Abstract
Interleukin-19 (IL-19) plays an important role in asthma by stimulating T helper type 2 (Th2) cytokine production. Interestingly, IL-4, a key Th2 cytokine, in turn up-regulates IL-19 expression in bronchial epithelial cells, so forming a positive feedback loop. In atopic dermatitis (AD), another Th2 disease closely related to asthma, IL-19 is up-regulated in the skin. We propose to use IL-4 transgenic (Tg) mice and human keratinocyte culture to delineate the molecular mechanisms involved in the up-regulation of IL-19 in AD. IL-19 is similarly up-regulated in the skin of IL-4 Tg mice as in human AD. Next we show that IL-4 up-regulates IL-19 expression in keratinocytes. Interestingly, the up-regulation was suppressed by a pan-Janus kinase (Jak) inhibitor, suggesting that the Jak–signal transducer and activator of transcription (Jak-STAT) pathway may be involved. Dominant negative studies further indicate that STAT6, but not other STATs, mediates the up-regulation. Serial 5′ deletion of the IL-19 promoter and mutagenesis studies demonstrate that IL-4 up-regulation of IL-19 in keratinocytes involves two imperfect STAT6 response elements. Finally, chromatin immunoprecipitation assay studies indicate that IL-4 increases the binding of STAT6 to its response elements in the IL-19 promoter. Taken together, we delineate the detailed molecular pathway for IL-4 up-regulation of IL-19 in keratinocytes, which may play an important role in AD pathogenesis.
Keywords: atopic dermatitis, interleukin-4, interleukin-19, keratinocyte, signal transducer and activator of transcription 6, signal transducer and activator of transcription 6 response elements
Introduction
Atopic dermatitis (AD), a chronic inflammatory skin disease, affects 10–20% of children and 1–3% of adults in developed countries.1,2 In addition, many of the AD patients also suffer from other atopic disorders, such as asthma and allergic rhinitis.3–5
The aetiology and pathogenesis of AD are not fully understood. It is generally accepted that the dysregulation of the immune response and disruption of the skin barrier function may contribute to the development and progression of this disease.6–8 We and others have shown that interleukin-4 (IL-4) plays an essential role in the pathophysiology of AD.9–12 Evidence supporting IL-4 as a key factor for AD includes (i) IL-4 is significantly up-regulated in the skin of AD patients; (ii) IL-4 is the key cytokine for the production of IgE, which is significantly up-regulated in the serum of AD patients; (iii) over-expressing IL-4 in the basal epidermis of IL-4 transgenic (Tg) mice resulted in chronic pruritic inflammatory skin lesions, which meet the clinical and histological diagnostic criteria for human AD;13,14 (iv) the clinical relevance of IL-4 in IL-4 Tg mice and human AD is validated by a recent clinical trial of an antibody to IL-4 receptor α, which provides statistically significant improvement in disease severity, per cent body involvement, and pruritus in patients affected by moderate to severe AD in only 4 weeks.15
Interleukin-19, a pro-inflammatory cytokine known to stimulate the production of T helper type 2 (Th2) cytokines,16–20 plays an essential role in the pathogenesis of asthma,21–24 another Th2 disease closely related to AD. Importantly, it was reported that IL-19 is dramatically up-regulated in the skin of AD patients.25 Recently it was reported that keratinocyte-specific knockout of SOCS3 resulted in severe skin inflammation, up-regulation of IL-19 in the skin, and high serum IgE levels.26 In addition, we found that IL-19 is among the few genes most significantly up-regulated by IL-4 in keratinocytes by screening hundreds of genes using PCR array.27 These findings led us to hypothesize that IL-4 from activated immune cells up-regulates IL-19 expression in keratinocytes, which in turn may up-regulate Th2 cytokines, so forming a positive feedback loop and finally leading to the development and exacerbation of AD.
Using IL-4 Tg mice that over-express IL-4 exclusively in the epidermis, we have demonstrated that IL-19 mRNA and protein levels are up-regulated in the skin of these mice as compared with wild-type mice. We have also established that IL-4 up-regulation of IL-19 in keratinocytes is at the transcriptional level and involves signal transducer and activator of transcription 6 (STAT6). Previously, Huang et al.22 demonstrated that IL-4 up-regulation of IL-19 expression in human bronchial epithelial cells involves a distal perfect STAT6 response element, whereas our data show clearly that in skin epidermal keratinocytes, such up-regulation involves the binding of STAT6 to different STAT6 sites.
Materials and methods
Materials
Interleukin-4, PCR SuperMix High Fidelity, EpiLife® Medium, Human Keratinocyte Growth Supplement, Dulbecco’s modified Eagle’s medium (DMEM) with or without phenol red, fetal bovine serum (FBS), charcoal-stripped FBS, penicillin/streptomycin, non-essential amino acids, sodium pyruvate, reverse transcriptase, Alexa Fluor 594 secondary antibody and Trizol reagent were obtained from Invitrogen (Carlsbad, CA); IL-19 antibodies (anti-human and anti-mouse) were bought from R&D Systems (Minneapolis, MN); SYBR Green PCR Master Mix and Effectene Transfection Reagent were from QIAGEN Inc. (Valencia, CA); trypsin-EDTA was purchased from Mediatech (Herndon, VA); the human IL-19 ELISA kit was from Assay Biotechnology (Sunnyvale, CA); KpnI, HindIII, T4 polynucleotide kinase, and T4 DNA ligase were bought from Fermentas (Pittsburgh, PA); Passive Lysis 5× Buffer, Dual-Luciferase Reporter Assay were obtained from Promega (Madison, WI); Janus kinase (Jak) inhibitor I was purchased from Calbiochem (La Jolla, CA). The DN-STAT6 was kindly given to us by Dr Steven M. Dubinett (University of California at Los Angeles, Los Angeles, CA);28 the DN-STAT3 is from Dr Toshio Hirano (Osaka University, Osaka, Japan);29 the DN-STAT5a was kindly provided by Dr Alice Mui (University of British Columbia, British Columbia, Canada);30 and the DN-STAT5b is a gift from Dr Li-yuan (Baylor College of Medicine, Houston, TX).31
Animals
The IL-4 Tg mice and wild-type mice (on C57BL/6 background) were kept at 25° with a 14-hr light/10-hr dark cycle. All experimental procedures were performed in accordance with the principles of the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Illinois at Chicago. Around 80–90 days after birth, skin lesions and normal skin tissues were collected from the ears of the IL-4 Tg mice and wild-type mice.
