Abstract
Two mechanisms have emerged as major regulators of membrane shape: BAR domain-containing proteins, which induce invaginations and protrusions, and nuclear promoting factors, which cause generation of branched actin filaments that exert mechanical forces on membranes. While a large body of information exists on interactions of BAR proteins with membranes and regulatory proteins of the cytoskeleton, little is known about connections between these two processes. Here, we show that the F-BAR domain protein pacsin2 is able to associate with actin filaments using the same concave surface employed to bind to membranes, while some other tested N-BAR and F-BAR proteins (endophilin, CIP4 and FCHO2) do not associate with actin. This finding reveals a new level of complexity in membrane remodeling processes.
Keywords: cryo-electron microscopy, F-BAR protein pacsin2, F-actin binding, membrane sculpting
Introduction
The members of the Bin-Amphiphysin-Rvs161/167 (BAR) domain superfamily of proteins have emerged as key players at the interface between the phospholipid bilayer and the actin cytoskeleton by connecting cytoskeleton-regulatory processes to their specific sites of action at the membrane 1,2. The BAR domains display affinity for curved membrane areas and/or induce membrane curvature upon membrane binding 3,4. Four types of BAR domains can be discerned with respect to the intrinsic curvature of their membrane binding sites: the N-terminal-BAR (N-BAR) and the extended-FCH-BAR (F-BAR) bind to membrane invaginations, whereas the inverse BAR (I-BAR) and the pinkbar proteins bind to the curved sites within membrane protrusions or flat membranes, respectively 5. Tip-to-tip and/or lateral interactions of dimeric BAR domain units result in long filaments that wind around membrane tubules in a spiral-like form that promote further tubule formation 4,6,7. Many BAR family members have an additional SH3 domain(s), which bind several proteins including nucleation promoting factors (NPFs), dynamin, and Wiscott–Aldrich syndrome protein (WASP/N-WASP protein family) 4,8–10 and thereby combine their membrane remodeling properties with activities of membrane fission and/or the pushing forces of the actin cytoskeleton 2,3,11. Pacsin2 (also termed syndapin 2 or FAP52) 12,13 is a member of the F-BAR domain protein subfamily. It is involved in clathrin-mediated endocytosis 14, vesicle budding from the trans-Golgi network 15, the biogenesis of caveolae 16, the formation/stabilization microspikes at the cell surface 17, and in regulation of cell spreading and migration 18. A unique feature of the F-BAR domain of pacsins is the 8-residue-long flexible loop (wedge loop) that protrudes from the concave side of the BAR domain and is involved in membrane binding 19,20. Liposome binding studies with pacsin1 revealed that the tubulation-inducing activity of the F-BAR domain alone is diminished in the full-length protein due to the autoinhibition by the C-terminal SH3 domain and the preceding linker 19,21. Interaction of the SH3 domain with the proline-rich region of binding partners (e.g. dynamin-1 and dynamin-2) promotes the membrane binding activity of the F-BAR domain 15. Here, we show that pacsin2 directly binds to actin filaments and demonstrate a new mode of interaction of this F-BAR protein, whereby the concave side of its F-BAR domain is not only the membrane binding site but can also be involved in protein interactions. In addition, we show that binding to F-actin is not a common feature of BAR domain proteins. The observed direct interactions between an F-BAR protein and F-actin open up new avenues of investigations of links between membrane remodeling and the actin cytoskeleton.
Results
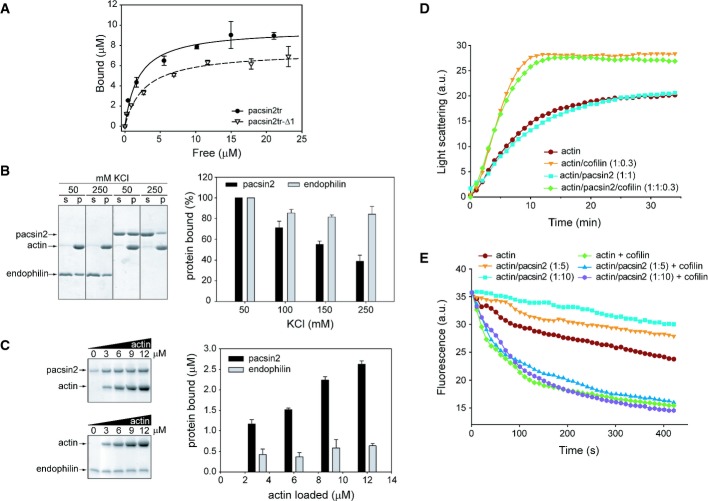
As shown by recent data, pacsin2 is involved in processes that require an active actin cytoskeleton 22. In order to assess the direct interaction of pacsin2 with actin filaments, as already shown for the N-BAR domain protein PICK1, we performed co-sedimentation assays. Both full-length pacsin2 as well as the C-terminally truncated pacsin2 variant pacsin2tr (residues 1–324) that mainly comprises the F-BAR domain were found to directly interact with F-actin (Fig 1A and B, Supplementary Fig S1). To assess the dissociation constant of the interaction between pacsin2 and F-actin, we used the C-terminally truncated pacsin2tr construct because of its higher stability at high protein concentrations compared to the full-length protein. Pacsin2tr was found to bind to F-actin with a dissociation constant of 1.92 ± 0.36 μM (Fig 1A). Since the wedge loop of pacsins was implicated in membrane binding (see below), we constructed a wedge loop mutant, both in the full-length (pacsin2-Δ1) and in the truncated pacsin2 variant (pacsin2tr-Δ1). The pacsin2tr-Δ1 mutant binds to F-actin with a similar dissociation constant (KD of 2.73 ± 0.56 μM) as the wild-type variant (Fig 1A). This suggests that the wedge loop has no specific role in the interaction between pacsin2 and F-actin.
