Abstract
The enzyme CTP synthase (CTPS) dynamically assembles into macromolecular filaments in bacteria, yeast, Drosophila, and mammalian cells, but the role of this morphological reorganization in regulating CTPS activity is controversial. During Drosophila oogenesis, CTPS filaments are transiently apparent in ovarian germline cells during a period of intense genomic endoreplication and stockpiling of ribosomal RNA. Here, we demonstrate that CTPS filaments are catalytically active and that their assembly is regulated by the non-receptor tyrosine kinase DAck, the Drosophila homologue of mammalian Ack1 (activated cdc42-associated kinase 1), which we find also localizes to CTPS filaments. Egg chambers from flies deficient in DAck or lacking DAck catalytic activity exhibit disrupted CTPS filament architecture and morphological defects that correlate with reduced fertility. Furthermore, ovaries from these flies exhibit reduced levels of total RNA, suggesting that DAck may regulate CTP synthase activity. These findings highlight an unexpected function for DAck and provide insight into a novel pathway for the developmental control of an essential metabolic pathway governing nucleotide biosynthesis.
Keywords: Ack, CTP synthase, cytoophidia, Drosophila oogenesis
Introduction
Metabolic enzymes can be dynamically regulated in response to nutrient availability and growth-promoting signals. Enzyme activity may be altered by transcriptional or post-transcriptional mechanisms such as covalent modifications (e.g. phosphorylation) or assembly into regulatory complexes. Recently, examples have emerged of transient assembly of metabolic enzymes into macromolecular structures 1,2, although, in general, the architecture and regulation of these assemblies is poorly understood.
One assembly that has been described in organisms from bacteria to mammals is comprised of the nucleotide biosynthetic enzyme CTP synthase (CTPS) 3,4. CTPS assembles into filaments dynamically in response to nutrient deprivation in yeast 5 although it has been unclear whether CTPS filaments are catalytically active. Consequently, the role of these structures in CTP biosynthesis has remained mysterious. CTPS filaments also occur in germline cells of the Drosophila ovary 5,6 where their function is also unknown. Here, we demonstrate that CTPS filaments form transiently during oogenesis and are catalytically active and that their assembly is regulated by a novel filament component, the non-receptor tyrosine kinase DAck. Our results establish a framework for understanding how the assembly of CTPS filaments provides temporal control over the production of CTP, an essential nucleotide and precursor for phospholipid biosynthesis, which is required during specific stages of oogenesis.
Results and Discussion
DACK-deficient female flies exhibit reduced fertility
Drosophila oogenesis depends on the production of “egg chambers” composed of 16-cell germline cysts (one oocyte and 15 supportive nurse cells) surrounded by a follicular epithelium. Egg chambers proceed through 14 morphologically defined stages of development over the course of 8 days to produce a mature egg 7. While the kinase DAck is important for Drosophila spermatogenesis 8, its role in oogenesis is unknown. We observed that female flies homozygous for a loss-of-function allele of DAck, DAck86 9, deposited eggs at a substantially slower rate than wild-type flies (Fig 1A), and expression of a wild-type DACK transgene in the DAck86 genetic background rescued this phenotype (Fig 1A and B). In contrast, transgenic expression of a kinase-dead DAck mutant (DAck-K156A) failed to rescue (Fig 1A and B), demonstrating that DAck kinase activity is critical for oogenesis.
Figure 1. DAck kinase activity is required for normal oogenesis in Drosophila.
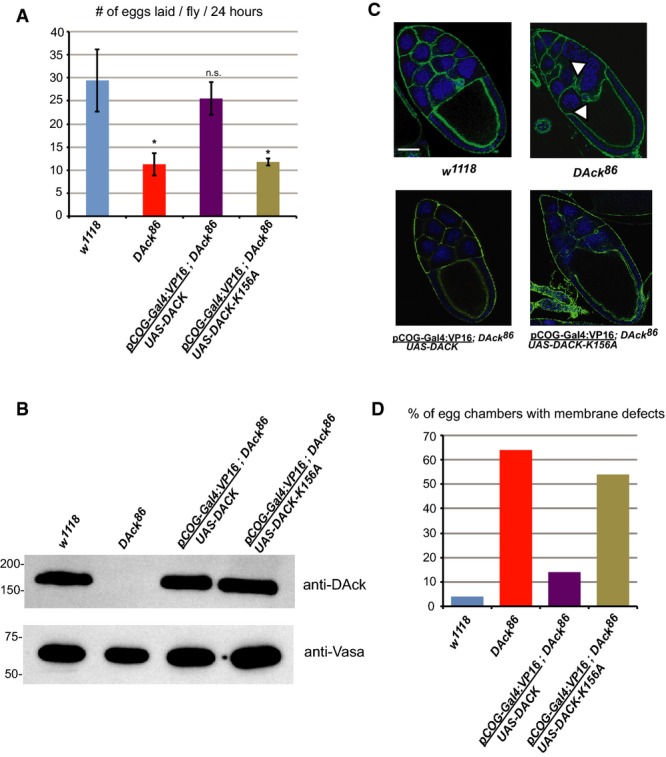
A Egg laying rates of w1118, DAck86, UAS-DACK/pCOG-Gal4:VP16;DAck86, and UAS-DACK-K156A/pCOG-Gal4:VP16;DAck86 flies. Error bars indicate standard deviation from three independent experiments. * denotes P-value < 0.01 as calculated by an unpaired, two-tailed Student’s t-test, and n.s. denotes not significantly different than w1118.
