Abstract
During muscle regeneration, the transcription factor Pax7 stimulates the differentiation of satellite cells (SCs) toward the muscle lineage but restricts adipogenesis. Here, we identify HDAC4 as a regulator of Pax7-dependent muscle regeneration. In HDAC4-deficient SCs, the expression of Pax7 and its target genes is reduced. We identify HDAC4-regulated Lix1 as a Pax7 target gene required for SC proliferation. HDAC4 inactivation leads to defective SC proliferation, muscle regeneration, and aberrant lipid accumulation. Further, expression of the brown adipose master regulator Prdm16 and its inhibitory microRNA-133 are also deregulated. Thus, HDAC4 is a novel regulator of Pax7-dependent SC proliferation and potentially fate determination in regenerating muscle.
Keywords: HDAC4, muscle regeneration, Pax7
Introduction
Skeletal muscle fulfills diverse functional demands and manages mechanical stress by modifying contractile and metabolic properties of myofibers and replacing those damaged via regenerative adult stem cells. The on-demand and activity-dependent muscle remodeling is achieved by elaborate reprogramming of muscle gene transcription 1. In myofibers, histone deacetylase 4 (HDAC4) is a critical factor that connects neural activity to the muscle transcriptional programs. Inactivation of HDAC4 suppresses denervation-induced muscle atrophy and synaptic gene expression while increases re-innervation 2–5. These findings highlight a central regulatory role of HDAC4 in activity-dependent muscle remodeling.
Muscle regeneration triggered by myofiber damage is another key adaptive program required for maintaining muscle integrity. Satellite cells (SCs), the principal muscle stem cells, are essential for damage-induced muscle regeneration 6,7. The paired box transcription factor Pax7 is the master regulator of SCs 8. Genetic ablation of Pax7 leads to a loss of SC pool and severely impaired muscle regeneration 9–11. Regeneration failure in Pax7 knockout (KO) mice is also accompanied by accumulation of lipids in muscle 9. Intriguingly, SCs have been shown to potentially develop into non-muscle lineages 12. Recent studies reveal a shared lineage of muscle and brown adipose tissue 13 and SCs have the potential to differentiate into brown adipocytes 14. In this program, microRNA-133 (miR-133), which targets and inhibits the master regulator of brown adipose tissue Prdm16, restricts SCs toward the muscle lineage 14. How Pax7 regulates SC activation and fate commitment during muscle regeneration remains incompletely understood. Here, we provide evidence that HDAC4 is a novel regulatory component in Pax7-dependent SC expansion and fate determination critical for adult muscle regeneration.
Results and Discussion
HDAC4 is required for satellite cell expansion in damaged muscle
We previously observed in muscle biopsy from an ALS patient that HDAC4 is localized not only to myofiber nuclei but also centrally located nuclei, a marker for newly regenerating fibers 15. Moreover, expression of HDAC4, similar to Pax7, progressively increased in tibialis anterior (TA) muscles damaged by cardiotoxin (CTX) (Fig 1A and B). These observations indicate that HDAC4 might be involved in muscle regeneration. To investigate the possibility, we generated satellite cell (SC)-specific HDAC4 knockout (KO) mice by crossing HDAC4Lox mice with a tamoxifen (Tmx)-inducible CreERT2 transgene that was placed at the endogenous Pax7 locus (Pax7-CreERT2). HDAC4Lox;Pax7-CreERT2 and control HDAC4Lox mice were treated with Tmx to activate Cre-mediated excision of HDAC4 in SCs 16, followed by CTX injection in TA and gastrocnemius (GA) muscles to activate SCs and muscle regeneration. The analyses of SCs purified by fluorescence-activated cell sorting (FACS) showed the efficiency of HDAC4 KO by this method is approximately 60–80% (Supplementary Fig S1A–C).
