects motility and invasion properties of cancer cells, we silenced MMP2 expression in KB cells. Silencing reduced MMP2 expression by more than 60% when compared to cells transfected with control siRNA (Fig. 5A). Consistent with reduced MMP-2, cell migration (Fig. 5B) and invasion (Fig. 5C) were both significantly reduced in KB-MMP-2-siRNA cells when compared to KB and KB-control-siRNA cell lines with unaffected MMP-2 expression.
Figure 5. MMP2 silencing inhibited motility and matrigel invasion in KB cells.
(A) MMP-2 protein levels in KB, KB-control-siRNA and KB-MMP2-siRNA cells using Western blot analysis. Protein levels were normalized to β-actin. (B) KB, KB-control-siRNA and KB-MMP2-siRNA cell motility as photographed and measured after 24 h. Growing KB cells were plated at 50 to 60% confluency in 6-well plates for 12 to 18 h before transfection. A swathe was scraped 72 hours after transfection and the efficacy of MMP-2 knockdown was confirmed by Western Blotting. Assays were performed in triplicate for each cell line and repeated 3 times. (C) KB, KB-control-siRNA and KB-MMP2-siRNA cell invasion using matrigel transwell analysis. For each cell line, 10 hpfs were randomly chosen, the number of invaded cells per field was counted and the total number of cells in ten fields was reported. The experiment was performed in four replicates (inserts) for each cell line and repeated 3 times with reproducible results. These values were analyzed by one-way ANOVA and significant differences were reported (P < 0.05). Data are expressed as mean ± SD.
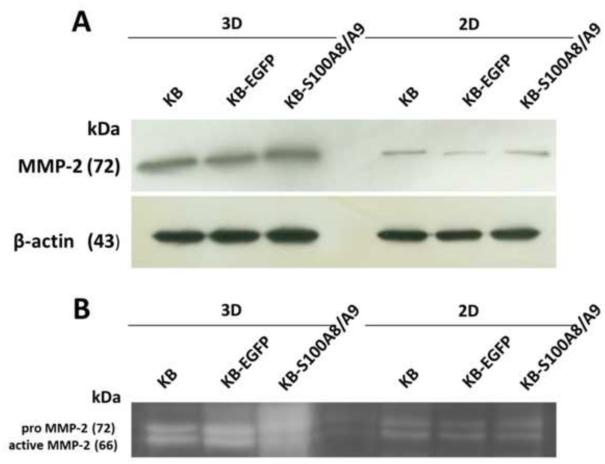
3.6. MMP2 expression and total cellular activity in 3D collagen matrix substrate cultures
To learn whether substrate tissue architecture would affect our results, we grew cells in 2D and 3D collagen matrix substrates and compared MMP-2 expression. MMP-2 protein levels in KB, KB-EGFP, and KB-S100A8/A9 cells were greater when grown in 3D than 2D collagen substrate cultures and were unaffected by the expression of S100A8/A9 (Fig. 6A). Similarly, there was no detectable difference in the proportion of pro MMP-2 to active MMP-2 in 2D and 3D cultures between any of the cell lines (Fig. 6B). Note that during electrophoresis, SDS activates the “cysteine switch”, dissociating Cys73 from bound zinc resulting in activation of the pro-MMP-2 (Snoek-van Beurden and Von den Hoff, 2005).
Figure 6. MMP2 expression and activity in 2D and 3D collagen cultures.
(A) MMP-2 protein levels in KB cells cultured in 2D and 3D collagen cultures using Western blot analysis. (B) Gelatinase activities in 2D and 3D collagen cultures as observed using zymography.
3.7. S100A8/A9 inhibition of MMP-2 expression and MMP2 or TIMP2 upstream methylation status
We determined whether DNA methylation status of the human MMP2 5’ upstream promoter region affects S100A8/A9-dependent MMP2 gene regulation. Most KB, KB-EGFP and KB-S100A8/A9 cells show MMP2 methylation (Fig. S2A). There is a weak unmethylated signal suggesting that some cells contain unmethylated MMP2 promoters (Fig. S2A). To better characterize the methylation status of the human MMP2 upstream region in KB-EGFP and KB-S100A8/A9 cells, a 221 bp BIS1 region with 26 CpG sites and a 230 bp BIS2 region with 6 CpG sites were chosen for further analysis (Chernov et al., 2009). In a total of six clones that represent the population of KB-EGFP and KB-S100A8/A9 cells, the frequency of methylation sites was similar in KB-EGFP and KB-S100A8/A9 cells (Fig. S2B). When compared to the BIS2 region, the BIS1 region of MMP2 was hypermethylated in KB-EGFP and KB-S100A8/A9 cells (Fig. S2B). Only a few methylated sites were observed in BIS2 region in either KB-EGFP or KB-S100A8/A9 cells (Fig. S2B).
