Abstract
DNA double-strand breaks (DSBs) activate a signaling pathway known as the DNA damage response (DDR) which via protein–protein interactions and post-translational modifications recruit signaling proteins, such as 53BP1, to chromatin flanking the lesion. Depletion of the SET8 methyltransferase prevents accumulation of 53BP1 at DSBs; however, this phenotype has been attributed to the role of SET8 in generating H4K20 methylation across the genome, which is required for 53BP1 binding to chromatin, prior to DNA damage. Here, we report that SET8 acts directly at DSBs during the DNA damage response (DDR). SET8 accumulates at DSBs and is enzymatically active at DSBs. Depletion of SET8 just prior to the induction of DNA damage abrogates 53BP1’s accumulation at DSBs, suggesting that SET8 acts during DDR. SET8’s occupancy at DSBs is regulated by histone deacetylases (HDACs). Finally, SET8 is functionally required for efficient repair of DSBs specifically via the non-homologous end-joining pathway (NHEJ). Our findings reveal that SET8’s active role during DDR at DSBs is required for 53BP1’s accumulation.
Keywords: 53BP1, DNA repair, H4K20, NHEJ, Pr-SET7, SET8
Introduction
DSBs are cytotoxic forms of DNA damage that can lead to oncogenic chromosomal rearrangements if improperly repaired 1. DSBs trigger a complex set of cellular responses collectively known as the DDR that impinges on transcription, DNA replication and cell cycle progression. Immediately after the detection of DSBs, the DDR pathway relaxes chromatin at these regions and promotes the stepwise accumulation of signaling and DNA repair proteins to chromatin flanking DSBs through post-translational modifications 2. One of the earliest events is the phosphorylation of a multitude of proteins, most notable among them is histone H2AX at serine 139 (known as γH2AX) 3, at DSBs by the ATM, ATR and/or the DNA-PK kinases. Spreading of γH2AX marked chromatin on either side of DSBs serves as a scaffold for anchoring large complexes of signaling and repair proteins that are detectable by immunofluorescence. The initial wave of protein phosphorylation is then followed by a wave of protein ubiquitylation by the RNF8 E3 ubiquitin ligase, which localizes to DSBs by binding to MDC1, a direct reader of γH2AX 4–6. The RNF8 E3 ligase then conjugates K48- and K63-linked ubiquitin chains onto histone and non-histone proteins at DSBs leading to recruitment of additional proteins, including the E3 ligase RNF168 7,8. Subsequent conjugation of K63-linked ubiquitin by RNF168 onto various proteins, including histones, leads to recruitment of downstream proteins, most notably BRCA1 and 53BP1 7,8. In addition to phosphorylation and ubiquitylation, damage-dependent post-translational modifications such as sumoylation, methylation and acetylation of histone and non-histone proteins at DSBs have also been reported 9. The DDR signaling cascade culminates in the selection of either NHEJ or homologous recombination (HR) for repair of each DSB.
Damage-induced chromatin binding of various signaling and repair proteins can require a combination of post-translational modifications. Several determinants of 53BP1 localization to chromatin flanking DSBs have been identified. 53BP1’s binding to DSBs strongly requires ubiquitin signaling as depletion of RNF8/RNF168 abrogates 53BP1 ionizing radiation-induced foci (IRIFs) 7,8. Moreover, H4K20 methylation is also necessary as demonstrated by the failure of the 53BP1 H4K20me1/2 binding mutant (D1521R) to form IRIFs 1. Two regions of 53BP1 mediate these binding events: first, its UDR domain interacts with K63-linked ubiquitin on H2A/H2AX K15 10 and second, its tandem Tudor domain binds H4K20me2 11. While the H2A/H2AX K15ub is placed specifically at DSBs by RNF168 10,12, the H4K20me2 is present abundantly across the genome according to mass spectrometry-based quantification 13,14. To make H4K20me2 accessible for 53BP1, RNF8 promotes proteolysis of competing H4K20me2 binding proteins such as L3MBTL1 15 and JMJD2A 16 at DSBs. While H4K20me2 may be globally abundant, local increases in H4K20me1, H4K20me2 and H4K20me3 at DSBs have also been observed 17. WHSC1/MMSET methyltransferase 17,18 may contribute to this increase; however, it is not clear whether WHSC1/MMSET is required for 53BP1’s accumulation 19,20.
SET8 is a monomethyltransferase that is required during normal cell cycle where it plays a role in replication, transcription and chromosome segregation 21. SET8’s substrates include both histone H4K20 and non-histone proteins. Although SET8 accumulates at UV laser-induced damage 22, SET8’s level did not increase significantly at endonuclease-induced DSBs as assessed by chromatin immunoprecipitation (ChIP) 17. Here, we report that SET8 is enzymatically active at DSBs and that SET8’s activity during the DDR is required for 53BP1 accumulation.