Cell culture
Primary human keratinocytes (Invitrogen) were cultured in EpiLife® Medium supplemented with Human Keratinocyte Growth Supplement (low calcium). Then the cells were switched to DMEM supplemented with 1% charcoal-dextran-treated FBS for 3 days to differentiate (high calcium). Immortalized human keratinocytes, HaCat cells (from Dr Lin Chen), were grown in DMEM supplemented with FBS (10%), non-essential amino acids, and penicillin/streptomycin. The cells were incubated in a humidified atmosphere of 5% CO2 at 37°. Fresh culture media were replaced every 48 h. Cells were treated with various concentrations of IL-4 in DMEM supplemented with 1% charcoal-dextran-treated FBS for 24 hr. Next, cells were washed twice with ice-cold PBS and were frozen at −80° until assays were performed.
RNA isolation and real-time RT-PCR analysis
Total RNA from cell culture was isolated using Trizol reagent according to the manufacturer’s instructions. The RT and real-time PCRs were performed as previously described.32 Briefly 1 μg of total RNA was reverse-transcribed and the final volume was 100 μl. Five microlitres and 10 μl of cDNA were used for GAPDH and IL-19, respectively. For each real-time PCR (25 μl total volume), cDNA was mixed with 2× SYBR Green PCR Master Mix containing the following primers. The mouse IL-19 primers used were 5′-ATC CTG TCC CTG GAG AAC CT-3′ and 5′-CTC TCC TGA TGG TCC TGG AA-3′; the mouse GAPDH primers used were 5′-AGA ACA TCA TCC CTG CAT CC-3′ and 5′-ACA CAT TGG GGG TAG GAA CA-3′; the human IL-19 primers used were 5′-AAG GAT CAT CAG GAG CCA AA-3′ and 5′-CAC TGC CTC TGT TCC TGA CA-3′; the human GAPDH primers used were 5′-ACA CCC ACT CCT CCA CCT TT-3′ and 5′-TGC TGT AGC CAA ATT CGT TG-3′. The PCR were carried out in duplicate, and the samples were analysed on a Stratagene Mx3000 real-time PCR machine (Santa Clara, CA).
Immunofluorescence confocal microscopy
Immunofluorescence microscopy on tissues was performed as described previously.33 Briefly, mouse ear tissues were fixed in 4% paraformaldehyde, followed by immersion overnight in 30% sucrose solution, and then embedded in OCT compound (Fisher Scientific, Pittsburgh, PA). Cryosections were incubated overnight at 4° with diluted anti-IL-19 antibody, followed by incubation with the Alexa Fluor-594 secondary antibody at room temperature for 1–2 hr (Invitrogen).
Immunofluorescence microscopy on cell cultures was performed as described previously.34 Briefly, HaCat cells were treated with either IL-4 (10 ng/ml) or vehicle in 1% charcoal-dextran-treated FBS medium on DB chamber slides (BD Biosciences, Bedford, MA) for 24 h. Next the cells were fixed with 4% paraformaldehyde solution for 20 min and permeabilized in PBS containing 0·1% Triton X-100 and 0·2% Tween-20 for 30 min at room temperature. The cells were incubated with anti-IL-19 overnight at 4° followed by incubation with an Alexa Fluor-594 secondary antibody at room temperature for 1–2 hr (Invitrogen).
The slides were mounted in Vectashield medium (Vector Laboratories, Inc., Burlingame, CA) containing a counterstain for DNA 4′,6-diamino-2-phenylindole and were examined using a Zeiss LSM 510 META confocal microscope (Oberkochen, Germany) equipped with a 63× water immersion objective lens.
ELISA
ELISA was performed according to the manufacturer’s instructions. Briefly 12-well plates were used. When cells are 60–80% confluent, they were treated with IL-4 (10 ng/ml) for 24 hr in 1 ml of DMEM (1% charcoal-stripped FBS). Then, 100 μl of cell medium samples and standards were loaded into each well of the 96-well plate, followed by primary antibody incubation, serial washes and avidin–horseradish peroxidase-linked secondary antibody. The reaction was developed with 3,3′,5,5′-tetramethylbenzidine and the absorbance was read at 450 nm on a Synergy HT Multi-Mode microplate reader (BioTek, Winooski, VT).
Generation of the IL-19 promoter–luciferase constructs
Different lengths of IL-19 promoter–luciferase reporters were generated using the sticky end PCR cloning technique.35 The human genomic BAC clone (BACPAC Resource Center Oakland, California; clone ID: RP11-237C22) contains the IL-19 promoter sequence described previously.36 The fragments were cloned into the KpnI and HindIII sites of the pGL3-Enhancer Vector (Promega, Madison, WI). The primers used in the subcloning are listed in Table 1.
Table 1.
