Abstract
Activating mutations in Wnt and EGFR/Ras signaling pathways are common in colorectal cancer (CRC). Remarkably, clonal co-activation of these pathways in the adult Drosophila midgut induces “tumor-like” overgrowths. Here, we show that, in these clones and in CRC cell lines, Dpp/TGF-β acts as a tumor suppressor. Moreover, we discover that the Iroquois/IRX-family-protein Mirror downregulates the transcription of core components of the Dpp pathway, reducing its tumor suppressor activity. We also show that this genetic interaction is conserved in human CRC cells, where the Iro/IRX proteins IRX3 and IRX5 diminish the response to TGF-β. IRX3 and IRX5 are upregulated in human adenomas, and their levels correlate inversely with the gene expression signature of response to TGF-β. In addition, Irx5 expression confers a growth advantage in the presence of TGF-β, but is selected against in its absence. Together, our results identify a set of Iro/IRX proteins as conserved negative regulators of Dpp/TGF-β activity. We propose that during the characteristic adenoma-to-carcinoma transition of human CRC, the activity of IRX proteins could reduce the sensitivity to the cytostatic effect of TGF-β, conferring a growth advantage to tumor cells prior to the acquisition of mutations in TGF-β pathway components.
Keywords: Dpp, EGFR/Ras, TGF-β, Wnt
Introduction
Colorectal cancer (CRC) is a slow progressing disease characterized by the transition from normal mucosa to adenoma and then further to carcinoma. Adenoma development usually starts with the constitutive activation of the Wnt signaling pathway primarily due to mutations in the negative regulator APC 1–3. Wnt pathway activation induces a massive hyperproliferation of the stem/progenitor compartment that leads to the formation of small, benign adenomas 2,4–6. In many cases, APC mutations are followed by oncogenic activation of K-RAS, which induces the development of large, aggressive adenocarcinomas 6–8.
CRC progression also requires further mutations in different signaling pathways. A large proportion of intestinal tumors show inactivating mutations in components of the TGF-β pathway 9,10. As TGF-β signaling exerts cytostatic effects on epithelial tumor cells, its downregulation may play an essential role in allowing tumor progression. Consistent with this finding, TGF-β signaling inactivation in mice mutant for APC induces the development of invasive adenocarcinomas 11,12. Around 50% of CRCs become resistant to TGF-β through inactivating mutations of the TGF-β receptors or the intracellular signaling proteins SMADs 9,10,13. Yet, the mechanisms accounting for the somatic loss of TGF-β responsiveness in CRC cases with no identifiable alterations in the pathway are not well understood 14–16.
In the adult Drosophila midgut, Wnt and EGFR/Ras signaling pathways regulate homeostasis and intestinal stem cell (ISC) proliferation 17–20, and alterations induced by the combined activation of these two pathways expand as aggressive intestinal tumor-like overgrowths that reproduce many hallmarks of human CRC 21,22. Here, we take advantage of this cancer model to show that Dpp signaling, Drosophila’s main TGF-β superfamily member, is downregulated in Apc-Ras clones due to the upregulation of Mirror (Mirr). Indeed, we show that Mirr, a member of the Iro/IRX complex protein, acts as a negative transcription regulator of many core components of the Dpp pathway. Moreover, we show that silencing of the Dpp pathway activity by Mirr is required for the growth of Apc-Ras clones. Finally, we also show that this genetic regulation may be conserved in mammals, where the Iro/IRX proteins IRX3 and IRX5 reduce the ability of human colon cancer cells to respond to TGF-β. We propose that IRX proteins may play an important role regulating TGF-β response during the initial steps of the adenoma-to-carcinoma transition, giving an advantage to tumor cells before specific mutations in the TGF-β pathway are acquired.