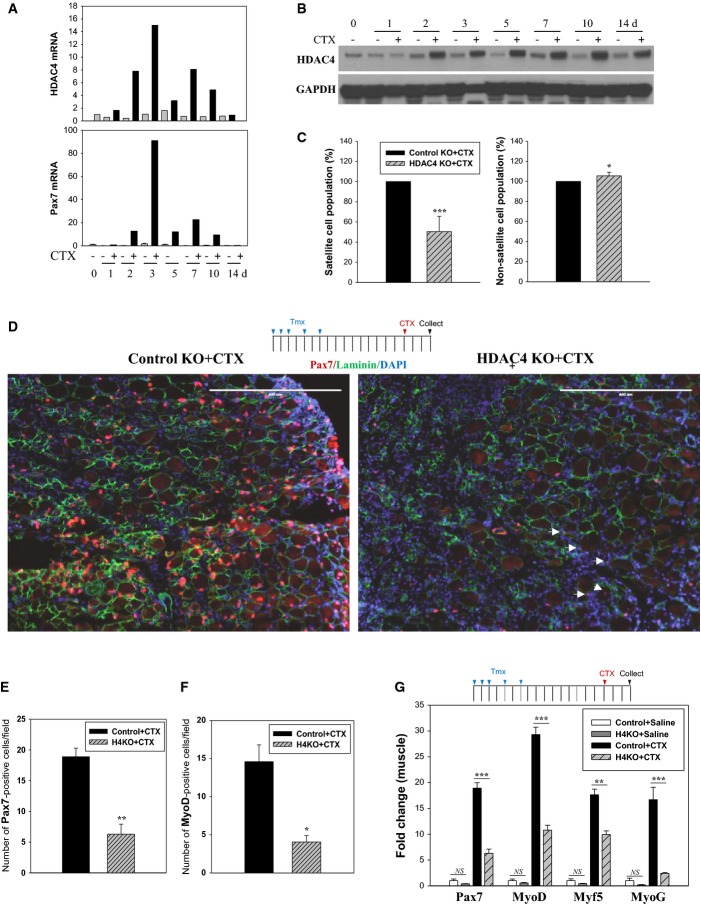
Figure 1. HDAC4 is required for SC expansion in damaged muscle.
A Expression of HDAC4 mRNA at different time points in damaged muscle. TA muscles in 8-week-old C57BL/6 male mice (eight mice) were untreated (0 day), treated with saline (left leg) or cardiotoxin (CTX) (right leg) and muscles were harvested at indicated time points. Columns, mean of PCR duplicates. n = 1 for each time point.
B Expression of HDAC4 protein in damaged muscle. TA muscles in 8-week-old C57BL/6 male mice (8 mice) were untreated (0 day), treated with saline (left leg) or CTX (right leg) and muscles were harvested at indicated time points. n = 1 for each time point.
C Effect of SC-specific HDAC4 KO on populations of activated SCs by FACS analysis. TA and GA muscles in mice treated with tamoxifen (Tmx) were injured by CTX treatment for 2–3 days. Columns, mean; bars, SD of nine (SCs) or four (non-zs) independent experiments. *P < 0.05, ***P < 0.001 versus control KO+CTX (unpaired Student’s t-test).
D–F Effects of HDAC4 KO on the number of activated SCs expressing Pax7 or MyoD. TA muscles in 7-week-old male mice were stained with indicated antibodies. Note that SCs expressing low levels of Pax7 were also counted as positive (white arrow). Field means a grid (250 × 250 μm). Approximately 40 grids per muscle were counted. Scale bar: 400 μm. Columns, mean; bars, SEM. n = 3 for each group. *P < 0.05, **P < 0.01 versus control KO+CTX (unpaired Student’s t-test).
G mRNA expression of genes in regenerating muscle. TA muscles in 7-week-old male mice were used. Columns, mean; bars, SEM. n = 4 for each group. **P < 0.01, ***P < 0.001; NS, not significant, P > 0.05 (unpaired Student’s t-test).