Primers used in the generation of the IL-19 promoter-luciferase constructs
| Primer name | Sequence |
|---|---|
| −2463 bp to +21 bp blunt end sense | 5′-GGA CAG GAT TTG AGG AGC TG-3′ |
| −2463 bp to +21 bp sticky end sense | 5′-GTA CGG ACA GGA TTT GAG GAG CTG-3′ |
| −2210 bp to +21 bp blunt end sense | 5′-ACC CAT GGG AGA AGC TCT TT-3′ |
| −2210 bp to +21 bp sticky end sense | 5′-GTA CAC CCA TGG GAG AAG CTC TTT-3′ |
| −986 bp to +21 bp blunt end sense | 5′-TGG TAG CTC CAA GTC TCA TCT TC-3′ |
| −986 bp to +21 bp sticky end sense | 5′-GTA CTG GTA GCT CCA AGT CTC ATC TTC-3′ |
| −582 bp to +21 bp blunt end sense | 5′-TAG AGC ACC AAC AGC CCC TA-3′ |
| −582 bp to +21 bp sticky end sense | 5′-GTA CTA GAG CAC CAA CAG CCC CTA-3′ |
| −247 bp to +21 bp blunt end sense | 5′-TTC TCC CAG AGG CAC AGT AAA-3′ |
| −247 bp to +21 bp sticky end sense | 5′-GTA CTT CTC CCA GAG GCA CAG TAA A-3′ |
| IL-19 promoter common blunt end antisense | 5′-AAT TTC ACC ATT GCA CTC CAG-3′ |
| IL-19 promoter common sticky end antisense | 5′-AGC TAA TTT CAC CAT TGC ACT CCA G-3′ |
Mutation of the IL-19 promoter
Mutation of the IL-19 promoter was performed using the QuikChange II site-directed mutagenesis kits (Stratagene) according to the manufacturer’s protocol. The –582 bp promoter was used as the template. Mutation was confirmed through DNA sequencing. The primers used to mutate the distal STAT site are 5′-GCC TAC TCT CTA ATG GAG TAG TGG AAA ATC CAT TTG CTT CTT TGT CTG TG-3′ and 5′-CAC AGA CAA AGA AGC AAA TGG ATT TTC CAC TAC TCC ATT AGA GAG TAG GC-3′; The primers used to mutate the proximal STAT site are 5′-GGG TCT AGA TTG GGA TCC TCT TGG ATG TAC CCA TCT TGA GTA TGT GAC CAT G-3′ and 5′-CAT GGT CAC ATA CTC AAG ATG GGT ACA TCC AAG AGG ATC CCA ATC TAG ACC C-3′.
Transfection of HaCat cells
Effectene transfection was performed according to the manufacturer’s instructions. Briefly, HaCat cells were seeded in 12-well plates and cultured until 60–80% confluency. Equal amounts of DNA were transfected per well. Eighteen to 19 h after transfection, the cells were treated with IL-4 (10 ng/ml) for another 24 hr. Transient expression of the reporter gene was quantified by a standard luciferase assay and was normalized against renilla luciferase. Luciferase activities were measured using a luminometer (Lumat LB 9507 luminometer; EG&G Berthold, Oak Ridge, TN).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed using a Sigma (St Louis, MO) Imprint® ChIP kit according to the manufacturer’s instructions. Anti-STAT6 (Millipore, Temecula, CA) was incubated with the samples at room temperature for 90 min on an orbital shaker. After cross-link-reversal and DNA purification, real-time PCR was carried out on the samples and inputs. The primers used are 5′-GCA CAT CTG CAC ATT CCA AG-3′ and 5′-GGG TTC TTA GAT CTC ACA CAG GA-3′. PCR products were purified and sequenced to confirm specificity.
Statistical analysis
Data were examined by the Student’s t-test (Figs 1, 2b and 5), one-way analysis of variance followed by the Tukey test (Figs 2a and 3a,b), or two-way analysis of variance followed by Bonferroni post-tests (Figs 3c and 4) using prism software (GraphPad Software, Inc., San Diego, CA). Values were considered statistically significant at P < 0·05.
Figure 1.
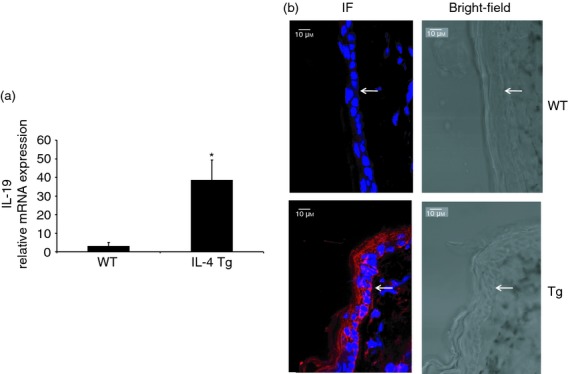
Interleukin-19 (IL-19) expression is up-regulated in the skin of IL-4 transgenic (Tg) mice. IL-19 expression was compared between wild-type mice and IL-4 Tg mice by real-time RT-PCR (a) and confocal microscopy (b). (a) Mouse GAPDH was used as the housekeeping gene, and values are expressed as the mean ± SEM (n = 5). *P < 0·05 versus wild-type (WT). (b) IL-19, red; nucleus, blue. Arrow denotes the dermal–epidermal border. Experiments were repeated three times.
Figure 2.
