Abstract
Interferon-γ (IFN-γ) is an integral and critical molecule of the immune system, with multiple functions, mostly related to the T helper type 1 (Th1) response to infection. It is critical for defence against mycobacterial infection and is of increasing interest in defence against fungi. In this article, we review the genetic and epigenetic variants affecting IFN-γ expression and investigate its role in disease, with an emphasis on fungal diseases such as invasive and chronic pulmonary aspergillosis. Over 347 IFN-γ gene variants have been described, in multiple ethnic populations. Many appear to confer a susceptibility to disease, especially tuberculosis (TB) and hepatitis, but also some non-infectious conditions such as aplastic anaemia, cervical cancer and psoriasis. Several epigenetic modifications are also described, increasing IFN-γ expression in Th1 lymphocytes and reducing IFN-γ expression in Th2 lymphocytes. Recombinant IFN-γ administration is licensed for the prophylaxis of infection (bacterial and fungal) in patients with the phagocyte functional deficiency syndrome chronic granulomatous disease, although the benefits appear limited. Interferon-γ therapy is given to patients with profound defects in IFN-γ and interleukin-12 production and appears to be beneficial for patients with invasive aspergillosis and cryptococcal meningitis, but the studies are not definitive. A high proportion of patients with chronic pulmonary aspergillosis are poor producers of IFN-γ in response to multiple stimuli and could also benefit from IFN-γ administration. The investigation and management of patients with possible or demonstrated IFN-γ deficiency in adulthood is poorly studied and could be greatly enhanced with the integration of genetic data.
Keywords: epigenetics, fungal disease, genetics, interferon-γ, tuberculosis
Introduction
Interferon-γ (IFN-γ; also known as type II interferon) is a cytokine that is critical in both innate and adaptive immunity in humans. It is a highly pleiotropic cytokine produced by many immune cells in response to interleukin-12 (IL-12) as well as to microbial stimuli such as zymosan, lipopolysaccharide and β-glucan, which acts to stimulate and modulate the immune response by modulating the production or activities of several cytokines and chemokines.1,2 It is also an important activator of macrophages and one of the key cytokines that distinguishes differentiated T cells as either T helper type 1 (Th1; IFN-γ-producing) or Th2.1,2 Because of the importance of IFN-γ in human immune responses, it is unsurprising that genetic and epigenetic variations within the IFN-γ gene are associated with a range of diseases. These genetic and epigenetic variations are reviewed here. Several genetic IFN-γ and IL-12 receptor defects are also described, but are not reviewed here. The currently under-studied role of IFN-γ genetic and epigenetic variation in fungal disease is also discussed.
IFN-γ and host immunity
Interferon-γ is important in the immune response to various pathogens. Recognition of these pathogens by Toll-like receptors or other receptors induces production of IL-12 by macrophages and dendritic cells, which in turn stimulates Th1 responses and production of IFN-γ.1,3 Thus IFN-γ has many important immunostimulatory and immunomodulatory effects.
Interferon-γ up-regulates antigen presentation by MHC class I and class II by increasing expression of the subunits as well as by increasing the expression and activity of the proteasome.4 Increased presentation by MHC increases the visibility of the pathogen to the host, and so increases the host ability to recognize and respond to the pathogen. Interferon-γ is also important in activation of macrophages to produce tumour necrosis factor-α, which then acts together with IFN-γ to increase macrophage phagocytosis and microbicidal activity, such as production of reactive nitrogen and oxygen species including superoxide radicals, nitric oxide and hydrogen peroxide.1,3,5 In addition, IFN-γ enhances lymphocyte recruitment and results in prolonged activation within the tissues, induces components of the complement cascade and the acute phase response, plays a role in IgG class switching, and has direct anti-viral effects.6,7 Interferon-γ is also key in controlling naive CD4 T-cell differentiation into Th1 effector T cells, critical mediators of cellular immunity against viral and intracellular bacterial infections.4
Production of IFN-γ is affected by various other members of the immune response, via the action of various transcription factors which activate or repress its transcription. Interleukin-12 enhances IFN-γ production via activation of signal transducer and activator of transcription 4 (STAT4) and subsequent increased expression of IFNG.8 Interleukin-18, IFN-α, IL-12 and IL-2 also promote IFN-γ production and can augment IL-12-induced IFN-γ production.4,9 Interleukin-21, IL-18 and IL-15 can act in synergy to enhance IFN-γ production by cells.9 In addition, IFN-γ strongly up-regulates its own expression.10 Transforming growth factor-β inhibits IFN-γ expression by inhibiting expression of the transcription factors T-bet and STAT4, which are important for IFN-γ expression.11 Transforming growth factor-β also induces phosphorylation of SMAD3, which then binds with SMAD4 forming a heterodimer that can bind to the IFNG promoter and repress transcription.12 Interleukin-6 potentiates expression of the suppressor of cytokine signalling-1, which then prevents the phosphorylation and subsequent activation of STAT1.13 As STAT1 influences IFN-γ expression by potentiating the expression T-bet, prevention of STAT1 activation prevents IFN-γ expression.13
Genetic variation in the IFN-γ gene
A number of studies have identified 419 variations in the IFN-γ gene (data from Ensembl website;14 Table 1, Fig. 1). These fall into different categories, described in Table 1. These variations may or may not affect the expression of the IFN-γ gene or function of the protein, depending on their location within the gene and on their effect on the DNA sequence (Fig. 2).
Table 1.
Types and number of variations within the interferon-γ (IFN-γ) gene
| Type of variation | Description | Number present in IFN-γ gene |
|---|---|---|
| Splice donor variant | A splice variant that changes the two-base region at the 5′ end of an intron | 1 |
| Gain of stop variant | A sequence variant whereby a premature stop codon is created, leading to a shortened transcript | 1 |
| Loss of stop variant | A sequence variant whereby at least one base of the stop codon is changed, resulting in an elongated transcript | 1 |
| Non-synonymous (Missense) variant | A sequence variant, that changes one or more bases, resulting in a different amino acid sequence but where the transcript length is preserved | 31 |
| Splice region variant | A sequence variant in which a change has occurred within the region of the splice site, either within one or three bases of the exon or three to eight bases of the intron | 7 |
| Synonymous variant | A sequence variant where there is no resulting change to the encoded amino acid or transcript length | 10 |
| 5′ UTR variant | A variant in the 5′ untranslated region (UTR). This is upstream of the gene | 3 |
| 3′ UTR variant | A variant in the 3′ untranslated region (UTR). This is downstream of the gene | 16 |
| Intron variant | A variant that occurs within an intron | 119 |
| Upstream gene variant | A sequence variant located 5′ (upstream) of a gene | 119 |
| Downstream gene variant | A sequence variant located 3′ (downstream) of a gene | 118 |
A codon is a group of three bases that code for one amino acid, or start/stop signal. Data from Ensembl website14.
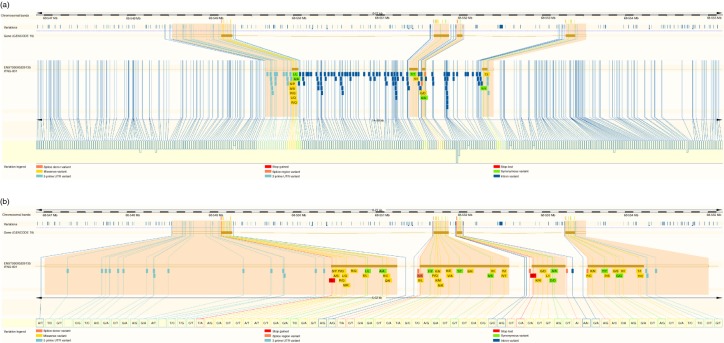
Figure 1.
Genetic variation within the interferon-γ gene (IFNG). (a) Variation within the IFN-γ gene, plus upstream and downstream regions; (b) variations within the exons and untranslated regions (UTRs), plus 20 bp either side of these. The variations track shows the variations present in the region (each line is a variation site). The exons (brown rectangles), introns (brown lines) and UTRs (unfilled rectangles) are shown. Variations within the exons and UTRs are highlighted with pale orange background throughout the figure, and are detailed using the variation legend shown. Figure adapted from Ensembl website14.
Figure 2.