Figure 1. Pacsin2 interacts with actin filaments, while endophilin does not.
A Filamentous actin was incubated with various amounts of pacsin2tr or pacsin2tr-Δ1. Upon ultracentrifugation, the amount of proteins in the pellet and supernatants was assessed by quantitative Western blotting. The exponential binding curves fitted for each set of data points are shown. Data represent mean values (± SE) of four independent experiments.
B Effects of KCl on co-sedimentation of endophilin BAR domain (endophilin) or full-length pacsin2 with F-actin. (s) and (p), supernatant and pellet fraction, respectively. Graph data are presented as percentage of binding compared with maximum (100%) pacsin2 binding at 50 mM KCl. Mean values (± SE) of three independent experiments are shown.
C Effects of increasing amounts of F-actin on co-sedimentation of endophilin BAR domain (endophilin) or full-length pacsin2. SDS–PAGE (left panel) and densitometric quantitation (right panel) of proteins found in the pellet fractions is shown. Graph data are presented as amount of protein bound to F-actin, from which amount of protein found in the pellet fraction without F-actin has been subtracted. Mean values (± SE) of three independent experiments are shown.
D Effects of pacsin2 on spontaneous polymerization of actin. Actin filaments were polymerized at 3 μM in the presence or absence of pacsin2 or cofilin (at molar ratios indicated in the figure). The results of a typical experiment are shown.
E Effects of pacsin2 on dilution-induced actin depolymerization. Actin filaments (4 μM) were depolymerized in the presence or absence of pacsin2 (at molar ratios as indicated in the figure) in the presence or absence of 100 nM cofilin. The plots of fluorescence intensity of pyrene-actin against time after dilution representing the results of a typical experiment are shown.
To further investigate the nature of pacsin2–F-actin binding and to ask whether other BAR domain-containing proteins might interact with actin filaments, we assayed selected N-BAR (endophilin) and F-BAR (CIP4 and FCHO2) proteins for binding to F-actin. For pacsin2 and endophilin, the specificity of the interaction with F-actin was tested in the co-sedimentation assay by either increasing the salt or the actin concentration (Fig 1B and C). In the first experiment, both proteins (pacsin2 and endophilin) co-sedimented with F-actin at 50 mM KCl (Fig 1B, Supplementary Fig S1). However, while there was only slight precipitation observed for pacsin2 when incubated in the absence of F-actin, relatively high self-precipitation of endophilin was observed at the same conditions (Supplementary Fig S1). Increasing the salt concentration to 250 mM KCl led to a reduction of the pacsin2–F-actin binding to approximately 40% as compared to that at 50 mM KCl (Fig 1B). This dependence on ionic strength indicates that pacsin2 most likely interacts with the negatively charged actin filaments via its positively charged concave surface. In contrast, co-sedimentation of endophilin with F-actin was not significantly reduced in the presence of high salt concentrations (85% at 250 mM KCl), suggesting an unspecific interaction.
In the second experiment, where actin concentration was systematically increased, both pacsin2 and endophilin showed some precipitation in the absence of F-actin (Fig 1C). Increasing amounts of F-actin led to an obvious enrichment of pacsin2 in the F-actin pellet fraction, while only a slight enrichment of endophilin in the F-actin pellet fraction was observed at all actin concentrations tested (Fig 1C), indicating that co-sedimentation of endophilin is actin independent. These results suggested that endophilin does not interact with F-actin, which was further confirmed by our EM studies (negative staining), where no decoration of actin filaments with endophilin was observed, while clear binding of pacsin2tr with actin filaments could be seen (Supplementary Fig S2). In addition, we found unspecific or no binding of FCHO2 and CIP4, respectively, to F-actin by using of co-sedimentation and EM studies (Supplementary Figs S3 and S4). Altogether, our results suggest that pacsin2 binds specifically to F-actin; however, binding of BAR domain-containing proteins to F-actin is not a common property of this protein superfamily.