B Immunoblot analysis of ovary extracts from w1118, DAck86, UAS-DACK/pCOG-Gal4:VP16;DAck86, and UAS-DACK-K156A/pCOG-Gal4:VP16;DAck86 flies using anti-DAck and anti-Vasa (loading control) antibodies.
C Stage 10 egg chambers from flies of the indicated genotypes stained with FITC-phalloidin to label the subcortical actin cytoskeleton (green) and Draq5 to label nuclei (blue). Single 0.2-μm confocal sections are shown. White arrowheads denote discontinuities in nurse cell plasma membranes. Scale bar, 20 μm.
D Quantitation of the membrane defect phenotype from stage 10 egg chambers (n = 50) of the indicated genotypes.
Egg chambers from DAck86 flies exhibited an apparent disruption in the continuity of the plasma membrane between adjacent nurse cells resulting in nurse cell fusion (arrowheads in Fig 1C). No plasma membrane defects were observed in the follicular epithelium. Transgenic re-expression of wild-type DAck but not DAck-K156A restored normal egg chamber morphology (Fig 1C and D), further demonstrating a key role for DAck activity in regulating germline cell membrane integrity.
Ack localizes to CTP synthase filaments
Kinases can localize to structural components within germ cells to regulate key developmental events. For example, Tec kinase localizes to ring canals between nurse cells during Drosophila oogenesis and failure of Tec recruitment leads to plasma membrane disruption and reduced fertility 10. We observed that DAck localized to single approximately 20-μm-long filamentous structures within the cytoplasm of each nurse cell in wild-type egg chambers (Fig 2A and Supplementary Movie S1). As expected, filamentous DAck staining was absent from DAck86 flies (Supplementary Fig S1A) and localization to filaments was restored upon re-expression of transgenic DAck, demonstrating specificity of the antibody. FLAG-DAck expressed in the female germline also localized to filaments (Supplementary Fig S1B), confirming this localization.
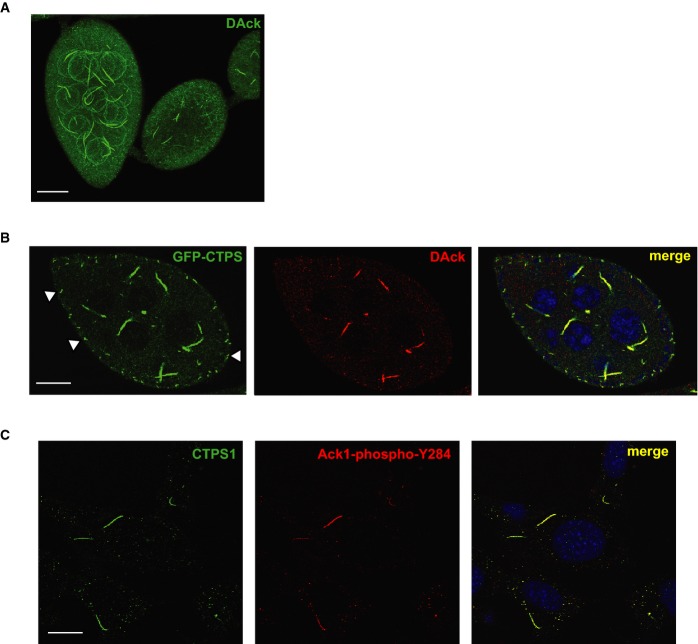
Figure 2. DAck/Ack1 localizes to CTPS filaments.
A Three-dimensional confocal projection of Drosophila egg chambers stained with an anti-DAck antibody. Spherical structures are non-specific staining of germ cell nuclear membranes. Scale bar, 25 μm.