Results and Discussion
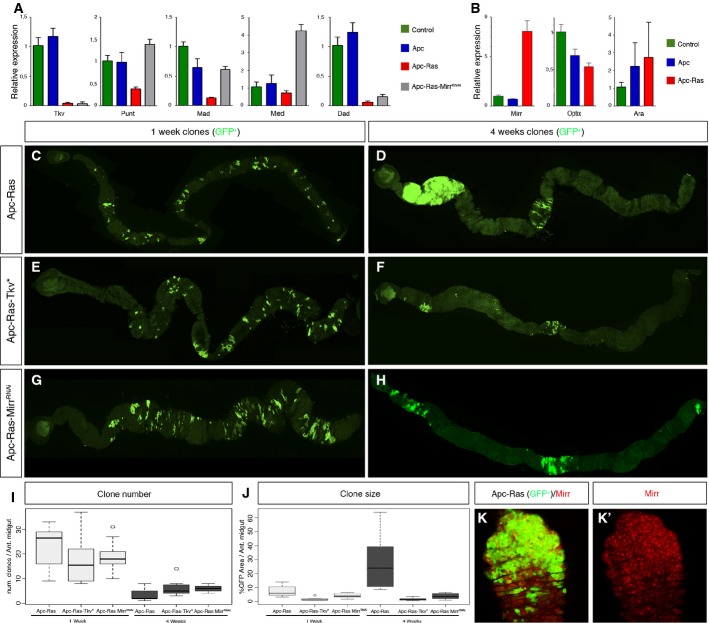
In human CRC, the activation of the Wg/Wnt and EGFR/Ras pathways is often followed by silencing of the TGF-β and/or BMP signaling pathways that act as tumor suppressors 10,14,16,23. Thus, we wondered whether this could be also the case in the tumor overgrowths caused by co-activation of the Wg/Wnt and EGFR/Ras signaling pathways in the adult Drosophila midgut. And if so, to use this system to unveil novel mechanisms that could account for the silencing of TGF-β and/or BMP signaling pathways in CRC cases with no identifiable mutations in components of both pathways. As a starting material, we generated clones of cells mutants for both Apc and Apc2 negative regulators of the Wg/Wnt pathway that over-expressed the oncogenic form UAS-RasV12, named Apc-Ras from now on, as previously described 21. Most of the Apc-Ras clones disappear over time, but the few that survive form tumor-like overgrowths 21. By flow cytometry, we sorted the GFP+ cells from wild-type, Apc, and Apc-Ras clones 4 weeks after clone induction and analyzed by qRT–PCR the expression of Dpp pathway components. Dpp binds to type I receptor thick veins (tkv) and type II receptor punt (put), allowing the activation of Mothers against Dpp (Mad). Activated Mad is then able to translocate the co-factor Medea (Med) into the nucleus to induce target gene expression. One of the target genes of the pathway is Daughters against Dpp (Dad), which acts as a negative pathway regulator. qRT–PCR analysis of GFP+ cells showed that Tkv, Put, Mad, Med, and Dad expression was downregulated in Apc-Ras clones when compared to wild-type or Apc clones (Fig 1A), suggesting that Dpp pathway activity could be reduced in Apc-Ras clones.
Figure 1. Dpp pathway suppresses Apc-Ras tumor growth and is regulated by Mirror.
A qRT–PCRs of the main core components of the Dpp pathway (tkv, put, Mad, and Med) and the target gene Dad 4 weeks after clone induction. Note that their expression is lower in Apc-Ras clones compared to wild-type or Apc clones.
B qRT–PCRs of Mirr, Optix, and Ara. Notice that Mirr is the only upregulated gene in Apc-Ras clones, but not in wild-type or Apc clones. Of note, qRT–PCR of Caup was negative in all conditions, and we discarded its expression in Apc-Ras clones by antibody staining (data not shown).
C–H Adult midguts showing Apc-Ras (C, D), Apc-Ras-Tkv* (E, F), and Apc-Ras-MirrRNAi (G, H) clones marked by GFP (green), 1 and 4 weeks after induction.
I Box-plot graph of the total number of clones in the anterior midgut.
J Box-plot graph of the clone area (GFP+) per anterior midgut area.
K Apc-Ras clones (green) stained with Mirr (red).
Data information: Graphs in (A, B) show a representative experiment (mean ± SEM of relative expression, n = 3). Box-plots in (I, J) show data from Table 1. The length of the box represents the distance between the 25th and 75th percentiles, the interior horizontal line represents the group median, and the whiskers extend to the group minimum and maximum values.
Source data are available online for this figure.
We then analyzed whether forced activation of the Dpp pathway could also modulate tumor initiation or growth in Drosophila. We generated Apc-Ras clones over-expressing the constitutively active form of the Dpp receptor TkvQ253 24 (Apc-Ras-Tkv* clones). One week after induction, Apc-Ras-Tkv* clones were similar to Apc-Ras clones in number, size, and distribution along the entire gut (Fig 1C, E, I, J and Table 1). However, 4 weeks after induction, Apc-Ras-Tkv* clones were noticeably smaller than Apc-Ras clones, albeit similar in number and distribution (Fig 1D, F, I, J and Table 1), indicating that over time, enforced Dpp pathway activity acts as a tumor suppressor.
Table 1.