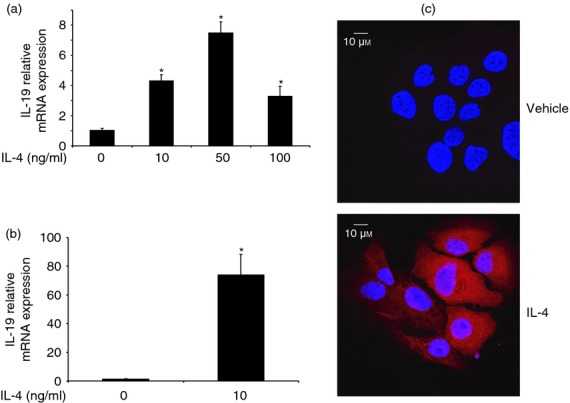
Interleukin-4 (IL-4) up-regulates the expression of IL-19 in primary keratinocytes and HaCat cells. (a) Primary keratinocytes were treated with different concentrations of IL-4 for 24 hr, and IL-19 expression was analysed by real-time RT-PCR. Human GAPDH was used as the housekeeping gene, and values are expressed as the mean ± SEM (n = 3). *P < 0·05 versus 0 ng/ml. HaCat cells were treated with IL-4 (10 ng/ml) for 24 hr and IL-19 expression was analysed by real-time RT-PCR (b). Values are expressed as the mean ± SEM (n = 4). *P < 0·05 versus 0 ng/ml. (c) IL-19, red; nucleus, blue; experiments were repeated three times.
Figure 5.
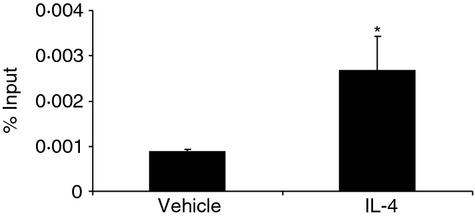
Interleukin-4 (IL-4) increases the binding of signal transducer and activator of transcription 6 (STAT6) to the IL-19 promoter. HaCat cells were treated with IL-4 (10 ng/ml) or vehicle for 24 hr. ChIP assay was performed to study the binding of STAT6 to the IL-19 promoter using an anti-STAT6 antibody. Data were quantified by real-time PCR and PCR products were sequenced. Values are expressed as the mean ± SEM (n = 3). *P < 0·05 versus vehicle.
Figure 3.
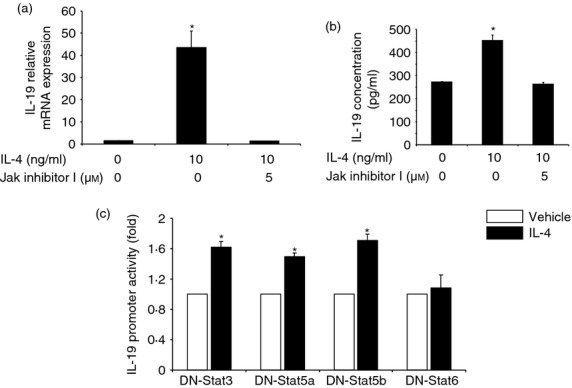
The Janus kinase–signal transducer and activator of transcription 6 (Jaks-STAT6) pathway is involved in interleukin-4 (IL-4) up-regulation of IL-19 in HaCat cells. (a) HaCat cells were treated with IL-4 and Jak inhibitor I (a pan-Jak inhibitor) for 24 hr and were subjected to real-time RT-PCR analysis (a) and ELISA (b). Values are expressed as the mean ± SEM (n = 4). *P < 0·05 versus control. (c) HaCat cells were co-transfected with an IL-19 promoter and with either dominant negative (DN)-STAT3, DN-STAT5a, DN-STAT5b or DN-STAT6 for 18 hr. The cells were then treated with either IL-4 or vehicle for another 24 hr. Promoter activity was quantified by a standard luciferase assay and was normalized against renilla luciferase. The experiment was repeated three times. Values are expressed as the mean ± SEM. *P < 0·05 versus vehicle.
Figure 4.
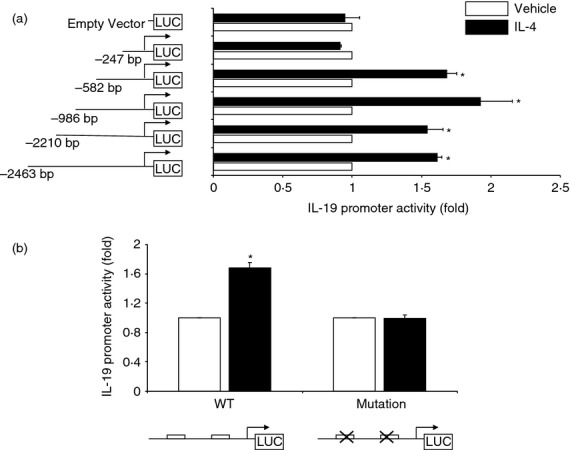
Localization of the interleukin-4 (IL-4) response elements in the IL-19 promoter. (a), HaCat cells were transfected with an equal amount of different IL-19 promoter reporter constructs. Eighteen hours after transfection, the cells were treated with IL-4 (10 ng/ml) for another 24 hr. The experiment was repeated six times. Values are expressed as the mean ± SEM. *P < 0·05 versus vehicle. (b), A 603 bp human IL-19 promoter was mutated at both of the two putative signal transducer and activator of transcription 6 (STAT6) sites. HaCat cells were transfected with an equal amount of the promoter reporter constructs. The experiment was repeated six times. Values are expressed as the mean ± SEM. *P < 0·05 versus vehicle.
Results
IL-19 is up-regulated in the IL-4 Tg mice
Interleukin-19 has been reported to be up-regulated in the skin of AD patients.25 To examine whether it is also up-regulated in the IL-4 Tg mice, an AD mouse model, we used real-time RT-PCR and immunofluorescence to study IL-19 expression in the skin of both wild-type and IL-4 Tg mice. As shown in Fig. 1, IL-19 is up-regulated at both the mRNA and protein levels in the IL-4 Tg mice compared with the wild-type mice, which is consistent with the findings documented in human AD.