B Egg chamber from CTPS-GFP protein trap fly line (CA06746) stained with an anti-DAck antibody and Draq5. A single 0.2-μm section is shown. White arrowheads denote filaments in follicle cells labeled with CTP synthase, but not DAck. Scale bar, 20 μm.
C MCF7 cells treated with 200 μM DON were fixed and stained with anti-CTPS1 and anti-phospho-Y284-Ack1 antibodies. Scale bar, 10 μm.
DAck filaments were reminiscent in appearance of cytoplasmic assemblies recently described in bacteria 11, yeast 5, flies 4,6, and mammalian cells 12 that are composed of the enzyme CTPS, which catalyzes the rate-limiting step in the production of cytidine nucleotides 13. Strikingly, DAck co-localized with GFP-tagged CTPS 14 filaments in germline cells of Drosophila egg chambers (Fig 2B). Interestingly, however, no filamentous DAck staining was observed in cells of the surrounding follicular epithelium, despite the presence of short CTPS filaments there (arrowheads in Fig 2B).
Pharmacological depletion of CTP by treatment with the CTPS inhibitors 6-diazo-5-oxo-L-norleucine (DON) or azaserine induces filament formation in metazoan cells and tissues 5,15. To determine whether filamentous localization of Ack is conserved in mammals, we immunostained DON-treated MCF7 cells with anti-CTPS1 and anti-phospho-Y284-Ack1 antibodies, revealing that co-localization of Ack and CTPS to filaments is conserved (Fig 2C).
Membrane defects in DAck mutants are linked to reduced CTPS activity
In addition to its role as an essential nucleotide, CTP plays a critical role as a precursor for the biosynthesis of phospholipids including phosphatidylcholine and phosphatidylinositol 16. Inhibition of CTPS decreases cellular CTP pools with a concomitant reduction in phospholipid biosynthesis 17,18. Furthermore, Cct1, the enzyme downstream of CTPS in the phosphatidylcholine biosynthetic pathway, is critical for Drosophila oogenesis 19. To determine whether the apparent membrane defects in DAck86 egg chambers reflect reduced phospholipid levels, we examined the distribution of the plasma membrane-enriched lipid phosphatidylinositol 4,5-bisphosphate (PIP2) using GFP fused to the pleckstrin homology domain of phospholipase Cδ (GFP-PH-PLCδ), a selective binder of PIP2 20. GFP-PH-PLCδ was expressed in the female germline of either wild-type or DAck86 flies, and isolated egg chambers from the two genotypes were pooled and analyzed by confocal microscopy. DAck immunostaining was used to differentiate wild-type from DAck86 mutant egg chambers within the same field in order to allow direct comparison of GFP signal intensities. We observed strikingly reduced intensity in DAck86 egg chambers compared to wild-type egg chambers (Fig 3A). These results demonstrate reduced phospholipid membrane composition in DAck86 flies perhaps due to reduced CTPS catalytic activity in this genotype.
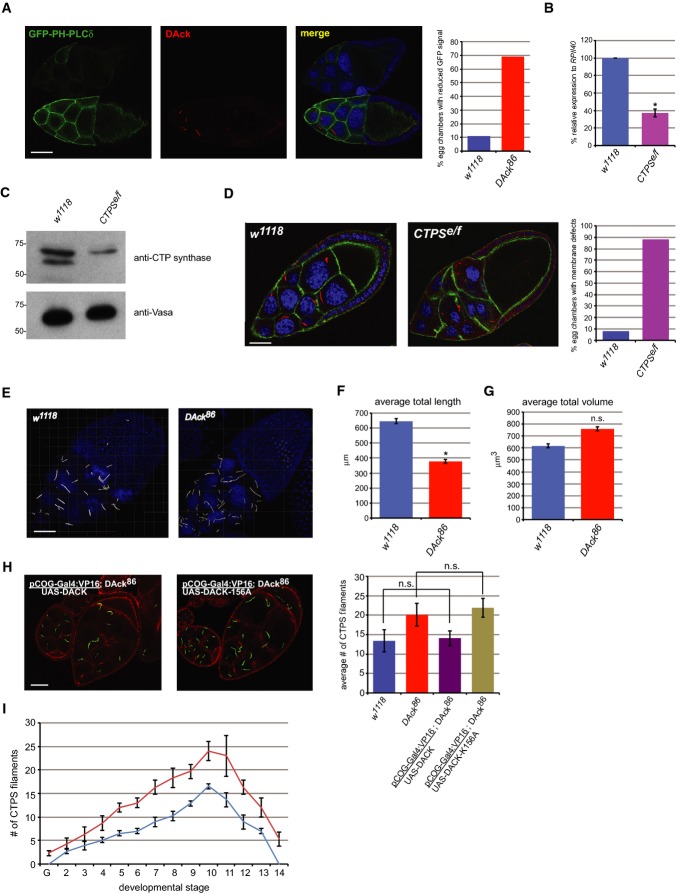
Figure 3. DAck regulates CTPS filament morphology.