Clone characterization: quantification of the number, size, and distribution of the clones of the different genotypes analyzed
| Number of clones (num clones ± SD) | Clone size (GFP+ area/domain area ± SD) | Clone distribution (GFP+ area/total area ± SD) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | W | n | A | M | P | A | M | P | A | M | P |
| Wild-type | 1 | 8 | 37 ± 10 | 2 ± 2 | 58 ± 13 | 3 ± 2 | 0.1 ± 0.1 | 3 ± 2 | 49 ± 10 | 0.3 ± 0.3 | 51 ± 10 |
| Apc-Ras | 1 | 10 | 23 ± 8 | 5 ± 3 | 23 ± 6 | 7 ± 4 | 8 ± 9 | 4 ± 2 | 52 ± 15 | 10 ± 9 | 38 ± 18 |
| Apc-Ras Tkv* | 1 | 10 | 17 ± 9 | 3 ± 3 | 25 ± 9 | 2 ± 1 | 5 ± 7 | 3 ± 2 | 30 ± 10 | 14 ± 19 | 55 ± 16 |
| Apc-Ras MirrRNAi | 1 | 9 | 19 ± 7 | 2 ± 2 | 25 ± 10 | 4 ± 2 | 2 ± 3 | 5 ± 3 | 45 ± 13 | 3 ± 6 | 52 ± 14 |
| Apc-Ras TkvDN | 1 | 6 | 23 ± 7 | 2 ± 2 | 31 ± 11 | 3 ± 1 | 3 ± 5 | 6 ± 4 | 32 ± 14 | 4 ± 7 | 64 ± 21 |
| Apc-Ras-TkvDN-MirrRNAi | 1 | 6 | 22 ± 7 | 2 ± 1 | 28 ± 16 | 5 ± 1 | 5 ± 4 | 7 ± 2 | 38 ± 9 | 6 ± 7 | 55 ± 8 |
| Wild-type | 4 | 10 | 25 ± 7 | 8 ± 4 | 24 ± 4 | 3 ± 1 | 4 ± 3 | 4 ± 2 | 38 ± 12 | 9 ± 7 | 53 ± 13 |
| Apc-Ras | 4 | 19 | 3 ± 2 | 2 ± 2 | 3 ± 3 | 26 ± 16 | 19 ± 25 | 3 ± 5 | 82 ± 22 | 11 ± 15 | 7 ± 14 |
| Apc-Ras Tkv* | 4 | 8 | 6 ± 3 | 6 ± 2 | 3 ± 1 | 2 ± 1 | 22 ± 17 | 0.8 ± 0.8 | 31 ± 17 | 57 ± 12 | 11 ± 8 |
| Apc-Ras MirrRNAi | 4 | 9 | 6 ± 1 | 3 ± 2 | 3 ± 2 | 4 ± 2 | 17 ± 17 | 0.6 ± 0.4 | 55 ± 25 | 35 ± 25 | 10 ± 6 |
| Apc-Ras TkvDN | 4 | 5 | 7 ± 2 | 2 ± 1 | 5 ± 3 | 36 ± 26 | 34 ± 21 | 1 ± 1 | 85 ± 9 | 13 ± 6 | 2 ± 1 |
| Apc-Ras-TkvDN-MirrRNAi | 4 | 5 | 4 ± 2 | 3 ± 1 | 4 ± 3 | 28 ± 15 | 19 ± 13 | 0.8 ± 0.5 | 88 ± 9 | 8 ± 7 | 4 ± 3 |
(A) anterior midgut, (M) Fe/Cu domain, (P) posterior midgut, (W) weeks after clone induction, (n) number of guts analyzed. Data from wild-type and Apc-Ras clones come from 21.
Source data are available online for this table.
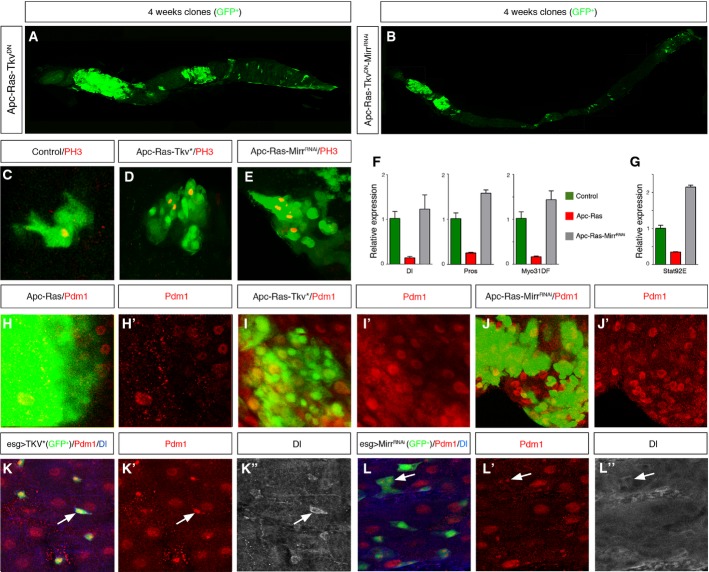
We next sought to determine the mechanism that allows the coordinated downregulation of Dpp pathway components in Apc-Ras clones. We explored the possibility that most, if not all, genes of the Dpp pathway could contain binding sites for a common negative regulator of transcription in their promoters. To address this issue, we scanned the promoter regions of Tkv, Put, Mad, and Med using the matScan software 25 in search for putative binding sites for known transcription factors. We identified a total of 70 transcription factors, yet only four of them hit at least three of the four promoter regions analyzed; these were the three components of the Iroquois/IRX complex araucan (ara), caupolican (caup) and mirror (mirr), and the homebox-containing gene optix. Of these, only Mirr was over-expressed in Apc-Ras clones, both transcriptionally (Fig 1B) and at the protein level (Fig 1K; Supplementary Fig S1A). Mirr usually acts as a transcriptional repressor 26,27, making it a bona fide candidate to be responsible for the downregulation of Dpp pathway components in Apc-Ras clones. Corroborating this hypothesis, over-expression of the RNAi of Mirr in an Apc-Ras background (Apc-Ras-MirrRNAi) recovered the expression of Put, Mad, and Med to at least wild-type levels (Fig 1A). Moreover, Apc-Ras-MirrRNAi clones were similar to Apc-Ras-Tkv* clones in size, number, and distribution both 1 and 4 weeks after clone induction (Fig 1G, H, I and J), albeit the effect of MirrRNAi was slightly weaker (Table 1). Furthermore, the negative effect of MirrRNAi in the growth of Apc-Ras clones was suppressed when co-expressed in the presence of a dominant-negative form of Tkv, TkvDN 28 (Apc-Ras-TkvDN-MirrRNAi clones) (Fig 2B; Table 1; Supplementary Fig S1B). As a control for this experiment, expression of the TkvDN transgene did not affect the growth of Apc-Ras clones (Apc-Ras-TkvDN clones) (Fig 2A; Table 1; Supplementary Fig S1B). Taken together, these results uncover Mirr as a novel tumor-promoting protein through its role as a negative transcriptional regulator of core Dpp pathway components.