During FACS analyses, we noted that the number of activated SCs was significantly reduced in HDAC4 KO mice, whereas non-SC cells were slightly increased (Fig 1C), suggesting that SC expansion might be impaired. To determine whether HDAC4 is required for injury-stimulated SC activation, we quantified Pax7-positive SCs in TA muscles treated with CTX. Immunostaining showed that the frequency of Pax7-positive cells as well as the staining intensity of Pax7 was much lower in HDAC4 KO compared to CTX-treated control muscles (Fig 1D and E, and Supplementary Fig S1D). MyoD-positive activated SCs were similarly reduced in HDAC4 KO muscle (Fig 1F). In contrast, numbers of quiescent SCs in uninjured muscle were not significantly different in control and HDAC4 KO muscle, indicating that HDAC4 regulates SC expansion during regeneration (Supplementary Fig S1E and F). Supporting this conclusion, RNA analyses revealed that expression of genes required for SC activation and muscle differentiation, including Pax7, MyoD, Myf5, and Myogenin (MyoG), were all markedly reduced in damaged HDAC4 KO muscles (Fig 1G). A modest reduction of these genes was detected in saline-treated HDAC4 KO muscle, likely due to mild muscle injury caused by the injection procedure, which generates local activation of SCs. Further, intraperitoneal injection of Tmx alone did not significantly reduce the expression of Pax7 and myogenic factors in uninjured muscle of HDAC4 KO mice (Supplementary Fig S2A). Together, these results show that HDAC4 inactivation impairs SC expansion induced by injury.
HDAC4 regulates the expression of Pax7 and its target genes in activated SCs
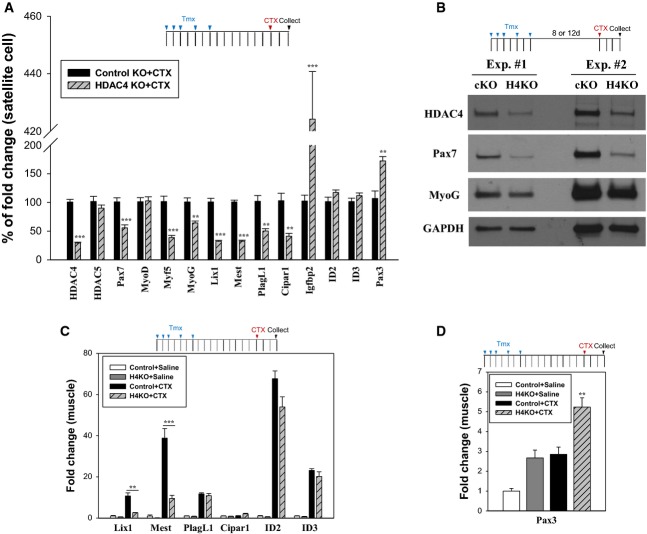
We next investigated whether HDAC4 regulates gene transcription program in SCs. SCs were isolated from CTX-injured control and HDAC4 SC KO muscles, cultured in growth medium (GM) and subjected to RNA analysis. We found that the expression of the SC-master transcription factor, Pax7, was significantly reduced in HDAC4 KO SCs (Fig 2A). Immunoblot analysis confirmed its reduction in HDAC4 KO SCs (Fig 2B). Supporting a Pax7 deficiency, the expression of Myf5, a well-established Pax7 target, was also reduced (Fig 2A). This reduction of Pax7 and Myf5 is specifically caused by the loss of HDAC4, as their expression was not changed in activated SCs or damaged TA muscles isolated from HDAC4lox/lox and HDAC4lox/lox; Pax7-CreERT2 mice without Tmx treatment (Supplementary Fig S2B and C). Importantly, the expression of MyoD was not affected in HDAC4 KO SCs (Fig 2A), consistent with a previous report that MyoD is not regulated by Pax7 17. MyoG, a downstream effector of muscle differentiation, was modestly decreased in HDAC4 KO SCs. To further determine the role of HDAC4 in Pax7-dependent gene transcription, we assessed a set of Pax7 target genes previously identified 17,18. As shown in Fig 2A, Pax7 target genes including Lix1, Mest, PlagL1, and Cipar1 were all reduced in HDAC4 KO SCs. In contrast, ID2 and ID3 were unchanged in HDAC4 KO SCs. Conversely, Igfbp2, a gene negatively regulated by Pax7 17, was increased in HDAC4 KO SCs. We further analyzed Pax7 target gene expression in regenerative TA muscle. In addition to Myf5 (Fig 1G), Lix1 and Mest were significantly reduced in regenerative HDAC4 KO muscle, whereas other Pax7 targets were unaffected (Fig 2C). Together, these results show that HDAC4 positively regulates Pax7 and a subset of Pax7 target genes in activated SCs.
