Abstract
Background & Aims
Cancer stem cells (CSCs) can contribute to hepatocellular carcinoma (HCC) progression and recurrence following therapy. The presence of tumor-associated macrophages (TAMs) in patients with HCC is associated with poor outcomes. It is not clear whether TAMs interact with CSCs during HCC development. We investigated whether TAMs affect the activities of CSCs in the microenvironment of human HCCs.
Methods
HCCs were collected from 17 patients during surgical resection and single cell suspensions were analyzed by flow cytometry. CD14+ TAMs were isolated from the HCC cell suspensions and placed into co-culture with HepG2 or Hep3B cells, and CSC functions were measured. The interleukin 6 (IL6) receptor was blocked with a monoclonal antibody (tocilizumab), and STAT3 was knocked down with small hairpin RNAs in HepG2 cells. Xenograft tumors were grown in NOD-SCID/Il2Rgnull mice from human primary HCC cells or HepG2 cells.
Results
CD44+ cells from human HCCs and cell lines formed more spheres in culture and more xenograft tumors in mice than CD44− cells, indicating that CD44+ cells are CSCs. Incubation of the CD44+ cells with TAMs promoted expansion of CD44+ cells, and increased their sphere formation in culture and formation xenograft tumors in mice. In human HCC samples, numbers of TAMs correlated with numbers of CD44+ cells. Of all cytokines expressed by TAMs, IL6 was increased at the highest level in human HCC co-cultures, compared with TAMs not undergoing co-culture. IL6 was detected in the microenvironment of HCC samples and induced expansion of CD44+ cells in culture. Levels of IL6 correlated with stages of human HCCs and detection of CSC markers. Incubation of HCC cell lines with tocilizumab or knockdown of STAT3 in HCC cells reduced the ability of TAMs to promote sphere formation by CD44+ cells in culture and growth of xenograft tumors in mice.
Conclusions
CD44+ cells isolated from human HCC tissues and cell lines have CSC activities in vitro and form a larger number of xenograft tumors in mice than CD44− cells. TAMs produce IL6, which promotes expansion of these CSCs and tumorigenesis. Levels of IL6 in human HCC samples correlate with tumor stage and markers of CSCs. Blockade of IL6 signaling with tocilizumab, a drug approved by the Food and Drug Administration for treatment of rheumatoid arthritis, inhibits TAM-stimulated activity of CD44+ cells. This drug might be used to treat patients with HCC.
Keywords: liver cancer, mouse model, IL6 receptor, cancer biology
Introduction
Hepatocellular carcinoma (HCC) is a leading cause of death among solid malignancies and has a rising incidence in the U.S.1, 2. HCC is linked to the incidence of liver disease, with viral hepatitis being one of the strongest risk factors 1. HCC outcomes are poor3, 4, and one medication, sorafenib, has marginal efficacy5. Cancer stem cells (CSC) may account for tumor recurrence following therapy and to tumor development and metastasis. CSCs may not be a fixed cell population and may exhibit plasticity regulated by tumor microenvironmental factors (reviewed6,7). Such regulation has been shown with colon cancer tumor associated fibroblasts and with breast cancer bone marrow mesenchymal stem cells8, 9.
The immune cell component has prognostic importance in HCC and other malignancies10–12. Effector T cells, T regulatory cells, and suppressive tumor associated macrophages (TAMs) all balance tumor immunity in HCC patients10, 13, 14. We and others have shown TAMs in HCC to be particularly important to this balance of immune response and overall prognosis. However the immune infiltrating cell influence on CSC functionality, particularly in human HCC, is not completely defined. We tested the hypothesis that HCC infiltrating TAMs promote HCC CSC function. We show that CD44+ CSCs are expanded by human HCC TAMs by their secretion of IL-6 and activation of STAT3 signaling in HCC cells. This effect was blocked by the IL-6 receptor blocking antibody, Tocilizumab, an FDA approved drug, highlighting a novel therapeutic strategy for targeting CSCs in HCC.
Materials and Methods
Human subjects
HCC tissues were obtained from patients undergoing surgical resection as described13. All patients gave written informed consent. The study was approved by and followed the University of Michigan IRB guidelines.
Additional Methods can be found online in the “Supplemental Information”.
Results
HCC cells express multiple potential CSC markers
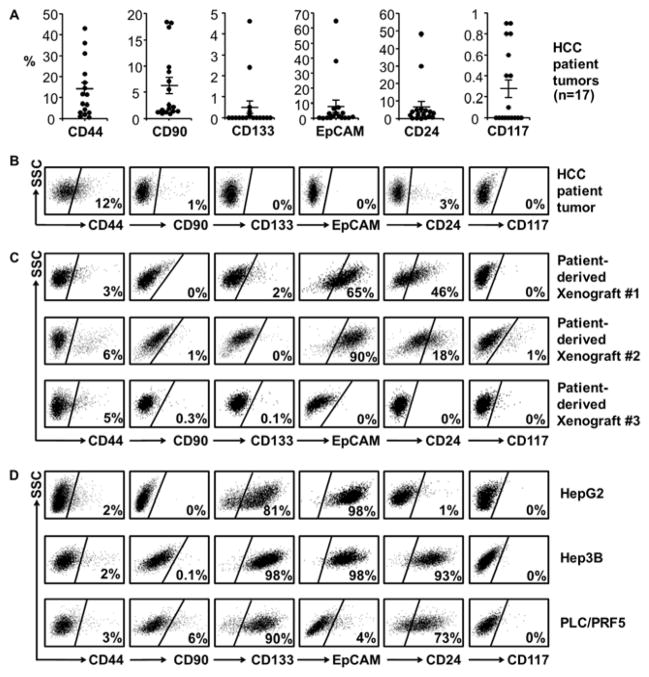
Many cell surface markers have been described to define human HCC CSCs, including CD24, CD44, CD90, CD117, CD133 and EpCAM15–17. To test whether these markers identify CSCs in a Western HCC patient cohort, surgically resected HCCs (n=17 patients, Supplementary Table 1) underwent single cell suspension and flow cytometry by gating on 7-AAD−CD45− (excluding non-viable and immune cells), and stained for putative CSC markers (Figure 1A–B). HCC patients had extremely low percentage (less than 1%) tumor cell populations of CD117, CD133 and EpCAM. However, CD24+, CD44+ and CD90+ cells were reproducibly observed with CD44+ being present in all HCCs. Cells which were co-positive for CD44 and other markers were rarely identified with occasionally CD44+/CD90+ and CD44+/CD24+ populations being noted in some tumors, however these were less than 1% of the total population (Supplementary Figure 1A). CD44, when determined by immunohistochemistry, was primarily in a membraneous pattern on HCC cells with some staining noted on stromal cells such as infiltrating leukocytes (Supplementary Figure 1B). Early passage HCC patient xenograft tumors and HCC cell lines were next examined for CSC markers (Figure 1C–D). All HCC patient-derived xenografts and cell lines consistently possessed a CD44+ population (2–6%), whereas CD90 (0–6%), CD133 (0–98%) and EpCAM (0–98%) populations varied in their presence considerably. Thus a small, discrete population of CD44+ HCC cells was reliably identified in all fresh HCC patient specimens, patient-derived xenograft tissues, and HCC cell lines.
Figure 1. CSC markers in human HCC.
(A) Flow cytometry analysis of putative CSC markers in primary HCC tumors (n=17 patients, mean±SEM). Results are percentage marker positive population in 7-AAD−CD45− (viable non-immune) cells. CD44+ percent was significantly higher than other markers (p < 0.05).
(B) Representative HCC patient FACS in (A).
(C) Representative patient-derived HCC xenografts FACS (n=3 patients) gated on mCD45−H2Kd− to exclude murine infiltrating cells.
(D) Representative FACS for CSC markers in HCC cell lines.
Results are the percentage positive population in viable cells relative to negative isotype controls.