We also determined whether the DNA methylation status of human TIMP-2, an endogenous inhibitor of matrix metalloproteinases (MMPs) (Ivanova et al., 2004), contributes to S100A8/A9-dependent MMP-2 gene regulation. A 329 bp BIS1 region with 16 CpG sites and a 220 bp BIS2 region with 12 CpG sites in human TIMP-2 upstream region were chosen for bisulfite sequencing analysis (Chernov et al., 2009). In contrast to hypermethylated BIS1, the 5’ end of the BIS2 region of the human TIMP-2 upstream region was less methylated, whereas the 3’ end of the region was highly methylated and similar in KB-EGFP and KB-S100A8/A9 cells (Fig. S3).
4. Discussion
S100A8/A9 plays a mechanistic role in cell cycle control of certain carcinoma cells, maintaining the control checkpoint at G2/M (Khammanivong et al., 2013). As we now report, the presence of this protein complex in carcinoma cells (KB) appears to modulate additional features of the cancer cell phenotype in vitro, including MMP-2-dependent motility and invasion through matrigel precoated membranes. S100A14 also regulates expression of MMP-2 in squamous cell carcinoma (Chen et al., 2012). Unlike S100A8/A9, which does not interact with G1/S controls, S100A14 functions to control MMP-2 through p53-dependent transcriptional regulation. Since KB cells expressing S100A8/A9 show slower growth than sham-transfected cells, we might have assumed cell cycle control to be a singular explanation for the phenotypic differences we observed. In the present study, however, we showed that expression of S100A8/A9 in KB and TR146 carcinoma cells selectively reduces production of MMP-2. Attributable to S100A8/A9, reduced MMP-2 also attenuates the invasive and migratory potential of the KB and TR146 carcinoma cells as studied in vitro.
This unexpected connection between S100A8/A9 and MMP-2 production and activity in vitro and the associated changes in cancer phenotype would appear to be relevant to human cancer. It has long been established that deregulated motile behavior contributes to pathological processes including tumor metastasis (Haemmerlin & Sträuli, 1981). Facilitating extracellular matrix or basement membrane invasion and migration, cells migrate from a primary tumor to blood vessels or lymphatics. To model and compare cell motility and invasive capacity among cell lines in vitro, cell migration and matrigel invasion assays are commonly used.
Tumor cell invasion and metastasis, characteristic of malignant phenotypes, requires regulated expression of MMPs (Vinihen et al., 2005; Pellikainen et al., 2004). Among the MMPs, MMP-2 and MMP-9 have well-characterized roles in the invasiveness and metastasis of cancer cells (de Vicente et al., 2005; Zhou et al., 2010; Roh et al., 2012). In our study, the production of S100A8/A9 selectively reduced the concomitant production of MMP-2 protein and associated enzymatic activity. Expression of other MMPs by the KB cells was not similarly affected by S100A8/A9. Using knock-in and knock-down approaches to change expression of S100A8/A9 in different carcinoma cell lines, we showed that expression of MMP-2 and the malignant phenotype were co-regulated in vitro. Silencing MMP2 increased motility and invasiveness of the KB cells, directly linking the modulation of these activities by S100A8/A9 to MMP-2 levels. Hence, S100A8/A9 reduces MMP-2 expression and activity, suggesting less degradation of ECM components, and a less malignant phenotype expressed by carcinoma cells.
Carcinoma cells in vitro produce their own two-dimensional extracellular matrix, which can be modified by MMP-2 to facilitate cell adhesion and migration (Lu et al., 2012; Wu et al., 2012; Chen et al., 2013). Cells grown in 3D conditions produce higher levels of MMPs than in 2D monolayers, reflecting the need to degrade the more complex, spatially constrained architecture of the extracellular matrix (Olsen et al., 2010; Sarkar & Yong, 2009). Over time, MMPs process and mature the extracellular matrix (Karamichos et al., 2007). The changing characteristics of the matrix are likely to signal for variation in MMP expression.