Next, we investigated whether pacsin2 has any effect on actin polymerization rates. To address this question, spontaneous polymerization of actin in the presence or absence of pacsin2tr was monitored by light scattering (Fig 1D). Interestingly, when compared to a control experiment with actin alone, pacsin2tr did not display an effect on actin polymerization either at low (1:1) or at higher (1:5) actin to pacsin2tr molar ratios (Fig 1D, Supplementary Fig S5A). In the polymerization assay, cofilin enhances the elongation rate of spontaneous actin polymerization by severing actin filaments and increasing the number of filaments ends 23. As expected, when using cofilin in our assay, actin polymerization was accelerated and the kinetics reached a plateau at about 15 min (Fig 1D). However, pacsin2 did not inhibit this activity, suggesting that binding of pacsin2 to F-actin differs from that of, for example, tropomyosin, which stabilizes F-actin and thus inhibits the actin severing activity of cofilin 24. To further explore this phenomenon, we analyzed the effects of pacsin2tr on the in vitro depolymerization kinetics of pyrene-labeled F-actin in the absence or presence of cofilin. As expected, fluorescence intensity decreased in function of time when pyrene-labeled F-actin was diluted in F-buffer (Fig 1E). The extent of depolymerization was similar when actin filaments were prepared by polymerization in the presence of pacsin2 at low actin to pacsin2 molar ratios (1:1 and 1:2) (Supplementary Fig S5B). However, when F-actin was mixed with pacsin2 at higher molar ratios (1:5 and 1:10, actin to pacsin2) (Fig 1E), a decrease in actin depolymerization rate was observed, indicating that pacsin2 is able to increase the stability of F-actin. When the F-actin and F-actin–pacsin2 complexes were subjected to dilution-induced depolymerization in the presence of cofilin, hardly any difference in the depolymerization rates was observed (Fig 1E), suggesting that pacsin2 is not able to reduce the activity of cofilin in this assay. In conclusion, pacsin2 increases the stability of actin filaments upon binding but does not seem to have nucleation and/or severing activity itself nor an effect on the activity of cofilin.
To investigate the competition between membrane and F-actin binding of pacsin2, a lipid co-flotation assay was performed. In the presence of liposomes, a significant amount of purified full-length pacsin2 was present in the lipid-containing fraction, whereas in the absence of liposomes, all pacsin2 resided in the dense fractions. However, in the presence of F-actin, the amount of pacsin2 in lipid-containing fraction was slightly diminished. Thus, these data suggest that binding of pacsin2 to liposomes is competed in the presence of F-actin (Supplementary Fig S6).
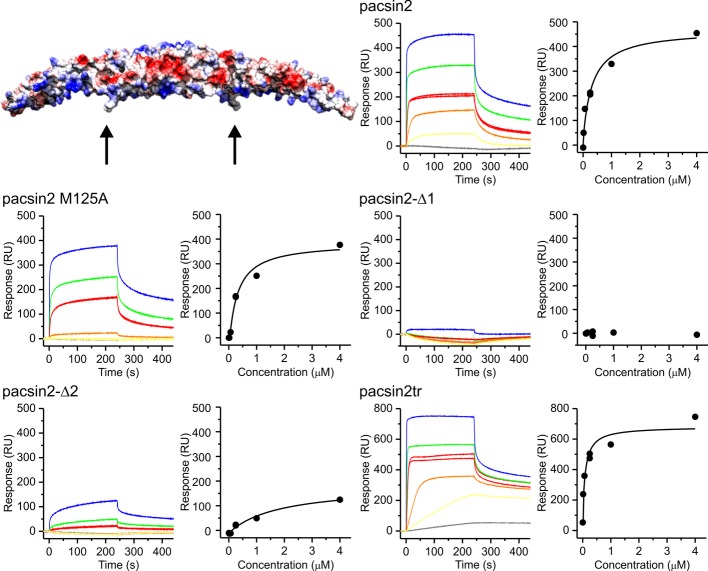
We used surface plasmon resonance (SPR), in order to quantitatively compare the interaction of pacsin2 and wedge loop mutants with membranes. We employed small unilamellar vesicles (SUVs) composed of negatively charged 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS). Pacsin2 did not interact with SUVs prepared from zwitterionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and these vesicles served as a control surface (for details see Materials and Methods). Pacsin2 exhibited considerable interactions with POPS SUVs with the estimated KD of 0.22 ± 0.1 μM (Fig 2). Two methionine residues (Met125 and Met126) located at the tip of the wedge loop were proposed to have a role in anchoring the protein to lipid membranes (Fig 2). Mutation of one of these residues, Met125Ala (pacsin2-M125A), weakened membrane interactions and increased the KD to 0.4 ± 0.21 μM. Removal of both methionines (pacsin2-Δ2) resulted in an almost tenfold decrease of affinity, with KD of 3.1 ± 2.7 μM. The pacsin2-Δ1 variant did not interact with SUVs at all, thereby confirming the prime importance of the wedge loop for membrane binding. We also assessed interactions of the C-terminally truncated pacsin2tr that lacks the SH3 domain and the preceding linker and as expected found it to bind to membranes slightly better than the wild-type, with the KD of 0.14 ± 0.05 μM.