CD44+ HCC cells have CSC properties
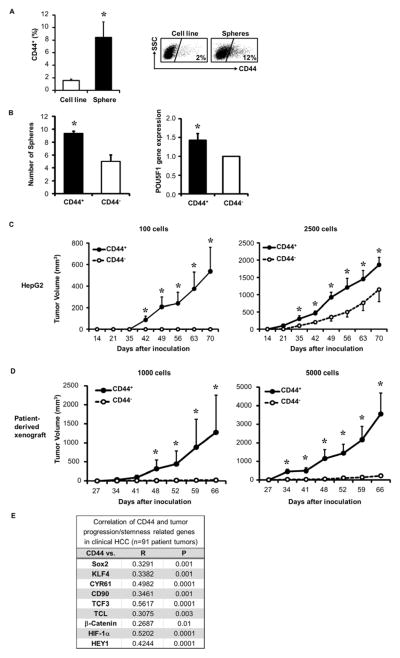
Since the CD44+ population was detected in all HCCs (Figure 1) and is a CSC marker in other malignancies18, 19, we examined whether the CD44+ subset was enriched for CSC function. CSCs have self-renewal capacity and form tumor spheres in serum-free and anchorage-independent conditions. HepG2 cells, when in sphere forming conditions, were able to generate spheres that could be passaged (Supplementary Figure 2A). HepG2 spheres were enriched for CD44+ cells more than 4 fold compared to conventional cultured HepG2 cells (Figure 2A) and had increased stem cell associated POU5F1 gene expression (previously known as OCT-4) and CD44 (Supplementary Figure 2B). We then sorted CD44+ and CD44− HepG2 cells and found that CD44+ cells formed almost two-fold more spheres and had higher POU5F1 gene expression than CD44− cells (Figure 2B). Likewise, HCC patient xenografts sorted for CD44+ cells demonstrated increased POU5F1 expression (Supplementary Figure 2C).
Figure 2. CD44+ HCC cells are enriched for CSC properties.
(A) HepG2 CD44+ cell enrichment on spheres at two weeks in sphere media, (n=3), and representative FACS.
(B) Sorting of CD44+ vs. CD44− HepG2 cells enriches sphere forming capability and POU5F1 expression by qRT-PCR (n=3).
(C–D) Tumorigenicity of 100–5000 sorted CD44+ vs. CD44− HepG2 cells (C) or HCC patient xenografts (D) following implantation into NSG mice (n=3–5 mice/group). One of three representative experiments or patients is shown in (C) and (D).
(E) Correlation between CD44 and stemness-related genes in clinical HCC tumors (n=91) using microarray database20. Spearman rank order correlation co-efficient (R) and P-value (P) were determined.
*p < 0.05. Data presented as mean±SEM.
To compare the in vivo tumor forming capacity of CD44+ and CD44− cells, limiting dilution tumor initiating assays of sorted CD44+ and CD44− HCC cells from HepG2 or HCC patient-derived xenograft tumors were performed in immune deficient NOD-SCID/IL-2Rgnull (NSG) mice (Figure 2C–D). CD44+ cells had higher tumorigencity than CD44− cells. As few as 100 CD44+ HepG2 cells and 1000 CD44+ cells from two HCC patient-derived xenografts showed a 50%–100% tumor formation rate, whereas no tumor was formed with the same number of CD44− cells. Although CD44− cells occasionally formed tumors at higher cell numbers, tumor volumes were smaller compared to those from CD44+ cells (Supplementary Figure 2D). Tumors formed from CD44+ cells had similar histology to the original unsorted (parental) HCC xenograft (Supplementary Figure 2E) and had similar stem cell marker expression percentages (Supplementary Figure 2F), demonstrating recapitulation and differentiation of the parental tumor heterogeneity by the CD44+ subset. A described HCC gene expression data set20 showed that CD44 expression correlated with tumor progression and stemness gene expression (Figure 2E). CSCs are resistant to chemotherapeutic agents and other cytotoxic agents. To test if CD44+ HCC CSCs exhibited this feature, HepG2 cells were exposed to the chemotherapeutic agent, cisplatin, and the CD44+ population was more resistant to cell death compared to the CD44− cells (Supplementary Figure 3A). CD44+ HepG2 cells were also more resistant to cell death after direct co-culture with activated effector T cells than CD44− cells (Supplementary Figure 3B). Altogether these data suggest that the CD44+ population exhibits the CSC properties of self-renewal, recapitulation of tumor heterogeneity, and resistance to cytotoxic environments.
HCC TAMs promote HCC CSCs expansion
CSC properties can be promoted by microenvironmental factors.8, 9, 21 To determine the relationship between CSCs and TAMs, freshly digested HCC patient tumor cells underwent flow cytometry. The quantity of CD44+ HCC cells was positively correlated with TAM (CD14+) quantity in HCC patients (Figure 3A). We once again evaluated an HCC patient gene expression dataset 20 and found a correlation between TAMs (CD14) and genes related to tumor progression and stemness (Figure 3B). Another gene expression data set that contained status of HCC clinical stage22 was examined and TAMs (CD68) was correlated with HCC stage (Supplementary Figure 4A) concordant to previous studies linking TAMs to HCC prognosis13, 14, 23. Since HCC TAMs and CD44+ CSCs were correlated, we evaluated whether HCC TAMs promoted HCC CSCs. TAMs were enriched from resected HCCs and co-cultured in dual-well chambers with HepG2 cells. The CD44+ HCC cell percentage increased 1.86±0.2 fold during TAM co-culture and TAMs enhanced stem cell related gene, POU5F1, expression in HepG2 cells (Figure 3C). Following co-culture, HepG2 cells also had greater sphere production compared to control, confirming the CSC promoting effects of TAMs (Figure 3C). Hep3B cells when co-cultured with TAMs had similar inductions of CD44+ subset expansion and increased sphere forming capacity (Supplementary Figure 5A–B). TAMs generated from donor blood CD14+ cells in co-culture with HepG2 cells also induced CD44+ subset expansion, increased POU5F1 expression, and increased sphere formation (Supplementary Figure 6A–C). Other genes upregulated during co-culture included CD44, the epithelial mesenchymal transition (EMT) signature genes Snail and Zeb1, hypoxia inducible gene HIF1α, Wnt signaling genes TCF7, β-catenin and c-myc, as well as Notch downstream target HEY1 (Supplementary Figure 6D).
Figure 3. TAMs promote HCC CSC expansion.
(A) Correlation of HCC TAM infiltration (Lin−CD11b+CD14+) and CD44+ HCC cells (Lin−CD44+) in HCC patients by FACS on fresh cell suspensions (n=17 patients, R=0.66, p<0.005, Spearman rank order correlation) and representative FACS shown.
(B) Correlation of CD14 and stemness-related genes in clinical HCC tumors (n=91, p<0.05) using a microarray database20. Spearman rank order correlation co-efficient (R) and P-value (P) were determined.
(C) Enrichment of CD44+ HCC cells by TAMs after three day transwell co-culture followed by FACS of HepG2 cells (n=5 HCC patients), POU5F1 expression by qRT-PCR (n=5 HCC patients), and sphere formation capacity (n=10). Control is HepG2 cells alone. Representative FACS and spheres assays (10x mag) are shown.
(D) Increased tumor growth following co-culture with TAMs. 1000 HepG2 cells following co-culture without or with TAMs were injected subcutaneously into opposing flanks of NSG mice, respectively, and tumor volume determined, n=5 mice/group, one of four representative experiments is shown.
(E) CD44+ subset is expanded in vivo in response to TAMs. HCC tumors were established in NSG mice by HepG2 cell IP injection. Once established, TAMs were IP injected and HCC cells three days later underwent FACS for CD44 (n=5 mice/group).
*p < 0.05. Data presented as mean±SEM.