Figure 2. HDAC4 regulates the expression of Pax7 and associated genes in SCs.
A mRNA expression of genes in SCs. Activated SCs isolated from 15-week-old female mice were cultured for 3 days in growth medium (GM). Medium was changed at 2 days after plating. Columns, mean; bars, SEM. n = 5 for each group. **P < 0.01, ***P < 0.001 versus control KO+CTX (unpaired Student’s t-test).
B Protein expression of HDAC4, Pax7, and MyoG in SCs. Activated SCs isolated from muscles in 13-week-old female (Exp. no. 1) or 10-week-old male (Exp. no. 2) mice were cultured for 2 days in GM.
C, D mRNA expression of potential Pax7-target genes and Pax3 in damaged muscle. cDNAs used in Fig 1G were used. Columns, mean; bars, SEM. n = 4 for each group. **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
HDAC4 regulates SC proliferation
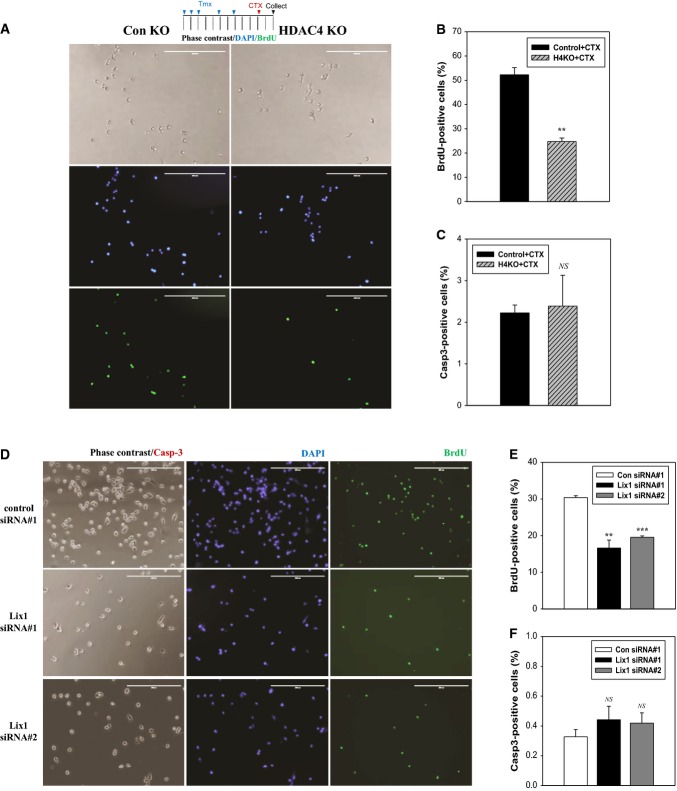
The reduction of Pax7-positive cells in injured HDAC4 KO muscle indicates a defect in SC expansion. To assess whether HDAC4 regulates SC proliferation, SCs were purified from CTX-treated control and HDAC4 SC KO muscle, cultured in GM, and pulse-labeled with BrdU. HDAC4 KO SCs showed greater than twofold reduction in proliferation (Fig 3A and B). Apoptosis, as indicated by active caspase-3, was similar between control and HDAC4 KO SCs (Fig 3C). To determine whether HDAC4 regulates differentiation, purified SCs were cultured in differentiation medium (DM) and assessed by the expression of myosin heavy chain. Both control and KO SCs underwent efficient differentiation although quantification revealed a modest but statistically significant reduction in HDAC4 KO SCs (Supplementary Fig S3A and B). Collectively, these results indicate that HDAC4 mainly regulates SC proliferation.
Figure 3. HDAC4 and HDAC4-regulated Lix1 regulate SC proliferation.
A–C Effects of HDAC4 KO on proliferation and apoptosis in SCs. Isolated SCs from 10-week-old male mice were cultured for 2 days in GM. Proliferation and apoptosis were monitored by staining cells with BrdU and caspase-3 antibodies, respectively. Approximately, 300 nuclei were measured per muscle. Scale bar: 200 μm. Columns, mean; bars, SEM. n = 3 for each group. **P < 0.01; NS, not significant, P > 0.05 (unpaired Student’s t-test).