A Decreased PIP2 levels in egg chambers of DAck86 flies. Expression of a UAS-GFP-PH-PLCδ transgene was driven in the female germline using nanos-Gal4:VP16 in both w1118 and DAck86 flies. Egg chambers from both genotypes were dissected and pooled and then stained with an anti-DAck antibody and imaged by confocal microscopy. A wild-type egg chamber is on the bottom and a DAck86 egg chamber is on top. Single 0.2-μm confocal sections are shown. Scale bar, 40 μm. Quantitation shows the percentage of stage 10 egg chambers (n = 50) of each genotype with reduced GFP-PH-PLCδ signal.
B Quantitative real-time PCR analysis of Drosophila CTP synthase (isoform C) expression using total RNA isolated from w1118 and CTPSe/f flies. The mean value, normalized to levels of RpII140 mRNA from three independent reactions, is shown. Error bars indicate standard deviation. * denotes P-value < 0.01 as calculated by an unpaired, two-tailed Student’s t-test.
C Western blot analysis of ovary lysates from w1118 and CTPSe/f flies using anti-CTP synthase and anti-Vasa antibodies.
D Immunostaining of egg chambers from w1118 and CTPSe/f flies with anti-CTP synthase antibody (red), FITC-phalloidin (green), and Draq5 (blue). Scale bar, 30 μm. Quantitation shows the percentage of stage 9 egg chambers (n = 50) of each genotype with membrane defects.
E Increased numbers and altered morphology of CTPS filaments in DAck86 flies. Stage 10 egg chambers dissected from CA06746 and CA06746;DAck86 flies were imaged by confocal microscopy. Z-stacks were subsequently volume-rendered using Imaris software. CTPS filaments are shown in white and nuclei in blue. Scale bar, 30 μm.
F Average sum of total lengths of CTPS filaments from stage 10 egg chambers (n = 5) from w1118 and DAck86 flies with indicated standard deviation. * denotes P-value < 0.05 as calculated by an unpaired, two-tailed Student’s t-test.
G Average sum of total volumes of CTPS filaments from stage 10 egg chambers (n = 5) from w1118 and DAck86 flies with indicated standard deviation. n.s. denotes not significantly different from w1118 as calculated by an unpaired, two-tailed Student’s t-test.
H The kinase activity of DAck is necessary for normal CTPS filament morphology. Egg chambers of the indicated genotypes were stained with anti-CTP synthase antibody (green) and rhodamine-phalloidin (red). Scale bar, 20 μm. Quantitation shows the average number of CTPS filaments in stage 9 egg chambers (n = 30) of each genotype. Error bars indicate standard deviation, and n.s. denotes not significant as calculated by an unpaired, two-tailed Student’s t-test.
I Numbers of CTPS filaments throughout the 14 developmental stages of oogenesis in w1118 (blue) and DAck86 (red) flies. The number of filaments per egg chamber of each developmental stage was determined by immunostaining with an anti-CTP synthase antibody. Shown is the average number observed from three independent experiments. Error bars indicate standard deviation. G = germarium.
If reduced phospholipid levels in DAck86 egg chambers are indeed due to reduced CTPS activity, then flies expressing reduced CTPS should exhibit similar phenotypes. We generated heteroallelic CTPS mutant flies (hereafter referred to as CTPSe/f), in which CTPS gene expression is reduced by 62% (Fig 3B) and CTPS protein levels are similarly reduced (Fig 3C). As expected, CTPS immunostaining is markedly decreased in CTPSe/f egg chambers (Fig 3D) as is GFP-PH-PLCδ staining at germ cell membranes (Supplementary Fig S2). These CTPS hypomorphic female flies are viable but exhibit reduced egg production (see Fig 4C). Moreover, discontinuities in the plasma membranes of germline cells and dramatic nurse cell fusion were observed, phenotypes highly similar to DAck86 (compare Fig 3D, right panel with Fig 1C, top right panel). These results support the idea that maintenance of adequate CTP levels is promoted by DAck and is required for germ cell membrane integrity and oogenesis.
Figure 4. Catalytically active CTPS forms filaments.