Figure 2. TkvDN is epistatic to Mirr.
A, B Adult midguts showing Apc-Ras-TkvDN (A) and Apc-Ras-TkvDN-MirrRNAi (B) clones marked by GFP (green) 4 weeks after clone induction. Note that Dpp pathway inactivation blocks the tumor suppressor effect of MirrRNAi.
C–E Wild-type (C), Apc-Ras-Tkv* (D), and Apc-Ras-MirrRNAi (E) GFP+ clones (green) stained with PH3 (red). Note the increased number of PH3+ cells in Apc-Ras-Tkv* and Apc-Ras-MirrRNAi clones compared to a wild-type clone of similar size.
F, G qRT–PCRs of Dl, Pros, and Myo31DF (F) and stat92e (G) showing that Apc-Ras-MirrRNAi clones restore the expression of these markers to wild-type levels. Graphs show a representative experiment (mean ± SEM of relative expression, n = 3).
H–J Apc-Ras (H), Apc-Ras-Tkv* (I), and Apc-Ras-MirrRNAi (J) GFP+ clones (green) marked with the EC marker Pdm1 (red).
K, L Expression of activated Tkv* (K) or MirrRNAi (L) in progenitor cells induces the expression of the EC marker Pdm1 (red) in ISCs, marked by Dl (blue).
Source data are available online for this figure.
We next asked how Dpp activity is able to block the growth of Apc-Ras clones. During normal homeostasis, Dpp promotes copper cell differentiation in the gastric region 29,30 and, under some circumstances, is able to restrict ISC proliferation 29,31. We ruled out a role of Dpp pathway imposing, directly or indirectly, a blockade on ISC proliferation because Apc-Ras-Tkv* and Apc-Ras-MirrRNAi clones showed a higher number of positive cells for the mitotic marker phosphohistone 3 (PH3) (Fig 2D and E) than control wild-type clones of similar size (Fig 2C). Alternatively, we considered whether Dpp activity could promote cell differentiation in Apc-Ras clones. In wild-type guts, ISCs divide generating another ISC and an enteroblast (EB), which differentiates toward an enteroendocrine cell (EE) or an enterocyte (EC) 32,33. We observed that both Apc-Ras-Tkv* and Apc-Ras-MirrRNAi clones showed a remarkable increase in the number of ECs compared to Apc-Ras clones (Fig 2H–J). qRT–PCR analysis of Apc-Ras-MirrRNAi clones showed that the expression of the ISC marker Delta (Dl), the EE marker Prospero (Pros), and the EC marker Myo31DF (Fig 2F) was restored to wild-type levels, as well as Stat92E, a key component of the Jak/Stat pathway and a differentiation inducer in the adult gut 34–36 (Fig 2G). We also observed that over-expression of TkvQ253 or MirrRNAi in normal epithelium progenitor cells (ISCs and EBs) resulted in the ectopic expression of the EC marker Pdm1 in the ISCs, identified by the expression of Dl (Fig 2K and L; Supplementary Fig S1B). Together, these results suggest that Dpp pathway activity might restrict tumor growth by inducing cell differentiation, imposing an EC or EE fate in cells that would otherwise develop into a mass of undifferentiated, proliferative cells 21.