To determine whether TAM-mediated effects effected tumor formation in vivo, 1000 HepG2 cells following transwell co-culture without or with TAMs were placed in the flanks of NSG mice. Indeed, HepG2 cells previously co-cultured with TAMs generated larger tumors in vivo compared to non-co-cultured HepG2 cells (Figure 3D). Using another in vivo model, TAMs were injected intraperitoneal (IP) into NSG mice with previously established HCC peritoneal tumors. HCC tumor cells following TAM injection showed an increased CD44+ population (Figure 3E). A similar effect was also noted in established orthotopic hepatic HCC tumors following TAM IP injection (Supplementary Figure 7A–B). Altogether, these data indicate that HCC TAMs promote HCC CSCs expansion in vitro and in vivo.
TAMs promote HCC CSC expansion through IL-6 signaling
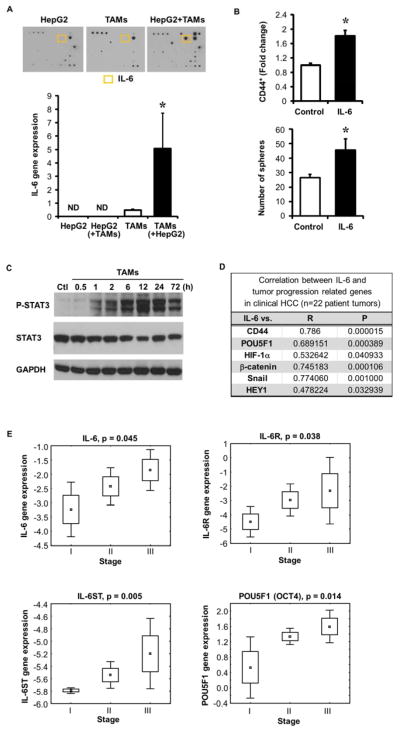
To determine how TAMs induced HCC CSC expansion, supernatants from HepG2/TAM co-cultures were examined using an array for 82 cytokines/chemokines (Figure 4A, top and Supplementary Table 3). IL-6 was the most significantly increased cytokine, whereas IL-6 was not detectable in isolated HepG2 or macrophage supernatants. ELISAs confirmed that HepG2 cells alone did not produce IL-6 and TAMs alone showed low IL-6 levels, whereas HepG2/TAM co-cultures had greater than a 10 fold increase in IL-6 (0.46 ng/mL) (Supplementary Figure 8A). Bulk HCC cells, rather than the CD44+ subset, primarily induced TAM IL-6 production (Supplementary Figure 8B). TAMs were the major IL-6 source as quantitative RT-PCR (Figure 4A, bottom) showed high IL-6 gene expression in co-cultured TAMs compared to TAMs or HepG2 cells alone.
Figure 4. TAM secreted IL-6 promotes CSC expansion in HCC.
(A) HepG2/TAMs transwell co-culture supernatants underwent cytokine antibody array (top, representative one of three experiments) while cells underwent evaluation for IL-6 by qRT-PCR and compared to HepG2 or TAMs alone (bottom, mean±SEM of relative gene expression, *p<0.05 vs. TAMs alone, n=3 experiments). IL-6 protein is highlighted in yellow. Cell type assessed indicated by ( ). ND is non-detectable.
(B) CD44+ cell enrichment and sphere forming capacity following three day IL-6 culture (10 ng/mL). CD44 determined by FACS and sphere formation determined at 2 weeks (n=3, *p<0.05 vs. vehicle control).
(C) Western blotting demonstrating increased phosphorylated STAT3 (P-STAT3) following exposure of HepG2 cells to TAMs vs. without TAMs (Ctl). One of three representative experiments is shown.
(D) Correlation of IL-6 with CD44 and other stem cell-related gene expression in clinical HCC tumors as determined by qRT-PCR (n=22 patients). Spearman rank order correlation co-efficient (R) and P-value (P) were determined.
(E) Correlation of IL-6, IL-6 receptors (IL-6R and IL-6ST), and stemness-related gene POU5F1 to HCC clinical stage using a microarray database22 (p<0.05) (n=60 patients). Spearman rank order P-value (p) was determined.
To further examine IL-6 promotion of HCC CSC expansion, recombinant human IL-6 was added to HepG2 or Hep3B cells and increased CD44+ cells were noted along with increased sphere formation (Figure 4B, Supplementary Figure 5C–D). IL-6 is known to signal via STAT324 and indeed, phospho-STAT3 (Tyr705) was elevated in HepG2 cells within one hour and was persistently elevated after three days of TAM co-culture (Figure 4C). Likewise, co-culture supernatants activated STAT3 when placed on HepG2 cells (Supplementary Figure 8C) suggesting that secreted factors contributed to sustained STAT3 activation.
To determine the clinical relationship between IL-6 and CSCs, HCC tumors (n=22 patients) were evaluated (Figure 4D). IL-6 mRNA was detected in 73% of HCC patients and was significantly correlated with expression of CD44 (standard), POU5F1, HIF-1α, β-catenin, Snail, and HEY1. Examination of a publicly available HCC patient gene expression dataset20 showed that IL-6 expression was correlated to stemness and tumor progression-related gene expression (Supplementary Figure 9A). Next we examined whether IL-6 expression and its receptor subtypes (IL-6R and IL-6ST) had prognostic implications in an HCC gene expression data set that included clinical stage22 and found these gene expression signatures correlated with HCC stage (Figure 4E). Using this same data set, expression of stemness related and EMT related genes (POU5F1, TWIST1, CTNNB1) were also increased with higher clinical stage (Figure 4E and Supplementary Figure 9B). Together these clinical and ex vivo data suggest a significant role for HCC TAM secreted IL-6 to promote CSC expansion in human HCC.