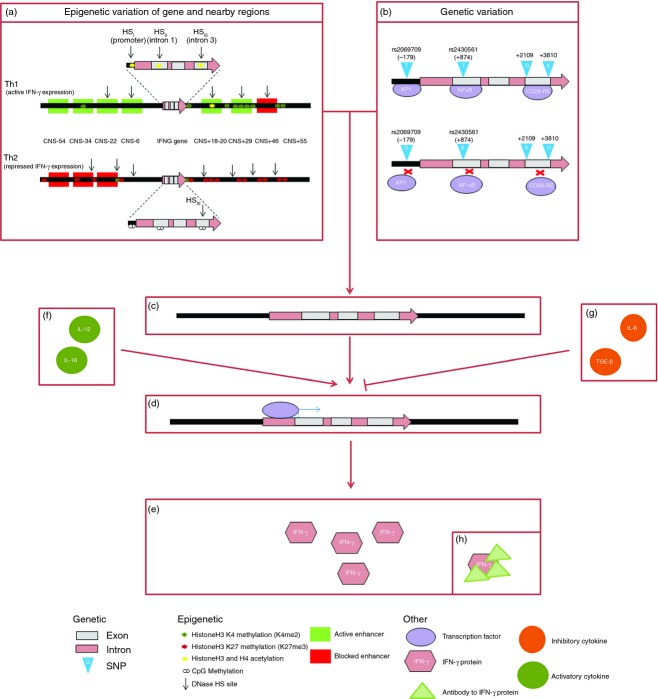
Differential mechanisms to impairment of interferon-γ responses. Epigenetic (a) and genetic (b) variations affect the chromatin structure and result in a specific pattern of variation at the DNA level of the interferon-γ gene (IFNG) and surrounding regions (c). Epigenetic variations include histone acetylation and H3K4 methylation, which activate gene expression, and DNA CpG methylation and histone H3K27 methylation, which repress gene expression. Genetic variation includes single nucleotide polymorphisms (SNPs) that affect the presence or functioning of a transcription factor binding site. Rs2430561 (+874T/A) is located in a nuclear factor-κB (NF-κB) binding site and NF-κB binds preferentially to the T allele. Rs2069709 (−179G/T) is located in the promoter. The T may create a putative activator protein 1 (AP-1) binding element or oestrogen-like response element. SNPs in intron 3 (+2109A/G and +3810G/A) may also affect transcription, by altering the binding of protein complexes such as CD28-RE. Binding of transcription factors (d) is affected by this genetic and epigenetic variation as well as by the presence or absence of the transcription factors themselves, and binding of different transcription factors activates or represses expression of the IFNG gene, to affect production of IFN-γ protein (e). Cytokines such as interleukin-12 (IL-12) and IL-18 can promote IFNG expression (f), while those such as IL-6 and transforming growth factor-β (TGF-β) can prevent expression (g), so altering the production of IFN-γ protein (e). This IFN-γ protein can be prevented from functioning by the presence of IFN-γ antibodies (h).
IFN-γ genetic variation and disease
Many variations within the IFN-γ gene have been shown to be associated with disease (Table 2).15–46 These associations may be related to expression of the IFN-γ gene. Gene expression is often affected by single nucleotide polymorphisms (SNPs) in either the promoter region or in nuclear factor-κB (NF-κB) binding regions. In the case of IFN-γ, there are two genetic variations that are known to affect expression. These are a polymorphic CA repeat region, where allele #2 (12 repeats) has been shown to result in high IFN-γ production when cells are stimulated,47 and a T/A SNP (rs2430561, +874T/A) in intron 1, where the T allele correlates to allele #2 of the CA repeat and high IFN-γ production.48 This T/A SNP is located in an NF-κB binding site and NF-κB binds preferentially to the T allele; the presence of the A allele reduces NF-κB binding, thereby reducing IFN-γ expression in response to stimuli.48 Studies have found that the rs2430561 SNP is associated with various diseases, including hepatitis20,41–43 and TB,19,21,27,34 in various populations (Table 2). Other, less well studied IFN-γ SNPs also appear to affect IFN-γ expression. Rs2069709 is a G to T transition at position −179 in the promoter region.49 It has been proposed that the T allele may create a putative activator protein 1 binding element or oestrogen-like response element, and electrophoretic mobility shift analysis has identified a unique complex that binds to the −179T variant but not to the −179G variant.49,50 Cells transfected with reporter complexes containing the T allele produce up to 13-fold more IFN-γ than those containing the G allele.49 This is true in T cells and peripheral blood mononuclear cells, but does not have the same effect in lamina propria cells, perhaps because of differences in oestrogen or CD2 signalling within these cells.49,50 This SNP was identified in HIV-infected individuals, and appears to affect AIDS progression.17,49 It is rare in Europeans (minor allele frequency 0·001), but is more common in African Americans (minor allele frequency 0·021).49 Two SNPs in intron 3 (+2109A/G and +3810G/A) of the IFN-γ gene may also affect transcription, by altering the binding of protein complexes, including CD28-RE, which itself binds to the transcription factors nuclear factor of activated T cells and to NF-κB.16 The +2109 G allele and +3810 A appear to form a DNA/protein complex that was not formed by the respective A and G alleles.16
Table 2.
Interferon-γ (IFN-γ) genetic associations with disease
| Variation | Association | Ethnicity | References |
|---|---|---|---|
| rs2430561 (+874A/T) | Paediatric tuberculosis, particularly in females | Han Chinese | 21 |
| rs2069718 (+3234C/T) | |||
| rs2069705/rs2430561/rs2069718 haplotype | |||
| rs3138557 (CA repeat) | Ossification of the posterior longitudinal ligament | Korean | 22 |
| rs2430561 (+874T/A) | Increased fatigue during the acute sickness response | Caucasian | 23 |
| rs2430561 (+874T/A) | Paroxysmal nocturnal haemoglobinuria | Han Chinese | 24 |
| rs3138557/rs2430561 haplotype | Number of vessels affected and severity in coronary heart disease | Korean | 25 |
| rs2430561 (+874T/A) | Cortical cataracts and posterior subcapsular cataracts | Indian | 26 |
| rs2430561 (+874T/A) | Mild pulmonary tuberculosis | Pakistani | 27 |
| rs2430561/IL10 rs1800870/IL6 | |||
| rs1800795 haplotype | Tuberculosis (mild or advanced) | Pakistani | 27 |
| rs2430561 (+874T/A) | Aplastic anaemia | Chinese | 28 |
| rs2430561 (+874T/A) | Response to immunosuppressive therapy in aplastic anaemia | Chinese | 28 |
| rs2069727 (A/G) | Acute lymphoblastic leukaemia in males | Welsh and Mexican | 29 |
| rs2430561 (+874T/A) | Protective against leprosy | Brazilian | 30 |
| rs11177074 (C/T) | Cervical cancer | Costa Rican | 31 |
| rs2069727/rs2069718/rs2430561/rs2069705 haplotype | Atopic dermatitis complicated by eczema herpeticum | Mixed African American and Caucasian | 32 |
| rs2430561 (+874T/A) | Chagas disease | Colombian | 33 |
| rs2430561 (+874T/A) | Tuberculosis (pulmonary and extra-pulmonary) | Brazilian | 34 |
| rs2430561 (+874T/A) | Cervical cancer | Swedish | 35 |
| rs2069705 | Systemic lupus erythematosus | Korean | 36 |
| rs2430561 (+874T/A) | Mediterranean spotted fever | Sicilian | 37 |
| TNFA rs1800629/IL6 rs1800795/IFNG rs2430561 haplotype | Spontaneous preterm birth | Brazilian | 38 |
| rs2430561 (+874T/A) | Cervical cancer | Indian | 39 |
| rs2430561 (+874T/A) | Psoriasis vulgaris | Polish | 40 |
| rs2069707 (−764C/G) | Recovery from hepatitis C virus infection | American | 41 |
| rs3138557 (CA repeat) rs2430561(+874T/A) | Chronic hepatitis B virus infection | Polynesian | 42 |
| rs2430561(+874T/A) | Liver cirrhosis in chronic hepatitis C | Taiwanese | 43 |
| rs2430561(+874T/A) rs3138557 (CA repeat) | IgA nephropathy | Italian | 44 |
| rs2430561 (+874T/A) rs3138557 (CA repeat) rs2430561(+874T/A) | Intrauterine hepatitis B virus infection | Chinese | 20 |
| rs2430561(+874T/A) | Tuberculosis | Sicilian | 19 |
| −1616G/A | Pulmonary tuberculosis | West African | 18 |
| +3234T/C | Pulmonary tuberculosis | West African | 18 |
| rs2069709 (−179G/T) | AIDS progression in HIV-positive individuals | African American | 17 |
| +2109A/G | Severe hepatic fibrosis in hepatic schistosomiasis | Sudanese | 16 |
| +3810G/A | Severe hepatic fibrosis in hepatic schistosomiasis | Sudanese | 16 |
| rs2430561(+874T/A) | Tuberculosis | Chinese | 15 |
| rs3138557 (CA repeat) | Tuberculosis | Chinese | 15 |
| rs3138557 (CA repeat) | Hepatitis E | Indian | 45 |
| rs3138557 (CA repeat) | Tuberculosis | Chinese | 46 |
| rs2430561(+874T/A) | Tuberculosis | Chinese | 46 |
Other SNPs in IFN-γ may affect its function. Of particular interest are SNPs within the exons of the gene, as these are the regions that are made into the final protein. Amino acid changing SNPs, called non-synonymous or missense SNPs, are more likely to have an effect than synonymous SNPs, and, depending on the location, may or may not affect the expression or function of the final protein. The effect of an SNP on protein function can be predicted using the PolyPhen-251 and SIFT52 programs. PolyPhen-2 predicts variation effects based on physical and comparative considerations, while SIFT predicts variation effects based on sequence homology and the physical–chemical similarity between the alternative amino acids. The non-synonymous variations in the IFN-γ gene, and their predicted effects on protein function, are shown in Table 3 (data from Ensembl website14). This table shows that there are many variations in the IFN-γ gene that are predicted to have deleterious or damaging effects on the IFN-γ protein function. In addition to non-synonymous SNPs, SNPs that cause gain or loss of ‘stop’ signals can result in a truncated or elongated protein, which may affect function. Two such SNPs exist in IFN-γ (Table 3).