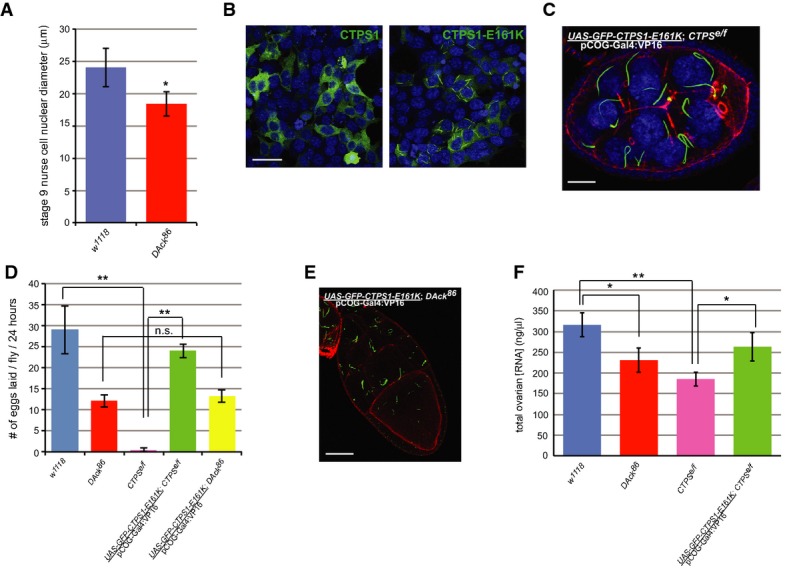
A Defective endoreplication in DAck86 egg chambers. Diameters of nurse cell nuclei were measured from stage 9 egg chambers (n = 30) of each genotype. Shown is the average diameter with error bars indicating standard deviation. * denotes P-value < 0.05 as calculated by an unpaired, two-tailed Student’s t-test.
B Localization of constitutively active CTPS1. HEK293 cells transfected with constructs encoding GFP-tagged human CTPS1 or CTPS1-E161K were imaged by confocal microscopy. Scale bar, 50 μm.
C Localization of human GFP-CTPS1-E161K (green) in Drosophila ovarian germline cells. A three-dimensional confocal projection from a stage 8 egg chamber of the indicated genotype stained with rhodamine-phalloidin (red) and Draq5 (blue). Scale bar, 20 μm.
D Egg laying rates from flies of the indicated genotypes. The average rate from three independent experiments is shown. Error bars indicate standard deviation. ** denotes P-value < 0.005 as calculated by an unpaired, two-tailed Student’s t-test, and n.s. denotes not significantly different from w1118.
E Localization of human GFP-CTPS1-E161K (green) in Drosophila ovarian germline cells. A 0.4-μm confocal section from a stage 10 egg chamber of the indicated genotype stained with rhodamine-phalloidin (red) is shown. Scale bar, 40 μm.
F Quantitation of total ovarian RNA from flies of the indicated genotypes. Equal volumes of ovarian tissue were dissected from each genotype, and total RNA was isolated and measured. The average concentration from three independent experiments is shown. Error bars indicate standard deviation. * denotes P-value < 0.05, and ** denotes P-value < 0.005 as calculated by an unpaired, two-tailed Student’s t-test.
DAck regulates CTP synthase filament morphology
Other macromolecular assemblies of metabolic enzymes exhibit highly regulated assembly that promotes catalytic activity 1. CTPS filaments were more numerous and appeared fragmented in DAck86 nurse cells compared to wild-type nurse cells (Fig 3E). Quantitation of three-dimensional image stacks confirmed that total filament length per egg chamber is indeed less in DAck86 flies (Fig 3F). However, due to the increased number of smaller, thinner filaments, total filament volume in wild-type and DAck86 egg chambers is comparable (Fig 3G), indicating that the morphological organization of the filaments is the most significant difference. Importantly, the morphology of CTPS filaments was dependent on DAck kinase activity, as expression of wild-type but not kinase-dead DAck-K156A rescued the altered CTPS filament morphology in the DAck86 genetic background (Fig 3H).
An analysis of filament number across the stages of oogenesis also revealed striking differences between the genotypes. Short CTPS foci are observed beginning in stage 2 egg chambers of wild-type flies that grow increasingly longer and proportionately wider through subsequent stages. At stage 11 and beyond, CTPS filaments began to disappear (Fig 3I), consistent with published results 6. However, CTPS protein was still present at equal levels in these late-stage egg chambers (Supplementary Fig S3), suggesting that the filaments are disassembled rather than degraded. By contrast, filaments were already observed in the germarium in DAck86 flies, earlier in the developmental timecourse than for wild-type flies (Fig 3I). Filaments in DAck86 flies also persisted inappropriately into late-stage egg chambers. These observations demonstrate a role for DAck in regulating the temporal assembly of filaments during oogenesis, perhaps linking the timing of filament assembly and disassembly to the morphological defects observed in DAck86 egg chambers.