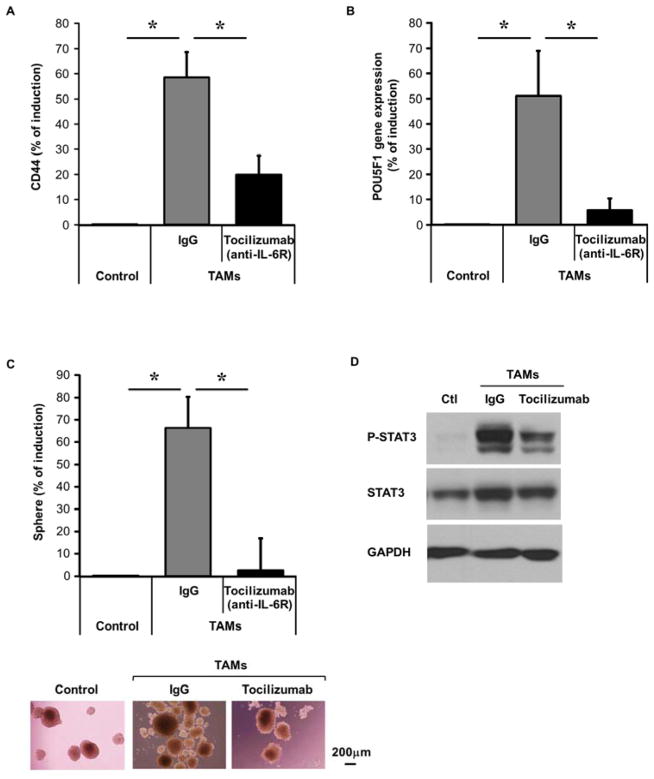
Blockade of IL-6/STAT3 signaling using Tocilizumab (anti-IL-6 receptor antibody) inhibits TAM promotion of HCC CSC expansion
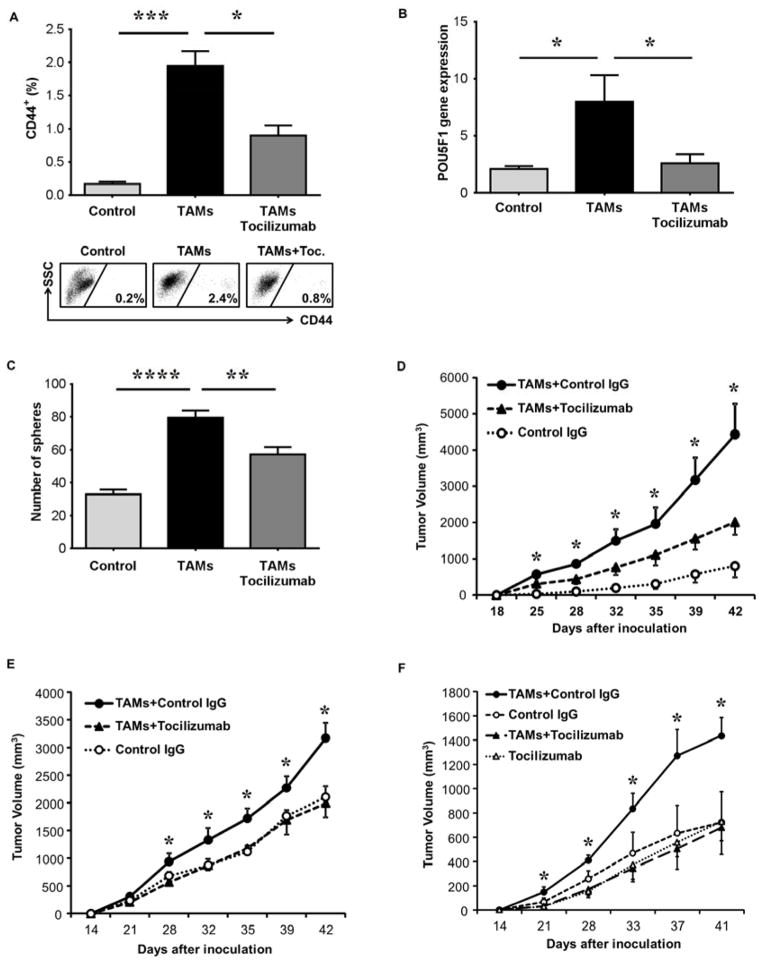
To validate that IL-6 signaling is essential for TAM-enhanced CSC function, we used Tocilizumab25, an FDA approved humanized anti-IL-6 receptor antibody, during co-culture assays. TAM-induced expansion of the HepG2 CD44+ population was inhibited by Tocilizumab (79.8±24.4%) (Figure 5A). Tocilizumab inhibited TAM-induced POU5F1 gene expression by 80±32.4% (Figure 5B) and sphere formation by 96±21% (Figure 5C). Additionally, Tocilizumab inhibited TAM-induced phospho-STAT3 levels during co-culture (Figure 5D) and when HepG2 cells were exposed to co-culture supernatants (Supplementary Figure 10A).
Figure 5. Tocilizumab (IL-6 receptor blockade) disrupts TAM-enhanced CSC expansion in HCC.
(A) Tocilizumab (5 μg/ml) inhibits TAM-induced CD44+ HepG2 cell expansion following three day co-culture with TAMs. CD44+ determined by FACS (n=5).
(B) Tocilizumab inhibits TAM-induced HepG2 POU5F1 gene expression as determined by qRT-PCR following co-culture as in (A) (n=5).
(C) TAM-induced sphere-forming capability is inhibited by Tocilizumab. Co-cultures performed as in (A) with HepG2 cells then undergoing sphere assay (n=8, representative photomicrographs 10x magnification).
(D) Western blot of HepG2 cells for phosphorylated STAT3 shows reduction by Tocilizumab (5 μg/mL) after co-culture with TAMS. One of three independent experiments is shown.
*p < 0.05. Data presented as mean±SEM.
We next evaluated whether Tociluzimab could sustainably decrease HCC tumor growth in vivo by inhibiting TAM-induced CSC function. TAMs were injected (IP) into NSG mice bearing HCC peritoneal tumors, with or without Tocilizumab, and tumors were isolated later to determine surface CD44 (Figure 6A) and POU5F1 gene expression (Figure 6B). Tocilizumab inhibited TAM-mediated CD44 HCC subset expansion and POU5F1 gene expression in this in vivo model. HCC CSC functional capacity due to TAMs was also inhibited by Tocilizumab as single cell suspensions from intraperitoneal tumors had reduced sphere formation (Figure 6C). Secondary in vivo tumor formation was also inhibited in Tocilizumab treated intraperitoneal tumors after re-implantation into NSG mice (Figure 6D). To further evaluate in vivo effects, Tocilizumab was utilized during TAMs/HepG2 co-cultures and subsequent in vivo HCC tumor formation was measured following flank implantation into NSG mice; Tocilizumab inhibited TAM-mediated tumor formation when used during the co-culture period (Figure 6E). Accordingly, HepG2 cells placed with TAMs in Matrigel® into NSG mice generated larger tumors compared to HepG2 cells alone, whereas TAM-enhanced tumor growth was completely blocked by Tocilizumab in vivo (Figure 6F). TAM-enhanced tumor growth was not due to endogenous murine macrophages since NSG mice are immunocompromised and no murine (F4/80+) macrophages were detected in multiple NSG tumor xenografts (Supplementary Figure 10B). These data indicate that human HCC TAMs promote HCC CSC expansion and tumor progression through IL-6 signaling. Furthermore, using multiple experimental in vivo HCC systems, these effects can be inhibited by the drug, Tocilizumab, suggesting a potential therapeutic target for CSCs in the HCC microenvironment.
Figure 6. Tocilizumab blocks TAM promotion of HCC tumor growth and stemness expansion in vivo.
(A) HepG2 cells were injected IP into NSG mice and six weeks later TAMs were injected IP (30 million cells) and three days later tumor nodules underwent FACS for CD44. Tocilizumab was administered (20 mg/kg IP) at time of TAM injection (n=5 mice per group, one of two representative experiments is shown and representative FACS.
(B) HepG2 and TAMs were injected IP into NSG mice as in (A) and tumor nodules underwent qRT-PCR for POU5F1 expression.
(C) Sphere assay was performed (1000 cells per group) on tumor nodule cells following experiment as described in (A), (n=5).
(D) Peritoneal tumors from (A) underwent single cell suspension and were injected subcutaneously (1000 cells/group) into NSG mice. Tumor volume was measured (n=5 mice/group). One of two representative experiments is shown.
(E) HepG2 cells, following co-culture (1000 cells) without or with TAMs and human IgG control antibody or TAMs and Tocilizumab (5μg/ml), were injected subcutaneously into NSG mice. Tumor volume was measured (n=4 mice). One of three independent experiments is shown.
(F) Tocilizumab blockade of TAM-induced tumor growth in vivo following flank co-implantation of HepG2 (5000 cells) and TAMs (10,000 cells) in matrigel into NSG mice. Tocilizumab (20 mg/kg IP every week) or IgG isotype control antibody was administered and tumor volume was measured (n=5 mice/group). One of two representative experiments is shown.
*p < 0.05, **p < 0.01, ****p < 0.0001. Data presented as mean±SEM.