Since carcinoma cells in vivo establish a 3D extracellular matrix (Kulasekara et al., 2009), we also compared MMP2 expression and MMP-2 activity in KB cells using 2D and 3D type I collagen matrix culture models (Fig. 6). Since the 3D model involved growth of KB cells in a type I collagen gel, we established 2D growth on a type I collagen substrate for comparison. On 2D and 3D collagen substrates, S100A8/A9 positive and negative (KB and KB-EGFP) KB cell lines showed similar MMP-2 expression and inactive and active pro-enzyme forms; MMP-2 levels appeared greater in cells grown in 3D cultures. In contrast, S100A8/A9-dependent down-regulation of MMP2 was observed only when the cancer cells produce their own 2D ECM. Since MMP2 expression by KB, KB-EGFP and KB-S100A8/A9 cells was similar in 2D and 3D collagen type I cultures, we would expect that native ECMs produced largely by the carcinoma cells would replicate the down-regulation seen in 2D cultures. Hence, an outside-in signal from the tumor stroma is suggested to regulate MMP-2 production in 2D or 3D conditions and the regulation by S100A8/A9 is indirect.
Indeed, we sought evidence for a direct mechanism whereby S100A8/A9 would affect promoter methylation of MMP2 or TIMP2. There was no significant difference in the MMP2 or TIMP2 promoter methylation status in KB cells in the presence and absence of S100A8/A9. The data suggest, therefore, that S100A8/A9 affects stromal synthesis and ECM fabrication or the selectivity of the outside-in signal that regulates MMP expression.
The invasiveness and migratory potential of carcinoma cells involves independently regulated expression of MMP2 and other factors to remodel tumor stroma (ECM). During tumorigenesis in vivo, carcinoma cells engage and direct stromal fibroblasts to cooperatively control the expression and characteristics of the stromal environment (Dolznig et al., 2011; Lee et al., 2011). When grown on type I collagen in 2D and 3D cultures, however, keratinocyte-releasable factors caused co-cultured fibroblasts to up-regulate MMP1 and MMP3 (Li et al., 2009). When co-cultured in 3D conditions on Matrigel matrix with breast cancer cells, fibroblasts increased production of S100A4 and MMP-2 and showed greater invasive phenotype (Olsen et al., 2010). Fibroblast MMPs and the composition of the stroma are also important determinants of the fate of tumor cells.
Understanding the molecular basis of cancer cell invasion and migration is crucial to the development of more successful and more targeted therapies. Metastasis is the final stage in tumor progression from a normal cell to a fully malignant cell and is the cause of 90% of all deaths from cancer (Yilmaz et al., 2007). Metastasis of cancer cells is a complex multistep process involving cell adhesion, motility, migration and invasion. Hence, interruption of one or more of these steps may limit direct tissue invasion and contribute to anti-metastatic therapy (Nam & Shom, 2009).
5. Conclusion
Taken together, our results demonstrate that intracellular S100A8/A9 was able to inhibit cancer cell migration and stromal invasion. The underlying mechanism requires that S100A8/A9 indirectly attenuate expression and activity of MMP-2 in the carcinoma lines studied. How this mechanism manifests in more complex, native 3D co-culture models and in vivo remains to be learned.