Catalytically active CTP synthase localizes to filaments
During mid-oogenesis, a massive supply of nucleotides is required for the synthesis of cellular DNA and RNA. Nurse cells in egg chambers of stages 2–10 undergo 10–12 cycles of endoreplication of nuclear DNA 21 producing an approximately 1,000-fold increase in their DNA content. Endoreplication is essential to support increased transcription and protein production for deposition into developing oocytes 22. Likewise, RNA production, particularly ribosomal RNA, is strikingly upregulated in nurse cells from these same stages in parallel with increases in ribosomal gene copy number 23. The presence of CTPS filaments in Drosophila ovarian germ cells at stages with a high demand for CTP, together with the observation that reduced CTPS expression is associated with defects in oogenesis, suggests that CTPS filaments are assembled to promote CTP biosynthesis and support endoreplication. Consistent with this model, the diameters of DAck86 nurse cell nuclei were significantly smaller than wild-type (Fig 4A), most likely as a result of defects in endoreplication due to reduced CTP production.
The catalytic activity of CTPS is controlled by both transcriptional and post-transcriptional mechanisms including phosphorylation 24–27. Indeed, filaments can be labeled with a phospho-specific antibody against CTPS phosphorylated on serine 36 6, a post-translational modification associated with increased enzymatic activity 25. An alternate method of enzyme regulation occurs via end product inhibition in which CTP inhibits the activity of CTPS itself 16. An evolutionarily conserved glutamate residue at position 161 (Supplementary Fig S4A) is critical for end product inhibition, as expression of a mutant form of CTP synthase (E161K) that is insensitive to feedback inhibition causes an increase in cellular CTP 17. We generated the E161K mutation in human CTPS1 (CTP synthase isoform 1) and expressed it in HEK293 cells as a GFP fusion protein. Remarkably, while wild-type GFP-tagged CTPS1 was primarily cytosolic, CTPS1-E161K localized to filaments (Fig 4B) that were morphologically similar to those in Drosophila egg chambers 5,6 or DON-treated mammalian cells 12,15. Thus, unlike the wild-type form, the constitutively active mutant human protein is prone to assemble into filaments.
The observation that a constitutively active mutant of CTPS spontaneously assembles into filaments supports the hypothesis that filaments are assembled for the purpose of nucleotide generation and that CTPS is catalytically active within them. We tested whether GFP-CTPS1-E161K could rescue the sterility of CTPSe/f female flies when expressed in the germline at endogenous levels (Supplementary Fig S4B). Remarkably, the active human enzyme assembled into filaments in the Drosophila ovary (Fig 4C) and rescued both the membrane integrity defects (Fig 4C) and the sterility of CTPSe/f female flies (Fig 4D). Based on these results, we conclude that Drosophila CTPS is likely catalytically active within filaments and that filament assembly promotes CTP synthesis.
Although transgenic expression of human GFP-CTPS1-E161K rescued the fertility defect of CTPSe/f mutant flies, it did not rescue the fertility of DAck86 mutant flies (Fig 4D). While this could be explained by additional, CTPS-independent roles for DAck during oogenesis, morphological analysis of transgenic GFP-CTPS1-E161K in both genetic backgrounds suggested an alternative possibility. CTPS1-E161K filaments in CTPSe/f mutant egg chambers appeared morphologically normal, while in the DAck86 mutant background, CTPS1-E161K filaments were more numerous and fragmented (Fig 4E) with a similar appearance as endogenous CTPS filaments in DAck86 egg chambers (Fig 3E). This suggests that CTPS1-E161K filaments remain dependent on DAck for normal assembly and function. Furthermore, it implies that filament assembly and enhancement of CTPS activity are separable events.
To more directly assess a role for DAck in regulating CTPS activity, we measured total RNA from wild-type and DAck86 mutant ovaries (Fig 4F). While DAck86 flies lay fewer eggs (Fig 1A), the proportion of early- to late-stage egg chambers from these flies is similar to wild-type flies, allowing for a direct comparison of RNA levels using equal volumes of tissue. Total ovarian RNA levels were decreased in DAck86 mutant flies compared to wild-type flies, consistent with an important role of DAck in RNA production. Importantly, CTPSe/f flies also exhibited reduced ovarian RNA levels, as expected, and this phenotype was rescued by transgenic expression of GFP-CTPS1-E161K (Fig 4F). Taken together, these data suggest that DAck plays a role in nucleotide biosynthesis by modulating CTPS filament organization which ultimately promotes CTPS enzymatic activity.