STAT3 signaling is required for TAM-induced HCC CSC expansion
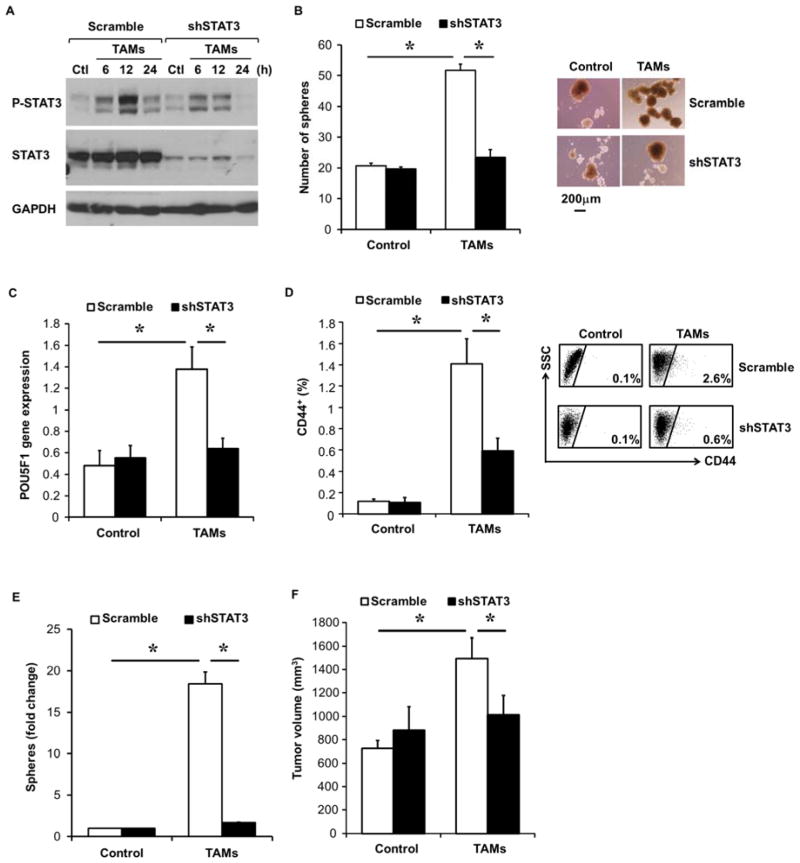
To determine whether STAT3 is required for TAM-mediated HCC CSC expansion, stable STAT3 knockdown in HepG2 cells was established using STAT3 shRNA expressing plasmid (Supplementary Figure 10C). TAMs increased phospho-STAT3 in control HepG2 cells expressing scramble shRNA, whereas STAT3 shRNA HepG2 cells had impaired phospho-STAT3 induction following TAM co-culture (Figure 7A). We next investigated whether STAT3 depletion would affect TAM-mediated CSC expansion. TAMs enhanced sphere formation in scramble shRNA HepG2 cells, but could not in STAT3 shRNA HepG2 cells (Figure 7B). Additionally, TAMs-mediated POU5F1 expression in HCC cells was blocked in STAT3 shRNA HepG2 cells compared to scramble shRNA HepG2 cells (Figure 7C).
Figure 7. STAT3 signaling is critical for TAM-induced CSC expansion in HCC.
(A) Disrupted HepG2 STAT3 activation following TAM co-culture in STAT3 shRNA HepG2 cells. Scramble shRNA or STAT3 shRNA HepG2 cells were co-cultured with TAMs for indicated time (hours). Phospho-STAT3 and total STAT3 protein levels were assessed by western blotting. One of three representative experiments is shown.
(B) Scramble shRNA or STAT3 shRNA HepG2 cells were co-cultured with TAMs for 3 days, and then collected for sphere assay (n=5). One representative photomicrograph (10x magnification) is shown.
(C) Co-cultures with scramble shRNA or STAT3 shRNA HepG2 cells and TAMs were performed as in (B) and POU5F1 gene expression determined by qRT-PCR (n=5).
(D) Inhibition of TAM-mediated CD44+ subset expansion in vivo following STAT3 knockdown. Scramble shRNA or STAT-3 shRNA HepG2 cells were injected IP into NSG mice and six weeks later TAMs (30 million) were IP injected and CD44 subset determined by FACS three days later (n=5 mice/group and representative FACS).
(E) STAT3 knockdown blocks TAM-induced spheres. Experiment performed as in (D) and HCC tumor single cells were placed into sphere assays (n=9–10 tumors/group).
(F) Loss of TAM-mediated tumor-promoting effects in STAT3 knockdown HepG2 cells. Scramble shRNA or STAT-3 shRNA HepG2 cells (1000) co-cultured without or with TAMs were injected subcutaneously into NSG mice. Tumor volume at 42 days after cell implantation is shown (n=5 mice). One of three representative experiments is shown.
*p < 0.05. Data presented as mean±SEM.
To examine whether STAT3 disruption could inhibit TAM-mediated promotion of HCC CSC capacity, HepG2 cells expressing scrambled or STAT3 shRNA were co-cultured with TAMs. STAT3 knockdown significantly inhibited TAM-mediated CD44+ subset expansion (Figure 7D). Similarly, TAM-mediated HCC CSC expansion was disrupted in STAT3 knockdown HCC cells in sphere assays (Figure 7E). Co-culturing scramble shRNA HepG2 cells with TAMs followed by implantation in NSG mice generated significantly larger tumors compared to scramble shRNA HepG2 cells alone, however, STAT3 shRNA HepG2 cells were disrupted in their ability to respond to TAM-induced tumor formation (Figure 7F). These data indicate that, similar to Tocilizumab, targeted STAT3 disruption significantly inhibited TAM-mediated promotion of HCC CSC expansion in vitro and in vivo.
Discussion
In this study, we examined the CSC profile of a Western HCC patient cohort, patient xenografts, and cell lines and defined CD44 as an important marker of HCC CSCs. We show that HCC CSCs are expanded by patient TAMs. A significant correlation between TAMs and CD44+ HCC cells in patient tumors along with stemness related gene expression was demonstrated. This was supported by CSC subset expansion after HCC cell exposure to TAMs in vitro. IL-6 secretion by TAMs accounts for this effect as IL-6 was the most highly induced cytokine in the HCC/TAM “niche” and independently stimulated the CD44+ expansion and CSC function. A clinical HCC dataset also supported a correlation between IL-6 and stem cell related gene expression.
The CD44+ subset was reproducibly demonstrated whereas other putative subsets were much less reliably demonstrated. CD90 and CD24 were identified in some primary tumor samples and may also represent populations enriched for stem cell capacity15, 16. Several reports have attempted to describe surface markers of HCC CSCs, including EpCAM, CD133, CD90, and CD2415, 16, 26. Nonetheless, we have shown the CD44 subset to have enriched CSC features in HCC. CD44 is a CSCs marker in other cancers and participates in key stem cell biology such as growth factor receptor stabilization (e.g. c-met) and microenvironmental niche homing27, 28. Thus CD44 served as a useful CSC marker in this study evaluating TAMs effect on HCC CSC expansion.
The CSC hypothesis contributes to recurrence following therapy, tumorigenesis, and EMT processes resulting in metastasis (review 6, 29). The CSC state may be induced by stromal factors in that colon cancer associated fibroblasts increase CSC expansion via wnt signaling,8 myeloid stem cells increase CSC expansion in human breast cancer xenografts,9 and myeloid derived suppressor cells support CSC expansion in ovarian cancer30. We and others have shown a diverse immune cell infiltrate exists in HCC stroma to include T effector cells, T regulatory cells (Treg), and TAMs which effect the balance of tumor immunity and also affect HCC patient outcome10, 13, 14. Therefore our study highlights TAMs as contributor to the CSC “niche” in a human hepatic cancer such as HCC and that this effect is controlled through IL-6 signaling.
