Figure 1. Identification of Cofilin pathway dysregulation in GBM.
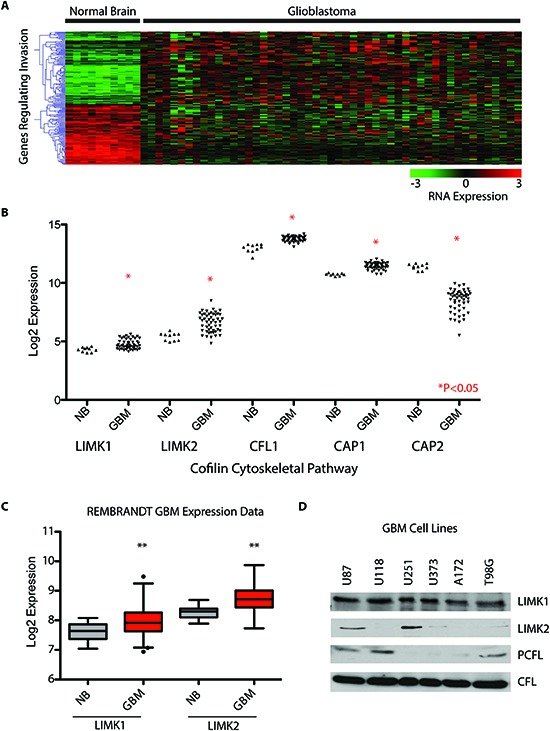
(A) 700 Probe sets were investigated representing 400 genes involved in migration and invasion. Using Sam-Pairwise analysis, a fold change of 1.5 was used, p<0.05 and a Q value of <0.01. 141 Genes were identified as significantly up or down regulated compared in mesenchymal glioblastoma (n=51) versus normal brain (n=10) (B) Invasion Pathway Analysis identified significant deregulation of the Cofilin Pathway (C) LIMK1 and LIMK2 which phosphorylate CFL are up-regulated in GBM using the REMBRANDT brain tumor data set. (D) CFL is upregulated in GBM and LIMK1 and 2 are present in GBM cells. *p<0.05, **p<0.01.
