Abstract
Macrophages detect bacterial infection through pattern recognition receptors (PRRs) localized at the cell surface, in intracellular vesicles or in the cytosol. Discrimination of viable and virulent bacteria from non-virulent bacteria (dead or viable) is necessary to appropriately scale the anti-bacterial immune response. Such scaling of anti-bacterial immunity is necessary to control the infection, but also to avoid immunopathology or bacterial persistence. PRR-mediated detection of bacterial constituents in the cytosol rather than at the cell surface along with cytosolic recognition of secreted bacterial nucleic acids indicates viability and virulence of infecting bacteria. The effector responses triggered by activation of cytosolic PRRs, in particular the RIG-I-induced simultaneous rapid type I IFN induction and inflammasome activation, are crucial for timely control of bacterial infection by innate and adaptive immunity. The knowledge on the PRRs and the effector responses relevant for control of infection with intracellular bacteria will help to develop strategies to overcome chronic infection.
Keywords: cytosolic pattern recognition receptors, interferon, RIG-I inflammasome activation, secreted bacterial nucleic acids
Introduction
The immune system has evolved to provide protection against infectious microorganisms. Physical barriers in the skin, the gastrointestinal, and the respiratory tracts provide a first line of defense to prevent invasion of bacteria that colonize body surfaces. Those bacteria that breach these barriers are not only confronted with a dense network of macrophages located within mucosal tissues in the skin, but also with macrophages from the liver or spleen that provide immune surveillance against circulating bacteria in the blood stream. Phagocytic cell populations such as granulocytes, monocytes, and tissue-resident macrophages are equipped with germ-line encoded pattern recognition receptors (PRR) that detect bacteria leading to the induction of innate immune responses (Akira & Takeda, 2004; Akira et al, 2006; Gao et al, 2008). PRRs recognize pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs), that is cell stress, damage, or cell death associated with infection. Because of bacterial immune evasion and existence of secretion systems in bacteria that deliver virulence factors into the cytosol, infected cells require a timely and coordinated activity of PRRs to combat bacterial infection. The PRRs and the signaling pathways involved in this aspect have been the matter of intense research efforts over the last years.
Binding and phagocytosis of bacteria followed by induction of inflammation are important for macrophage control of bacterial dissemination in tissues or blood. Even before phagocytosis, some bacteria such as Helicobacter pylori deliver degradation products of cell wall peptidoglycan into the cytosol of epithelial cells and thereby activate cytosolic PRR-induced signaling (Girardin et al, 2003a; Viala et al, 2004). PRRs are not exclusively expressed by macrophages, but are also found on organ-resident non-immune cell populations such as endothelial and epithelial cells, which allow these cells to engage in immune surveillance and induction of inflammation (Knolle et al, 1997; Viala et al, 2004; Irving et al, 2014). Phagocytic elimination of bacteria or bacterial debris is mainly performed by macrophages and granulocytes, but also by epithelial cells (Travassos et al, 2010; Irving et al, 2014).
While the rapid containment of bacterial infection and potent induction of innate immune responses are relevant to control local infection and prevent it from spreading further, it is equally important to prevent immune pathology resulting from overshooting immunity (Blander & Sander, 2012). Here, we review the current knowledge on the key PRRs and signaling pathways that contribute to the scaling of immune responses against intracellular bacterial infection. The activation of PRRs localized in different subcellular compartments and their cross talk will be reviewed with particular reference to their role in cytosolic recognition of bacterial nucleic acids for generation of protective immunity. Since a wealth of knowledge has been collected on the immune response against Gram-positive intracellular Listeria monocytogenes, we will mainly discuss the principles of anti-bacterial immunity against this pathogen but also refer to other important pathogenic bacterial species.