Supplementary Material
6. Acknowledgements
This work was supported by NIH/NIDCR R01DE021206 to MCH and by CAPES 6177/11-5 to EJS. These studies were performed by EJS to satisfy partial requirements for the PhD degree at the Piracicaba School of Dentistry, University of Campinas, SP, Brazil.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- Chernov AV, Sounni NE, Remacle AG, Strongin AY. Epigenetic control of the invasion-promoting MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J Biol Chem. 2009;284:12727–34. doi: 10.1074/jbc.M900273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champaiboon C, Sappington KJ, Guenther BD, Ross KF, Herzberg MC. Calprotectin S100A9 calcium-binding loops I and II are essential for keratinocyte resistance to bacterial invasion. J Biol Chem. 2009;13:7078–90. doi: 10.1074/jbc.M806605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xiao Z, Meng Y, Zhao Y, Han J, Su G, Chen B, Dai J. The enhancement of cancer stem cell properties of MCF-7 cells in 3D collagen scaffolds for modeling of cancer and anti-cancer drugs. Biomaterials. 2012;33:1437–44. doi: 10.1016/j.biomaterials.2011.10.056. [DOI] [PubMed] [Google Scholar]
- Chen JS, Huang XH, Wang Q, Huang JQ, Zhang LJ, Chen XL, Lei J, Cheng ZX. Sonic hedgehog signaling pathway induces cell migration and invasion through focal adhesion kinase/AKT signaling-mediated activation of matrix metalloproteinase (MMP)-2 and MMP-9 in liver cancer. Carcinogenesis. 2012;34:10–9. doi: 10.1093/carcin/bgs274. [DOI] [PubMed] [Google Scholar]
- Chen H, Yuan Y, Zhang C, Luo A, Ding F, Ma J, Yang S, Tian Y, Tong T, Zhan Q, Liu Z. Involvement of S100A14 protein in cell invasion by affecting expression and function of matrix metalloproteinase (MMP)-2 via p53-dependent transcription regulation. J Biol Chem. 2012;287:17109–19. doi: 10.1074/jbc.M111.326975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman N, Stanley MA. Expression of the myelomonocytic antigens CD36 and L1 by keratinocytes in squamous intraepithelial lesions of the cervix. Hum Pathol. 1994;25:73–79. doi: 10.1016/0046-8177(94)90174-0. [DOI] [PubMed] [Google Scholar]
- De Vicente JC, Fresno MF, Vilalain L, Vega JA, Hernández Vallejo G. Expression and clinical significance of matrix metalloproteinase-2 and matrix metalloproteinase-9 in oral squamous cell carcinoma. Oral Oncol. 2005;41:283–93. doi: 10.1016/j.oraloncology.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Dolznig H, Rupp C, Puri C, Haslinger C, Schweifer N, Wieser E, Kerjaschki D, Garin-Chesa P. Modeling colon adenorcarcinomas in vitro a 3D co-culture system induces cancer-relevant pathways upon tumor cell and stromal fibroblast interaction. Am J Pathol. 2011;179:487–501. doi: 10.1016/j.ajpath.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberley ED, Murphy LC, Watson PH. S100 proteins and their influence on pro-survival pathways in cancer. Biochem Cell Biol. 2004;82:508–15. doi: 10.1139/o04-052. [DOI] [PubMed] [Google Scholar]
- Farias LC, Gomes CC, Rodrigues MC, de Castro WH, Lacerda JC, Ferreira EF, Gomez RS. Epigenetic regulation of matrix metalloproteinase expression in ameloblastoma. BMC Clin Pathol. 2012;12:11. doi: 10.1186/1472-6890-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung LF, Lo AK, Yuen PW, Liu Y, Wang XH, Tsao SW. Differential gene expression in nasopharyngeal carcinoma cells. Life Sci. 2000;67:923–36. doi: 10.1016/s0024-3205(00)00684-6. [DOI] [PubMed] [Google Scholar]
- Gerlach RF, Demacq C, Jung K, Tanus-Santos JE. Rapid separation of serum does not avoid artificially higher matrix metalloproteinase (MMP-9) level in serum versus plasma. Clin Biochem. 2007;40:119–23. doi: 10.1016/j.clinbiochem.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–18. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- Heizmann CW, Fritz G, Schafer BW. S100 proteins: structure, functions and pathology. Front Biosci. 2002;7:d1356–68. doi: 10.2741/A846. [DOI] [PubMed] [Google Scholar]