Table 3.
Non-synonymous and loss/gain of stop single nucleotide polymorphisms within the interferon-γ gene
| SNP ID | Location (Chr:bp) | Alleles | Type | AA change | AA position | SIFT | PolyPhen |
|---|---|---|---|---|---|---|---|
| COSM942843 | 12:68551973 | C/A | Stop gain | E/* | 61 | N/A | N/A |
| COSM549550 | 12:68549134 | T/A | Stop lost | */L | 167 | N/A | N/A |
| rs372093951 | 12:68549141 | A/G | Missense | S/P | 165 | tolerated | benign |
| rs369578383 | 12:68549144 | C/A | Missense | A/S | 164 | deleterious | benign |
| rs201359065 | 12:68549155 | C/T | Missense | R/Q | 160 | tolerated | benign |
| rs374634889 | 12:68549161 | A/T | Missense | L/Q | 158 | tolerated | possibly damaging |
| rs377736305 | 12:68549179 | C/T | Missense | R/Q | 152 | deleterious | possibly damaging |
| rs150875052 | 12:68551842 | C/T | Missense | V/I | 73 | tolerated | benign |
| rs76012457 | 12:68551993 | C/T | Missense | G/D | 54 | tolerated | benign |
| rs371849964 | 12:68553385 | G/A | Missense | T/I | 4 | deleterious | probably damaging |
| COSM942837 | 12:68549155 | C/T | Missense | R/Q | 160 | tolerated | benign |
| COSM356728 | 12:68549164 | A/T | Missense | M/K | 157 | tolerated | benign |
| COSM549549 | 12:68549203 | G/A | Missense | S/L | 144 | tolerated | benign |
| COSM1476858 | 12:68549246 | G/A | Missense | R/C | 130 | deleterious | probably damaging |
| COSM549548 | 12:68549249 | G/T | Missense | Q/K | 129 | deleterious | probably damaging |
| COSM1363841 | 12:68551694 | G/A | Missense | S/L | 122 | deleterious | probably damaging |
| COSM942839 | 12:68551724 | C/T | Missense | R/Q | 112 | tolerated | benign |
| COSM1210288 | 12:68551729 | C/A | Missense | K/N | 110 | tolerated | benign |
| COSM1582160 | 12:68551730 | T/A | Missense | K/M | 110 | tolerated | benign |
| COSM1476859 | 12:68551735 | G/C | Missense | N/K | 108 | tolerated | benign |
| COSM1210289 | 12:68551752 | T/C | Missense | K/E | 103 | tolerated | possibly damaging |
| COSM942840 | 12:68551754 | A/G | Missense | V/A | 102 | tolerated | benign |
| COSM942841 | 12:68551796 | C/T | Missense | S/N | 88 | tolerated | benign |
| COSM942842 | 12:68551865 | C/A | Missense | R/I | 65 | tolerated | benign |
| COSM694678 | 12:68551865 | C/G | Missense | R/T | 65 | tolerated | benign |
| COSM942844 | 12:68551983 | C/A | Missense | K/N | 57 | deleterious | benign |
| COSM942845 | 12:68552003 | G/T | Missense | L/I | 51 | tolerated | probably damaging |
| COSM942846 | 12:68553283 | A/C | Missense | F/C | 38 | deleterious | possibly damaging |
| COSM194534 | 12:68553291 | C/A | Missense | K/N | 35 | deleterious | probably damaging |
| COSM1705908 | 12:68553320 | G/A | Missense | P/S | 26 | tolerated | benign |
| COSM942847 | 12:68553338 | C/T | Missense | G/S | 20 | tolerated | benign |
| COSM240197 | 12:68553353 | C/T | Missense | V/I | 15 | tolerated | benign |
| COSM1363842 | 12:68553388 | T/C | Missense | Y/C | 3 | tolerated | possibly damaging |
AA, amino acid.
Data from Ensembl14.
Epigenetic variation
Epigenetic modifications are extremely flexible and often reversible inheritable changes that can affect the accessibility of DNA for gene expression without affecting the DNA sequence itself. DNA is stored wrapped around cylinder-like structures called histones to form chromatin fibres. Epigenetic modifications of these histones or of the DNA itself can affect the structure of the chromatin fibre, making the DNA within it more or less accessible to the DNA binding proteins that are required to initiate the process of transcription, consequently affecting production of RNA and protein. Modifications that result in compacted (closed) chromatin lead to gene silencing, while those that result in relaxed (open) chromatin allow for gene expression.
Histone modification occurs predominantly at the N-terminal ‘tails’ of histones, which can undergo enzymatic post-translational modification including methylation, acetylation, phosphorylation, ubiquitination and sumoylation [addition of a small ubiquitin-like modifier (SUMO) protein].53 The effect of these varies depending on their location within a gene, and which residue is affected, but generally, histone acetylation causes activation of a gene and increased gene expression, while the effect of histone methylation is dependent on position; histone methylation at positions H3K4, H3K36, H3K79 results in activation and histone methylation at positions H3K9, H3K27, H4K20 results in silencing of a gene.53 The effects of phosphorylation, ubiquitination and sumoylation are less clearly defined, however, ubiquitination may increase transcriptional elongation and sumoylation appears to antagonize ubiquitination and acetylation to repress transcription.53
At the DNA level, the major epigenetic modification is CpG methylation, where the cytosine nucleotide of a CpG dinucleotide (CpG island) is methylated. This prevents recruitment of methyl-sensitive DNA binding proteins, preventing the initiation of transcription, and also generates an inaccessible chromatin structure.54 Together, these effects prevent gene expression and silence the gene. DNase hypersensitivity sites are areas of chromatin with increased sensitivity to an enzyme called DNase I. These areas of chromatin are highly accessible to DNA binding proteins, resulting in increased transcription and gene expression.
Epigenetic modifications occur naturally (e.g. during development and cellular differentiation), but can also be influenced by environmental factors including diet,55 smoking56 and microbial infections.57,58 The epigenetic modifications caused by these environmental exposures may have disease contributing effects, and may be one explanation for the disease discordance observed in identical twins as they age.59,60 Many diseases have been shown to involve aberrant epigenetic profiles, including cancer, atherosclerosis and osteoarthritis.60–63 Environmental exposure may contribute to these. For example, it has been proposed that early stage nutrition can affect CpG methylation levels, in turn affecting susceptibility to chronic disease as an adult,55 and smoking may cause promoter hypermethylation and silencing of p16, a tumour suppressor gene, possibly increasing susceptibility to oral cancer.56
Epigenetic variation and the IFN-γ gene
There is much evidence that the IFN-γ gene is subject to epigenetic modification, and that this modification is flexible and reversible. The epigenetic modifications are controlled by various transcription factors, including t-bet and GATA3, and those modifications that occur at promoter regions or conserved non-coding sequence sites are particularly important as these areas are involved in gene expression.64,65 In particular, much work has been completed investigating the differing patterns of epigenetic modification of the IFN-γ gene in Th1 and Th2 cells.
Interferon-γ is one of the key cytokines that distinguishes differentiated T cells as either Th1 (produce IFN-γ) or Th2 (do not produce IFN-γ). Differentiation of T cells involves various epigenetic changes in a 100-kb region surrounding the IFN-γ gene itself.66 These epigenetic modifications include gain or loss of histone modifications and changes in DNase hypersensitivity sites and CpG dinucleotide methylation, which activate the IFN-γ gene in Th1 cells and silence the IFN-γ gene in Th2 cells.66,67
Th1 cells produce IFN-γ. Within these cells, the IFN-γ gene shows histone H4 acetylation and histone H3 lysine 4 (H3K4) methylation and DNase hypersensitivity sites that are not present in naive T cells, including strong sites within the conserved non-coding sequence regions and the promoter.65–67 In addition, the CpG methylation seen in specific sites within the IFN-γ gene in naive T cells is largely lost during Th1 differentiation such that Th1 cells show reduced CpG methylation.66 The promoter is completely non-methylated.66 These changes in methylation and DNase hypersensitivity are associated with an open chromatin structure and increased production of IFN-γ.66 Like Th1 cells, natural killer cells produce IFN-γ. The IFN-γ gene within these cells also shows histone H4 acetylation and histone H3K4 methylation.67
Th2 cells do not produce IFN-γ. Within these cells, the IFN-γ gene shows histone H3K27 di-methylation and tri-methylation and no hyperacetylation.65,66 These modifications repress gene expression. As with Th1 cells, DNase hypersensitivity sites that are not present in naive T cells are observed, but these are different to the pattern in Th1 cells and fall adjacent to but not within conserved non-coding sequence sites.66 In contrast to Th1 differentiation, during Th2 differentiation the CpG methylation of naive T cells is largely maintained and the promoter becomes hypermethylated.66 These modifications silence gene expression.
IFN-γ epigenetic variation and disease
Interferon-γ gene epigenetics has been investigated in a growing number of conditions over recent years, and IFN-γ methylation has now been implicated in diseases from asthma to periodontitis (Table 4).68–74 Some diseases are associated with decreased (hypo-) methylation of the IFN-γ gene, others with increased (hyper-) methylation.
Table 4.