The regulation of metabolic enzyme activity by the reversible formation of macromolecular assemblies is an emerging paradigm in biology with only a few other examples 1,2. Here, we provide new evidence that CTPS filaments are catalytically active based on the constitutive activity and dramatic filament-forming ability of the E161K mutant both in mammalian cells and when expressed in germ cells of the Drosophila ovary. Active CTPS filaments form in a stage-specific manner during oogenesis to support the high demand for CTP for rapid membrane expansion and RNA production in developing germ cells. Our data suggest that a mechanism utilized by single-celled organisms during nucleotide deprivation 5 has been integrated with tyrosine kinase signaling pathways in a tissue that requires transiently high levels of CTPS activity during development. Our results support a model in which the catalytic activity of DAck regulates the morphology and subsequently the enzymatic activity of CTP synthase filaments, suggesting that the assembly of a proper filamentous structure is key for promoting CTPS catalytic activity. As we have not been able to detect phosphorylation of CTPS by Ack (Supplementary Fig S5), DAck likely phosphorylates an as yet unidentified substrate that is essential for linking individual CTPS filaments into large, bundled, catalytically active assemblies. Thus, elucidation of the upstream signaling events that control Ack function and identification of Ack substrates in germline cells of the ovary will be informative for understanding how dynamic CTPS assembly is controlled.
Materials and Methods
Antibodies
Anti-DAck antibody (JCD2) was provided by J. Dixon. Anti-CTP synthetase 1/2 (y-88), anti-CTPS1 (C-13), anti-Vasa (d-260), anti-myc (9E10), anti-β-tubulin (H-235), and anti-phospho-tyrosine (PY20) antibodies were from Santa Cruz Biotechnology. Anti-GFP antibody (JL-8) was from Clontech. Anti-FLAG antibody (M2) was from Sigma-Aldrich. Anti-phospho-Y284-Ack1 antibody (ab74091) was from Abcam.
Fly stocks
Stocks were maintained and all crosses performed at 25°C. w1118 flies were used as a control in all experiments. DAck86 flies were provided by N. Harden. The CTP synthase GFP protein trap line, CA06746, was provided by A. Spradling. CTPsyne01207/TM6b and CTPsynf01941/TM6b were generated by Exelixis and maintained by the Harvard stock center. CTPSe/f flies were generated by crossing CTPsyne01207/TM6b and CTPsynf01941/TM6b. UAS-GFP-PH-PLCδ flies were provided by L. Cooley. Expression of UAS-DACK, UAS-FLAG-DACK, and UAS-GFP-CTPS1-E161K transgenes in germline cells of the ovary was driven with pCOG-Gal4:VP16.
Fertility analysis was performed essentially as described 28. Briefly, equal numbers of 3- to 4-day-old virgin females of the indicated genotype were crossed to an equal number of w1118 males at 25°C. Eggs were collected on grape juice agar plates and counted at 24 h.
Acknowledgments
We thank N. Harden (Simon Fraser University), A. Spradling (Carnegie Institute), J. Dixon (UC San Diego), L. Graves (UNC, Chapel Hill), B. Turk (Yale), and L. Cooley (Yale) for fly stocks and reagents and D. Wiest (Fox Chase Cancer Center) for critical reading of the manuscript. This work was supported by the American Cancer Society (PF-11-068-01-TBE) to T.I.S., National Institutes of Health Grants R01 GM083025 to J.R.P., R01 HD065800 to A.M.O., and P30 CA006927 to Fox Chase Cancer Center, and by pilot funding from Fox Chase Cancer Center.