Results
SET8 is required during DDR for 53BP1 binding to chromatin flanking DSBs
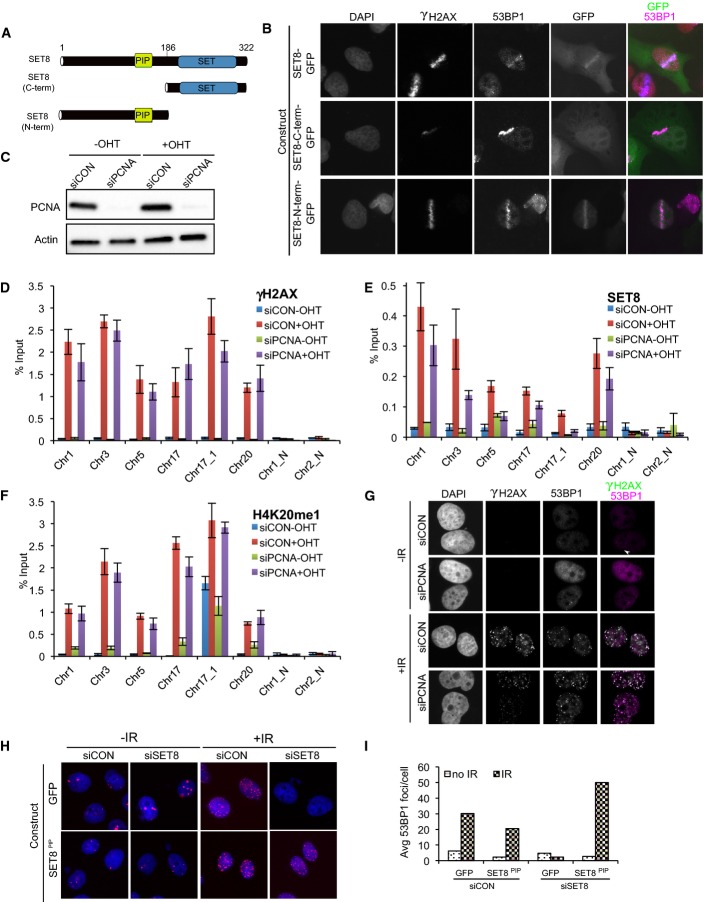
SET8 was shown to accumulate at UV laser-induced damage; however, laser micro-irradiation may also generate many types of DNA lesions depending on the laser power and wavelength, so it is unclear whether SET8 localization to laser micro-irradiation foci was mediated by DSBs or by other types of DNA damages. To address this question, we used an inducible endonuclease system that generates DSBs at defined sites in the genome 23. To determine which AsiSI recognition sequences are cut in U2OS cells, we performed ChIP followed by high-throughput sequencing using anti-γH2AX antibody on cells either treated or not with tamoxifen. We observed clear γH2AX peaks at approximately 72 AsiSI sites in tamoxifen-treated cells but not in untreated cells (data not shown). From these experiments, we selected 6 sites that are cut in a tamoxifen-dependent manner and 2 sites that have AsiSI recognition sequences but were negative for γH2AX. We performed ChIP using antibodies against SET8 and H4K20me1 and quantified their enrichment at the selected AsiSI cut sites 2 h after tamoxifen treatment. We observed a significant level of increase in SET8 and H4K20me1 compared to its levels at these sites prior to treatment with tamoxifen (Fig 1A; P < 0.01 for both SET8 and H4K20me1, paired t-test).
Figure 1. SET8 localizes to DSBs.
A Endogenous SET8 and H4K20me1 accumulate at DSBs in a damage-dependent manner. ChIP-qPCR for SET8 and H4K20me1 was carried on using U2OS cells stably expressing the tamoxifen (4-OHT)-inducible AsiSI endonuclease system. Results are normalized to the input. P < 0.05 (paired t-test for difference in SET8 and H4K20me1 at DSBs in −OHT and +OHT conditions).
B Western blot showing the SET8 knockdown efficiency after siRNA treatment. Whole-cell extracts from U2OS cells treated either with control or siRNA against SET8 (two subsequent siRNA transfections in 48 h) were immunoblotted and probed with the indicated antibodies. Actin was used as a loading control.
C Levels of γH2AX at DSBs are similar in siCON and siSET8 cells (P = 0.5; paired t-test for difference in γ-H2AX in −OHT and +OHT conditions). ChIP-qPCR was done on the same batch of cells as shown in (B).
D, E Increases in H4K20me1 and 53BP1 at DSBs are abrogated in SET8-depleted cells. ChIP-qPCR experiment was performed with anti-53BP1 and anti-H4K20me1 antibodies. Data are shown relative to input (P < 0.05; paired t-test for difference in H4K20me1 and 53BP1 at DSBs in −OHT and +OHT conditions).
F Abrogation of 53BP1 IRIFs upon knockdown of endogenous SET8 cannot be rescued by catalytic deficient SET8. Immunofluorescence for 53BP1 in SET8-depleted U2OS cells (two subsequent siRNA transfections in 48 h) after expression of GFP or siRNA-resistant SET8 form or siRNA-resistant SET8 catalytic dead mutant followed by IR treatment.
G Quantification of rescue experiments shown in (F).
H SET8 colocalizes with 53BP1. U2OS cells expressing GFP-SET8 were sensitized with BrdU and subjected to laser micro-irradiation. Cells were fixed and immunostained for γH2AX and 53BP1 30 min after laser irradiation. GFP-SET8 was visualized by GFP fluorescence.
I Quantification of the extent of colocalization of SET8 with 53BP1 shown in (H).
To test whether SET8 is required for increases in H4K20me1 and 53BP1 at DSBs, we performed ChIP with anti-H4K20me1 and anti-53BP1 antibodies after depleting SET8 with siRNAs (Fig 1B). We also performed ChIP with an anti-γH2AX antibody to control for potential differences in levels of DSBs in the cell population treated with non-targeting (siCON) or SET8 siRNA (siSET8). Although the levels of γH2AX IRIF formation in siSET8 cells are noticeably higher than in siCON cells in both unchallenged and IR-treated cells (Supplementary Fig S1), we observed no significant difference in the accumulation of γH2AX at AsiSI sites in siCON or siSET8 cells induced with tamoxifen (Fig 1C; P = 0.6, Mann–Whitney U-test). H4K20me1 and 53BP1 accumulated at DSBs as expected in siCON cells (Fig 1D and E); however, its increase at DSBs was abrogated in siSET8 cells (Fig 1D and E; P < 0.01 for both H4K20me1 and 53BP1, paired t-test) despite insignificant differences in γH2AX levels at DSBs in siCON and siSET8 cells (Fig 1C). The depletion of SET8 was specific since a siRNA-resistant full-length form of SET8 rescued 53BP1 IRIFs (Fig 1F and G). Moreover, a catalytic dead point mutant (R265G) form of SET8 failed to rescue 53BP1 IRIFs (Fig 1F and G). To determine the extent of SET8 co-localization with 53BP1 within the same cell, U2OS cells expressing GFP-SET8 were sensitized in BrdU-containing medium, micro-irradiated using a UV laser and processed for immunofluorescence. We observed that SET8-GFP accumulated and co-localized with 53BP1 in the majority of micro-irradiated cells (Fig 1H and I). We conclude that SET8 methyltransferase accumulates at DSBs and promotes H4K20me1 and 53BP1 accumulation at these sites.