The clinical significance of TAM IL-6 in promoting CSC expansion is supported by the blockade of TAM-promoted CSC expansion using an FDA approved drug for the treatment of rheumatoid arthritis, Tocilizumab, a potent inhibitor of the IL-6 receptor25. STAT3 signaling in HCC cells was accordingly critical as STAT3 knock down disrupted TAM-promoted CSC expansion in vitro and in vivo. Overall, these data suggest that targeting the IL-6/STAT3 signaling pathway to inhibit CSC function has important therapeutic implications for the treatment of HCC. IL-6 and STAT3 signaling are important for HCC development in murine models, with hepatic associated macrophages representing a major paracrine IL-6 source during HCC progression and autocrine IL-6 contributing significantly in HCC initiation from HCC progenitor cells31–34. In line with this possibility, paracrine IL-6 from TAMs appeared to be the major source of IL-6 in our study of established human HCCs though we can’t completely exclude an additional autocrine IL-6 effect in HCC patients. However TAMs elicit significant CSC functional expansion in vitro and in vivo which is disrupted by the anti-IL-6 receptor drug, Tocilizumab. Indeed, chronic hepatitis is a known risk factor for HCC development and in this setting IL-6 levels are increased1, 35. IL-6 and STAT3 signaling can increase CSC properties in breast, glioblastoma, and colorectal cancers36–39. Additionally, a recent report in an Eastern HCC patient cohort has shown that STAT3-mediated signaling was important for increasing HCC EMT following enrichment of CD24+ cells in HCC cell lines after cisplatin exposure16. These studies suggest IL-6 and STAT3 signaling may serve as important therapeutic targets in HCC.
Pharmaceutical development aimed at disrupting IL-6 and STAT3 signaling in cancer to inhibit CSCs is proceeding40. We have identified that STAT3 signaling in HCC cells following TAM interaction is critical to increased CSC expansion and increased tumor growth. Therefore therapies targeting this pathway, beside those designed to disrupt IL-6 itself, are worthy of additional pre-clinical evaluation in controlling HCC CSC function. Our study shows for the first time that Tociluzimab, and thus targeting the IL-6 receptor or STAT3 signaling, may provide future treatment efficacy in HCC and are thus important for clinical study. Furthermore, our study highlights the importance of targeting the immune microenvironment as a mechanism to inhibit CSC function in HCC.
Supplementary Material
Supplemental Table 1. HCC Patient Clinical Characteristics
Indicated baseline clinical characteristics of HCC patient tumors used for analysis following resection. BCLC (Barcelona Clinic Liver Cancer Staging), AFP (alphafetoprotein, ng/mL), VI (vascular invasion). Etiology of liver disease, if any, is provided.
Supplemental Table 2. Primers for qRT-PCR.
Sense and anti-sense sequences for the indicated human genes are listed.
Supplemental Table 3. Human cytokine antibody arrays in HepG2/TAMs co-culture Cytokines in the conditioned media of HepG2 alone, monocytes alone, or HepG2/TAMs co-cultures in transwell were measured by immunoblot as described in experimental procedures. The table shows relative densitometry of cytokine array results. Plus (+) indicates detectable cytokines, more (+) correlates with higher level and five plus (+++++) is the highest level. ND indicates non-detectable cytokines. Data are representative of three experiments.
(A) Representative flow cytometry plots of HCC patient tumor processed as described in Figure 1A and plotted for CD24+ or CD90+ vs. CD44+.
(B) Representative IHC staining for CD44 on human HCC tumors (left panels) and paired surrounding cirrhotic or non-cirrhotic liver (right panels). Large arrows point to CD44+ HCC tumor cells with membranous staining and small arrow heads indicate CD44+ stromal cells which were primarily infiltrating leukocytes (40x magnification).
(A) Primary HepG2 sphere cells form 2nd and 3rd generation spheres when plated in ultra-low attachment wells in sphere medium for 2 weeks.
(B) Conventional cultured HepG2 cells (white bar) or HepG2 sphere cells (black bar) underwent qRT-PCR analysis for POU5F1 and CD44 (standard isoform) and showed higher stemness associated gene expression. Data is representative for one of three experiments.
(C) qRT-PCR for stem cell gene POU5F1 expression was performed following cell sorting of CD44+ and CD44− cells from HepG2 cells. Data is representative for one of three experiments.
(D) Representative photograph of gross tumor size following implantation of CD44+ vs. CD44− sorted HCC cells from HCC patient xenografts in NSG mice.
(E) Hematoxylin and eosin staining of representative xenografted tumors formed from parental HCC tumor or from tumors following sorting for CD44+ cells (40x magnification).
(F) Sorted CD44+ cells from HCC patient derived xenografts were implanted into NSG mice as described in Figure 2. Subsequent tumors were subjected to FACS for indicated cell surface markers. One of three representative experiments is shown.
(A) HepG2 cells were treated without or with cisplatin (10μg/ml) for 3 days and percentage of CD44+ HepG2 cells were quantified by FACS.
(B) HepG2 cells were co-cultured in transwells with CD3+ T cells isolated from healthy donor PBMCs and activated by anti-CD3 and anti-CD28 antibodies and percentage of CD44+ cells was determined by FACS.
Representative dot plots from one of three independent experiments are shown.
Correlation of TAM levels as determined by CD68 gene expression to HCC clinical stage using a microarray database 7. Spearman correlation and P-value (P) were determined (n=60 patients).
(A) Enrichment of CD44+ HCC cells by TAMs after three day transwell co-culture followed by FACS of Hep3B cells (n=4 experiments),
(B) Sphere formation capacity following co-culture of Hep3B cells with TAMs as in (A). (n=4 experiments).
(C–D) CD44+ cell enrichment and sphere forming capacity following three day culture of Hep3B cells with IL-6 (20 ng/mL). CD44 determined by FACS and sphere formation determined at 2 weeks (n=4 experiments).
Control is Hep3B cells alone. *p< 0.05 vs. control.
(A) Enrichment of CD44+ HCC cells by TAMs. HepG2 cells were plated in the bottom of the transwell plates, whereas TAMs (n=11 donors) were added in the upper chamber. After 3-day co-culture, HepG2 cells were collected for FACS analysis of CD44. Representative FACS is shown.
(B) Increased expression of stemness related genes in HepG2 cells following co-culture with TAMs. qRT-PCR was performed for POU5F1 gene expression comparing HepG2 cells co-cultured without or with TAMs (n=5 donors).
(C) Increased sphere-forming capacity in HepG2 cells following co-culture with TAMs. HepG2 cells were co-cultured with TAMs as in (A), and then were collected for sphere assay (n=5). Representative photomicrographs (10x magnification) are shown.
(D) Increased expression of CSC related genes in HepG2 cells following co-culture with TAMs. qRT-PCR was performed for CD44 (standard isoform), Snail, Zeb1, TCF7, β-catenin, c-myc, Hif-1α, and HEY1 comparing HepG2 cells co-cultured without or with TAMs (n=5).
*p < 0.05. Data presented as mean ± SEM.
HCC tumors were established in NSG mice by HepG2 cell liver orthotopic injection (1 million cells). Once established, TAMs were injected intraperitoneal (ip) and HCC tumors were processed into single cell suspensions three days later and underwent FACS for CD44 (A) and RT-PCR for POU5F1 expression (B) (n=3 mice). Representative FACS plot is shown. *p < 0.05. Data presented as mean±SEM.
(A) IL-6 is increased in supernatant of HepG2 and TAMs co-culture as determined by ELISA (n=3, *p< 0.05 vs. other conditions). ND=non-detectable.
(B) HepG2 cells (Bulk) or HepG2 cells sorted by FACSAria for CD44+ cells were placed into co-culture transwells with TAMs and ELISA performed for IL-6 after 48 hours. Controls are Bulk HepG2 cells or TAMs alone (non-co-cultured). Data presented as mean ± SEM, n=3, *p < 0.05 vs. TAMs alone.
(C) Co-cultures with TAMs and HepG2 cells performed followed by collection of supernatant and placement onto HepG2 cells for indicated times and STAT3 activation determined by western blot. Representative of three experiments is shown.
(A) Positive correlation of IL-6 and tumor progression-related genes in clinical HCC tumors (n=91, p<0.05) using a microarray database 6. Spearman rank order correlation co-efficient (R) and P-value (P) were determined.
(B) A microarray database 7 was used to examine the correlation of the stemness-related gene CTNNB1 and EMT-related gene TWIST1 expression to HCC clinical stage (p<0.05). Spearman rank order P-value (p) was determined (n=60 patients).