D–F Effects of Lix1 knockdown (KD) on proliferation and apoptosis in SCs. Pooled SCs from muscles treated with CTX for 2 days in 8-week-old C57BL/6 male mice (10 mice) were transfected with Lix1 siRNAs and cultured for 2 days in GM. Medium was changed at 1 day after plating. Approximately 1,500 nuclei were counted per well. Scale bar: 200 μm. Columns, mean; bars, SEM. n = 3 for triplicate wells. **P < 0.01, ***P < 0.001 versus control siRNA no. 1 (unpaired Student’s t-test).
Lix1 is required for Pax7-dependent SC proliferation
Although ID3 was proposed as a Pax7 target required for SC proliferation 19, its expression in SCs or regenerative muscle was not affected by HDAC4 inactivation (Fig 2A and C). Our analysis of a limited set of Pax7 target genes has identified Lix1 (limb expression 1) and Mest (mesoderm-specific transcript) as two genes whose expressions were strongly suppressed in both HDAC4 KO SCs and muscles (Fig 2A and C). To test the possibility that they might be the factors that promote SC proliferation, we knocked down Lix1 or Mest in SCs isolated from CTX-activated muscle. These siRNAs effectively suppressed Lix1 or Mest expression (Supplementary Fig S3C and D). We found that Lix1 KD significantly reduced SC proliferation without inducing apoptosis (Fig 3D–F), whereas Mest KD had no effect (Supplementary Fig S3E). Expression of Pax7 and myogenic factors were unaffected by Lix1 KD (Supplementary Fig S3C). These results indicate that Lix1 is a Pax7 target gene required for efficient SC proliferation.
HDAC4 is required for efficient damage-induced muscle regeneration
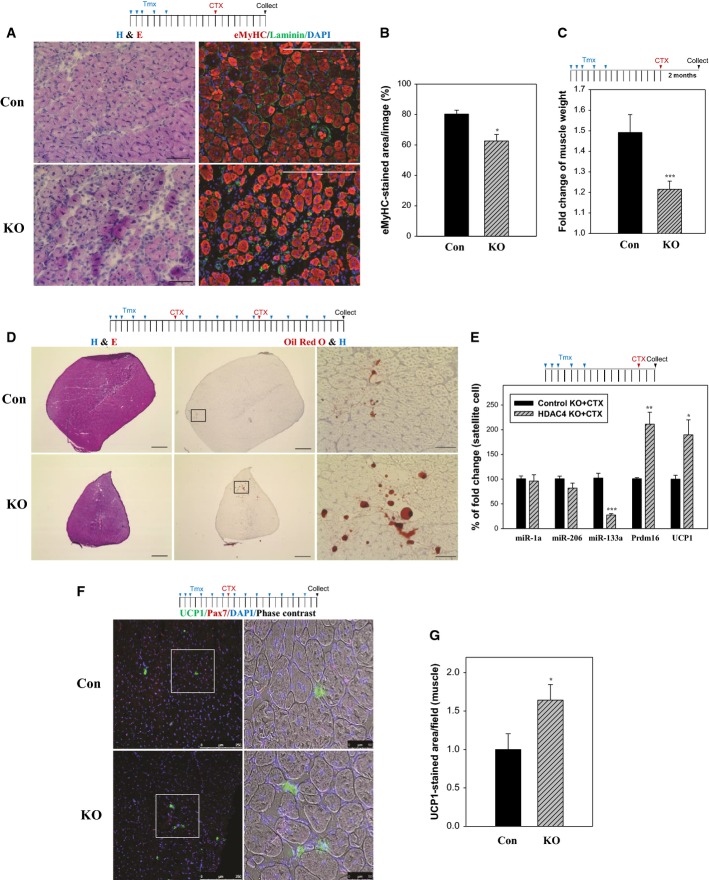
We next determined whether muscle regeneration is impaired in HDAC4 SC KO mice. At 8 days post-CTX treatment, control muscles underwent proper regeneration as illustrated by relatively uniform myofibers that are positive for embryonic myosin (eMyHC) and centrally localized nuclei (Fig 4A). In HDAC4 SC KO muscle, the regenerating fibers appeared to be smaller and interspersed with infiltrated cells. Quantification using eMyHC staining confirmed a reduction of regenerative fibers in the HDAC4 KO mice (Fig 4B). At 5 weeks post-CTX treatment, HDAC4 KO muscles showed reduced muscle size, whereas the overall muscle structure appeared to be normal (Supplementary Fig S4A), indicating delayed muscle regeneration. Supporting these results, regeneration-associated muscle hypertrophy was significantly inhibited in HDAC4 KO TA muscle (Fig 4C). In Pax7 SC-specific KO animals, the muscle regeneration defect becomes more apparent when Cre-mediated recombination (driven by Tmx-inducible Cre) was sustained by continuous administration of Tmx and muscle was injured by repeated cycles of CTX injection 9,10. Adapting this experimental condition, we found that muscle regeneration was indeed more profoundly impaired in HDAC4 SC KO muscles, as indicated by a significant reduction in muscle size (Fig 4D). In SC-specific Pax7 KO mice, regeneration failure is accompanied by accumulation of lipids in muscle 9. Analysis of HDAC4 SC KO muscles also revealed aberrant increase in neutral lipid stained by Oil Red O (Fig 4D and Supplementary Fig S4B). Further analysis showed that Oil Red O-stained cells were located between regenerating myofibers (Supplementary Fig S4C), indicating an expansion of interstitial adipocytes in HDAC4 SC KO muscle. Collectively, these data show that HDAC4 SC KO impairs muscle regeneration.