IL-4 stimulates IL-19 expression in keratinocytes
Since IL-19 is up-regulated in the skin of human AD and IL-4 Tg mice, we asked the question of whether the high levels of IL-19 are directly caused by the over-expression of IL-4. Towards that end, we used primary human keratinocytes and a human keratinocyte cell line, HaCat cells. The cells were treated with IL-4 for 24 hr, and IL-19 expression was examined by real-time RT-PCR. Indeed, IL-4 stimulates IL-19 expression in both the primary keratinocytes and HaCat cells (Fig. 2)27 although we noticed an exaggerated response of HaCat cells to IL-4 stimulation. Since primary keratinocytes are very refractory to transfection, we used HaCat cells in our study of the signal transduction pathway for IL-4 regulation of IL-19 in keratinocytes. Interleukin-19 protein expression is also up-regulated by IL-4 (Fig. 2c). The data clearly suggest that the high levels of IL-19 in the skin of human AD and IL-4 Tg mice are mediated by the over-expression of IL-4 in the skin.
The Jak-STAT6 pathway is involved in IL-4 up-regulation of IL-19 in HaCat cells
The Jak-STAT pathway is a major signal transduction pathway for IL-4 signalling in keratinocytes,9,37–40 so we first examined whether Jaks play any roles here. As shown in Fig. 3(a,b), Jak inhibitor I (a pan-Jak inhibitor), which inhibits all the Jak family members, completely abolished the up-regulation of IL-19 at the mRNA and protein levels, indicating that the Jak-STAT pathway may be involved in the regulation of IL-19 in human keratinocytes. With the understanding that Jaks are essential in IL-4 regulation of IL-19, we proceeded to the next logical step by examining whether STATs are also involved, and if so, which STAT plays the dominant role. Previously we reported that IL-4 stimulates the phosphorylation of STAT3 and STAT6 among STAT family members in keratinocytes.9 We co-transfected HaCat cells with an IL-19 promoter–luciferase construct and with dominant negative (DN)-STAT3, DN-STAT5a, DN-STAT5b or DN-STAT6. Eighteen to 19 hr after transfection, the cells were treated with IL-4 for another 24 hr and IL-19 promoter activity was examined by luminometry. As shown in Fig. 3(c), DN-STAT6, but not other DNs, completely eliminated IL-4 stimulation of IL-19 promoter activity, supporting the notion that IL-4 regulation of IL-19 is indeed through the Jak-STAT6 pathway.
Localization of the IL-4 responsive element in the IL-19 promoter
The final and most critical step for investigating IL-4 up-regulation of IL-19 expression is to locate the promoter area where IL-4 confers its activity. Towards that end, we generated different lengths of IL-19 promoters and transfected HaCat cells with these promoters. The cells were co-transfected with a renilla luciferase expression vector, allowing for normalization of transfection efficiency. As shown in Fig. 4(a), serial 5′ deletion of the IL-19 promoter revealed that the region between −582 and −247 bp upstream of the transcription start site is essential for IL-4 up-regulation of IL-19. Analysis of this region did not find any perfect STAT6 site (5′-TTC and GAA-3′ spaced by four nucleotides). However, we found two corresponding imperfect STAT6 sites (aaaTTTattgGAAactact and tcttTTCtggCAAcatctt). This finding is in sharp contrast to the up-regulation of IL-19 gene in human bronchial epithelial cells, where the IL-4 responsive site was reported to be between –2259 bp –and –2145 bp.22 It is well known that STAT proteins may bind to imperfect sites, especially when two imperfect STAT sites are adjacent.41–44 In addition, we previously demonstrated that STAT3 binds to two imperfect STAT sites in the promoter of the α2-macroglobulin gene.34 Although mutation of either of the two sites failed to generate conclusive data, mutation of the two elements simultaneously, as shown in Fig. 4(b), led to total elimination of IL-4 action, which indicates that the two imperfect STAT6 sites are essential for IL-4 up-regulation of IL-19 in keratinocytes.
To further validate whether IL-4 affects the activated STAT6 binding to this area (−582 to −247 bp), we used a monoclonal antibody to immunoprecipitate STAT6. The ChIP assay showed clearly that IL-4 increased STAT6 binding to the IL-19 promoter (Fig. 5). DNA sequencing of the PCR products confirmed that STAT6 physically binds to the area demonstrated by the serial 5′ deletion study.
Discussion
Interleukin-19, an IL-10 subfamily member, is expressed in myeloid cells, keratinocytes and bronchial epithelial cells.45–47 It signals through Jak1/Tyk2-STAT1/STAT3 and is usually associated with pro-inflammatory responses in innate immunity.45 Extensive research has shown that IL-19 plays an important role in the pathogenesis of asthma,21–24 probably through its ability to stimulate the production of Th2 cytokines.16–20 Interestingly, IL-19 is also up-regulated in the skin of patients with AD, another Th2 disease closely related to asthma.25
Previously, Huang et al.22 reported that IL-4 up-regulates IL-19 expression in airway epithelia through STAT6 binding to a perfect STAT6 site (TTC and GAA spaced by four nucleotides) −2259 bp in the IL-19 promoter based on the ChIP studies. We found that the STAT6 response elements involved in IL-4 regulation of IL-19 in keratinocytes are different from that reported in bronchial epithelial cells. It is possible that IL-19 regulation involves different cis-acting elements in different cell types. On the other hand, without additional functional studies, the data from the ChIP study alone cannot conclude that the perfect STAT6 response element is essential for IL-4 regulation of IL-19. In our study on keratinocytes, after removing the perfect STAT6 site from the IL-19 promoter, we still observed the stimulation of IL-19 by IL-4. The serial 5′ deletion study finally located an area between −582 and −247 bp, which appears to be essential for IL-4 up-regulation of IL-19 in keratinocytes. However, we did not find any perfect STAT6 site in this area. Although STAT6 prefers the classical perfect STAT6 site, it is known that STAT6 could also bind to other less than perfect sites.41–44 Carefully analysing this area, we found two imperfect STAT6 sites, mutation of either of which generated inconclusive results. However, after we mutated both of these sites, we found that IL-4 up-regulation of IL-19 is totally abolished, suggesting that two STAT6 homodimers may bind to the less than perfect STAT6 sites, forming a relatively stable protein–DNA complex, possibly interacting with other adjacent transcription factors and co-activators. Future studies may further investigate the relationship between these two imperfect STAT6 sites with the distal perfect STAT6 sites, which may play a less important role in the assembly of the general transcription machinery.