Interferon-γ epigenetic modification and disease
| Modification of interferon-γ gene | Association | References |
|---|---|---|
| Hypermethylation in effector T cells | Asthma (in discordant asthmatic twins) | 76 |
| Hypermethylation of the promoter in blood DNA | Diisocyanate induced occupational asthma | 71 |
| Hypomethylation | Increased diastolic blood pressure in elderly subjects | 72 |
| Hypomethylation of the promoter in T cells | Biliary atresia | 73 |
| Hypomethylation in bile duct cells | Biliary atresia | 74 |
| Hypomethylation of the promoter | Gingival biopsy samples of sites of chronic periodontitis | 77 |
| Hypomethylation | Samples of inflamed dental pulp, compared with healthy dental pulp | 78 |
| Hypomethylation in peripheral T cells | Inflammatory bowel disease patients requiring surgery, compared with non-surgical patients | 68 |
| Hypomethylation of the promoter in blood DNA | Increasing job seniority in chemical plant workers | 69 |
| Hypomethylation of the promoter | Severe acute graft-versus-host disease | 70 |
Interferon-γ methylation, and consequently IFN-γ gene expression in humans, has been shown to be modified by various microbial factors. HIV causes hypermethylation and silencing of the IFN-γ gene, possibly as a method to evade the immune response,58 while hypomethylation and activation of the IFN-γ gene is found in Epstein–Barr virus-transformed B cells.57 It has been suggested that low-level microbial exposure during early life can reduce demethylation of the IFN-γ gene in naive T cells, reducing activation of this gene and leading to an increased risk of allergic disease.75
Asthma in humans appears to be associated with increased methylation and consequent decreased expression of the IFN-γ gene. Effector T cells from discordant asthmatic twins show increased methylation and decreased expression of the IFN-γ gene, compared with their non-asthmatic twin.76 T-cell function is also reduced.76 Similarly, hypermethylation of the IFN-γ promoter has also been found in blood DNA from workers with diisocyanate-induced occupational asthma, suggesting that, in these subjects, diisocyanate exposure may have caused increased methylation of the IFN-γ gene, leading to increased production of IFN-γ and the development of asthma.71 Interferon-γ promoter methylation status was found to be a sensitive and specific method for identifying diisocyanate asthma workers.71
Other diseases are associated with decreased methylation and increased expression of the IFN-γ gene. Diastolic blood pressure is negatively associated with methylation of the IFN-γ gene (as blood pressure increases, methylation decreases), as shown by longitudinal measurements of DNA methylation in elderly subjects.72 People with high diastolic blood pressure have hypomethylation of the IFN-γ gene. The IFN-γ promoter is also hypomethylated in T cells and bile duct cells from patients with biliary atresia, together with the expected increased gene expression.73,74 Similar promoter hypomethylation and increased gene expression have been observed in gingival biopsy samples of sites of chronic periodontitis.77 Reduced methylation was also found in samples of inflamed dental pulp, when compared with healthy dental pulp.78
Interferon-γ antibodies
Antibodies to IFN-γ may be found in a few apparently normal individuals, 2–3% in the Netherlands, with slightly higher rates in older adults.79 Most anti-IFN-γ antibodies are IgG class, but they may or may not be functional. Production of functional anti-IFN-γ antibodies is more common in those of Asian descent, and is closely linked to certain HLA class II types.80 Over 100 individuals with neutralizing anti-IFN-γ antibodies and serious infection have been described, mostly in Asia, but not exclusively. It is probably more common than has been realized.81
Interferon-γ antibodies and disease
The most reported infections associated with anti-IFN-γ antibodies are disseminated or severe non-tuberculous mycobacterial infections, Mycobacterium tuberculosis, salmonellosis, varicella zoster reactivation, disseminated Penicillium marneffei infection, histoplasmosis, cryptococcosis, listeriosis and meliodosis,81–83 Unusual skin conditions also appear to be common, notably Sweet syndrome (neutrophilic dermatosis), but also erythema nodosum, pustular psoriasis and exanthematous pustulosis.83 Therapy with rituximab (anti-CD20) appears to be effective in blocking anti-IFN-γ antibody production.84
Assessment of IFN-γ deficits
Interferon-γ production is currently assessed using a whole blood stimulation assay, in which whole blood is stimulated with a variety of stimuli including zymosan, lipopolysaccharide, β-glucan, bacillus Calmette–Guérin and IL-12. The level of IFN-γ is measured after stimulation and this level of IFN-γ production is compared with the level of production by a control sample, so that impaired responses can be identified. As well as being used to investigate the inherent ability of an individual’s cells to produce IFN-γ, the IFN-γ release assays have been developed for use in the diagnosis of TB, as an alternative to the tuberculin skin test.85 Two commercial IFN-γ release assays are available; the ELISA, QuantiFERON-TB Gold Intube, and the ELISPOT, T-SPOT.TB. Both involve measuring IFN-γ release by T cells stimulated by M. tuberculosis antigens, which are present in infected individuals but not uninfected individuals or in the bacillus Calmette–Guérin vaccine.85 T cells that respond to these antigens should therefore only be present in infected individuals, and so only cells from infected individuals should release IFN-γ upon stimulation. However, as has been discussed, differences in expression, either constitutively, or in response to stimulation, can be caused by genetic and epigenetic variations within the IFN-γ gene, independent of the stimulus. This has not been rigorously assessed for TB diagnostic assays as an explanation for false negatives.
IFN-γ and mycobacterial infection
Interferon-γ is key in the immune response to M. tuberculosis. Interferon-γ production following recognition of this pathogen is important in macrophage activation and phagocytosis and results in inhibition of growth and death of the mycobacteria.5 Reduced IFN-γ production, and the resulting reduced Th1 response has been particularly associated with TB, and levels of IFN-γ and IL-12 increase during anti-TB treatment.86 In addition, adjuvant therapy with IFN-γ may be beneficial in TB patients.87 TB had been associated with mutations in the IFN-γ gene in many different ethnic groups.15,18,19,21,27,34,46 Reduced IFN-γ production and reduced Th1 responses have also been observed in non-tuberculosis mycobacterial infections such as those caused by Mycobacterium malmoense or Mycobacterium avium.88,89 Genetic associations with the IFN-γ gene have not yet been identified in these groups; however, associations with the IFN-γ receptor have been described, as have associations with anti-IFN-γ antibodies.90
IFN-γ and fungal infection
In addition to a role in bacterial infections, IFN-γ may also be important in defence against fungal infections. Cryptococcosis is usually caused by Cryptococcus neoformans, occasionally by Cryptococcus gattii, the latter notably in non-immunocompromised patients.91 Following inhalation, the usual manifestation of disease is meningitis, although pneumonia92 and other forms of disseminated cryptococcosis, notably skin and bone disease, also occur. Patients who fail to mount a directed IFN-γ response in cryptococcal meningitis are much more likely to die than those who do, irrespective of antifungal therapy.93 Aspergillosis is caused by the fungus Aspergillus, usually Aspergillus fumigatus.94 In the majority of individuals, inhaled A. fumigatus spores are cleared without causing disease, however, in immunocompromised individuals, A. fumigatus can cause an acute and severe disease called invasive aspergillosis (IA), and in overtly immunocompetent individuals A. fumigatus can cause chronic pulmonary aspergillosis (CPA).94
Invasive aspergillosis is a serious invasive fungal infection that occurs predominantly in the lung but also occasionally in other sites such as the paranasal sinuses, or postoperatively, and which can disseminate if untreated.95 It occurs in a wide variety of immunocompromised patients, including those undergoing organ transplants, critically ill patients, those receiving high-dose corticosteroid therapy, in liver failure and during neutropenia. Unless diagnosed early, it is associated with extremely high morbidity and mortality, although outcomes have improved in recent years with earlier diagnosis and better antifungal agents, notably voriconazole.95 Invasive aspergillosis is closely associated with profound neutropenia, monocytopenia and thrombocytopenia, or a blunted immune response (usually mediated by corticosteroid therapy). In addition, genetic susceptibility also plays a role and several mutations in donor or recipient following haematopoietic stem cell transplantation have been shown to affect susceptibility (e.g. ref. 96).
Chronic pulmonary aspergillosis is a serious and debilitating progressive lung condition that involves the formation of a cavity or cavities within the lung, with progressive fibrosis and consequent reduction in lung function and quality of life.94,97 A fungal ball, or aspergilloma, may be present.94 If untreated, CPA can be fatal and has a ≥ 50% 5-year mortality.98 Long-term treatment with expensive antifungal drugs is required to prevent deterioration; however, even with therapy, many patients do not improve but instead either deteriorate or remain stable, and morbidity and relapse remain high.99 Unlike in IA, CPA patients have overtly normal immune systems without deficient numbers of immune cells, and the reasons behind development of this disease are unclear. While patients almost invariably have some previous lung disease, such as pulmonary TB or chronic obstructive pulmonary disease, they do not generally have a clinical history of recurrent infection.100 In addition, the majority of patients with these underlying diseases do not develop CPA. It is likely that an immunogenetic deficiency is involved, and some genes have indeed been implicated (e.g. ref. 101). However, these associations do not explain all of the cases of CPA.