Figure 2. SPR analysis of pacsin2 and its variants interactions with POPS SUVs.
Raw sensorgrams are shown together with equilibrium binding analysis for each tested protein. The presented sensorgrams were doubly referenced as specified in Materials and Methods. The concentrations used were 2.6 nM (gray), 15.6 nM (yellow), 62.5 nM (orange), 250 nM (red in duplicate), 1 μM (green), and 4 μM (blue). The model of pacsin2 is presented with surface presentation colored by Coulomb potential. The wedge loop encompassing residues 122–126 (sequence HKQMM) substituted with G in pacsin2-Δ1 is indicated by an arrow. Met125, which was mutated to Ala, is located at the tip of the wedge loop (arrow).
Extensive binding of pacsin2tr to F-actin was observed by negative stain EM (Supplementary Fig S2C and D). To obtain higher resolution information, cryo-EM (Fig 3A and B) was used for three-dimensional reconstruction. The Iterative Helical Real Space Reconstruction approach 25 was used for three-dimensional reconstruction, starting with a solid cylinder as an initial reference and using 31,856 segments. A sorting of the image segments was then done to remove those that were partially or more lightly decorated by pacsin2tr, leaving 5,620 segments as extensively decorated. These were used for the final three-dimensional reconstruction (Fig 3C). Fitting a model of the actin filament 26 into this volume suggested a resolution of approximately 12 Å. Since the actin filament was not significantly changed by the binding of pacsin2tr at the available resolution, the additional density due to pacsin2tr was quite clear. Interestingly, the additional density was found to form two strands, each one associated with a long-pitch helical strand within F-actin, thus resembling the binding of tropomyosin bound to actin filaments as observed previously 27 (Fig 3D).
Figure 3. Three-dimensional reconstruction of F-actin with bound pacsin2.
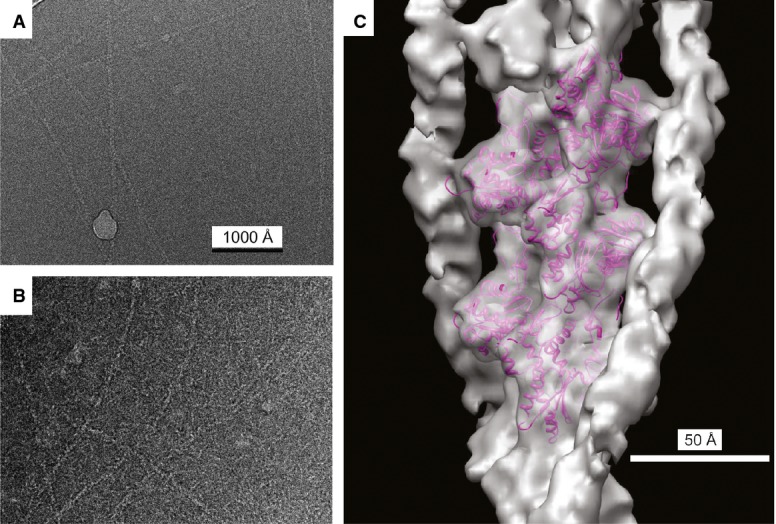
A Electron cryo-micrograph of naked, unstained frozen/hydrated F-actin.
B Electron cryo-micrograph of unstained frozen/hydrated F-actin decorated with pacsin2. Magnification settings are same as in (A).
C The surface of the reconstructed volume is shown in gray, with an atomic model of F-actin 26 shown as magenta ribbons. The remaining density running along each actin strand is pacsin2.
To map/fit pacsin2 into the additional electron density, we crystallized F-BAR domain of pacsin2 28 and solved its structure to 2.57 Å, with final refinement statistics of Rwork = 0.184 and Rfree = 0.222 (Supplementary Fig S7, Supplementary Table S1). Not surprisingly, it was found to be most similar to mouse pacsin1 (pdb code 2X3W) with r.m.s.d. between equivalent Cα atoms of 1.07 Å 21. Even by having a crystal structure of pacsin2, interpreting the additional density present in the three-dimensional reconstruction in terms of molecular models for pacsin was impaired by the substoichiometric binding of pacsin to F-actin and the fact that no regular pattern of binding (e.g. 1:2 or 1:3) was observed in averaged power spectra from the pacsin-decorated actin filaments.