(A) Supernatants obtained from TAM and HepG2 co-cultures were placed onto HepG2 cells with IgG or Tocilizumab (anti-IL-6 receptor, 5 μg/ml) for 30 minutes. Phosphor-STAT3 levels were determined by western blotting. Representative of three experiments is shown.
(B) Representative photomicrographs of F4/80 immunohistochemistry (40x magnification) showing absence of murine macrophages in NSG mice for subcutaneous (SQ) HCC xenografts, intraperitoneal (IP) HCC xenografts, and in orthotopic (liver) HCC xenografts. Positive control is small intestine from immune intact (C57BL/6) mouse.
(C) Stable knockdown of STAT3 in HepG2 cells following transduction with lentivirus expressing human STAT3 shRNA or scramble shRNA. Stable cell lines were established by puromycin (2 μg/ml) selection with knockdown of STAT3 shRNA confirmed by western blotting.
Acknowledgments
Grant Support: NIH grants CA151414 and the GI SPORE Career Development Award (THW) and NIH grants 5P30CA46592-24 and CA152470 (WZ)
Footnotes
Conflict of Interest: No conflicts of interest to disclose.
Author Contributions:
Study concept and design: SW, DMS, WZ, THW
Acquisition of data: SW, EZ, LV, AS, GL
Analysis and interpretation of data: SW, IK, AS, GL, DMS, WZ, THW
Drafting the manuscript: SW, WZ, THW
Critical revision of the manuscript: SW, DMS. WZ, THW
Statistical analysis: SW, IK, THW
Obtained funding: WZ and THW
Administrative, technical, or material support: WZ and THW
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–27. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Davila JA, Petersen NJ, McGlynn KA. The continuing increase in the incidence of hepatocellular carcinoma in the United States: an update. Ann Intern Med. 2003;139:817–23. doi: 10.7326/0003-4819-139-10-200311180-00009. [DOI] [PubMed] [Google Scholar]
- 3.Cho CS, Gonen M, Shia J, Kattan MW, Klimstra DS, Jarnagin WR, D’Angelica MI, Blumgart LH, DeMatteo RP. A novel prognostic nomogram is more accurate than conventional staging systems for predicting survival after resection of hepatocellular carcinoma. J Am Coll Surg. 2008;206:281–91. doi: 10.1016/j.jamcollsurg.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Vauthey JN, Lauwers GY, Esnaola NF, Do KA, Belghiti J, Mirza N, Curley SA, Ellis LM, Regimbeau JM, Rashid A, Cleary KR, Nagorney DM. Simplified staging for hepatocellular carcinoma. J Clin Oncol. 2002;20:1527–36. doi: 10.1200/JCO.2002.20.6.1527. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 6.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–2. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 7.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–96. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, Sprick MR, Kemper K, Richel DJ, Stassi G, Medema JP. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–76. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 9.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, Korkaya H, Heath A, Dutcher J, Kleer CG, Jung Y, Dontu G, Taichman R, Wicha MS. Breast Cancer Stem Cells Are Regulated by Mesenchymal Stem Cells through Cytokine Networks. Cancer Res. 2011;71:614–24. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu J, Xu D, Liu Z, Shi M, Zhao P, Fu B, Zhang Z, Yang H, Zhang H, Zhou C, Yao J, Jin L, Wang H, Yang Y, Fu YX, Wang FS. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology. 2007;132:2328–39. doi: 10.1053/j.gastro.2007.03.102. [DOI] [PubMed] [Google Scholar]
- 11.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 12.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–74. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 13.Wu K, Kryczek I, Chen L, Zou W, Welling TH. Kupffer cell suppression of CD8+ T cells in human hepatocellular carcinoma is mediated by B7-H1/programmed death-1 interactions. Cancer Res. 2009;69:8067–75. doi: 10.1158/0008-5472.CAN-09-0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuang DM, Zhao Q, Peng C, Xu J, Zhang JP, Wu C, Zheng L. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med. 2009;206:1327–37. doi: 10.1084/jem.20082173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW, Lam CT, Poon RT, Fan ST. Significance of CD90(+) Cancer Stem Cells in Human Liver Cancer. Cancer Cell. 2008;13:153–66. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, Reid LM, Minato H, Honda M, Kaneko S, Tang ZY, Wang XW. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–24. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Sole M, Tovar V, Alsinet C, Ramos AH, Barretina J, Roayaie S, Schwartz M, Waxman S, Bruix J, Mazzaferro V, Ligon AH, Najfeld V, Friedman SL, Sellers WR, Meyerson M, Llovet JM. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reim F, Dombrowski Y, Ritter C, Buttmann M, Hausler S, Ossadnik M, Krockenberger M, Beier D, Beier CP, Dietl J, Becker JC, Honig A, Wischhusen J. Immunoselection of breast and ovarian cancer cells with trastuzumab and natural killer cells: selective escape of CD44high/CD24low/HER2low breast cancer stem cells. Cancer Res. 2009;69:8058–66. doi: 10.1158/0008-5472.CAN-09-0834. [DOI] [PubMed] [Google Scholar]
- 22.Iizuka N, Oka M, Yamada-Okabe H, Nishida M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, Hamada K, Nakayama H, Ishitsuka H, Miyamoto T, Hirabayashi A, Uchimura S, Hamamoto Y. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361:923–9. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- 23.Oishi N, Kumar MR, Roessler S, Ji J, Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, Qin LX, Yamashita T, Woo HG, Kim YJ, Kaneko S, Tang ZY, Thorgeirsson SS, Wang XW. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56:1792–803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–8. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 25.Maini RN, Taylor PC, Szechinski J, Pavelka K, Broll J, Balint G, Emery P, Raemen F, Petersen J, Smolen J, Thomson D, Kishimoto T. Double-blind randomized controlled clinical trial of the interleukin-6 receptor antagonist, tocilizumab, in European patients with rheumatoid arthritis who had an incomplete response to methotrexate. Arthritis Rheum. 2006;54:2817–29. doi: 10.1002/art.22033. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, Reid LM, Minato H, Honda M, Kaneko S, Tang ZY, Wang XW. EpCAM-Positive Hepatocellular Carcinoma Cells Are Tumor-Initiating Cells With Stem/Progenitor Cell Features. Gastroenterology. 2008 doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002;16:3074–86. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–74. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 29.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 30.Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, Vatan L, Szeliga W, Mao Y, Thomas DG, Kotarski J, Tarkowski R, Wicha M, Cho K, Giordano T, Liu R, Zou W. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39:611–21. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda S, Hikiba Y, Sakamoto K, Nakagawa H, Hirata Y, Hayakawa Y, Yanai A, Ogura K, Karin M, Omata M. Ikappa B kinasebeta/nuclear factor-kappaB activation controls the development of liver metastasis by way of interleukin-6 expression. Hepatology. 2009;50:1851–60. doi: 10.1002/hep.23199. [DOI] [PubMed] [Google Scholar]
- 32.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–4. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 33.Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, Shalapour S, Seki E, Yost SE, Jepsen K, Frazer KA, Harismendy O, Hatziapostolou M, Iliopoulos D, Suetsugu A, Hoffman RM, Tateishi R, Koike K, Karin M. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384–96. doi: 10.1016/j.cell.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porta C, De Amici M, Quaglini S, Paglino C, Tagliani F, Boncimino A, Moratti R, Corazza GR. Circulating interleukin-6 as a tumor marker for hepatocellular carcinoma. Ann Oncol. 2008;19:353–8. doi: 10.1093/annonc/mdm448. [DOI] [PubMed] [Google Scholar]
- 36.Lin L, Fuchs J, Li C, Olson V, Bekaii-Saab T, Lin J. STAT3 signaling pathway is necessary for cell survival and tumorsphere forming capacity in ALDH(+)/CD133(+) stem cell-like human colon cancer cells. Biochem Biophys Res Commun. 2011;416:246–51. doi: 10.1016/j.bbrc.2011.10.112. [DOI] [PubMed] [Google Scholar]
- 37.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–92. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, Wu Z, Gonen M, Mulvey LA, Bessarabova MO, Huh SJ, Silver SJ, Kim SY, Park SY, Lee HE, Anderson KS, Richardson AL, Nikolskaya T, Nikolsky Y, Liu XS, Root DE, Hahn WC, Frank DA, Polyak K. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korkaya H, Liu S, Wicha MS. Regulation of cancer stem cells by cytokine networks: attacking cancer’s inflammatory roots. Clin Cancer Res. 2011;17:6125–9. doi: 10.1158/1078-0432.CCR-10-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28:4006–12. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. HCC Patient Clinical Characteristics
Indicated baseline clinical characteristics of HCC patient tumors used for analysis following resection. BCLC (Barcelona Clinic Liver Cancer Staging), AFP (alphafetoprotein, ng/mL), VI (vascular invasion). Etiology of liver disease, if any, is provided.