As with mycobacterium infection, Th1 responses and macrophages are important in Aspergillus infection. Various studies have suggested that Th1 responses (e.g. IFN-γ, tumour necrosis factor-α, IL-15) are beneficial during infection with A. fumigatus, whereas uncontrolled Th2 responses (e.g. IL-4, IL-13) are detrimental.102,103 Recent reports indicate a role for IFN-γ in immune tolerance to A. fumigatus, acting via indoleamine 2,3-deoxygenase and culminating in inhibition of Th17 cell responses and control of inflammation and allergy in Aspergillus-related infections.104 Interferon-γ is therefore important in resistance to CPA and IA. Production of IFN-γ in response to standard stimuli is impaired in IA105 as well as in CPA.106,107 Both CPA and IA patients have been treated with recombinant IFN-γ with benefit;94,108 CPA patients have stable or improved disease when IFN-γ is given in combination with itraconazole, and a replacement dose of rIFN-γ (50 μg subcutaneously three times per week at night) can make a substantial difference to patients. In addition to its use in aspergillosis, exogenous IFN-γ therapy has proved beneficial in other patients with a range of invasive fungal infections (Ochroconis gallopava, Alternaria malorum, Pyrenochaeta romeroi, Davidiella tassiana and Candida albicans) after kidney transplantation.108 Several cases of disseminated invasive fungal infections that were refractory to conventional antifungal drug therapy were rapidly cured with IFN-γ therapy.108
Clinical trials of gIFN treatment in infection and fungal diseases
Recombinant IFN-γ was originally licensed for infection prophylaxis in patients with chronic granulomatous disease following a randomized controlled trial (RCT) showing a significant reduction of infections and severity of infection in those receiving rIFN-γ.109 A 12-month double-blind, placebo-controlled RCT of rIFN-γ was suggestive of benefit in HIV-positive patients with low CD4 cell counts, with reduced incidence of mucosal Candida, herpes simplex virus and cytomegalovirus infections and a trend towards increased survival (28% compared with 18%).110 The advent of combination antiretroviral therapy curtailed further evaluations. Also in AIDS, an RCT of rIFN-γ at 100 and 200 µg three times weekly added to antifungal therapy showed important trends in improvement in cryptococcal meningitis, with more rapid cerebrospinal fluid sterilization (32–36% rIFN-γ recipients versus 13% recipients (P = 0·072) and reduction in cryptococcal antigen (12- to 24-fold versus eightfold decrease, respectively) at 2 weeks.111 At 10 weeks, improved combined mycological and clinical success was seen in the rIFN-γ recipients (26% versus 8%; P = 0·078). Unfortunately the study was under-powered. A follow-up study comparing two and six doses of 100 μg of rIFN-γ in cryptococcal meningitis all treated with amphotericin B and flucytosine, showed faster organism clearance in both rIFN-γ groups compared with those not receiving rIFN-γ.112 No differences in mortality were seen. In pulmonary TB, an RCT comparing the addition of rIFN-γ given by nebulizer to anti-tuberculous therapy showed a significant difference in the rate of clearance of M. tuberculosis from the sputum smear at 4 weeks (P = 0·03) anti-tuberculous therapy alone.113 Both nebulized and subcutaneous rIFN-γ significantly reduced fever, wheeze, and night sweats at 4 weeks. Some open studies of rIFN-γ are suggestive of benefit in invasive aspergillosis,108,114–116 but no RCTs have been published. In contrast to these encouraging data, other RCTs have been negative. A large RCT in pulmonary fibrosis was stopped early with lack of benefit.117 In Chinese patients with chronic hepatitis B, rIFN-γ showed no benefit.118 Aerosolized rIFN-γ was ineffective in mild to moderate cystic fibrosis.119
IFN-γ genetic and epigenetic variation and fungal disease
As discussed, IFN-γ appears to be important in the immune response to fungi; in aspergillosis in particular, Th1 (IFN-γ-producing) responses appear beneficial in CPA, while uncontrolled Th2 responses are detrimental,102,103 and impaired IFN-γ responses are associated with aspergillosis, including CPA.102,103,105,107 Immune cells from aspergillosis patients have deficient IFN-γ production, and patients benefit from treatment with recombinant IFN-γ. The production of IFN-γ that is measured by this assay can be affected by genetic and epigenetic variations within the IFN-γ gene. It is likely that the deficient responses observed in cells from CPA patients are a result of genetic or epigenetic factors within the DNA encoding the IFN-γ gene.
Identification of IFN-γ SNPs affecting expression, either constitutively or in response to stimuli, may be useful as indicators for IFN-γ treatment in aspergillosis patients or those with other fungal disease. In addition, identification of variations that are associated with aspergillosis or other fungal diseases may be useful as genetic markers of susceptibility to these diseases and could help to identify at risk individuals. Therefore, genetic studies of IFN-γ and its role in fungal diseases such as aspergillosis would be invaluable.
In addition, although there is no evidence yet as to whether the IFN-γ is epigenetically altered in patients with aspergillosis or other fungal diseases, it is likely that epigenetic changes that reduce expression of IFN-γ, such as hypermethylation of the gene or promoter, may increase susceptibility to this disease. Investigation and identification of these would also be invaluable.
Conclusions
The IFN-γ gene is subject to both genetic and epigenetic variations, some of which have been associated with gene expression and with disease. IFN-γ therapy is given to patients with profound defects in IFN-γ and IL-12 production. A high proportion of patients with CPA are poor producers of IFN-γ in response to multiple stimuli and IFN-γ therapy appears to be beneficial for patients with IA and CPA. The investigation and management of patients with possible or demonstrated IFN-γ deficiency in adulthood are poorly studied and could be greatly enhanced with the integration of genetic data. Variation in the IFN-γ gene may be important in fungal disease, including aspergillosis, particularly in CPA and IA, and genetic and epigenetic studies investigating IFN-γ in aspergillosis would be useful tools to elucidate a possible role for this variation in both susceptibility to aspergillosis and in identification and stratification of patients who would benefit from IFN-γ therapy.
Glossary
- CPA
chronic pulmonary aspergillosis
- IA
invasive aspergillosis
- IFN-γ
interferon γ
- IL
interleukin
- MAF
minor allele frequency
- NF-κB
nuclear factor-κB
- RCT
randomised control trial
- rIFN-γ
recombinant interferon-γ
- SNP
single nucleotide polymorphism
- STAT
signal transducer and activator of transcription
- SUMO
small ubiquitin-like modifier
- TB
tuberculosis
- Th1
T helper type 1
Disclosures
The authors have no competing interests.
References
- Watford WT, Moriguchi M, Morinobu A, O’Shea JJ. The biology of IL-12: coordinating innate and adaptive immune responses. Cytokine Growth Factor Rev. 2003;14:361–8. doi: 10.1016/s1359-6101(03)00043-1. [DOI] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-γ: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–89. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Robinson CM, O’Dee D, Hamilton T, Nau GJ. Cytokines involved in interferon-γ production by human macrophages. J Innate Immunol. 2010;2:56–65. doi: 10.1159/000247156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn JR, Wilson CB. Regulation of interferon-γ during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- Denis M, Gregg EO, Ghandirian E. Cytokine modulation of Mycobacterium tuberculosis growth in human macrophages. Int J Immunopharmacol. 1990;12:721–7. doi: 10.1016/0192-0561(90)90034-k. [DOI] [PubMed] [Google Scholar]
- Hill N, Sarvetnick N. Cytokines: promoters and dampeners of autoimmunity. Curr Opin Immunol. 2002;14:791–7. doi: 10.1016/s0952-7915(02)00403-x. [DOI] [PubMed] [Google Scholar]
- Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-γ. Annu Rev Immunol. 1997;15:749–95. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Fitz L, Ryan M, et al. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J Exp Med. 1989;170:827–45. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strengell M, Matikainen S, Siren J, Lehtonen A, Foster D, Julkunen I, Sareneva T. IL-21 in synergy with IL-15 or IL-18 enhances IFN-γ production in human NK and T cells. J Immunol. 2003;170:5464–9. doi: 10.4049/jimmunol.170.11.5464. [DOI] [PubMed] [Google Scholar]