In order to identify/map residues that are proximal in the pacsin2/F-actin complex, we performed cross-linking experiments, using a zero-length cross-linker 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), followed by mass spectrometric analysis. After cross-linking, a band of about 90 kDa appeared on SDS–PAGE, indicating the formation of specific cross-links between one actin subunit (42 kDa) and one pacsin2 subunit (52 kDa) (Supplementary Fig S8). To identify the sites of intermolecular cross-links, this 90 kDa band was excised from SDS–PAGE and analyzed by high-resolution LC-MS/MS after trypsin treatment. A representative MS/MS spectrum of a specific cross-link product, which allowed the identification of a linkage between Lys62 of pacsin2 and Asp25 of actin is depicted in Supplementary Fig S9. In total, 9 cross-links were identified for the pacsin2/F-actin complex (Supplementary Table S2). The major cross-linking products involve two clusters of lysine residues (53, 64, and 101; 143, 147, and 150) mapping on the concave surface of the pacsin2 BAR domain (Fig 4A) and the aspartic acid residues (24 and 25) and glutamic acid residues (99 and 100) on actin subunits, mapping to the long-pitch helical strand on F-actin (Fig 4B). The first lysine cluster is located in the pacsin2 dimerization domain and the second is close to the proline residue 144 that introduces the specific kink in the F-BAR of pacsin2, causing diverse orientations of the tails in the pacsin protein family (Supplementary Fig S10). The distances between the two lysine clusters in the F-BAR domain of pacsin (47–68 Å) correspond well with the distance between the clusters identified on actin protomers (58 Å), which correspond to the long-pitch helical strand on F-actin. Furthermore, the identification of these cross-links agrees with the electrostatic nature of the pacsin2–F-actin interaction, already observed in co-sedimentation assays.
Figure 4. Mapping of residues obtained by XL-MS analysis of pacsin2:F-actin complex.
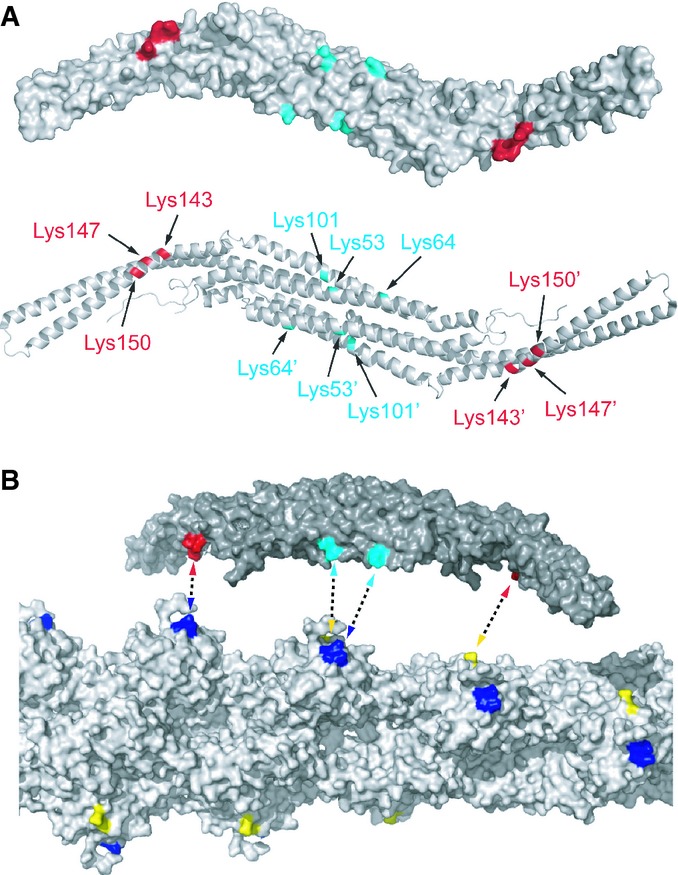
A Lysine residues 53, 64, and 101 (depicted in cyan) and lysine residues 143, 147, and 150 (depicted in red) belonging to first and second cluster, respectively, are localized on the concave surface of the pacsin2 F-BAR domain. Model of pacsin2 F-BAR domain represented as surface (top) or cartoon (bottom) is shown. Prime (′) is used as distinguisher between residues belonging to individual subunits of pacsin2 dimer.
B Aspartic acid residues 24 and 25 (depicted in yellow) and glutamic acid residues 99 and 100 (depicted in blue) on actin subunits map to the long-pitch helical strand on F-actin. Dashed double arrows indicate possible ways of cross-linking. The distances between the two lysine clusters in the F-BAR domain of pacsin (47–68 Å) correspond with the distance between the clusters identified on actin protomers (58 Å). Model of pacsin2 F-BAR domain (top) and F-actin (bottom) is represented as solvent accessible surface.
Discussion
While the BAR superfamily proteins sculpt membrane through the curvature of their BAR domains, actin polymerization provided by nucleation promoting factors (NPFs) and the Arp2/3 complex generates a pushing force against the membrane. BAR domain proteins are therefore often found at the membrane/cytoskeleton interface in protein assemblies that display a complex regulation 22. A study by Rocca et al 29 on a protein complex involving the N-BAR domain protein PICK1 gives a first notion of a direct binding between a BAR domain and F-actin; however, molecular details about this interaction were not reported 29. Here, we show for the first time that the concave membrane binding face of an F-BAR domain also directly interacts with actin filaments, as supported by several lines of evidence. (i) The co-sedimentation assays revealed a salt dependence and thereby suggested the electrostatic nature of the interaction between pacsin2 and actin. (ii) The XL-MS experiments identified clusters of residues on the concave side of pacsin2. (iii) The co-flotation assay revealed some, albeit weak, competition between negatively charged liposomes and F-actin for binding to the F-BAR domain of pacsin2.