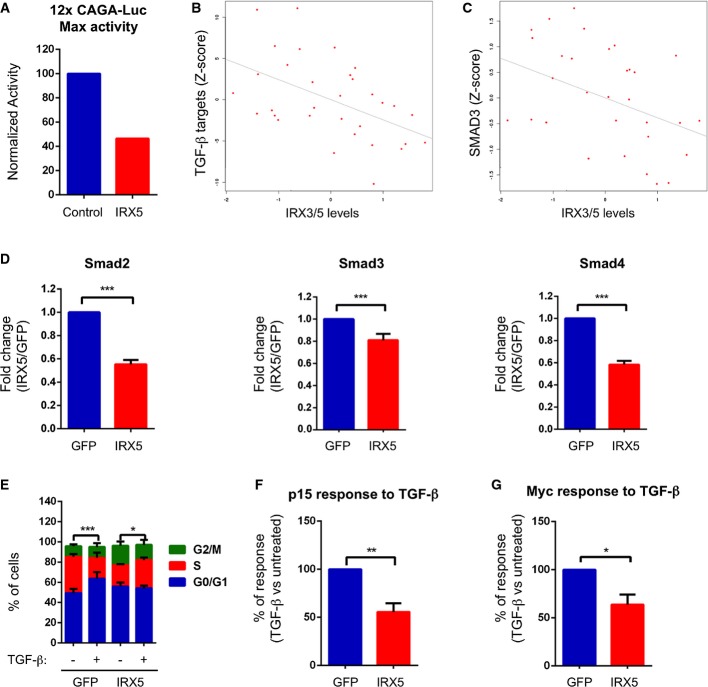
The above results lead us to hypothesize that Iro/IRX homologue proteins might play a similar role by reducing the ability of tumor cells to respond to TGF-β during the transition from adenoma to carcinoma in the human colon. The mammalian Iro/IRX complex is composed by six genes, IRX1 to IRX6, found in two clusters of three genes each 37. Despite the fact that all Iro/IRX proteins share high homology in the homeodomain, there is no direct relationship between any Drosophila Iro and any vertebrate IRX proteins. However, previous comparative analysis of the transcriptional profiles of human colorectal adenoma samples identified IRX3 as one of the most upregulated transcription factors compared to healthy tissue 38. Furthermore, we found that IRX3 and IRX5 were upregulated in adenomas arising in APC mutant mice compared to normal mucosa samples (Supplementary Fig S2A). These observations led us to explore the impact of IRX3 and IRX5 on the activity of the TGF-β pathway in mammalian cells. Knockdown of IRX3 and IRX5 in two different human cell lines induced downregulation of SMAD3 (Supplementary Fig S2B). We identified IRX optimal binding sites in the proximal SMAD3 promoter suggesting direct regulation. SW837 is the only well-established CRC cell line that shows a transcriptional response to TGF-β 39. These cells were derived from a late-stage CRC and expressed undetectable levels of IRX3 and IRX5. We thus co-transfected this cell line with the TGF-β reporter 12XCAGA-Luc 40 together with IRX3 or IRX5 expression vectors and measured luciferase levels in TGF-β-treated versus untreated cells. These assays showed that IRX3 or IRX5 over-expression induced a 4-fold reduction in reporter activity compared to cells transfected with a control vector, with a 47% of the absolute maximal activation in IRX5-over-expressing cells (Fig 3A).
Figure 3. Irx3 and Irx5 regulate the response to TGF-β in human cells.
A Graph of a representative experiment showing maximum luciferase activity upon TGF-β treatment of cells transfected with an empty vector or the Irx5-expressing vector.
B Negative correlation between TGF-β targets and IRX3/IRX5 expression in the cohort of adenoma samples from the GSE8671 dataset (Spearman’s correlation = −0.47507, P-value = 0.0065275, n = 32).
C Negative correlation between SMAD3 and IRX3/IRX5 expression in the same dataset as in (B) (Spearman’s correlation = −0.43218, P-value = 0.014199).
D qRT–PCRs of Smad2, Smad3, and Smad4 in LSTBRII cells expressing GFP or IRX5. Cells were induced with doxycycline for 24 h, and the levels of Smad2, Smad3, and Smad4 were assessed by qRT–PCR. Data are presented as mean fold change over GFP cells ± SEM (n = 4) (***P-value < 0.001, two-tailed t-test).
E Cell cycle profile of LSTBRII cells expressing GFP or IRX5 in response to TGF-β. Cells were induced with doxycycline and treated with TGF-β for 72 h. Data are presented as % of cells in G0/G1, S, or G2/M from the total population ± SEM (n = 6) (***P-value < 0.001; *P-value < 0.05, two-way ANOVA). Comparisons refer to cells in S phase.
F, G p15/Cdkn2b (F) or cMyc (G) response to TGF-β treatment of LSTBRII cells expressing GFP or IRX5. Cells were induced with doxycycline for 24 h and then treated with TGF-β for an additional 24 h. Results are expressed considering the transcriptional upregulation of p15/Cdkn2b (F) or the transcriptional downregulation of cMyc (G) of GFP cells (+TGF-β versus –TGF-β) as 100%. Data are presented as mean % of response ± SEM (n = 4) (*P-value < 0.05, **P-value < 0.01, two-tailed t-test).
Source data are available online for this figure.