- Hardy KJ, Sawada T. Human γ interferon strongly upregulates its own gene expression in peripheral blood lymphocytes. J Exp Med. 1989;170:1021–6. doi: 10.1084/jem.170.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JT, Martin SL, Xia L, Gorham JD. TGF-β1 uses distinct mechanisms to inhibit IFN-γ expression in CD4+ T cells at priming and at recall: differential involvement of Stat4 and T-bet. J Immunol. 2005;174:5950–8. doi: 10.4049/jimmunol.174.10.5950. [DOI] [PubMed] [Google Scholar]
- Yu J, Wei M, Becknell B, et al. Pro- and antiinflammatory cytokine signaling: reciprocal antagonism regulates interferon-γ production by human natural killer cells. Immunity. 2006;24:575–90. doi: 10.1016/j.immuni.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Diehl S, Anguita J, Hoffmeyer A, Zapton T, Ihle JN, Fikrig E, Rincón M. Inhibition of Th1 differentiation by IL-6 is mediated by SOCS1. Immunity. 2000;13:805–15. doi: 10.1016/s1074-7613(00)00078-9. [DOI] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Li L, Zhu X. Polymorphism of the interferon-γ gene and risk of tuberculosis in a southeastern Chinese population. Hum Immunol. 2008;69:129–33. doi: 10.1016/j.humimm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Chevillard C, Moukoko CE, Elwali NE, et al. IFN-γ polymorphisms (IFN-γ +2109 and IFN-γ +3810) are associated with severe hepatic fibrosis in human hepatic schistosomiasis (Schistosoma mansoni. J Immunol. 2003;171:5596–601. doi: 10.4049/jimmunol.171.10.5596. [DOI] [PubMed] [Google Scholar]
- An P, Vlahov D, Margolick JB, Phair J, O’Brien TR, Lautenberger J, Winkler CA. A tumor necrosis factor-α-inducible promoter variant of interferon-γ accelerates CD4+ T cell depletion in human immunodeficiency virus-1-infected individuals. J Infect Dis. 2003;188:228–31. doi: 10.1086/376455. [DOI] [PubMed] [Google Scholar]
- Cooke GS, Campbell SJ, Sillah J, et al. Polymorphism within the interferon-γ/receptor complex is associated with pulmonary tuberculosis. Am J Respir Crit Care Med. 2006;174:339–43. doi: 10.1164/rccm.200601-088OC. [DOI] [PubMed] [Google Scholar]
- Lio D, Marino V, Serauto A, et al. Genotype frequencies of the +874T→A single nucleotide polymorphism in the first intron of the interferon-γ gene in a sample of Sicilian patients affected by tuberculosis. Eur J Immunogenet. 2002;29:371–4. doi: 10.1046/j.1365-2370.2002.00327.x. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhu QR, Gu SQ, Fei LE. Relationship between IFN-γ gene polymorphism and susceptibility to intrauterine HBV infection. World J Gastroenterol. 2006;12:2928–31. doi: 10.3748/wjg.v12.i18.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Jiao WW, Feng WX, Wu XR, Xiao J, Miao Q, et al. IFNG polymorphisms are associated with tuberculosis in Han Chinese pediatric female population. Mol Biol Rep. 2013;40:5477–82. doi: 10.1007/s11033-013-2647-7. [DOI] [PubMed] [Google Scholar]
- Kim KT, Kim DH, Chung JY, Lee S, Joo J, Nah SS, Song HY, Kim HJ. Association of interferon γ polymorphism with ossification of the posterior longitudinal ligament in the Korean population. Immunol Invest. 2012;41:876–87. doi: 10.3109/08820139.2012.714437. [DOI] [PubMed] [Google Scholar]
- Piraino B, Vollmer-Conna U, Lloyd AR. Genetic associations of fatigue and other symptom domains of the acute sickness response to infection. Brain Behav Immun. 2012;26:552–8. doi: 10.1016/j.bbi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY, Yang XJ, Yang SS, Wang W, Tian YY, Cao FL, Zhou J. Association analysis of cytokine polymorphisms and plasma level in Northern Chinese Han patients with paroxysmal nocturnal hemoglobinuria. Chin Med J (Engl) 2012;125:1576–80. [PubMed] [Google Scholar]
- Kim HJ, Kang SW, Chung JH, Kim SJ, Choe BK. Polymorphisms of the interferon γ gene and coronary artery disease in the Korean population. Mol Biol Rep. 2012;39:5425–32. doi: 10.1007/s11033-011-1342-9. [DOI] [PubMed] [Google Scholar]
- Manne M, Gunde S, Kondreddy RK, Thurlapati N, Tirunilai P. Association of IFN-g+874(T/A) polymorphism with female patients of age-related cataracts. Oman J Ophthalmol. 2012;5:32–6. doi: 10.4103/0974-620X.94764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A, Hasan Z, Dawood G, Hussain R. Differential combination of cytokine and interferon- γ +874 T/A polymorphisms determines disease severity in pulmonary tuberculosis. PLoS ONE. 2011;6:e27848. doi: 10.1371/journal.pone.0027848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Zeng F, Zhang JY, Mu XY, Meng WT, Ma HB, Liu T. Association of the interferon-γ single nucleotide polymorphism +874(T/A) with response to immunosuppressive therapy in patients with severe aplastic anemia. Blood Cells Mol Dis. 2010;45:313–6. doi: 10.1016/j.bcmd.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Morrison BA, Ucisik-Akkaya E, Flores H, Alaez C, Gorodezky C, Dorak MT. Multiple sclerosis risk markers in HLA-DRA, HLA-C, and IFNG genes are associated with sex-specific childhood leukemia risk. Autoimmunity. 2010;43:690–7. doi: 10.3109/08916930903567492. [DOI] [PubMed] [Google Scholar]
- Cardoso CC, Pereira AC, Brito-de-Souza VN, et al. IFNG +874 T→A single nucleotide polymorphism is associated with leprosy among Brazilians. Hum Genet. 2010;128:481–90. doi: 10.1007/s00439-010-0872-x. [DOI] [PubMed] [Google Scholar]
- Wang SS, Gonzalez P, Yu K, et al. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PLoS ONE. 2010;5:e8667. doi: 10.1371/journal.pone.0008667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DY, Gao PS, Grigoryev DN, et al. Human atopic dermatitis complicated by eczema herpeticum is associated with abnormalities in IFN-γ response. J Allergy Clin Immunol. 2011;127:965–73.e1–5. doi: 10.1016/j.jaci.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres OA, Calzada JE, Beraun Y, Morillo CA, Gonzalez A, Gonzalez CI, Martín J. Role of the IFNG +874T/A polymorphism in Chagas disease in a Colombian population. Infect Genet Evol. 2010;10:682–5. doi: 10.1016/j.meegid.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallinoto AC, Graca ES, Araujo MS, Azevedo VN, Cayres-Vallinoto I, Machado LF, Ishak MO, Ishak R. IFNG +874T/A polymorphism and cytokine plasma levels are associated with susceptibility to Mycobacterium tuberculosis infection and clinical manifestation of tuberculosis. Hum Immunol. 2010;71:692–6. doi: 10.1016/j.humimm.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Ivansson EL, Juko-Pecirep I, Gyllensten UB. Interaction of immunological genes on chromosome 2q33 and IFNG in susceptibility to cervical cancer. Gynecol Oncol. 2010;116:544–8. doi: 10.1016/j.ygyno.2009.10.084. [DOI] [PubMed] [Google Scholar]
- Kim K, Cho SK, Sestak A, Namjou B, Kang C, Bae SC. Interferon-γ gene polymorphisms associated with susceptibility to systemic lupus erythematosus. Ann Rheum Dis. 2010;69:1247–50. doi: 10.1136/ard.2009.117572. [DOI] [PubMed] [Google Scholar]
- Forte GI, Scola L, Misiano G, et al. Relevance of γ interferon, tumor necrosis factor α, and interleukin-10 gene polymorphisms to susceptibility to Mediterranean spotted fever. Clin Vaccine Immunol. 2009;16:811–5. doi: 10.1128/CVI.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura E, Mattar R, de Souza E, Torloni MR, Goncalves-Primo A, Daher S. Inflammatory cytokine gene polymorphisms and spontaneous preterm birth. J Reprod Immunol. 2009;80:115–21. doi: 10.1016/j.jri.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Kordi Tamandani MK, Sobti RC, Shekari M, Mukesh M, Suri V. Expression and polymorphism of IFN-γ gene in patients with cervical cancer. Exp Oncol. 2008;30:224–9. [PubMed] [Google Scholar]
- Baran W, Szepietowski JC, Mazur G, Baran E. IFN-γ promoter gene polymorphism in psoriasis vulgaris. Biomarkers. 2008;13:52–8. doi: 10.1080/13547500701610273. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yang H, Borg BB, et al. A functional SNP of interferon-γ gene is important for interferon-α-induced and spontaneous recovery from hepatitis C virus infection. Proc Natl Acad Sci USA. 2007;104:985–90. doi: 10.1073/pnas.0609954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott W, Gane E, Winship I, Munn S, Tukuitonga C. Polymorphism in intron 1 of the interferon-γ gene influences both serum immunoglobulin E levels and the risk for chronic hepatitis B virus infection in Polynesians. Immunogenetics. 2007;59:187–95. doi: 10.1007/s00251-006-0184-4. [DOI] [PubMed] [Google Scholar]
- Dai CY, Chuang WL, Hsieh MY, et al. Polymorphism of interferon-γ gene at position +874 and clinical characteristics of chronic hepatitis C. Transl Res. 2006;148:128–33. doi: 10.1016/j.trsl.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Schena FP, Cerullo G, Torres DD, et al. Role of interferon-γ gene polymorphisms in susceptibility to IgA nephropathy: a family-based association study. Eur J Hum Genet. 2006;14:488–96. doi: 10.1038/sj.ejhg.5201591. [DOI] [PubMed] [Google Scholar]