Our cryo-EM study of F-actin decorated with BAR domain of pacsin2 showed that its binding occurs along the long-pitch strands of the actin filament, reminiscent of tropomyosin interaction with F-actin. Furthermore, F-actin binding assays suggest that pacsin2 is an actin-side binding protein that does increase the stability of actin filaments, but does not protect them from cofilin-mediated depolymerization activity. Pacsins can recruit N-WASP to membranes and trigger local actin polymerization in vivo in an Arp2/3-complex-dependent manner 30. In this respect, it will be interesting to see whether Arp2/3 recruitment can also be initiated by pacsin2 when bound to F-actin, thereby providing an additional mode of actin regulation.
There are several structural differences between N-BAR and F-BAR domain proteins 6,9,20,29,31–33 which suggest that these subfamilies could differ in the binding to F-actin, with F-BAR domain proteins being more suited for the interaction. However, an exception is the N-BAR domain of PICK1, which was shown to interact directly with actin filaments 29. This interaction was found to be almost completely abolished by substitution of two adjacent lysine residues constituting the F-actin binding motif in pan1 34 with concomitant inhibition of PICK1–lipid vesicle interactions. Structure based sequence alignment of N-BAR domain proteins (Supplementary Fig S11) revealed that this double lysine motif is partially conserved within the N-BAR domain subfamily, suggesting that other N-BAR domain proteins might interact with F-actin, too. However, as shown in our study, the N-BAR protein endophilin, which also displays this motif, does not interact with the F-actin, implying that in PICK1 additional residues play a role in the interaction with F-actin. In the structure based sequence alignment of the F-BAR domain protein family, this double lysine motif of PICK1 is not conserved 35 at the same position in the structure.
We investigated the conservation of surface residues (lysine clusters) of pacsin2 identified by our XL-MS analysis of F-actin:pacsin2 complex. Interestingly, we found that these residues form a distinct spatial distribution of positive charges on the pacsin2 and are not conserved throughout the F-BAR domain proteins (Supplementary Fig S12). Our analysis suggests that a specific pattern of positively charged residues is required for F-actin binding, as opposed to less stringent requirements of charge distribution for membrane binding of F-BAR domains in general. This is corroborated by our finding that two other F-BAR domain proteins (CIP4 and FCHO2) do not interact with actin filaments. In conclusion, binding to actin filaments seems to be a specific feature of some BAR domain proteins and further studies are needed to identify sequence and structural determinants that confer distinctive actin binding modes of N-BAR and F-BAR domain proteins.
The finding that the F-BAR domain of pacsin2 is not only associated with a membrane system but can also directly bind to actin filaments raises the question of the significance of this interaction in the cell biological context. Based on our study and experimental data from the literature, we propose three scenarios where the pacsin–actin interaction may play a significant role: regulation of cytoskeletal dynamics, sequestration of membrane-dissociated pacsin, and recruitment of pacsin to active sites at the membrane.
Materials and Methods
F-actin co-sedimentation assay
Actin co-sedimentation assay was essentially done as described in Kostan et al 36 with slight modifications. Briefly, samples of purified rabbit skeletal muscle actin (12 μM) in 20 μl of G-buffer (5 mM Tris–HCl pH 8, 0.2 mM CaCl2, 0.2 mM ATP, 2 mM β-mercapthoethanol) were polymerized by addition of 1/10 volume of 10× F-buffer (50 mM Tris, 0.5 M KCl, 20 mM MgCl2, 20 mM ATP, pH 8), and incubation for 30 min at room temperature. For determining dissociation constants, increasing concentrations of pacsin2tr or pacsin2tr-Δ1 (0–100 μM) in 20 μl of 1× F-buffer were added to polymerized actin samples and incubated for another 30 min. For co-sedimentation with increasing amounts of salts, pacsin2 or endophilin (both at 12 μM) in 20 μl of 1× F-buffer, supplemented with or without KCl to final concentration of 50, 150, 250, or 450 mM, was added to polymerized actin samples and incubated for another 45 min. For co-sedimentation with increasing amounts of actin, pacsin2, endophilin, or CIP4 (at 12, 12 and 4 μM, respectively) in 20 μl of 1× F-buffer was mixed with 20 μl of 1× F-buffer containing 0, 6, 12, 18, 24 μM pre-polymerized actin and incubated for another 45 min. Actin filaments and proteins bound were sedimented by centrifugation (217,000 × g, 30 min, 20°C), and equivalent volumes of the supernatant and pellet fractions were analyzed by SDS–PAGE and/or immunoblotting using an StrepMAB-Classic mouse antibody (IBA) to Strep-tag II, and either goat anti-mouse IgG conjugated to horseradish peroxidase (Pierce) and detection by CL-XPosure films (Pierce) or IRDye 680LT Goat anti-mouse (LI-COR) and detection by the ODYSSEY Infrared imaging System (LI-COR). Film exposures were scanned and quantified using the ImageQuant 5.1 software package (Molecular Dynamics). For determination of dissociation constants, the amount of pacsin2tr or pacsin2tr-Δ1 bound to F-actin was fit to a single rectangular hyperbola using Prism 4 (GraphPad Software).