Although the depletion of SET8 abrogates 53BP1 accumulation at DSBs 11 (Fig 1F and G), this might be an indirect consequence of a global reduction of pre-existing H4K20me upon SET8 depletion (Fig 1B) rather than due to the abrogation of SET8’s activity on H4K20 or some other unknown substrate(s) specifically at DSBs. To test whether damage-dependent SET8 accumulation at DSBs is required for 53BP1 foci formation, we used the auxin-inducible degron (AID) system that allows degradation of a target protein within few hours 24 (Fig 2A). An siRNA-resistant form of SET8 was fused to GFP-AID and expressed in TIR1 expressing DLD-1 cells 25. Treatment with auxin led to efficient degradation of SET8-GFP-AID protein in 6 h (Fig 2B). AID-GFP-SET8 expressing cells transfected with siRNA against SET8 were able to rescue 53BP1 IRIFs, indicating that SET8-AID form is functional (Fig 2C).
Figure 2. SET8’s activity during DDR is required for 53BP1 foci formation.
A Illustration of the auxin-inducible degron (AID) system.
B DLD1 cell line stably expressing TIR1 and dox-inducible siRNA-resistant AID-GFP-SET8 construct was treated with either control or SET8-targeting siRNA for 48 h. AID-GFP-SET8 was degraded by treating the cells with auxin for 6 h, after depleting the DMEM medium of dox. Cells were irradiated with 10 Gy and allowed to recover for 1 h. Whole-cell extracts were immunoblotted with the indicated antibodies to access the knockdown and AID-GFP-SET8 degradation efficiency. Blots for H4K20me1 and actin were quantified using ImageJ.
C Immunofluorescence for 53BP1 in DLD1-TIR1 cells expressing AID-GFP-SET8. Cells were irradiated with 10 Gy of ionizing radiation and fixed after 1 h.
D Quantification of experiment shown in (C). Median number of 53BP1 IRIFssiSET8 + IR = 31, median number of 53BP1 IRIFssiSET8 + IR + auxin = 8; P = 2.2 × 10−64, Mann-Whitney U-test.
In IR-treated siCON cells, treatment with auxin resulted in non-significant reduction in 53BP1 IRIFs (median number of 53BP1 IRIFs siCON+IR = 20, median number of 53BP1 IRIFs siCON+IR+auxin = 19; P = 0.1, Mann–Whitney U-test) (Fig 2C and D), indicating that auxin does not interfere with formation of 53BP1 IRIFs. In contrast, in IR-treated siSET8 cells, treatment with auxin resulted in a significant reduction in 53BP1 IRIFs (median number of 53BP1 IRIFs siSET8+IR = 31, median number of 53BP1 IRIFs siSET8+IR+auxin = 8; P = 2.2 × 10−64, Mann–Whitney U-test) (Fig 2C and D). To determine the extent to which the reduction in 53BP1 IRIF in siSET8 + IR + auxin condition can be attributed to the reduction in the global levels of H4K20me1 that reduced upon auxin-mediated depletion of the ectopic AID-GFP-SET8, we quantified the H4K20me1 Western blots and normalized it to the levels of actin in each of the conditions (Fig 2C; bottom panel). Upon treatment of cells with auxin, levels of bulk H4K20me1 reduced by 30% in IR-treated siCON cells and by 23% in IR-treated siSET8 cells. Thus, the striking 90% reduction in 53BP1 IRIFs in siSET8 + IR + auxin cells cannot be explained by the reduction in the bulk H4K20me1 due to auxin-mediated depletion of ectopic SET8. These results strongly suggest that SET8’s activity during the DDR, through the methylation of H4K20 (Fig 1C) or of some unknown substrate(s), is required for recruitment of 53BP1 to DSBs.
SET8 binding to DSBs can occur independently of PCNA
To identify the region required for localization of SET8 at DSBs, we fused the N-terminal fragment, containing the PIP box, and C-terminal fragment containing only the catalytic domain of SET8 to GFP (Fig 3A) and performed laser micro-irradiation experiments. The C-terminus of SET8 that contains the methyltransferase domain localized to the nucleus but failed to co-localize with γH2AX and 53BP1 (Fig 3B; Supplementary Fig S2). In contrast, the N-terminus of SET8 did localize to laser lines, similar to the full-length form (Fig 3B). We conclude that SET8’s N-terminal half is sufficient for its localization to laser micro-irradiation mediated DNA damage.
Figure 3. SET8 can localize to and promote 53BP1’s accumulation at DSBs independently of PCNA.
A Schematic of SET8 protein and the truncation forms.
B The N-terminus of SET8 is sufficient for DSB localization. U2OS cells were transfected with the indicated constructs.
C Immunoblots showing knockdown efficiency in whole-cell extracts derived from U2OS-ER-AsiSI cells transfected with non-targeting or PCNA-targeting siRNAs.
D–F ChIP-qPCR experiments were performed in U2OS-ER-AsiSI cell line transfected with the indicated siRNAs using anti-γH2AX, anti-SET8 and anti-H4K20me1 antibodies. P ≫ 0.05 (paired t-test) for difference in γH2AX levels at DSBs in siCON and siPCNA treated with OHT. P = 0.002 (paired t-test) for reduction in SET8 levels in siPCNA cells compared to siCON cells. P ≫ 0.05 (paired t-test) for reduction in H4K20me1 in siPCNA cells compared to siCON cells. Mean proportional reduction in SET8 levels at DSBs in siPCNA cells treated with OHT compared to siCON cells treated with OHT is 0.61 with a standard deviation of 0.26.