- Arora R, Saha A, Malhotra D, Rath P, Kar P, Bamezai R. Promoter and intron-1 region polymorphisms in the IFNG gene in patients with hepatitis E. Int J Immunogenet. 2005;32:207–12. doi: 10.1111/j.1744-313X.2005.00512.x. [DOI] [PubMed] [Google Scholar]
- Tso HW, Ip WK, Chong WP, Tam CM, Chiang AK, Lau YL. Association of interferon γ and interleukin 10 genes with tuberculosis in Hong Kong Chinese. Genes Immun. 2005;6:358–63. doi: 10.1038/sj.gene.6364189. [DOI] [PubMed] [Google Scholar]
- Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott PJ, Hutchinson IV. In vitro production of IFN-γ correlates with CA repeat polymorphism in the human IFN-γ gene. Eur J Immunogenet. 1999;26:1–3. doi: 10.1046/j.1365-2370.1999.00122.x. [DOI] [PubMed] [Google Scholar]
- Pravica V, Perrey C, Stevens A, Lee JH, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human IFN-γ gene: absolute correlation with a polymorphic CA microsatellite marker of high IFN-γ production. Hum Immunol. 2000;61:863–6. doi: 10.1016/s0198-8859(00)00167-1. [DOI] [PubMed] [Google Scholar]
- Bream JH, Ping A, Zhang X, Winkler C, Young HA. A single nucleotide polymorphism in the proximal IFN-γ promoter alters control of gene transcription. Genes Immun. 2002;3:165–9. doi: 10.1038/sj.gene.6363870. [DOI] [PubMed] [Google Scholar]
- Gonsky R, Deem RL, Bream JH, Young HA, Targan SR. An IFNG SNP with an estrogen-like response element selectively enhances promoter expression in peripheral but not lamina propria T cells. Genes Immun. 2006;7:342–51. doi: 10.1038/sj.gene.6364305. [DOI] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;76:7.20.1–7.20.41. doi: 10.1002/0471142905.hg0720s76. Chapter 7: Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression – belts, braces, and chromatin. Cell. 1999;99:451–4. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–8. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- von Zeidler SV, Miracca EC, Nagai MA, Birman EG. Hypermethylation of the p16 gene in normal oral mucosa of smokers. Int J Mol Med. 2004;14:807–11. doi: 10.3892/ijmm.14.5.807. [DOI] [PubMed] [Google Scholar]
- Bonilla V, Sobrino F, Lucas M, Pintado E. Epstein–Barr virus transformation of human lymphoblastoid cells from patients with fragile X syndrome induces variable changes on CGG repeats size and promoter methylation. Mol Diagn. 2003;7:163–7. doi: 10.1007/BF03260033. [DOI] [PubMed] [Google Scholar]
- Mikovits JA, Young HA, Vertino P, et al. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the γ interferon (IFN-γ) promoter and subsequent downregulation of IFN-γ production. Mol Cell Biol. 1998;18:5166–77. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Bressler J, Beaudet AL. Epigenetics and human disease. Annu Rev Genomics Hum Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- Poulsen P, Esteller M, Vaag A, Fraga MF. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007;61:38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–40. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- Lund G, Andersson L, Lauria M, et al. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147–54. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- Im GI, Choi YJ. Epigenetics in osteoarthritis and its implication for future therapeutics. Expert Opin Biol Ther. 2013;13:713–21. doi: 10.1517/14712598.2013.764410. [DOI] [PubMed] [Google Scholar]
- Aune TM, Collins PL, Collier SP, Henderson MA, Chang S. Epigenetic activation and silencing of the gene that encodes IFN-γ. Front Immunol. 2013;4:112. doi: 10.3389/fimmu.2013.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-γ loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-γ. Nat Immunol. 2007;8:732–42. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci USA. 2005;102:17095–100. doi: 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsky R, Deem RL, Landers CJ, Derkowski CA, Berel D, McGovern DP, Targan SR. Distinct IFNG methylation in a subset of ulcerative colitis patients based on reactivity to microbial antigens. Inflamm Bowel Dis. 2011;17:171–8. doi: 10.1002/ibd.21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Sartori S, Tarantini L, Motta V, Rota F, Costa G. Epigenetic effects of shiftwork on blood DNA methylation. Chronobiol Int. 2010;27:1093–104. doi: 10.3109/07420528.2010.490065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RM, Suarez-Alvarez B, Salvanes R, et al. DNA methylation dynamics in blood after hematopoietic cell transplant. PLoS ONE. 2013;8:e56931. doi: 10.1371/journal.pone.0056931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang B, Bernstein DI, Lummus ZL, Ying J, Boulet LP, Cartier A, Gautrin D, Ho SM. Interferon-γ promoter is hypermethylated in blood DNA from workers with confirmed diisocyanate asthma. Toxicol Sci. 2013;133:218–24. doi: 10.1093/toxsci/kft079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeeff SE, Baccarelli AA, Halonen J, et al. Association between blood pressure and DNA methylation of retrotransposons and pro-inflammatory genes. Int J Epidemiol. 2013;42:270–80. doi: 10.1093/ije/dys220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Zhao R, Zheng S. Changes in epigenetic regulation of CD4+ T lymphocytesin biliary atresia. Pediatr Res. 2011;70:555–9. doi: 10.1203/PDR.0b013e318232a949. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Eauclaire SF, Mugnier M, et al. DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology. 2011;53:905–14. doi: 10.1002/hep.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuillermin PJ, Ponsonby AL, Saffery R, Tang ML, Ellis JA, Sly P, Holt P. Microbial exposure, interferon γ gene demethylation in naive T-cells, and the risk of allergic disease. Allergy. 2009;64:348–53. doi: 10.1111/j.1398-9995.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- Runyon RS, Cachola LM, Rajeshuni N, et al. Asthma discordance in twins is linked to epigenetic modifications of T cells. PLoS ONE. 2012;7:e48796. doi: 10.1371/journal.pone.0048796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Crivello A, Offenbacher S, Moretti A, Paquette DW, Barros SP. Interferon-γ promoter hypomethylation and increased expression in chronic periodontitis. J Clin Periodontol. 2010;37:953–61. doi: 10.1111/j.1600-051X.2010.01616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso FP, Viana MB, Sobrinho AP, Diniz MG, Brito JA, Gomes CC, Moreira PR, Gomez RS. Methylation pattern of the IFN-γ gene in human dental pulp. J Endod. 2010;36:642–6. doi: 10.1016/j.joen.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Sauerborn M, van de Vosse E, Delawi D, van Dissel JT, Brinks V, Schellekens H. Natural antibodies against bone morphogenic proteins and interferons in healthy donors and in patients with infections linked to type-1 cytokine responses. J Interferon Cytokine Res. 2011;31:661–9. doi: 10.1089/jir.2010.0075. [DOI] [PubMed] [Google Scholar]
- Chi CY, Chu CC, Liu JP, et al. Anti-IFN-γ autoantibodies in adults with disseminated nontuberculous mycobacterial infections are associated with HLA-DRB1*16:02 and HLA-DQB1*05:02 and the reactivation of latent varicella-zoster virus infection. Blood. 2013;121:1357–66. doi: 10.1182/blood-2012-08-452482. [DOI] [PubMed] [Google Scholar]
- Browne SK. Anticytokine autoantibody-associated immunodeficiency. Annu Rev Immunol. 2014;32:635–57. doi: 10.1146/annurev-immunol-032713-120222. [DOI] [PubMed] [Google Scholar]
- Kampitak T, Suwanpimolkul G, Browne S, Suankratay C. Anti-interferon-γ autoantibody and opportunistic infections: case series and review of the literature. Infection. 2011;39:65–71. doi: 10.1007/s15010-010-0067-3. [DOI] [PubMed] [Google Scholar]
- Chan JF, Trendell-Smith NJ, Chan JC, Hung IF, Tang BS, Cheng VC, Yeung CK, Yuen KY. Reactive and infective dermatoses associated with adult-onset immunodeficiency due to anti-interferon-γ autoantibody: Sweet’s syndrome and beyond. Dermatology. 2013;226:157–66. doi: 10.1159/000347112. [DOI] [PubMed] [Google Scholar]
- Browne SK, Zaman R, Sampaio EP, et al. Anti-CD20 (rituximab) therapy for anti-IFN-γ autoantibody-associated nontuberculous mycobacterial infection. Blood. 2012;119:3933–9. doi: 10.1182/blood-2011-12-395707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HW, Hur M. Interferon-γ release assays for the diagnosis of latent tuberculosis infection: an updated review. Ann Clin Lab Sci. 2013;43:221–9. [PubMed] [Google Scholar]