Supplemental Table 2. Primers for qRT-PCR.
Sense and anti-sense sequences for the indicated human genes are listed.
Supplemental Table 3. Human cytokine antibody arrays in HepG2/TAMs co-culture Cytokines in the conditioned media of HepG2 alone, monocytes alone, or HepG2/TAMs co-cultures in transwell were measured by immunoblot as described in experimental procedures. The table shows relative densitometry of cytokine array results. Plus (+) indicates detectable cytokines, more (+) correlates with higher level and five plus (+++++) is the highest level. ND indicates non-detectable cytokines. Data are representative of three experiments.
(A) Representative flow cytometry plots of HCC patient tumor processed as described in Figure 1A and plotted for CD24+ or CD90+ vs. CD44+.
(B) Representative IHC staining for CD44 on human HCC tumors (left panels) and paired surrounding cirrhotic or non-cirrhotic liver (right panels). Large arrows point to CD44+ HCC tumor cells with membranous staining and small arrow heads indicate CD44+ stromal cells which were primarily infiltrating leukocytes (40x magnification).
(A) Primary HepG2 sphere cells form 2nd and 3rd generation spheres when plated in ultra-low attachment wells in sphere medium for 2 weeks.
(B) Conventional cultured HepG2 cells (white bar) or HepG2 sphere cells (black bar) underwent qRT-PCR analysis for POU5F1 and CD44 (standard isoform) and showed higher stemness associated gene expression. Data is representative for one of three experiments.
(C) qRT-PCR for stem cell gene POU5F1 expression was performed following cell sorting of CD44+ and CD44− cells from HepG2 cells. Data is representative for one of three experiments.
(D) Representative photograph of gross tumor size following implantation of CD44+ vs. CD44− sorted HCC cells from HCC patient xenografts in NSG mice.
(E) Hematoxylin and eosin staining of representative xenografted tumors formed from parental HCC tumor or from tumors following sorting for CD44+ cells (40x magnification).
(F) Sorted CD44+ cells from HCC patient derived xenografts were implanted into NSG mice as described in Figure 2. Subsequent tumors were subjected to FACS for indicated cell surface markers. One of three representative experiments is shown.
(A) HepG2 cells were treated without or with cisplatin (10μg/ml) for 3 days and percentage of CD44+ HepG2 cells were quantified by FACS.
(B) HepG2 cells were co-cultured in transwells with CD3+ T cells isolated from healthy donor PBMCs and activated by anti-CD3 and anti-CD28 antibodies and percentage of CD44+ cells was determined by FACS.
Representative dot plots from one of three independent experiments are shown.
Correlation of TAM levels as determined by CD68 gene expression to HCC clinical stage using a microarray database 7. Spearman correlation and P-value (P) were determined (n=60 patients).
(A) Enrichment of CD44+ HCC cells by TAMs after three day transwell co-culture followed by FACS of Hep3B cells (n=4 experiments),
(B) Sphere formation capacity following co-culture of Hep3B cells with TAMs as in (A). (n=4 experiments).
(C–D) CD44+ cell enrichment and sphere forming capacity following three day culture of Hep3B cells with IL-6 (20 ng/mL). CD44 determined by FACS and sphere formation determined at 2 weeks (n=4 experiments).
Control is Hep3B cells alone. *p< 0.05 vs. control.
(A) Enrichment of CD44+ HCC cells by TAMs. HepG2 cells were plated in the bottom of the transwell plates, whereas TAMs (n=11 donors) were added in the upper chamber. After 3-day co-culture, HepG2 cells were collected for FACS analysis of CD44. Representative FACS is shown.
(B) Increased expression of stemness related genes in HepG2 cells following co-culture with TAMs. qRT-PCR was performed for POU5F1 gene expression comparing HepG2 cells co-cultured without or with TAMs (n=5 donors).
(C) Increased sphere-forming capacity in HepG2 cells following co-culture with TAMs. HepG2 cells were co-cultured with TAMs as in (A), and then were collected for sphere assay (n=5). Representative photomicrographs (10x magnification) are shown.
(D) Increased expression of CSC related genes in HepG2 cells following co-culture with TAMs. qRT-PCR was performed for CD44 (standard isoform), Snail, Zeb1, TCF7, β-catenin, c-myc, Hif-1α, and HEY1 comparing HepG2 cells co-cultured without or with TAMs (n=5).
*p < 0.05. Data presented as mean ± SEM.
HCC tumors were established in NSG mice by HepG2 cell liver orthotopic injection (1 million cells). Once established, TAMs were injected intraperitoneal (ip) and HCC tumors were processed into single cell suspensions three days later and underwent FACS for CD44 (A) and RT-PCR for POU5F1 expression (B) (n=3 mice). Representative FACS plot is shown. *p < 0.05. Data presented as mean±SEM.
(A) IL-6 is increased in supernatant of HepG2 and TAMs co-culture as determined by ELISA (n=3, *p< 0.05 vs. other conditions). ND=non-detectable.
(B) HepG2 cells (Bulk) or HepG2 cells sorted by FACSAria for CD44+ cells were placed into co-culture transwells with TAMs and ELISA performed for IL-6 after 48 hours. Controls are Bulk HepG2 cells or TAMs alone (non-co-cultured). Data presented as mean ± SEM, n=3, *p < 0.05 vs. TAMs alone.
(C) Co-cultures with TAMs and HepG2 cells performed followed by collection of supernatant and placement onto HepG2 cells for indicated times and STAT3 activation determined by western blot. Representative of three experiments is shown.
(A) Positive correlation of IL-6 and tumor progression-related genes in clinical HCC tumors (n=91, p<0.05) using a microarray database 6. Spearman rank order correlation co-efficient (R) and P-value (P) were determined.
(B) A microarray database 7 was used to examine the correlation of the stemness-related gene CTNNB1 and EMT-related gene TWIST1 expression to HCC clinical stage (p<0.05). Spearman rank order P-value (p) was determined (n=60 patients).
(A) Supernatants obtained from TAM and HepG2 co-cultures were placed onto HepG2 cells with IgG or Tocilizumab (anti-IL-6 receptor, 5 μg/ml) for 30 minutes. Phosphor-STAT3 levels were determined by western blotting. Representative of three experiments is shown.
(B) Representative photomicrographs of F4/80 immunohistochemistry (40x magnification) showing absence of murine macrophages in NSG mice for subcutaneous (SQ) HCC xenografts, intraperitoneal (IP) HCC xenografts, and in orthotopic (liver) HCC xenografts. Positive control is small intestine from immune intact (C57BL/6) mouse.
(C) Stable knockdown of STAT3 in HepG2 cells following transduction with lentivirus expressing human STAT3 shRNA or scramble shRNA. Stable cell lines were established by puromycin (2 μg/ml) selection with knockdown of STAT3 shRNA confirmed by western blotting.