G 53BP1 IRIFs are unaffected after PCNA depletion. Immunostaining of U2OS cells transfected with non-targeting or PCNA-targeting siRNAs. Cells were allowed to recover for 1 h after irradiation (10 Gy) prior to staining.
H, I Abrogation of 53BP1 IRIFs by depletion of endogenous SET8 can be rescued with a siRNA-resistant form of SET8 containing a PCNA interacting mutation. Immunofluorescence of U2OS cells treated with non-targeting or SET8-targetting siRNAs co-transfected with plasmids encoding GFP or siRNA-resistant SET8 with PCNA binding mutant.
Recruitment of SET8 to UV laser micro-irradiation-induced DNA damage requires PCNA 22. SET8 interacts with PCNA on chromatin via its PIP box motif 22,26–29. As the N-terminal region of SET8 that is sufficient to localize to laser micro-irradiation sites (Fig 3B) contains a PIP box, we directly tested for PCNA’s role in recruiting SET8 at AsiSI-induced DSBs by performing ChIP in PCNA-depleted cells (Fig 3C; Supplementary Fig S1B). In cells treated with control siRNA, AsiSI-sensitive sites were enriched for γH2AX, SET8 and H4K20me1 after tamoxifen treatment (Fig 3D–F). In PCNA-depleted cells, SET8 enrichment at DSBs was reduced by approximately 61% (Fig 3E) and H4K20me1 levels reduced by approximately 20% (Fig 3F), suggesting that SET8 recruitment to DSBs can also occur independently of PCNA. To test whether the observed reduction in SET8 at DSBs upon PCNA depletion is sufficient to abrogate SET8’s role during DDR, we tested for defects in 53BP1 IRIFs in PCNA-depleted cells; however, we found that 53BP1 IRIFs were unaffected in PCNA-depleted cells (Fig 3G). As a corollary to these experiments, we tested whether an siRNA-resistant form of SET8 containing mutations in the PIP box motif (F184A/Y185A) that does not interact with PCNA 29 could rescue 53BP1 foci formation. Both the wild-type and PIP box mutant forms of SET8 restored the accumulation of 53BP1 at DSBs (Fig 3H and I), suggesting that interaction with PCNA is not required for this activity. Altogether, our results suggest that SET8’s recruitment to and its role in promoting 53BP1’s accumulation at DSBs can occur independently of PCNA.
HDACs negatively regulate SET8’s occupancy at DSBs
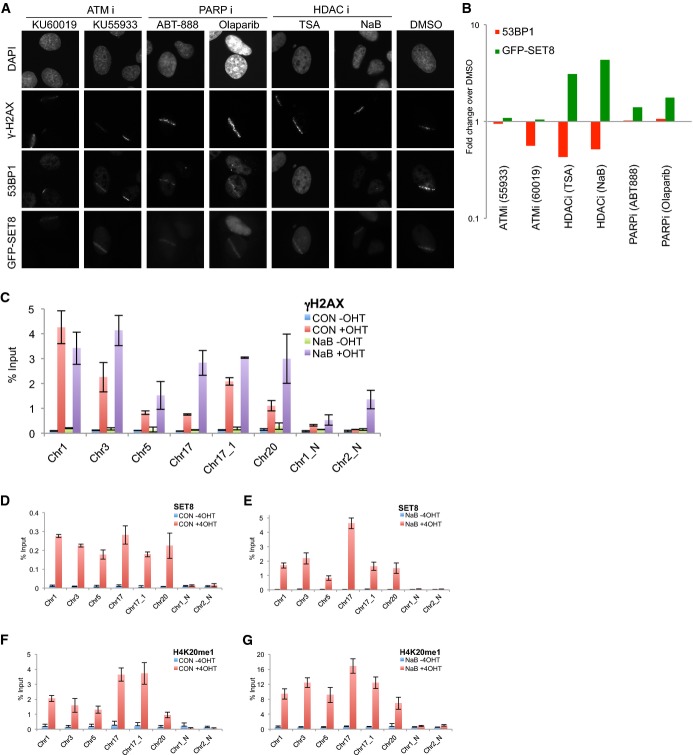
To identify pathways that mediate SET8 recruitment to DSBs, we focused on proteins that influence 53BP1’s recruitment to DSBs such as ATM 1 and HDACs 20,30. Additionally, recruitment of SET8 to chromatin is dependent on the activity of poly-ADP ribosylase polymerase I (PARP1) 31. In parallel, we treated GFP-SET8 expressing cells with two ATM inhibitors (KU-55933 and KU-60119), two HDAC Class I and II inhibitors (trichostatin and sodium butyrate) and two PARP inhibitors (olaparib and ABT-888) and then micro-irradiated cells. The number of cells with discernable recruitment of 53BP1 or SET8-GFP to DNA damage was divided by the number of cells with γH2AX positive signal to determine the percent positive cells. 53BP1’s recruitment to laser lines was reduced by approximately 50% in cells treated with ATMi; however, no discernible effect on SET8’s recruitment to laser-induced damage was observed under these conditions (Fig 4A and B). Treatment of cells with PARP1 inhibitors did not affect 53BP1, but SET8’s recruitment to laser lines increased by approximately 20% (Fig 4A and B). In contrast, inhibiting activity of HDAC (Class I and II) reduced steady-state levels of 53BP1 at DSBs by approximately 50% in agreement with previous reports 20,30 while unexpectedly increasing the intensity and the proportion of cells with SET8 micro-irradiation laser lines by three times (Fig 4A and B). To test whether inhibiting HDACs affects SET8’s occupancy at DSBs, we performed the ChIP assay after AsiSI-induced DSBs with anti-γH2AX, anti-SET8 and anti-H4K20me1 antibodies. Levels of γH2AX increased by 25% in NaB-treated +OHT cells compared to untreated +OHT cells (Fig 4C); however, the levels of SET8 (Fig 4D and E) increased by almost 20-fold, consistent with the increase in intensity of GFP-SET8 in the micro-irradiation experiments (Fig 4A and B). Congruent with increase in SET8 at DSBs in HDACi-treated cells, levels of H4K20me1 increased by fourfold under this condition (Fig 4F and G). We conclude that HDACs (Class I and II) counteract SET8’s occupancy at DSBs.