Figure 4. HDAC4 inactivation impairs muscle regeneration and increases adipocytes in regenerating muscle.
A, B Defective regeneration in HDAC4 SC KO damaged muscle. TA muscles in 10-week-old male mice were stained with H&E for histology or with embryonic myosin (eMyHC) to mark regenerating fibers. Scale bar: 50 (left) and 200 μm (right). Columns, mean; bars, SEM. n = 3 for each group. *P < 0.05, ***P < 0.001 versus control KO (unpaired Student’s t-test).
C Reduced hypertrophy in HDAC4 SC KO muscles. TA muscles in 9- to 10-week-old mice were used. Fold change is expressed as CTX-treated muscle/saline-treated contralateral muscle in each mouse. Columns, mean; bars, SD. n = 4 for control KO and n = 5 for HDAC4 KO. ***P < 0.001 versus control KO (unpaired Student’s t-test).
D Delayed muscle regeneration and increased lipid accumulation in SC-specific HDAC4 KO muscle. TA muscles in 8-week-old male mice were stained with H&E or with Oil Red O and H to mark lipid and nuclei, respectively. Scale bar: 500 (left and middle) and 50 μm (right).
E RNA expression of miR-1a, miR-133a, miR-206, Prdm16 and UCP1 in HDAC4 KO SCs. Total RNAs prepared in Fig 2A were used. Columns, mean; bars, SEM. n = 5 for each group. *P < 0.05, **P < 0.01, ***P < 0.001 versus control KO+CTX (unpaired Student’s t-test).
F, G Increased brown adipogenesis in HDAC4 SC KO regenerating muscle. TA muscles in 12-week-old male mice were stained with indicated antibodies. Scale bar: 250 (left) and 50 μm (right). Columns, mean; bars, SEM. n = 4 for control KO and n = 3 for HDAC4 KO. *P < 0.05 versus control KO+CTX (unpaired Student’s t-test).
HDAC4 inactivation increases brown adipocytes in regenerative muscle and deregulates miR-133
SC has the potential to develop into brown adipose tissue, a fate that is regulated by miR-133 14. miR-133 targets and inhibits Prdm16, the master transcription factor for brown fat adipogenesis 14,20,21. Inhibition of miR-133 can reprogram SCs toward the brown adipose lineage 14. To investigate whether HDAC4 regulates miR-133 expression, we assessed mature miR-133a and two other muscle microRNAs, miR-1a and miR-206, both of which repress Pax7 expression and SC proliferation 22. As shown in Fig 4E, expression of miR-133a, but not miR-1a or miR-206, was reduced in HDAC4 KO SCs. Consistent with a reduction in miR-133a, a significant induction of Prdm16 and a brown fat marker, UCP1, were observed in HDAC4 KO SCs. Supporting these findings, UCP1-positive cells were increased in HDAC4 KO muscle (Fig 4F and G). Collectively, these results indicate that HDAC4 restricts the expression of the miR-133–Prdm16 axis important for the brown adipocyte lineage determination.