Author contributions
TIS, AMO, and JRP designed experiments, TIS, KPS, SVT, and EN performed experiments and analyzed data, and TIS, AMO, and JRP wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo WC, Calise SJ, von Muhlen CA, Satoh M, Chan EK. Molecular cell biology and immunobiology of mammalian rod/ring structures. Int Rev Cell Mol Biol. 2014;308:35–74. doi: 10.1016/B978-0-12-800097-7.00002-6. [DOI] [PubMed] [Google Scholar]
- Liu JL. The enigmatic cytoophidium: compartmentation of CTP synthase via filament formation. BioEssays. 2011;33:159–164. doi: 10.1002/bies.201000129. [DOI] [PubMed] [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL. Intracellular compartmentation of CTP synthase in Drosophila. J Genet Genomics. 2010;37:281–296. doi: 10.1016/S1673-8527(09)60046-1. [DOI] [PubMed] [Google Scholar]
- King RC. Ovarian Development in Drosophila melanogaster. New York: Academic Press; 1970. [Google Scholar]
- Abdallah AM, Zhou X, Kim C, Shah KK, Hogden C, Schoenherr JA, Clemens JC, Chang HC. Activated Cdc42 kinase regulates Dock localization in male germ cells during Drosophila spermatogenesis. Dev Biol. 2013;378:141–153. doi: 10.1016/j.ydbio.2013.02.025. [DOI] [PubMed] [Google Scholar]
- Zahedi B, Shen W, Xu X, Chen X, Mahey M, Harden N. Leading edge-secreted Dpp cooperates with ACK-dependent signaling from the amnioserosa to regulate myosin levels during dorsal closure. Dev Dyn. 2008;237:2936–2946. doi: 10.1002/dvdy.21722. [DOI] [PubMed] [Google Scholar]
- Lu N, Guarnieri DJ, Simon MA. Localization of Tec29 to ring canals is mediated by Src64 and PtdIns(3,4,5)P3-dependent mechanisms. EMBO J. 2004;23:1089–1100. doi: 10.1038/sj.emboj.7600127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo WC, Satoh M, Kasahara H, Terada N, Hamazaki T, Chan JY, Yao B, Tamayo S, Covini G, von Mühlen CA. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS ONE. 2011;6:e29690. doi: 10.1371/journal.pone.0029690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman I. Enzymatic amination of uridine triphosphate to cytidine triphosphate. J Biol Chem. 1956;222:765–775. [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Zhang J, Tastan OY, Deussen ZA, Siswick MY, Liu JL. Glutamine analogs promote cytoophidium assembly in human and Drosophila cells. J Genet Genomics. 2011;38:391–402. doi: 10.1016/j.jgg.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Chang YF, Carman GM. CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog Lipid Res. 2008;47:333–339. doi: 10.1016/j.plipres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander DB, O’Brien DJ, Gorman JA, Carman GM. Effect of CTP synthetase regulation by CTP on phospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:18992–19001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- Hatch GM, McClarty G. Regulation of cardiolipin biosynthesis in H9c2 cardiac myoblasts by cytidine 5′-triphosphate. J Biol Chem. 1996;271:25810–25816. doi: 10.1074/jbc.271.42.25810. [DOI] [PubMed] [Google Scholar]
- Gupta T, Schupbach T. Cct1, a phosphatidylcholine biosynthesis enzyme, is required for Drosophila oogenesis and ovarian morphogenesis. Development. 2003;130:6075–6087. doi: 10.1242/dev.00817. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126:293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod JJ, Jacobs-Lorena M, Crippa M. Changes in rate of RNA synthesis and ribosomal gene number during oogenesis of Drosophila melanogaster. Dev Biol. 1977;57:393–402. doi: 10.1016/0012-1606(77)90224-x. [DOI] [PubMed] [Google Scholar]
- Chang YF, Martin SS, Baldwin EP, Carman GM. Phosphorylation of human CTP synthetase 1 by protein kinase C: identification of Ser(462) and Thr(455) as major sites of phosphorylation. J Biol Chem. 2007;282:17613–17622. doi: 10.1074/jbc.M702799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TS, O’Brien DJ, Carman GM. Phosphorylation of CTP synthetase on Ser36, Ser330, Ser354, and Ser454 regulates the levels of CTP and phosphatidylcholine synthesis in Saccharomyces cerevisiae. J Biol Chem. 2003;278:20785–20794. doi: 10.1074/jbc.M301394200. [DOI] [PubMed] [Google Scholar]
- Higgins MJ, Graves PR, Graves LM. Regulation of human cytidine triphosphate synthetase 1 by glycogen synthase kinase 3. J Biol Chem. 2007;282:29493–29503. doi: 10.1074/jbc.M703948200. [DOI] [PubMed] [Google Scholar]
- Han GS, Sreenivas A, Choi MG, Chang YF, Martin SS, Baldwin EP, Carman GM. Expression of human CTP synthetase in Saccharomyces cerevisiae reveals phosphorylation by protein kinase A. J Biol Chem. 2005;280:38328–38336. doi: 10.1074/jbc.M509622200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson GS, Guarnieri DJ, Simon MA. Src64 is required for ovarian ring canal morphogenesis during Drosophila oogenesis. Development. 1998;125:2883–2892. doi: 10.1242/dev.125.15.2883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.