In addition to the STAT6 sites, the human IL-19 promoter also contains two nuclear factor-κB sites, two CAAT enhancer binding protein sites, and a total of seven putative keratinocyte growth factor binding motifs, which may link its role to skin diseases.17 Interleukin-17 signals through nuclear factor-κB to up-regulate IL-19 in human airway epithelial cells, synergistically enhancing IL-4 effects.22 Interestingly, IL-19 has been shown to play a role in psoriasis,48–50 but different from the Th2–IL-19 positive feedback loop hypothesis, the suggested mechanism involves crosstalk between IL-19 from keratinocytes and keratinocyte growth factor from CD8+ T cells.17,48
In conclusion, we have elucidated the detailed signal transduction pathway for IL-4 up-regulation of IL-19 in keratinocytes, which may play an important role in the pathogenesis of AD. We show that the IL-4 regulation of IL-19 in keratinocytes involves two imperfect STAT6 sites, indicating that imperfect STAT6 sites could play critical roles in gene regulation among several perfect potential STAT6 sites.
Acknowledgments
We would like to thank Dr Steven M. Dubinett for the DN-STAT6, Dr Toshio Hirano for the DN-STAT3, Dr Alice Mui for the DN-STAT5a, Dr Li-yuan Yu-Lee for the DN-STAT5b, and Dr. Ke Ma for technical support in confocal microscopy. This work is in part supported by the Albert H. and Mary Jane Slepyn Endowed Fellowship Fund (LB), the Lawrence S. Chan, MD Endowed Skin Research Fund (LB), and the Dr Orville J. Stone Endowed Professorship in Dermatology Fund (LSC).
Glossary
- AD
atopic dermatitis
- ChIP
chromatin immunoprecipitation
- DN
dominant negative
- FBS
fetal bovine serum
- IF
immunofluorescence
- Jak
Janus kinase
- Stat
signal transducer and activator of transcription
- TBS
tris-buffered saline
- Tg
transgenic
Disclosures
The authors declare that there are no conflicts of interest.
References
- Saito H. Much atopy about the skin: genome-wide molecular analysis of atopic eczema. Int Arch Allergy Immunol. 2005;137:319. doi: 10.1159/000086464. [DOI] [PubMed] [Google Scholar]
- DaVeiga SP. Epidemiology of atopic dermatitis: a review. Allergy Asthma Proc. 2012;33:227. doi: 10.2500/aap.2012.33.3569. [DOI] [PubMed] [Google Scholar]
- Kapoor R, Menon C, Hoffstad O, Bilker W, Leclerc P, Margolis DJ. The prevalence of atopic triad in children with physician-confirmed atopic dermatitis. J Am Acad Dermatol. 2008;58:68. doi: 10.1016/j.jaad.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Simpson EL, Eichenfield LF, Ellis CN, Mancini AJ, Paller AS. Current issues in atopic comorbidities and preventing the atopic march. Semin Cutan Med Surg. 2012;31:S6. doi: 10.1016/j.sder.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Scheinmann P, Pham TN, Karila C, de Blic J. Allergic march in children, from rhinitis to asthma: management, indication of immunotherapy. Arch Pediatr. 2012;19:330. doi: 10.1016/j.arcped.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Eichenfield LF, Ellis CN, Mancini AJ, Paller AS, Simpson EL. Atopic dermatitis: epidemiology and pathogenesis update. Semin Cutan Med Surg. 2012;31:S3. doi: 10.1016/j.sder.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Noh G, Lee J. Atopic dermatitis and cytokines: the immunoregulatory and therapeutic implications of cytokines in atopic dermatitis – part II: negative regulation and cytokine therapy in atopic dermatitis. Recent Pat Inflamm Allergy Drug Discov. 2012;6:248. doi: 10.2174/187221312802652802. [DOI] [PubMed] [Google Scholar]
- Lee J, Noh G, Lee S, Youn Y, Rhim J. Atopic dermatitis and cytokines: recent patents in immunoregulatory and therapeutic implications of cytokines in atopic dermatitis – part I: cytokines in atopic dermatitis. Recent Pat Inflamm Allergy Drug Discov. 2012;6:222. doi: 10.2174/187221312802652820. [DOI] [PubMed] [Google Scholar]
- Bao L, Shi VY, Chan LS. IL-4 regulates chemokine CCL26 in keratinocytes through the Jak1, 2/Stat6 signal transduction pathway: implication for atopic dermatitis. Mol Immunol. 2012;50:91. doi: 10.1016/j.molimm.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Chen L, Martinez O, Overbergh L, Mathieu C, Prabhakar BS, Chan LS. Early up-regulation of Th2 cytokines and late surge of Th1 cytokines in an atopic dermatitis model. Clin Exp Immunol. 2004;138:375. doi: 10.1111/j.1365-2249.2004.02649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Marble DJ, Agha R, Peterson JD, Becker RP, Jin T, Li J, Chan LS. The progression of inflammation parallels the dermal angiogenesis in a keratin 14 IL-4-transgenic model of atopic dermatitis. Microcirculation. 2008;15:49. doi: 10.1080/10739680701418416. [DOI] [PubMed] [Google Scholar]