In this study, we provide evidence that HDAC4 is required for efficient muscle regeneration induced by injury. The characterization of HDAC4 KO has revealed a significant reduction in Pax7 expression in SCs (Fig 2A and B). Indeed, subsets of Pax7 target genes were significantly mis-regulated in HDAC4 KO SCs (Fig 2A). Surprisingly, siRNA knockdown of HDAC4 in purified and activated SCs in vitro did not cause a reduction in Pax7 expression (Supplementary Fig S5A). These results indicate that regulation of Pax7 by HDAC4 is indirect and requires an intact muscle microenvironment in vivo. Further supporting an indirect mechanism, HDAC4 knockdown in C2C12 myoblasts did not affect acetyl-histone H4 (K8) levels in the promoter regions of Pax7 or Lix1 (Supplementary Fig S5B). It is of interest to note that HDAC4 can regulate cytokine production 2, which might influence the inflammatory program important for muscle regeneration 23. How HDAC4 in SCs communicates with other components of the muscle regeneration program to affect Pax7 expression awaits further studies.
The differential requirement of Pax7 target genes on HDAC4 could reflect the up-regulation of Pax3 in HDAC4 KO SCs and regenerating TA muscle (Fig 2A and D). This finding indicates that HDAC4 selectively regulates Pax7, but not Pax3, expression in SCs. Pax3 is highly related to Pax7 and can regulate similar sets of genes 17,18. Thus, the elevated Pax3 might partially compensate for Pax7 and maintain the expression of common targets. The differential dependence of Pax7 target genes on HDAC4 led us to focus on Lix1 and Mest, which are down-regulated in both HDAC4 KO SCs and damaged muscles, as candidates that mediate Pax7-dependent SC proliferation. We found that knockdown of Lix1, but not Mest, suppressed the proliferation of activated SCs (Fig 3E and Supplementary Fig S3E). This finding suggests that Pax7-dependent SC proliferation involves Lix1. The biochemical function of Lix1 is largely unknown. Its Drosophila orthologous, Lowfat (Lft), is a component of the FAT-signaling pathway that regulates planar cell polarity (PCP) 24. Interestingly, PCP plays an important role in SC expansion 25. Whether Lix1 and HDAC4 regulate SC proliferation via the PCP pathway requires further investigation.
Supporting its role as a positive regulator of Pax7, HDAC4 KO led to defects in SC proliferation and muscle regeneration. Similar to Pax7 KO mice, regenerative muscles in HDAC4 KO mice also showed aberrant lipid accumulation (Fig 4D). Interestingly, we found that HDAC4 KO in SCs reduces expression of miR-133a, a negative regulator of the brown fat transcription factor Prdm16. Consistent with this finding, the expression of Prdm16 and a marker for brown adipocytes, UCP1, were both elevated in HDAC4 KO SCs (Fig 4E). An increase in UCP1-positive cells was also observed in HDAC4 KO regenerating muscle (Fig 4F and G). These findings are consistent with a previous report that miR-133 suppresses SC differentiation to brown adipocytes 14. Our study therefore suggests that HDAC4 is required for maintaining miR-133a in SCs and thereby inhibits SC differentiation toward the brown adipose lineage. Interestingly, miR-133 over-expression can increase Pax7 levels in primary preadipocytes 14, indicating that miR-133 positively regulates Pax7 expression. Accordingly, HDAC4 could increase Pax7 expression in SCs via miR-133. Given the instructive role of miR-133 in brown adipose determination 14,20,21, this scenario implies a regulatory role of HDAC4 in SC fate determination. The physiological relevance of SC plasticity is not known. By directing some activated SCs toward an adipose lineage, this arrangement could potentially provide metabolic advantages to regenerative muscle. Since HDAC4 is regulated by multiple physiological and pathological conditions, its activity in SCs might influence the composition of muscle fibers and adipose tissue in regenerative muscle. Further studies are required to determine how HDAC4 regulates miR-133 expression and its impact on SC fate determination in response to patho-physiological conditions.