- de Oliveira LR, Peresi E, Golim MA, et al. Analysis of toll-like receptors, iNOS and cytokine profiles in patients with pulmonary tuberculosis during anti-tuberculosis treatment. PLoS ONE. 2014;9:e88572. doi: 10.1371/journal.pone.0088572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XF, Yang ZW, Li J. Adjunctive therapy with interferon-γ for the treatment of pulmonary tuberculosis: a systematic review. Int J Infect Dis. 2011;15:e594–600. doi: 10.1016/j.ijid.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Sahrbacher U, Naumann L, Reischl U, Scholmerich J, Gluck T. Reduced TH-1 cytokine release in an adult patient with chronic relapsing Mycobacterium malmoense infection. Infection. 2007;35:282–6. doi: 10.1007/s15010-007-4101-z. [DOI] [PubMed] [Google Scholar]
- Vankayalapati R, Wizel B, Samten B, et al. Cytokine profiles in immunocompetent persons infected with Mycobacterium avium complex. J Infect Dis. 2001;183:478–84. doi: 10.1086/318087. [DOI] [PubMed] [Google Scholar]
- Storgaard M, Varming K, Herlin T, Obel N. Novel mutation in the interferon-γ-receptor gene and susceptibility to mycobacterial infections. Scand J Immunol. 2006;64:137–9. doi: 10.1111/j.1365-3083.2006.01775.x. [DOI] [PubMed] [Google Scholar]
- Chen SC, Korman TM, Slavin MA, et al. Antifungal therapy and management of complications of cryptococcosis due to Cryptococcus gattii. Clin Infect Dis. 2013;57:543–51. doi: 10.1093/cid/cit341. [DOI] [PubMed] [Google Scholar]
- Harris JR, Lindsley MD, Henchaichon S, et al. High prevalence of cryptococcal infection among HIV-infected patients hospitalized with pneumonia in Thailand. Clin Infect Dis. 2012;54:e43–50. doi: 10.1093/cid/cir903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis JN, Casazza JP, Stone HH, Meintjes G, Lawn SD, Levitz SM, Harrison TS, Koup RA. The phenotype of the Cryptococcus-specific CD4+ memory T-cell response is associated with disease severity and outcome in HIV-associated cryptococcal meningitis. J Infect Dis. 2013;207:1817–28. doi: 10.1093/infdis/jit099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DW, Riniotis K, Dobrashian R, Sambatakou H. Chronic cavitary and fibrosing pulmonary and pleural aspergillosis: case series, proposed nomenclature change, and review. Clin Infect Dis. 2003;37:S265–80. doi: 10.1086/376526. [DOI] [PubMed] [Google Scholar]
- Patterson TF, Kirkpatrick WR, White M, et al. Invasive aspergillosis. Disease spectrum, treatment practices, and outcomes. I3 Aspergillus Study Group. Medicine (Baltimore) 2000;79:250–60. doi: 10.1097/00005792-200007000-00006. [DOI] [PubMed] [Google Scholar]
- de Boer MG, Jolink H, Halkes CJ, van der Heiden PL, Kremer D, Falkenburg JH, van de Vosse E, van Dissel JT. Influence of polymorphisms in innate immunity genes on susceptibility to invasive aspergillosis after stem cell transplantation. PLoS ONE. 2011;6:e18403. doi: 10.1371/journal.pone.0018403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shair K, Atherton GT, Harris C, Ratcliffe L, Newton PJ, Denning DW. Long-term antifungal treatment improves health status in patients with chronic pulmonary aspergillosis; a longitudinal analysis. Clin Infect Dis. 2013;57:828–35. doi: 10.1093/cid/cit411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HS, Jeon K, Um SW, Suh GY, Chung MP, Kim H, Kwon OJ, Koh WJ. Clinical characteristics and treatment outcomes of chronic necrotizing pulmonary aspergillosis: a review of 43 cases. Int J Infect Dis. 2010;14:e479–82. doi: 10.1016/j.ijid.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Felton TW, Baxter C, Moore CB, Roberts SA, Hope WW, Denning DW. Efficacy and safety of posaconazole for chronic pulmonary aspergillosis. Clin Infect Dis. 2010;51:1383–91. doi: 10.1086/657306. [DOI] [PubMed] [Google Scholar]
- Smith NL, Denning DW. Underlying conditions in chronic pulmonary aspergillosis including simple aspergilloma. Eur Respir J. 2011;37:865–72. doi: 10.1183/09031936.00054810. [DOI] [PubMed] [Google Scholar]
- Smith NL, Hankinson J, Simpson A, Bowyer P, Denning DW. A prominent role for the IL1 pathway and IL15 in susceptibility to chronic cavitary pulmonary aspergillosis. Clin Microbiol Infect. 2014;20:O480–8. doi: 10.1111/1469-0691.12473. [DOI] [PubMed] [Google Scholar]
- Murdock BJ, Shreiner AB, McDonald RA, Osterholzer JJ, White ES, Toews GB, Huffnagle GB. Coevolution of TH1, TH2, and TH17 responses during repeated pulmonary exposure to Aspergillus fumigatus conidia. Infect Immun. 2011;79:125–35. doi: 10.1128/IAI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersuk GM, Underhill DM, Zhu L, Marr KA. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J Immunol. 2006;176:3717–24. doi: 10.4049/jimmunol.176.6.3717. [DOI] [PubMed] [Google Scholar]
- Lass-Florl C, Roilides E, Loffler J, Wilflingseder D, Romani L. Minireview: host defence in invasive aspergillosis. Mycoses. 2013;56:403–13. doi: 10.1111/myc.12052. [DOI] [PubMed] [Google Scholar]
- Armstrong-James DP, Turnbull SA, Teo I, Stark J, Rogers NJ, Rogers TR, Bignell E, Haynes K. Impaired interferon-γ responses, increased interleukin-17 expression, and a tumor necrosis factor-α transcriptional program in invasive aspergillosis. J Infect Dis. 2009;200:1341–51. doi: 10.1086/605931. [DOI] [PubMed] [Google Scholar]
- Kelleher P, Goodsall A, Mulgirigama A, Kunst H, Henderson DC, Wilson R, Newman-Taylor A, Levin M. Interferon-γ therapy in two patients with progressive chronic pulmonary aspergillosis. Eur Respir J. 2006;27:1307–10. doi: 10.1183/09031936.06.00021705. [DOI] [PubMed] [Google Scholar]
- Döffinger R, Harris C, Lear S, Barcenas-Morales G, Newton P, Alachkar H, Kumararatne DS, Denning DW. 2012. Reduced IFN-g and IL-17 production in chronic pulmonary aspergillosis (CPA) Poster presented at 5th Advances Against Aspergillosis, Istanbul, Turkey.
- Armstrong-James D, Teo IA, Shrivastava S, Petrou MA, Taube D, Dorling A, Shaunak S. Exogenous interferon-γ immunotherapy for invasive fungal infections in kidney transplant patients. Am J Transplant. 2010;10:1796–803. doi: 10.1111/j.1600-6143.2010.03094.x. [DOI] [PubMed] [Google Scholar]
- The International Chronic Granulomatous Disease Cooperative Study Group. A controlled trial of interferon γ to prevent infection in chronic granulomatous disease. N Engl J Med. 1991;324:509–16. doi: 10.1056/NEJM199102213240801. [DOI] [PubMed] [Google Scholar]
- Riddell LA, Pinching AJ, Hill S, et al. A phase III study of recombinant human interferon γ to prevent opportunistic infections in advanced HIV disease. AIDS Res Hum Retroviruses. 2001;17:789–97. doi: 10.1089/088922201750251981. [DOI] [PubMed] [Google Scholar]
- Pappas PG, Bustamante B, Ticona E, et al. Recombinant interferon- γ 1b as adjunctive therapy for AIDS-related acute cryptococcal meningitis. J Infect Dis. 2004;189:2185–91. doi: 10.1086/420829. [DOI] [PubMed] [Google Scholar]
- Jarvis JN, Meintjes G, Rebe K, et al. Adjunctive interferon-γ immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. AIDS. 2012;26:1105–13. doi: 10.1097/QAD.0b013e3283536a93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson R, Condos R, Tse D, et al. Immunomodulation with recombinant interferon-γ1b in pulmonary tuberculosis. PLoS ONE. 2009;4:e6984. doi: 10.1371/journal.pone.0006984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar A, Rodriguez G, Rolston KV, et al. High-dose caspofungin combination antifungal therapy in patients with hematologic malignancies and hematopoietic stem cell transplantation. Bone Marrow Transplant. 2007;39:157–64. doi: 10.1038/sj.bmt.1705559. [DOI] [PubMed] [Google Scholar]
- Safdar A, Rodriguez G, Ohmagari N, Kontoyiannis DP, Rolston KV, Raad II, Champlin RE. The safety of interferon-γ-1b therapy for invasive fungal infections after hematopoietic stem cell transplantation. Cancer. 2005;103:731–9. doi: 10.1002/cncr.20883. [DOI] [PubMed] [Google Scholar]
- Armstrong-James D, Teo I, Herbst S, et al. Renal allograft recipients fail to increase interferon-γ during invasive fungal diseases. Am J Transplant. 2012;12:3437–40. doi: 10.1111/j.1600-6143.2012.04254.x. [DOI] [PubMed] [Google Scholar]
- King TE, Jr, Albera C, Bradford WZ, et al. Effect of interferon γ-1b on survival in patients with idiopathic pulmonary fibrosis (INSPIRE): a multicentre, randomised, placebo-controlled trial. Lancet. 2009;374:222–8. doi: 10.1016/S0140-6736(09)60551-1. [DOI] [PubMed] [Google Scholar]
- Lau JY, Lai CL, Wu PC, Chung HT, Lok AS, Lin HJ. A randomised controlled trial of recombinant interferon-γ in Chinese patients with chronic hepatitis B virus infection. J Med Virol. 1991;34:184–7. doi: 10.1002/jmv.1890340310. [DOI] [PubMed] [Google Scholar]
- Moss RB, Mayer-Hamblett N, Wagener J, et al. Randomized, double-blind, placebo-controlled, dose-escalating study of aerosolized interferon γ-1b in patients with mild to moderate cystic fibrosis lung disease. Pediatr Pulmonol. 2005;39:209–18. doi: 10.1002/ppul.20152. [DOI] [PubMed] [Google Scholar]