- Ivanova T, Vinokurova S, Petrenko A, Eshilev E, Solovyova N, Kisseljov F, Kisseljova N. Frequent hypermethylation of 5' flanking region of TIMP-2 gene in cervical cancer. Int J Cancer. 2004;108:882–6. doi: 10.1002/ijc.11652. [DOI] [PubMed] [Google Scholar]
- Karamichos D, Brown RA, Mudera V. Collagen stiffness regulates cellular contraction and matrix remodeling gene expression. J Biomed Mater Res A. 2007;83:887–94. doi: 10.1002/jbm.a.31423. [DOI] [PubMed] [Google Scholar]
- Ke Z, Lin H, Fan Z, Cai TQ, Kaplan RA, Ma C, Bower KA, Shi X, Luo J. MMP-2 mediates ethanol-induced invasion of mammary epithelial cells over-expressing ErbB2. Int J Cancer. 2006;119:8–16. doi: 10.1002/ijc.21769. [DOI] [PubMed] [Google Scholar]
- Khammanivong A, Wang C, Sorenson BS, Ross KF, Herzberg MC. S100A8/A9 (Calprotectin) negatively regulates G2/M cell cycle progression and growth of squamous cell carcinoma. PLoS One. 2013;9:e69395. doi: 10.1371/journal.pone.0069395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Shukunami C, Morioka Y, Matsumoto N, Takahashi R, Oh J, Atsumi T, Umezawa A, Kudo A, Kitayama H, et al. Dual effects of the membrane-anchored MMP regulator RECK on chondrogenic differentiation of ATDC5 cells. J Cell Sci. 2007;120:849–857. doi: 10.1242/jcs.03388. [DOI] [PubMed] [Google Scholar]
- Kong JP, Ding F, Zhou CN, Wang XQ, Miao XP, Wu M, Liu ZH. Loss of myeloid-related proteins 8 and myeloid-related proteins 14 expression in human esophageal squamous cell carcinoma correlates with poor differentiation. World J Gastroenterol. 2004;10:1093–7. doi: 10.3748/wjg.v10.i8.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara KK, Lukandu OM, Neppelberg E, Vintermyr OK, Johannessen AC, Costea DE. Cancer progression in associated with increased expression of basement membrane proteins in three-dimensional in vitro models of human oral cancer. Arch Oral Biol. 2009;54:924–31. doi: 10.1016/j.archoralbio.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Lee HO, Mullins SR, Franco-Barraza J, Valianou M, Cukierman E, Cheng JD. FAP-overexpressing fibroblasts produce an extracellular matrix that enhances invasive velocity and directionality of pancreatic cancer cells. BMC Cancer. 2011;13:245. doi: 10.1186/1471-2407-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Moeen Rezakhanlou A, Chavez-Munoz C, Lai A, Ghahary A. Keratinocyte-releasable factors increased the expression of MMP1 and MMP3 in co-cultured fibroblasts under both 2D and 3D culture conditions. Mol Cell Biochem. 2009;332:1–8. doi: 10.1007/s11010-009-0167-5. [DOI] [PubMed] [Google Scholar]
- Lu CC, Yang JS, Chiang JH, Hour MJ, Amagaya S, Lu KW, Lin JP, Tang NY, Lee TH, Chung JG. Inhibition of invasion and migration by newly synthesized quinazolinone MJ-29 in human oral cancer CAL 27 cells through suppression of MMP-2/9 expression and combined down-regulation of MAPK and AKT signaling. Anticancer Res. 2012;32:2895–903. [PubMed] [Google Scholar]
- Nam KS, Shon YH. Suppression of metastasis of human breast cancer cells by chitosan oligosaccharides. J Microbiol Biotechnol. 2009;19:629–33. doi: 10.4014/jmb.0811.603. [DOI] [PubMed] [Google Scholar]
- Neth P, Ries C, Karow M, Egea V, Ilmer M, Jochum M. The Wnt signal transduction pathway in stem cells and cancer cells: influence on cellular invasion. Stem Cell Rev. 2007;3:18–29. doi: 10.1007/s12015-007-0001-y. [DOI] [PubMed] [Google Scholar]
- Nisapakultorn K, Ross KF, Herzberg MC. S100A8/A9 expression in vitro by oral epithelial cells confers resistance to infection by Porphyromonas gingivalis. Infect Immun. 2001;69:4242–7. doi: 10.1128/IAI.69.7.4242-4247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen CJ, Moreira J, Lukanidin EM, Ambartsumian NS. Human mammary fibroblasts stimulate invasion of breast cancer cells in a three-dimensional culture and increase stroma development in mouse xenografts. BMC Cancer. 2010;19:444. doi: 10.1186/1471-2407-10-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlichenko LS, Radisky DC. Matrix metalloproteinases stimulate epithelial-mesenchymal transition during tumor development. Clin Exp Metastasis. 2008;25:593–600. doi: 10.1007/s10585-008-9143-9. [DOI] [PubMed] [Google Scholar]