Surface plasmon resonance
The surface plasmon resonance experiments (SPR) were performed using Biacore T100 and sensor chip L1 (Biacore, GE Healthcare) 37. Proteins were diluted in the SPR running buffer composed of 20 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.1 mg/ml BSA, pH 8. All experiments were performed at 25°C. Small unilamellar vesicles (SUVs) were prepared from 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) or 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS; both from Avanti Polar Lipids) by using sonication as described 38. The SUVs were injected over the surface of L1 sensor chip in the running buffer at a flow rate of 2 μl/min. The POPS SUVs were captured to the second flow cell, while the POPC SUVs were injected over the first flow cell and served as a control for nonspecific binding. The immobilization level for both SUVs was kept constantly around 1,200 RU. After lipid injection, the surface was stabilized with two 15 s injections of 10 mM NaOH. Wild-type pacsin2 and mutants were injected across the lipid surface at different concentrations, ranging from 2.6 nM to 4 μM. The proteins were injected for 4 min at a flow rate 10 μl/min, and the dissociation was monitored for additional 4 min. Between each protein injection, the surface was regenerated with two 30 s injections of 40 mM octylglucopyranoside and fresh lipid surface was prepared for each protein injection. Sensorgrams were doubly referenced for the nonspecific binding to the POPC LUVs and buffer injections and then analyzed with the Biacore T100 Evaluation Software. Equilibrium dissociation constants (KD) were determined by plotting the binding levels at the end of the injection, which for most protein injections reached steady-state levels, versus the concentration of the injected protein and fitting the data to the equation:
where Req is steady-state binding level, C is the concentration of proteins, and Rmax is the maximal response. The average KDs and standard deviations were determined from three independent titrations for each protein.
Cryo-electron microscopy
Rabbit skeletal muscle G-actin (10 μM) was polymerized in FEM-buffer (15 mM MOPS-buffer, pH 7.2, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP) for 2 h at room temperature. F-actin (4 μM) in FEM-buffer was incubated with pacsin2tr (20 μM) for 5–20 min at room temperature. The samples were frozen using a Vitrobot Mark IV (FEI) plunge freezer. Droplets (2.5–3 μl) were applied to glow discharged C-flat grids in 100% humidity at 20°C, with a blotting time of 3–3.5 s. Cryo-EM was done on a Tecnai F20 microscope with a field emission source, operated at 200 keV at a magnification of 50,000×. Images were recorded on film with defocus values from 1.9 to 4.1 μm. The micrographs were scanned on a Nikon Coolpix 8000 at a raster of 1.25 Å/pixel. They were decimated to 2.5 Å/pixel after filaments were extracted from the images using the helixboxer routine within EMAN 39. The SPIDER software package 40 was used for most other steps in the image processing, with the addition of routines for IHRSR 25. Boxes of length 200 pixels (500 Å) were used for sorting and three-dimensional reconstruction. Images were multiplied by the theoretical CTF function, which is the simple application of a Weiner filter in the limit of very low SNR. After the reconstruction was generated, it was corrected for the fact that the images were multiplied by the CTF twice (once by the microscope, once by us) by dividing by the sum of the squared CTFs.
Accession numbers
Coordinates of crystal form I were deposited to the Protein Data Bank under accession code: 4bne.
Acknowledgments
We wish to thank Joanne McCarthy and Stéphanie Monaco at ESRF for their assistance at the ID14-1 and ID14-2 beamlines. Many thanks to Gwyndaf Evans (DIAMOND) for helping in the initial phasing with SHARP. Maurizio Polentarutti (Elettra) is acknowledged for help with pressure cell and xenon derivatization of crystals. IT was a recipient of a Long Term Postdoctoral Fellowship from the European Molecular Biology Organisation, Heidelberg, Germany. In the early stage of this work, IT was supported by a TRIL fellowship from the International Centre of Theoretical Physics, Trieste, Italy. JK was supported by the Federal Ministry of Economy, Family and Youth through ‘Laura Bassi Centre of Expertise’ initiative project Number 253275 and by Austrian Science Fund (FWF) Project P22276. This work was supported by NIH GM081303 (to EHE). CS was supported by the University of Vienna. GA and VH were supported by the Slovenian Research Agency.