Despite having a functional TGF-β pathway, SW837 cells did not show a cytostatic response to TGF-β (Supplementary Fig S2C), implying that this cell line developed additional mechanisms to evade the tumor suppressive effects of TGF-β. In contrast, virtually all CRC cell lines display impaired TGF-β responses due to the acquisition of mutations in either TGF-β type II receptor (TGFBR2) or the intracellular mediator SMAD4 (reviewed in 41). To study the effect of TGF-β in CRC, we used Ls174T cells. This cell line bears inactivating mutations in TGFBR2 in homozygosis, yet we generated inducible clones in which the expression of wild-type TGFBR2 could be controlled at will by the presence or absence of doxycycline (LSTGFBR2). Addition of TGF-β to parental Ls174T cells (LS) resulted neither in phosphorylation of intracellular SMADs nor in transcriptional activation of reporter constructs driven by SMAD-binding sites (Supplementary Fig S2D and E). Re-expression of wild-type receptor in Ls174T cells restored TGF-β signaling as shown by robust phosphorylation of SMADs and activation of the SMAD-binding reporter 12XCAGA-Luc (Supplementary Fig S2D and E). In the presence of TGF-β, LSTGFBR2 cells arrested in the G1 phase of the cell cycle (Supplementary Fig S2F). This response is reminiscent of that observed in other epithelial tumor cells in which TGF-β drives cell cycle arrest by decreasing levels of c-MYC, which in turn releases the expression of the cell cycle inhibitor p15-CDKN2B 42. By genomewide expression profiling using microarrays, we identified genes regulated by TGF-β in LSTGFBR2 cells (Supplementary Table S1). This analysis confirmed that many of the genes downregulated corresponded to cell cycle regulators including c-MYC 42, whereas upregulated genes included the cell cycle inhibitor CDKN2B 42. We next interrogated a set of transcriptomic profiles of human adenomas and used the average expression of the TGF-β upregulated genes (twofold, P < 0.05, n = 242 genes) as a surrogate to identify tumors displaying distinct activity of the TGF-β pathway. This analysis revealed that IRX3 and IRX5 expression correlated inversely with the expression levels of the TGF-β program (R = −0.47507, P < 0.0065275) (Fig 3B). IRX3 and IRX5 expression also correlated inversely with SMAD3 expression (R = −0.43218, P < 0.014199) (Fig 3C). We obtained equivalent results using a second independent cohort of adenoma samples (Supplementary Fig S3A and B).
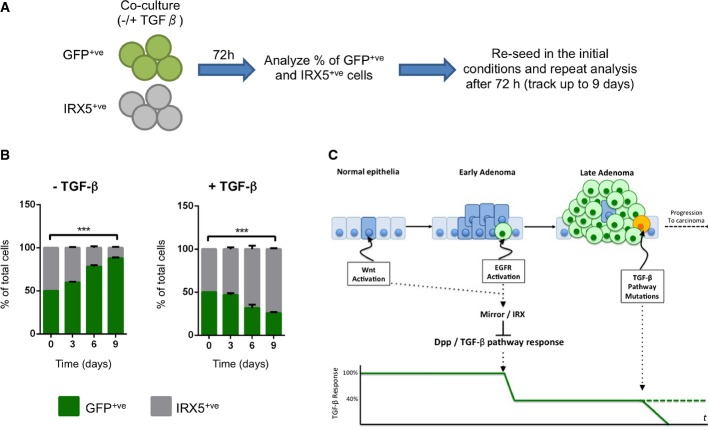
The above correlative data suggest that IRX activity can modulate TGF-β signaling in human colonic tumors. To test this possibility, we expressed IRX5 in LS174T cells while we reconstituted wild-type TGFBR2 expression. We confirmed that IRX5 decreased the levels of SMAD2, SMAD3 and SMAD4 mRNA (Fig 3D). As shown above, addition of TGF-β to control LSTGFBR2 cells induced arrest into the G1 phase of the cell cycle. IRX5 restrained this response (Fig 3E) including the modulation of c-MYC and p15-CDKN2B levels by TGF-β signaling (Fig 3F and G). To reinforce this observation, we performed competition assays by co-culturing control and IRX5-expressing cells (Fig 4A). In the absence of TGF-β, control cells outcompeted IRX5-expressing cells (Fig 4B). In sharp contrast, supplementation with TGF-β blocked proliferation of control cells, which were outcompeted by IRX5-expressing cells over the first week of culture (Fig 4B). Overall, these data show that the transcription factors IRX3 and IRX5 downmodulate TGF-β signaling in human CRC cells. This effect is reminiscent of that triggered by Drosophila IRX homologue in fly midgut tumors.
Figure 4. Irx proteins may play an essential role during the CRC adenoma-to-carcinoma transition.