- Sehra S, Yao Y, Howell MD, Nguyen ET, Kansas GS, Leung DY, Travers JB, Kaplan MH. IL-4 regulates skin homeostasis and the predisposition toward allergic skin inflammation. J Immunol. 2010;184:3186. doi: 10.4049/jimmunol.0901860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LS, Robinson N, Xu L. Expression of interleukin-4 in the epidermis of transgenic mice results in a pruritic inflammatory skin disease: an experimental animal model to study atopic dermatitis. J Invest Dermatol. 2001;117:977. doi: 10.1046/j.0022-202x.2001.01484.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Lin SX, Agha-Majzoub R, Overbergh L, Mathieu C, Chan LS. CCL27 is a critical factor for the development of atopic dermatitis in the keratin-14 IL-4 transgenic mouse model. Int Immunol. 2006;18:1233. doi: 10.1093/intimm/dxl054. [DOI] [PubMed] [Google Scholar]
- Beck LA, Thaci D, Hamilton JD, Ren H, Rocklin R, Ming J, Graham N, Radin A. Systemic treatment of patients with severe atopic dermatitis (AD) with an anti IL-4Rα mAb (REGN668/SAR231893) results in rapid and sustained improvements in disease signs and symptoms. J Invest Dermatol. 1893;133:S178. [Google Scholar]
- Azuma YT, Matsuo Y, Nakajima H, Yancopoulos GD, Valenzuela DM, Murphy AJ, Karow M, Takeuchi T. Interleukin-19 is a negative regulator of innate immunity and critical for colonic protection. J Pharmacol Sci. 2011;115:105. doi: 10.1254/jphs.10r02cr. [DOI] [PubMed] [Google Scholar]
- Gallagher G. Interleukin-19: multiple roles in immune regulation and disease. Cytokine Growth Factor Rev. 2010;21:345. doi: 10.1016/j.cytogfr.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Kim S, Miska KB, McElroy AP, Jenkins MC, Fetterer RH, Cox CM, Stuard LH, Dalloul RA. Molecular cloning and functional characterization of avian interleukin-19. Mol Immunol. 2009;47:476. doi: 10.1016/j.molimm.2009.08.027. [DOI] [PubMed] [Google Scholar]
- .Hsing CH, Hsu CC, Chen WY, Chang LY, Hwang JC, Chang MS. Expression of IL-19 correlates with Th2 cytokines in uraemic patients. Nephrol Dial Transplant. 2007;22:2230. doi: 10.1093/ndt/gfm179. [DOI] [PubMed] [Google Scholar]
- Oral HB, Kotenko SV, Yilmaz M, Mani O, Zumkehr J, Blaser K, Akdis CA, Akdis M. Regulation of T cells and cytokines by the interleukin-10 (IL-10)-family cytokines IL-19, IL-20, IL-22, IL-24 and IL-26. Eur J Immunol. 2006;36:380. doi: 10.1002/eji.200425523. [DOI] [PubMed] [Google Scholar]
- Liao SC, Cheng YC, Wang YC, et al. IL-19 induced Th2 cytokines and was up-regulated in asthma patients. J Immunol. 2004;173:6712. doi: 10.4049/jimmunol.173.11.6712. [DOI] [PubMed] [Google Scholar]
- Huang F, Wachi S, Thai P, Loukoianov A, Tan KH, Forteza RM, Wu R. Potentiation of IL-19 expression in airway epithelia by IL-17A and IL-4/IL-13: important implications in asthma. J Allergy Clin Immunol. 2008;121:1415. doi: 10.1016/j.jaci.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C, Park SH, Daley E, Emson C, Louten J, Sisco M, de Waal MR, Grunig G. Interleukin-19: a constituent of the regulome that controls antigen presenting cells in the lungs and airway responses to microbial products. PLoS ONE. 2011;6:e27629. doi: 10.1371/journal.pone.0027629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace E, Scafidi V, Di Bona D, et al. Increased expression of IL-19 in the epithelium of patients with chronic rhinosinusitis and nasal polyps. Allergy. 2012;67:878. doi: 10.1111/j.1398-9995.2012.02842.x. [DOI] [PubMed] [Google Scholar]
- Kunz S, Wolk K, Witte E, et al. Interleukin (IL)-19, IL-20 and IL-24 are produced by and act on keratinocytes and are distinct from classical ILs. Exp Dermatol. 2006;15:991. doi: 10.1111/j.1600-0625.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- Uto-Konomi A, Miyauchi K, Ozaki N, et al. Dysregulation of suppressor of cytokine signaling 3 in keratinocytes causes skin inflammation mediated by interleukin-20 receptor-related cytokines. PLoS ONE. 2012;7:e40343. doi: 10.1371/journal.pone.0040343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Shi VY, Chan LS. IL-4 up-regulates epidermal chemotactic, angiogenic, and pro-inflammatory genes and down-regulates antimicrobial genes in vivo and in vitro: relevant in the pathogenesis of atopic dermatitis. Cytokine. 2013;61:419. doi: 10.1016/j.cyto.2012.10.031. [DOI] [PubMed] [Google Scholar]