Author contributions
Crystal structure determination: IT. Biochemical experiments: JK, US, CS, JS. Electron microscopy experiments: AO. Surface plasmon resonance experiments: VH. Cell cultures, transfections, and immunofluorescence microscopy: JM, MN, IV. XL-MS experiments: JZ. Co-flotation experiments: YS. BI analysis: OIC. Experimental design, interpretation, and writing of manuscript: IT, JK, US, GA, JM, MN, IV, YS, JZ, V-PL, EHE, KDjC.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Aspenstrom P. Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int Rev Cell Mol Biol. 2009;272:1–31. doi: 10.1016/S1937-6448(08)01601-8. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Gautreau A. Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol. 2012;22:141–150. doi: 10.1016/j.tcb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Pykalainen A, Boczkowska M, Zhao H, Saarikangas J, Rebowski G, Jansen M, Hakanen J, Koskela EV, Peranen J, Vihinen H, et al. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat Struct Mol Biol. 2011;18:902–907. doi: 10.1038/nsmb.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- Wang Q, Kaan HY, Hooda RN, Goh SL, Sondermann H. Structure and plasticity of endophilin and sorting nexin 9. Structure. 2008;16:1574–1587. doi: 10.1016/j.str.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Daumke O, Roux A, Haucke V. BAR domain scaffolds in dynamin-mediated membrane fission. Cell. 2014;156:882–892. doi: 10.1016/j.cell.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M, Takenawa T, Yokoyama S. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J Biol Chem. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- Merilainen J, Lehto V-P, Wasenius V-M. FAP52, a novel, SH3 domain-containing focal adhesion protein. J Biol Chem. 1997;272:23278–23284. doi: 10.1074/jbc.272.37.23278. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148:1047–1062. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels MM, Dong J, Leibig W, Westermann P, Qualmann B. Complexes of syndapin II with dynamin II promote vesicle formation at the trans-Golgi network. J Cell Sci. 2006;119:1504–1516. doi: 10.1242/jcs.02877. [DOI] [PubMed] [Google Scholar]
- Senju Y, Itoh Y, Takano K, Hamada S, Suetsugu S. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J Cell Sci. 2011;124(Pt 12):2032–2040. doi: 10.1242/jcs.086264. [DOI] [PubMed] [Google Scholar]
- Shimada A, Takano K, Shirouzu M, Hanawa-Suetsugu K, Terada T, Toyooka K, Umehara T, Yamamoto M, Yokoyama S, Suetsugu S. Mapping of the basic amino-acid residues responsible for tubulation and cellular protrusion by the EFC/F-BAR domain of pacsin2/Syndapin II. FEBS Lett. 2010;584:1111–1118. doi: 10.1016/j.febslet.2010.02.058. [DOI] [PubMed] [Google Scholar]
- de Kreuk BJ, Nethe M, Fernandez-Borja M, Anthony EC, Hensbergen PJ, Deelder AM, Plomann M, Hordijk PL. The F-BAR domain protein PACSIN2 associates with Rac1 and regulates cell spreading and migration. J Cell Sci. 2011;124(Pt 14):2375–2388. doi: 10.1242/jcs.080630. [DOI] [PubMed] [Google Scholar]
- Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci USA. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomann M, Wittmann JG, Rudolph MG. A hinge in the distal end of the PACSIN 2 F-BAR domain may contribute to membrane-curvature sensing. J Mol Biol. 2010;400:129–136. doi: 10.1016/j.jmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci USA. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Su X, Nguyen V, Roberts K, Li X, Takakura A, Plomann M, Zhou J. Polycystin-1 regulates actin cytoskeleton organization and directional cell migration through a novel PC1-Pacsin 2-N-Wasp complex. Hum Mol Genet. 2014;23:2769–2779. doi: 10.1093/hmg/ddt672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- Behrmann E, Muller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro I, Nikki M, Glumoff T, Lehto V-P, Djinovic Carugo K. Crystallization and phasing of focal adhesion protein 52 from Gallus gallus. Acta Crystallogr D. 2004;60:539–541. doi: 10.1107/S090744490302907X. [DOI] [PubMed] [Google Scholar]
- Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels MM, Qualmann B. Syndapins integrate N-WASP in receptor-mediated endocytosis. EMBO J. 2002;21:6083–6094. doi: 10.1093/emboj/cdf604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 2010;21:340–349. doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Masuda M, Mochizuki N. Structural characteristics of BAR domain superfamily to sculpt the membrane. Semin Cell Dev Biol. 2010;21:391–398. doi: 10.1016/j.semcdb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostan J, Gregor M, Walko G, Wiche G. Plectin isoform-dependent regulation of keratin-integrin alpha6beta4 anchorage via Ca2+/calmodulin. J Biol Chem. 2009;284:18525–18536. doi: 10.1074/jbc.M109.008474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besenicar M, Macek P, Lakey JH, Anderluh G. Surface plasmon resonance in protein-membrane interactions. Chem Phys Lipids. 2006;141:169–178. doi: 10.1016/j.chemphyslip.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Hodnik V, Anderluh G. Capture of intact liposomes on biacore sensor chips for protein-membrane interaction studies. Methods Mol Biol. 2010;627:201–211. doi: 10.1007/978-1-60761-670-2_13. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li YH, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.