Materials and Methods
Mouse procedures
HDAC4Lox mice were provided by Dr. E. Olson 5. Pax7CreERT2 mice were purchased from Jackson Laboratory (B6;129-Pax7tm2.1(cre/ERT2)Fan/J). Wild-type C57BL/6 mice were also from Jackson Laboratory. Conditional strains contain HDAC4Lox; Pax7-CreERT2 (KO) or HDAC4Lox (control). Tamoxifen (Tmx; Sigma; T5648) treatment for Cre recombinase activation was performed by intraperitoneal injection to both control and HDAC4 KO mice (200 μl of 10 mg/ml in corn oil). Mouse gender, age, numbers, and Tmx treatment are indicated in each figure legend. To induce muscle degeneration/regeneration, mice were anesthetized and injected with 50 μl of 10 μM cardiotoxin (Sigma; C9759) in one site of TA muscle or in two sites of GA muscle. All mice were housed at the Duke University mouse facilities in accordance with the IACUC.
Acknowledgments
We thank Dr. Eric Olson for the generous gift of HDAC4-flox mouse. We also thank Mr. Thomas Fitzpatrick for quantification image data, Dr. Jae-woo Lee for MF20 antibody, and Ms. Minsi Zhang for discussions. This work was supported by 276662 (MDA) to MCC, AR060042 (NIAMS, NIH) to CMF and AR055613 (NIAMS, NIH) to TPY.
Author contributions
MCC designed and performed experiments and wrote the manuscript; SR conducted mouse genotyping, immunostaining, and quantification of data. RH performed ChIP experiments and analyzed data. BW helped with the animal works. MK provided technical assistances and edited the manuscript. CMF provided critical reagents, discussions and edited the manuscript. TPY supervised the project and edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Choi MC, Cohen TJ, Barrientos T, Wang B, Li M, Simmons BJ, Yang JS, Cox GA, Zhao Y, Yao TP. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell. 2012;47:122–132. doi: 10.1016/j.molcel.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Waddell DS, Barrientos T, Lu Z, Feng G, Cox GA, Bodine SC, Yao TP. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci USA. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther S, Kim J, Kostin S, Lepper C, Fan CM, Braun T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell. 2013;13:590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettor R, Milan G, Franzin C, Sanna M, De Coppi P, Rizzuto R, Federspil G. The origin of intermuscular adipose tissue and its pathophysiological implications. Am J Physiol Endocrinol Metab. 2009;297:E987–E998. doi: 10.1152/ajpendo.00229.2009. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013;17:210–224. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons BJ, Cohen TJ, Bedlack R, Yao TP. HDACs in skeletal muscle remodeling and neuromuscular disease. Handb Exp Pharmacol. 2011;206:79–101. doi: 10.1007/978-3-642-21631-2_5. [DOI] [PubMed] [Google Scholar]
- Nishijo K, Hosoyama T, Bjornson CR, Schaffer BS, Prajapati SI, Bahadur AN, Hansen MS, Blandford MC, McCleish AT, Rubin BP, et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23:2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol Biol Cell. 2009;20:3170–3177. doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Guo X, Li Y, Sun K, Lu L, Jiang L, Fu X, Zhu H, Sun H, Wang H, et al. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012;11:231–241. doi: 10.1016/j.stem.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol. 2012;14:1330–1335. doi: 10.1038/ncb2612. [DOI] [PubMed] [Google Scholar]
- Liu W, Bi P, Shan T, Yang X, Yin H, Wang YX, Liu N, Rudnicki MA, Kuang S. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9:e1003626. doi: 10.1371/journal.pgen.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, Wang DZ. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li Y, Wu Y, Wang L, Wang X, Du J. Interleukin-6/signal transducer and activator of transcription 3 (STAT3) pathway is essential for macrophage infiltration and myoblast proliferation during muscle regeneration. J Biol Chem. 2013;288:1489–1499. doi: 10.1074/jbc.M112.419788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Kucuk B, Irvine KD. Drosophila lowfat, a novel modulator of fat signaling. Development. 2009;136:3223–3233. doi: 10.1242/dev.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand F, Jones AE, Seale V, Scime A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.