A Schematic representation of cell competition assay. The same number of doxycycline-induced GFP or IRX5 LSTBRII cells was co-cultured in the presence or absence of TGF-β. After 72 h, cells were analyzed by FACS to quantify the % of GFP and IRX5 cells in the culture. Cells were then re-seeded and kept in the same culture conditions as before (either with or without TGF-β). This analysis was repeated at 6 and 9 days from the initial seeding.
B Cell competition assay of IRX5 and GFP LSTBRII cells. Data are presented as % of GFP and IRX5 cells ± SEM (n = 3) (***P-value < 0.001, two-way ANOVA).
C Proposed model in which expression of IRX genes in early adenomas would reduce the ability of tumor cells to respond to TGF-β, precluding its tumor suppressor activity. This reduced response would allow tumor cells to survive long enough to accumulate additional mutations.
Source data are available online for this figure.
We show that overcoming the suppressor effect of a TGF-β homologue is an essential step for the Wg/Wnt and EGFR/Ras signaling pathways to generate intestine tumors in Drosophila through the action of an Iro/IRX transcriptional repressor. Paralleling the observations in Drosophila, our experiments indicate that IRX3 and IRX5 lowers sensitivity to TGF-β signaling in mammalian cells. Based on these results, we propose a model where the synergy between Wnt and EGFR/Ras pathway activation would not only provide intestinal tumor cells with a proliferative advantage, but would also render them more resistant to the cytostatic and pro-apoptotic effect of TGF-β through the activation of IRX3 and/or IRX5 expression (Fig 4C). Perhaps, expression of IRX proteins allows early tumor cells to thrive in a TGF-β-rich context such as the inflammatory environment characteristic of most adenomatous lesions. Furthermore, transcriptional downregulation may modulate the activity of the pathway before specific mutations have time to accumulate, which fits well with the development of human CRC as a multistep process, characterized by the accumulation of mutations in different oncogenes and tumor suppressor genes. Moreover, we demonstrate that high levels of IRX expression confer a growth advantage in the presence of TGF-β, whereas in the absence of TGF-β, control cells overcompete IRX-expressing cells. We speculate that in this context, carcinoma cells that acquire mutations in the TGF-β pathway would then have selective pressure to reduce the level of IRX expression. Finally, our results show the utility of Drosophila as a model system for human diseases 21,43–45 to identify new genetic regulations.
Materials and Methods
Clone generation
MARCM clones were generated by a 1-h heat shock at 37°C of 2-to 5-day-old females and were marked by the progenitor cell marker escargot (esg) Gal4 line driving the expression of UAS GFP.
Staining and antibodies
Stainings were performed using standard protocols 21. Antibodies are described in Supplementary Methods.
Clone gene expression analysis
Drosophila midguts were dissected 4 weeks after induction. GFP+ cells were isolated by fluorescence-activated cell sorting (FACS), and RNA was extracted and amplified as described 46. Gene expression levels were assessed using Power Sybr Green quantitative PCR (Applied Biosystems).
Cell lines, transfections, and treatments
Human adenocarcinoma cell lines were cultured under standard conditions, and transfections were performed using PEI (polyethylenimine) reagent (Polysciences Inc.). Treatments are detailed in Supplementary Methods.
Acknowledgments
We are thankful to IRB Barcelona functional genomics facility, Cell separation Unit, and to Bloomington stock center for fly stocks. We also thank A. Calon and members of Casanova and Llimargas laboratories for discussions, E. Lonardo and D. Tauriello for mouse RNA samples, E. Espinet for LSTBR2 RNA samples, R.Gomis for MDA-MB-231 cells, Y. Rivera for technical help, and M. Furriols, M. Rahman, D. Shaye, and T. Stracker for comments on the manuscript. O.M. is a recipient of a FPU fellowship from the MEC, and F.M.B holds an IRB/PhD La Caixa fellowship. This work was supported by the MICINN (BFU2010-16016 and BFU2011-23479) to A.C.