- Pellikainen JM, Ropponen KM, Kataja VV, Kellokoski JK, Eskelinen MJ, Kosma VM. Expression of matrix metalloproteinase (MMP)-2 and MMP-9 in breast cancer with a special reference to activator protein-2, HER2, and prognosis. Clin Cancer Res. 2004;15:7621–7628. doi: 10.1158/1078-0432.CCR-04-1061. [DOI] [PubMed] [Google Scholar]
- Roesch Ely M, Nees M, Karsai S, Mägele I, Bogumil R, Vorderwülbecke S, Ruess A, Dietz A, Schnölzer M, Bosch FX. Transcript and proteome analysis reveals reduced expression of calgranulins in head and neck squamous cell carcinoma. Eur J Cell Biol. 2005;84:431–44. doi: 10.1016/j.ejcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Roh MR, Zheng Z, Kim HS, Kwon JE, Jeung HC, Rha SY, Chung KY. Differential expression patterns of MMPs and their role in the invasion of epithelial premalignant tumors and invasive cutaneous squamous cell carcinoma. Exp Mol Pathol. 2012;92:236–42. doi: 10.1016/j.yexmp.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Yong VW. Inflammatory cytokine modulation of matrix metalloproteinase expression and invasiveness of glioma cells in a 3-dimensional collagen matrix. J Neurooncol. 2009;91:157–64. doi: 10.1007/s11060-008-9695-1. [DOI] [PubMed] [Google Scholar]
- Sewell DA, Yuan CX, Robertson E. Proteomic signatures in laryngeal squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec. 2007;69:77–84. doi: 10.1159/000097406. [DOI] [PubMed] [Google Scholar]
- Schröpfer A, Kammerer U, Kapp M, Dietl J, Feix S, Anacker J. Expression pattern of matrix metalloproteinases in human gynecological cancer cell lines. BMC Cancer. 2010;10:553. doi: 10.1186/1471-2407-10-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek-van Beurden PA, Von den Hoff JW. Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. Biotechniques. 2005;38:73–83. doi: 10.2144/05381RV01. [DOI] [PubMed] [Google Scholar]
- Sorenson BS, Khammanivong A, Guenther BD, Ross KF, Herzberg MC. IL-1 receptor regulates S100A8/A9-dependent keratinocyte resistance to bacterial invasion. Mucosal Immunol. 2012;5:66–75. doi: 10.1038/mi.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugizov S, Berline J, Herrera R, Penaranda ME, Nakagawa M, Palesky J. Inhibition of human papillomavirus type 16 E7 phosphorylation by the S100 MRP-8/14 protein complex. J Virol. 2005;79:1099–112. doi: 10.1128/JVI.79.2.1099-1112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpeenniemi-Hujanen T. Gelatinases (MMP-2 and -9) and their natural inhibitors as prognostic indicators in solid cancers. Biochimie. 2005;87:287–97. doi: 10.1016/j.biochi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Vihinen P, Ala-aho R, Kahari VM. Matrix metalloproteinases as therapeutic targets in cancer. Curr Cancer Drug Targets. 2005;5:203–20. doi: 10.2174/1568009053765799. [DOI] [PubMed] [Google Scholar]
- Wang J, Cai Y, Xu H, Zhao J, Xu X, Han YL, Xu ZX, Chen BS, Hu H, Wu M, et al. Expression of MRP14 gene is frequently down-regulated in Chinese human esophageal cancer. Cell Res. 2004;14:46–53. doi: 10.1038/sj.cr.7290201. [DOI] [PubMed] [Google Scholar]
- Wu X, Chen H, Gao Q, Bai J, Wang X, Zhou J, Qiu S, Xu Y, Shi Y, Wang X, et al. Downregulation of JWA promotes tumor invasion and predicts poor prognosis in human hepatocellular carcinoma. Mol Carcinog. 2012 doi: 10.1002/mc.21981. doi: 10.1002/mc.21981. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Yilmaz M, Christofori G, Lehembre F. Distinct mechanisms of tumor invasion and metastasis. Trends Mol Med. 2007;13:535–41. doi: 10.1016/j.molmed.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Zhou CX, Gao Y, Johnson NW, Gao J. Immunoexpression of matrix metalloproteinase-2 and matrix metalloproteinase-9 in the metastasis of squamous cell carcinoma of the human tongue. Aust Dent J. 2010;55:385–9. doi: 10.1111/j.1834-7819.2010.01258.x. [DOI] [PubMed] [Google Scholar]
- Zucker S, Vacirca J. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev. 2004;23:101–117. doi: 10.1023/a:1025867130437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.