- Cui X, Zhang L, Luo J, Rajasekaran A, Hazra S, Cacalano N, Dubinett SM. Unphosphorylated STAT6 contributes to constitutive cyclooxygenase-2 expression in human non-small cell lung cancer. Oncogene. 2007;26:4253. doi: 10.1038/sj.onc.1210222. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yamanaka Y, Nakae K, et al. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651. [PMC free article] [PubMed] [Google Scholar]
- Mui AL, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J. 1996;15:2425. [PMC free article] [PubMed] [Google Scholar]
- Luo G, Yu-Lee L. Transcriptional inhibition by Stat5. Differential activities at growth-related versus differentiation-specific promoters. J Biol Chem. 1997;272:26841. doi: 10.1074/jbc.272.43.26841. [DOI] [PubMed] [Google Scholar]
- Bao L, Tessier C, Prigent-Tessier A, Li F, Buzzio OL, Callegari EA, Horseman ND, Gibori G. Decidual prolactin silences the expression of genes detrimental to pregnancy. Endocrinology. 2007;148:2326. doi: 10.1210/en.2006-1643. [DOI] [PubMed] [Google Scholar]
- Shi VY, Bao L, Chan LS. Inflammation-driven dermal lymphangiogenesis in atopic dermatitis is associated with CD11b+ macrophage recruitment and VEGF-C up-regulation in the IL-4-transgenic mouse model. Microcirculation. 2012;19:567. doi: 10.1111/j.1549-8719.2012.00189.x. [DOI] [PubMed] [Google Scholar]
- Bao L, Devi S, Bowen-Shauver J, Ferguson-Gottschall S, Robb L, Gibori G. The role of interleukin-11 in pregnancy involves up-regulation of α2-macroglobulin gene through janus kinase 2-signal transducer and activator of transcription 3 pathway in the decidua. Mol Endocrinol. 2006;20:3240. doi: 10.1210/me.2006-0296. [DOI] [PubMed] [Google Scholar]
- Zeng G. Sticky-end PCR: new method for subcloning. Biotechniques. 1998;25:206. doi: 10.2144/98252bm05. [DOI] [PubMed] [Google Scholar]
- Chen PJ, Wei CC, Wang C, Chen FW, Hsu YH, Chang MS. Promoter analysis of interleukin 19. Biochem Biophys Res Commun. 2006;344:713. doi: 10.1016/j.bbrc.2006.03.200. [DOI] [PubMed] [Google Scholar]
- Bao L, Zhang H, Chan LS. The involvement of the JAK-STAT signaling pathway in chronic inflammatory skin disease atopic dermatitis. JAKSTAT. 2013;2:e24137. doi: 10.4161/jkst.24137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi C, Fairchild HR, Madonna S, Scarponi C, De Pita O, Leung DY, Howell MD. IL-4 and IL-13 negatively regulate TNF-α- and IFN-γ-induced β-defensin expression through STAT-6, suppressor of cytokine signaling (SOCS)-1, and SOCS-3. J Immunol. 2007;179:984. doi: 10.4049/jimmunol.179.2.984. [DOI] [PubMed] [Google Scholar]
- Travagli J, Letourneur M, Bertoglio J, Pierre J. STAT6 and Ets-1 form a stable complex that modulates Socs-1 expression by interleukin-4 in keratinocytes. J Biol Chem. 2004;279:35183. doi: 10.1074/jbc.M403223200. [DOI] [PubMed] [Google Scholar]
- Takeda K, Tanaka T, Shi W, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Borner C, Woltje M, Hollt V, Kraus J. STAT6 transcription factor binding sites with mismatches within the canonical 5′-TTC..GAA-3′ motif involved in regulation of δ- and µ-opioid receptors. J Neurochem. 2004;91:1493. doi: 10.1111/j.1471-4159.2004.02846.x. [DOI] [PubMed] [Google Scholar]
- Kraus J, Borner C, Hollt V. Distinct palindromic extensions of the 5′-TTC..GAA-3′ motif allow STAT6 binding in vivo. FASEB J. 2003;17:304. doi: 10.1096/fj.02-0482fje. [DOI] [PubMed] [Google Scholar]
- Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- Hofmann SR, Rosen-Wolff A, Tsokos GC, Hedrich CM. Biological properties and regulation of IL-10 related cytokines and their contribution to autoimmune disease and tissue injury. Clin Immunol. 2012;143:116. doi: 10.1016/j.clim.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Sabat R. IL-10 family of cytokines. Cytokine Growth Factor Rev. 2010;21:315. doi: 10.1016/j.cytogfr.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Li HH, Lin YC, Chen PJ, et al. Interleukin-19 upregulates keratinocyte growth factor and is associated with psoriasis. Br J Dermatol. 2005;153:591. doi: 10.1111/j.1365-2133.2005.06665.x. [DOI] [PubMed] [Google Scholar]
- Romer J, Hasselager E, Norby PL, Steiniche T, Thorn CJ, Kragballe K. Epidermal overexpression of interleukin-19 and -20 mRNA in psoriatic skin disappears after short-term treatment with cyclosporine a or calcipotriol. J Invest Dermatol. 2003;121:1306. doi: 10.1111/j.1523-1747.2003.12626.x. [DOI] [PubMed] [Google Scholar]
- Otkjaer K, Kragballe K, Funding AT, Clausen JT, Noerby PL, Steiniche T, Iversen L. The dynamics of gene expression of interleukin-19 and interleukin-20 and their receptors in psoriasis. Br J Dermatol. 2005;153:911. doi: 10.1111/j.1365-2133.2005.06800.x. [DOI] [PubMed] [Google Scholar]