Author contributions
OM, FB, and AMS performed experiments. CSA performed the bioinformatic analysis. JC, EB, and ES contributed to the conceptual development of the project and to data interpretation. AC conceived and designed the experiments, interpreted the data, and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Morin PJ, Sparks AB, Sparks AB, Korinek V, Korinek V, Barker N, Barker N, Clevers H, Clevers H, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Batlle E, Henderson JT, Beghtel H, van den Born MMW, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Tomita N, Mondem T, Ohue M, Yana I, Takami K, Yamamoto H, Yagyu T, Kikkawa N, Shimano T. A detailed analysis of the role of K-ras gene mutation in the progression of colorectal adenoma. Br J Cancer. 1997;75:341–347. doi: 10.1038/bjc.1997.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen K-P, Alberici P, Fsihi H, Gaspar C, Breukel C, Franken P, Rosty C, Abal M, Marjou El F, Smits R, et al. APC and oncogenic KRAS Are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology. 2006;131:1096–1109. doi: 10.1053/j.gastro.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. New Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- Muñoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, Sozmen EG, Madison BB, Pozzi A, Moon RT, et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–9844. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- Grady WM, Rajput A, Myeroff L, Liu DF, Kwon K, Willis J, Markowitz S. Mutation of the type II transforming growth factor-beta receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Res. 1998;58:3101–3104. [PubMed] [Google Scholar]
- Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Brattain MG, Chang J, Kim S-J, et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- Levy L, Hill C. Alterations in components of the TGF-β superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Edgar BA. EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development. 2009;136:483–493. doi: 10.1242/dev.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo M-J, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero JB, Stefanatos RK, Myant K, Vidal M, Sansom OJ. Non-autonomous crosstalk between the Jak/Stat and Egfr pathways mediates Apc1-driven intestinal stem cell hyperplasia in the Drosophila adult midgut. Development. 2012;139:4524–4535. doi: 10.1242/dev.078261. [DOI] [PubMed] [Google Scholar]
- Cordero JB, Stefanatos RK, Scopelliti A, Vidal M, Sansom OJ. Inducible progenitor-derived Wingless regulates adult midgut regeneration in Drosophila. EMBO J. 2012;31:3901–3917. doi: 10.1038/emboj.2012.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell O, Merlos-Suárez A, Campbell K, Barriga FM, Christov CP, Miguel-Aliaga I, Batlle E, Casanova J, Casali A. Conserved mechanisms of tumorigenesis in the Drosophila adult midgut. PLoS ONE. 2014;9:e88413. doi: 10.1371/journal.pone.0088413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhao R, Huang P, Yang F, Quan Z, Xu N, Xi R. APC loss-induced intestinal tumorigenesis in Drosophila: roles of Ras in Wnt signaling activation and tumor progression. Dev Biol. 2013;378:122–140. doi: 10.1016/j.ydbio.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol Mech Dis. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Blanco E, Messeguer X, Smith TF, Guigó R. Transcription factor map alignment of promoter regions. PLoS Comput Biol. 2006;2:e49. doi: 10.1371/journal.pcbi.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilioni A, Craig G, Hill C, McNeill H. Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc Natl Acad Sci USA. 2005;102:14671–14676. doi: 10.1073/pnas.0502480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu MJ, González-Pérez E, Ajuria L, Samper N, González-Crespo S, Campuzano S, Jiménez G. Mirror represses pipe expression in follicle cells to initiate dorsoventral axis formation in Drosophila. Development. 2012;139:1110–1114. doi: 10.1242/dev.076562. [DOI] [PubMed] [Google Scholar]
- Haerry TE, Khalsa O, O’Connor MB, Wharton KA. Synergistic signaling by two BMP ligands through the SAX and TKV receptors controls wing growth and patterning in Drosophila. Development. 1998;125:3977–3987. doi: 10.1242/dev.125.20.3977. [DOI] [PubMed] [Google Scholar]
- Guo Z, Driver I, Ohlstein B. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J Cell Biol. 2013;201:945–961. doi: 10.1083/jcb.201302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Qi Y, Jasper H. Dpp signaling determines regional stem cell identity in the regenerating adult Drosophila gastrointestinal tract. Cell Rep. 2013;4:10–18. doi: 10.1016/j.celrep.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Han L, Shi L, Lin X. Trachea-derived dpp controls adult midgut homeostasis in Drosophila. Dev Cell. 2013;24:133–143. doi: 10.1016/j.devcel.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2005;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2005;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe K, Lee W-C, Micchelli CA. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev Biol. 2010;338:28–37. doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R. Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of drosophila intestinal stem cells. J Mol Cell Biol. 2010;2:37–49. doi: 10.1093/jmcb/mjp028. [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Modolell J. Iroquois genes: genomic organization and function in vertebrate neural development. Curr Opin Genet Dev. 2002;12:403–408. doi: 10.1016/s0959-437x(02)00317-9. [DOI] [PubMed] [Google Scholar]
- Sabates-Bellver J, van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA, Bujnicki JM, Menigatti M, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- Ijichi H, Ikenoue T, Kato N, Mitsuno Y, Togo G, Kato J, Kanai F, Shiratori Y, Omata M. Systematic analysis of the TGF-beta-Smad signaling pathway in gastrointestinal cancer cells. Biochem Biophys Res Commun. 2001;289:350–357. doi: 10.1006/bbrc.2001.5988. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz SD, Dawson DM, Willis J, Willson JKV. Focus on colon cancer. Cancer Cell. 2002;1:233–236. doi: 10.1016/s1535-6108(02)00053-3. [DOI] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massagué J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer. 2005;5:626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- Apidianakis Y, Rahme LG. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech. 2011;4:21–30. doi: 10.1242/dmm.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nat Rev Cancer. 2013;13:172–183. doi: 10.1038/nrc3461. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Roca E, Garcia-Albéniz X, Rodriguez-Mulero S, Gomis RR, Kornacker K, Auer H. Accurate expression profiling of very small cell populations. PLoS ONE. 2010;5:e14418. doi: 10.1371/journal.pone.0014418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.