Figure 4. HDACs negatively regulate SET8’s occupancy at DSBs.
A Representative images of U2OS cells transfected with GFP-SET8 and treated with the indicated inhibitors. Cells were allowed to recover for 1 h post-micro-irradiation prior to staining. Prior to micro-irradiation, cells were incubated with inhibitors as follows: TSA (1.3 μM, 16 h) or NaB (5 mM, 16 h), KU55933 (10 μM, 1 h), KU60019 (10 μM, 1 h), ABT-888 (10 μM, 1 h) and Olaparib (0.5 μM, 2 h).
B Quantification of results shown in (A). P-values were computed using Fisher’s exact test for difference in proportion of 53BP1- or SET8-GFP-positive cells in γH2AX laser-positive cells under the indicated treatments relative to DMSO. Statistical significance for reduction of 53BP1 at laser micro-irradiation: PTSA = 7 × 10−43; PNaB = 3 × 10−32. Statistical significance for increase of SET8 at laser micro-irradiation: PTSA = 6 × 10−7; PNaB = 4 × 10−12.
C–G γH2AX (C), SET8 (D, E) and H4K20me1 (F, G) ChIP-qPCR in U2OS-ER-AsiSI cells untreated (C, D and F) or treated with 5 mM NaB for 16 h (C, E and G). P << 0.05 (paired t-test for difference in SET8 and H4K20me1 levels in +OHT untreated cells and +OHT NaB-treated cells).
SET8 promotes DNA double-strand break repair via the NHEJ pathway
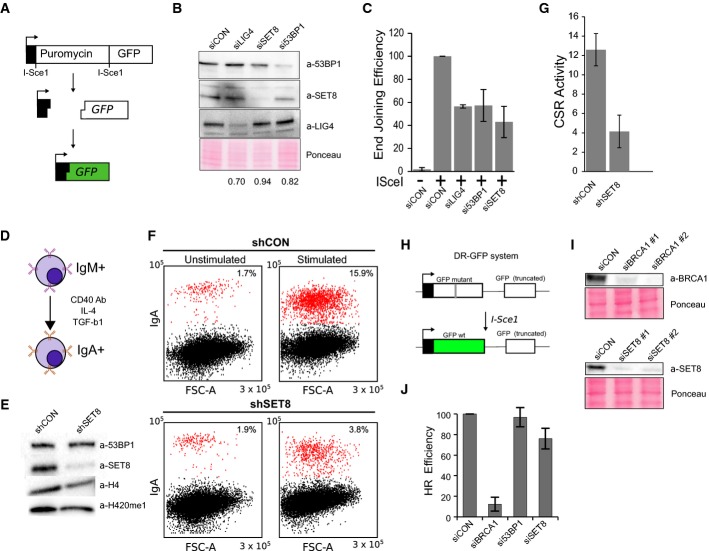
To test whether SET8 plays a functional role in DSB repair in vivo, we assessed the effect of SET8 depletion using a reporter-based NHEJ assay 32 (Fig 5A). Depletion of LIG4, a gene essential for canonical NHEJ, resulted in an approximately 50% reduction of GFP+ cells (Fig 5B and C). The depletion of 53BP1 or SET8, but not a non-targeting control, resulted in a similar decrease confirming that SET8 promotes NHEJ (Fig 5B and C). To substantiate the role of SET8 in NHEJ repair of DSBs using an assay that involves repair of natural DSBs, we performed the CH12F3-2 B-cell-based class switch recombination (CSR) assay 33 (Fig 5D). Upon activation of B cells, multiple breaks arise at the IgH locus and 53BP1 is essential for long-range end joining resulting in the switching of the antibody isotype 33. Upon stimulation, ∼16% of the B cells infected with a control shRNA underwent class switching from IgM to IgA; however, class switching was reduced to approximately 4% in SET8-depleted cells (Fig 5E–G). To test whether SET8 contributes to the homologous recombination (HR) directed repair of DSBs, we employed the previously described HR reporter assay (DR-GFP) 34 (Fig 5H). Expression of I-SceI endonuclease generates a site-specific DSB within the mutated GFP gene, which when repaired by gene conversion results in a functional GFP; extent of HR can thus be quantified using flow cytometry. We used siRNAs against BRCA1 as positive control and confirmed that the repair efficiency of these cells were severely abrogated (> 90%) (Fig 5I). Depletion of SET8, however, only partially decreased the HR efficiency (∼15%) (Fig 5J). Together, these results demonstrate that SET8 plays a role in DSB repair, particularly in the NHEJ pathway.
Figure 5. SET8 promotes NHEJ-dependent DSB repair.
A Illustration of the NHEJ assay 32.
B Whole-cell extracts from cells treated with the indicated siRNA were probed with the indicated antibodies to confirm knockdown efficiencies. The percent knockdown compared to control is indicated.
C GFP+ cells indicating NHEJ activity is plotted as a percent of the rejoining activity from cells treated with control siRNA. Results are the mean and standard deviation of 3 independent experiments.
D Class switch recombination (CSR) assay: CH12F3-2 B cells are stimulated with IL-4, TGF-β and anti-CD4 antibodies, and extent of CSR is measured by the cell surface expression of IgA.
E Whole-cell extracts from CH12-F3 cells infected with the indicated shRNAs were probed with the indicated antibodies to confirm knockdown efficiency.
F Representative flow cytometric profiles of shRNA-transduced CH12F3-2 cells unstimulated and stimulated with a cocktail of cytokines for 1 day and stained with anti-IgA antibody. Forward scatter is shown on the x-axis and the intensity of the IgA stain is shown on the y-axis.
G The proportion of IgA+ cells were determined by flow cytometry and plotted as the percent of cells that underwent class switching. Data are expressed as the mean ± standard deviation from three biological replicates from two independent shRNA infections.
H DR-GFP assay for HR. U2OS cells with chromosomally integrated cassette containing a full-length but mutant GFP gene containing the I-SceI site next to a truncated GFP. After induction of break within the mutant GFP, cells that have successfully carried out HR-directed gene conversion using the truncated GFP as a template are quantified by flow cytometry.
I Whole-cell extracts from DR-GFP U2OS treated with the indicated siRNAs were probed with the indicated antibodies to confirm knockdown efficiency.
J Quantification of proportion of GFP-positive cells after induction of I-SceI. Data are expressed (relative to the GFP-positive cells in the cells treated with I-SceI and siCON) as mean and standard deviation from three biological replicates from two independent siRNA infections.
Discussion
53BP1 plays a central role in DSB repair as it contributes to NHEJ directed repair by acting as a scaffold for the accumulation of other proteins such as RIF1 35–39 and PTIP 40 to inhibit end resection, the committing step of HR. Furthermore, 53BP1 underlies pathological DNA repair, tumorigenesis and sensitivity to chemotherapeutic agents such as PARP inhibitors in BRCA1-deficient cells, and therefore, it is important to identify all the determinants of its occupancy at DSBs. The determinants of 53BP1’s binding to chromatin flanking DSBs appear to be complex; H4K20 methylation 11, RNF8-dependent degradation of competing H4K20me readers 15,16, H4K16 deacetylation 20,30, and RNF168-mediated H2AK15 ubiquitylation 10 have all been suggested to play a role in this process. The results presented here show that SET8 is another factor that is also required for 53BP1’s accumulation at DSBs.
Depletion of SET8 via siRNAs or shRNAs results in significant reduction of genomewide levels of H4K20me1 and H4K20me2 prior to DNA damage, which make it difficult to determine whether SET8’s activity on H4K20 or some other unknown substrate(s) during the DDR is required for supporting 53BP1 function. To overcome this limitation, we employed the auxin degron system 24 that allows exogenous SET8 to be degraded within few hours so that genomewide levels of H4K20 methylation are not greatly perturbed (Fig 2C; bottom panel). Our finding that depletion of SET8 just prior to irradiation is sufficient to abolish 53BP1 IRIFs strongly supports our assertion that SET8 is actively required during DDR to recruit 53BP1.
Although PCNA regulates SET8’s localization to UV laser micro-irradiation-induced DNA damage, there appears be a PCNA-independent pathway for SET8 and 53BP1 localization to DSBs (Fig 3). We note it might be possible that during S-phase, the role of PCNA might be important. In agreement with previous reports 20,30, 53BP1’s recruitment to DSBs is negatively regulated by HDAC inhibition (Fig 4) and may be due to H4K16ac, which increases upon HDAC inhibition 41 and which interferes with the interaction between 53BP1 and H4K20me2. In contrast, DNA damage-induced SET8 occupancy and consequently the levels of H4K20me1 at DSBs are dramatically increased upon HDAC inhibition (Fig 4). This result is unexpected as in vitro experiments have shown that SET8 affinity and methylation activity at K20 on histone H4 with K16ac are significantly lower than on histone H4 without K16ac 42–44. The cause of this discrepancy is unclear but may have to do with differences in in vitro versus in vivo conditions such as presence of additional constraints imposed by chromatin state 45.
Using both artificially induced DSBs as well as a system using naturally arising DSBs in the B-cell class switch recombination process, we have demonstrated that SET8 is functionally required for repair specifically via the NHEJ pathway (Fig 5). SET8 depletion significantly altered the fidelity, efficiency or progression of NHEJ-dependent repair but only slightly affected HR. The affect on HR might be related to previously demonstrated role of SET8 during S-phase 46,47.
Our results are consistent with findings reported in a recent paper 48, demonstrating that SET8 accumulates at DSBs independently of PCNA and that accumulation of H4K20me and 53BP1 at DSBs depend on SET8. However, the approach used in their paper is significantly different from ours. In their paper, ChIP were performed at a single DSB at an artificial site in the genome after expression of the I-SceI endonuclease for 24–36 h, while in our system, the ChIP experiments were done 2 h after induction of the AsiSI endonuclease at multiple native, that is, unaltered sites in the genome. Furthermore, while Tuzon et al 48 claim that SET8 acts during DDR and that SET8-dependent increase in de novo H4K20me at DSBs is necessary for 53BP1 foci formation, no formal evidence was provided for this assertion. By using a degron system that allows depletion of SET8 within few hours without perturbing the bulk H4K20me1 (Fig 2), we have provided strong evidence that SET8 acts during DDR and that the abrogation of 53BP1’s foci in the absence of SET8 is due to loss of SET8’s role during DDR. It remains formally possible that non-histone targets of SET8 might also be important to recruit 53BP1 to DSBs; however, the most straightforward interpretation of our results is that active recruitment of SET8 to DSBs results in the local increase of methylated H4K20, which in turn permits the recruitment of 53BP1. Studies, including the report by Tuzon et al 48, that use siRNA- or shRNA-mediated depletion of SET8 cannot address this question unequivocally as the prolonged period of treatment required for efficient knockdown leads to eradication of the abundant pre-damage levels of H4K20me, making it difficult to determine whether the cause of abrogation of 53BP1’s accumulation at DSBs is the loss of pre-damage activity of SET8 in generating global H4K20me or the damage-dependent activity of SET8 in methylating H4K20 and/or some other target.
The finding that SET8 is required for 53BP1 accumulation during DDR, however, raises a question regarding how 53BP1 might accumulate during all the stages of the cell cycle. SET8 is a cell cycle regulated protein that is rapidly degraded during the onset of the S-phase reaching very low 22,26,27,29, yet detectable 29 and functional 46,47 levels, during S-phase. The apparent absence of SET8 during the S-phase might lead to the expectation that 53BP1 foci may not be able to form during the S-phase; however, experimentally this is not observed 49. It is possible that while the bulk of SET8 is degraded during the S-phase of the cell cycle, the residual SET8 is sufficient for its role in DDR. Furthermore, as SET8 is actively translated during the S-phase 29, it is also possible that the turnover of SET8 is prevented during DDR. This question needs to be addressed in the future.
In conclusion, we have identified SET8 as a direct and an active component of the DDR, which contributes to the accumulation of 53BP1 at DSBs. Given that 53BP1 underlies toxic genomic rearrangements in BRCA1-deficient cells, our work suggests that pharmacological inhibition of SET8 might be a viable therapeutic option for cancers driven by BRCA1 deficiency.
Materials and Methods
Cell culture
CH12F3-2 cells (a generous gift from Daniel Durocher, Toronto) were cultured in RPMI medium (Invitrogen) supplemented with 10% FBS (Invitrogen), 5% NCTC 109 medium (SIGMA), 1× GlutaMAX (Invitrogen) and 50 μM β-mercaptoethanol. U2OS cells were cultivated in High Glucose DMEM with GlutaMAX or McCoy’s medium supplemented with 10% fetal bovine serum (FBS) in 37°C under humidified atmosphere with 5% CO2. Cells were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. The pBABE HA-AsiSI-ER 23 plasmid was transfected into U2OS cells using Lipofectamine, and selection was performed using 1 μg/ml puromycin. Cells were treated with 300 nM OHT (4-hydroxytamoxifen) for 2 h.
Laser micro-irradiation
U2OS cells were seeded on 25-mm coverslips in a 6-well dish, transfected with 1 μg of the plasmids indicated in the figure legends using Lipofectamine 2000 (Invitrogen) and incubated with 10 μM BrdU (Invitrogen) for 12–24 h. Cells were micro-irradiated using a PALM MicroBeam Laser Capture microscope (Zeiss). Briefly, the coverslips were transferred to a magnetic incubation chamber (Chamlide) and overlaid with 1 ml medium. Coverslips were mounted on the microscope and irradiated with a pre-determined pattern using 20% cutting speed, 55% focus and 30% power through a 40× objective. The time required to complete the irradiation pattern was approximately 8 min. Cells were allowed to recover at 37°C for 30 min before being processed for immunofluorescence. It was empirically determined that these settings did not produce a detectable DNA damage response in unsensitized cells (data not shown). Prior to micro-irradiation, cells were incubated with inhibitors as follows: TSA (1.3 μM, 16 h) or NaB (5 mM, 16 h), KU55933 (10 μM, 1 h), KU60019 (10 μM, 1 h), ABT-888 (10 μM, 1 h) and Olaparib (0.5 μM, 2 h).
Immunofluorescence and microscopy
Cells were seeded on 0.17-mm glass coverslips and treated as indicated in the figure legends. Cells were rinsed 2 × with PBS, pre-extracted with CSK (10 mM HEPES-KOH pH 6.8, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2) for 5 min at 4°C, fixed with 3.7% paraformaldehyde in PBS for 15 min at room temperature and washed 4 × with PBS. Cells were permeabilized by incubation in PBS containing 0.25% Triton X-100 (v/v) for 15 min at room temperature and rinsed 1 × with PBS. Samples were blocked with antibody dilution buffer (5% BSA, 0.1% Triton X-100 in PBS) for approximately 1 h and incubated with the indicated primary antibody overnight at 4°C or at room temperature for 2 h. Secondary antibodies were incubated for 45 min at room temperature. Coverslips were mounted with Vectashield + DAPI (Vector Laboratories) or Prolong Diamond containing DAPI. The fluorescence from GFP fusion proteins was imaged from the intrinsic GFP fluorescence remaining after processing. Images were acquired using a Zeiss Axio Observer Z1 microscope using a 40× oil immersion objective and standard FITC, TRITC and Cy5 filter sets.
Class switching assay
To induce class switching, 200,000 transduced CH12F3-2 cells were seeded into 24-well plate and treated with 1 ng/ml TGF-β1 (Preprotech), 10 ng/ml IL-4 (Preprotech) and 1 μg/ml agonist anti-CD40 (BD). Cells were stained with a PE-conjugated anti-IgA antibody (eBiosciences) and analyzed using a FACSCantoII flow cytometer (BD Biosciences) and Flowing Software.
CH12 transductions
To produce virus, subconfluent HEK293T cells were grown in a 10-cm dish and transfected with 5 μg shRNA plasmid, and 2.5 μg each pSVC MV-IN VSVG and pMLV gag-pol pC19-3N (gifts from Dan Durocher, Toronto) using Lipofectamine 2000 (Invitrogen). After 24-h incubation at 37°C, the medium was changed to DMEM containing 20 mM HEPES-KOH pH 7.4, further incubated for 2.5 days at 34°C and filtered through a 0.45-μm filter. 2.5 ml filtered viral supernatant was added to 200,000 CH12F3-2 cells in a 6-well plate (Falcon), centrifuged for 90 min at 1,500 g and incubated at 37°C for approximately 5 h. 2.5 ml CH12F3-2 medium was added, and cells were incubated overnight at 37°C. Cells were progressively selected with 2, 4 and 8 μg/ml puromycin where each selection was carried out for approximately 4 days.
In vivo NHEJ Assay
EJ5 reporter cells were grown in 6-cm dishes and co-transfected with 250 pmol of the indicated siRNA for 2 days and re-transfected with 250 pmol siRNA, 4 μg pCMV3xnlsI-SceI or phCMV-1 I-SceI (non-functional control) and 0.5 μg pCAG Ds-Red (Addgene) as a transfection control. After 72 h, cells were washed 2 × with PBS and analyzed using a FACSCantoII flow cytometer (BD Biosciences) and Flowing Software.
Plasmids
siRNA-resistant plasmids encoding SET8 (322aa version) and SET8 (R268G) were described earlier 29 and were a gift from Eric Julien (Montpellier). SET8 was subcloned into pcDNA5/frt/to-GFP vector (Invitrogen). siRNA-resistant form of SET8-GFP was cloned into pcDNA5-PLK-AID, which was a generous gift from Andrew Holland (Johns Hopkins).
Antibodies
We used the following antibodies: mouse anti-γH2AX (ab81299, Abcam), mouse anti-H4K20me1 (C15200147 (MAb-147-100), Diagenode), rabbit anti-SET8 (2996, Cell Signaling Technology), rabbit anti-LIG4 (ab80514, Abcam), rabbit anti-53BP1 (NB100-305, Novus Biologicals) and anti-beta actin (ab8227, Abcam).
Chromatin immunoprecipitation
Chromatin was sonicated to an average size of 200–400 bp. Forty microlitre Dynabeads Protein G beads (Dynabeads, Invitrogen) were pre-bound with 3–10 μg antibody and added to the chromatin. After 16 h incubation at 4°C on a rotating wheel, beads were collected by DynaMag magnet. Beads were washed three times with Sonication buffer [50 mM Hepes pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 0.5 mM PMSF, Protease inhibitor cocktail (Roche)], three times with wash buffer A (Sonication buffer supplemented with 500 mM NaCl), three times with wash buffer B (20 mM Tris pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 0.5 mM PMSF, Roche protease inhibitor cocktail) and TE buffer pH 8.0 (10 mM Tris–HCl pH 8.0, 1 mM EDTA pH 8.0 supplemented with 50 mM NaCl). 200 μl of elution buffer (50 mM Tris pH 8.0, 1 mM EDTA, 1% SDS, 50 mM NaHCO3) was added to the beads for 20 min at 65°C. Eluates were purified after reverse crosslink using QIAquick PCR Purification kit (Qiagen). qPCRs were done using KAPA SYBR FAST qPCR kit according to the manufacturer’s instructions. The following primers were used: chr1: 5′-AAGACGGTTAGGTGCATGGAG-3′, chr3: 5′-AGGCCCTTTCCTCAAGAATGG-3′, chr5: 5′-TTCAGACTCAGTCAAGGCAAGG-3′, chr17: 5′-TAACCGACCTAGAGTGCACATG-3′, chr17_1: 5′-ACACAGTCCCCTCAAGAAGATC-3′, chr20: 5′-ATCAATACTGGCTCTCGCTACG-3′, chr1_N: 5′-TCATCCTACACAGAAGCCACTG-3′, chr2: 5′-GACTGCCTTGTAAACAGTGCTAC-3′.
RNA interference
Sequences of siRNAs used in this work are as follows: CON: 5′-UAAGGCUAUGAAGAGAUAC-3′, SET8 si #1: 5′-GCAACTAGAGAGACAAATC-3′, SET8 si #2: 5′-GATTGAAAGTGGGAAGGAA-3′, SET8 si #3: 5′-GUGGAUGCAACUAGAGAGACA-3′, 53BP1 si #1: 5′-GAAGGACGGAGUACUAAUA-3′, 53BP1 si #2: 5′-GCACACUUGUCACUCGUGU-3′, LIG4: 5′-AAGCCAGACAAAAGAGGUGAA-3′.
Inducible degron experiment
DLD-1 cell line stably expressing TIR1-9xMyc and siRNA-resistant AID-GFP-SET8 was transfected with siRNAs against SET8 or non-targeting control for 48 h. Cells were then treated with 50 mM of auxin (IAA, Sigma) for 6 h prior to IR exposure (10 Gy) and processed for immunostaining using antibody against 53BP1 after 1-h recovery.
Acknowledgments
This work was supported by funding from the Ontario Institute for Cancer Research to NNB. Authors wish to thank Eric Julien (Montpellier) for reagents and to Daniel Durocher (Toronto), Steve Jackson (Cambridge) and Michael Huen (Hong Kong) for feedback on the manuscript.
Author contributions
SD, JT and NNB conceived study and designed experiments. SD, JT and SL performed experiments. SD, JT and NNB analyzed results. NNB with input from SD and JT wrote Manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Smeenk G, van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu Rev Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Díaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J Biol Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu I, Ciccia A, Lewis SM, Elledge SJ. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci USA. 2011;108:13130–13134. doi: 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlerode AJ, Guan Y, Rajendran A, Ura K, Schotta G, Xie A, Shah JV, Scully R. Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PLoS ONE. 2012;7:e49211. doi: 10.1371/journal.pone.0049211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao KY, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol. 2013;5:157–165. doi: 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, Cleveland DW. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci USA. 2012;109:E3350–E3357. doi: 10.1073/pnas.1216880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell. 2010;40:22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Xu R, Zhu L, Cheng X, Wang Z, Manis J, Shipp MA. BAL1 and its partner E3 ligase, BBAP, link Poly(ADP-ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8. Mol Cell Biol. 2013;33:845–857. doi: 10.1128/MCB.00990-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem. 2013;288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkáč J, Cook MA, Rosebrock AP, Munro M, Canny MD, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L, Martínez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27:639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- Miller KM, Jackson SP. Histone marks: repairing DNA breaks within the context of chromatin. Biochem Soc Trans. 2012;40:370–376. doi: 10.1042/BST20110747. [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179:1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Spektor T, Kong X, Congdon LM, Wu S, Schotta G, Yokomori K, Rice JC. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Rep. 2014;8:430–438. doi: 10.1016/j.celrep.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaekers CH, van den Beucken T, Bristow RG, Chiu RK, Durocher D, Wouters BG. RNF8-independent Lys63 poly-ubiquitylation prevents genomic instability in response to replication-associated DNA damage. PLoS ONE. 2014;9:e89997. doi: 10.1371/journal.pone.0089997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.