Subcellular localization of PRRs
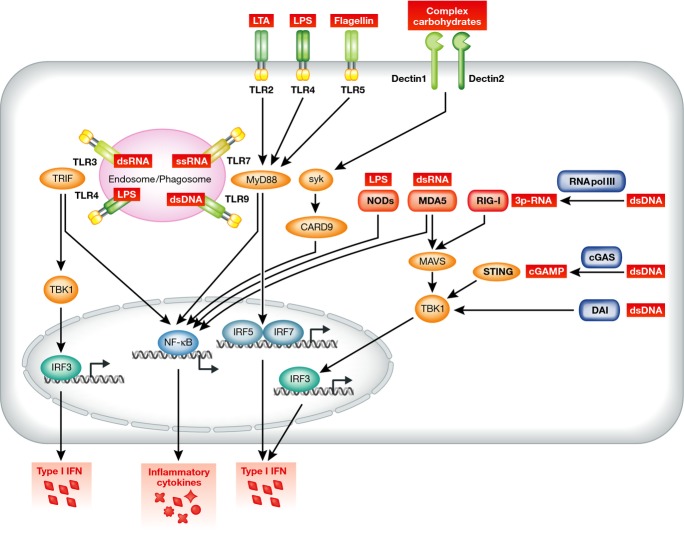
Closely interlinked networks of germ-line encoded PRRs exist to facilitate recognition of bacteria localized within different subcellular compartments in phagocytic immune cells (Fig 1). Membrane-anchored PRRs detecting PAMPs are located at the cell surface and within endosomal/phagosomal compartments. Toll-like receptors (TLRs) as well as C-type lectin receptors belong to this category of PRRs that overlook both, the cell surface and the endosomal compartments (Fig 1). Ligand binding and TLRs activation induce signaling via the adapter molecules MyD88 and/or TRIF to trigger the production of type I interferon (IFN) and inflammatory cytokines through the transcription factors NF-κB and interferon response factors (IRF) 3, 5, and 7 (Table 1 and Fig 1) (Blasius & Beutler, 2010; Kawai & Akira, 2010). The relative importance of type I IFN for pathogen control or regulation of immune responses will be dealt with later on in this review. Dectin receptors signal through the adapter molecules Syk and CARD9 to induce expression of inflammatory cytokines via NF-κB (Taylor et al, 2007; Saijo et al, 2010) (Table 1 and Fig 1). The sanctity of the cytosol is the result of most sensitive detection of any signs of infection through PRRs. Although the replication strategies of bacteria and viruses are fundamentally different, cytosolic PRRs have evolved a surveillance network that covers these demands. Two distinct classes of PRR families can be distinguished, that is the NOD receptors (NOD1 and NOD2) detecting structural elements of bacteria cell walls (Girardin et al, 2003a,b) and the RNA-sensing RIG-I-like helicase receptors (RIG-I and MDA5) (Table 1 and Fig 1). Upon ligand binding, cytosolic NOD receptors signal through NF-κB for induction of pro-inflammatory cytokines (Bertin et al, 1999; Ogura et al, 2001). Upon recognition of non-self RNA in the cytosol, RIG-I and MDA5 signal via the adapter molecule MAVS and TBK1 to activate interferon responsive factor 3 (IRF3) and thereby production of type I IFN and inflammatory cytokines. Recently, the nucleotidyl transferase cGAS has been identified that generates the second messengers cyclic-diadenylate monophosphate and cyclic-diguanylate monophosphate (cGAMP) in response to recognition of foreign DNA to trigger type I IFN induction via STING (Xiao & Fitzgerald, 2013). Interestingly, STING also serves as a receptor recognizing cGAMP directly released from cytosolic bacteria (Woodward et al, 2010). The RNA polymerase III transcribes foreign DNA into RIG-I ligands, namely 3p-RNA (Loo & Gale, 2011), which allows for RIG-I-dependent sensing of these RNA polymerase intermediates (Ablasser et al, 2009).
Figure 1. Recognition of PAMPs by pattern recognition receptors (PRRs).
Pattern recognition receptors are classified into three subgroups according to their localization in the cell. Toll-like receptors are located on plasma membranes at the cell surface and also within endosomal compartments. Certain TLRs are involved in recognition of proteins, lipids, and lipoproteins such as TLR1, TLR2, TLR4-6, and TLR11, while others detect the presence of nucleic acids, such as TLR3, TLR7, TLR8, and TLR9. Furthermore, C-type lectins constitute a separate family of membrane-anchored PRRs such as Dectin-1 and Dectin-2 that contribute to recognition of complex carbohydrate structures. Together, these receptors recognize a wide range of pathogen-associated molecular patterns and thus ensure that bacteria can be sensed in a synergistic fashion by different PRRs. Finally, sensing of pathogens within the cytosol is mediated by the family of NOD receptors and NLRs that include RIG-like helicases and other RNA-sensing receptors.
Table 1.
Localization and ligand properties of PRRs
| PRRs | Subcellular localization | Ligands recognized | Signaling pathways/transcription factors |
|---|---|---|---|
| Toll-like receptors | |||
| TLR2 | Cell surface/endosome | Lipoteichoic acid | MyD88, NF-κB |
| TLR3 | Endosome | Double-stranded RNA | TRIF, NF-κB/IRF3 |
| TLR4 | Cell surface/endosome | LPS | TRIF/MyD88/Mal/NF-κB/IRF3 |
| TLR5 | Cell surface | Flagellin | MyD88, NF-κB |
| TLR7/8 | Endosome | Single-stranded RNA | MyD88, NF-κB/IRF7 |
| TLR9 | Endosome | Double-stranded DNA | |
| C-type Lectins | |||
| Dectin-1 | Cell surface | β-glucan | Syk, CARD9, NF-κB |
| Dectin-2 | Cell surface | α-mannans | Syk, CARD9, NF-κB |
| NLR | |||
| NOD | Cytosol | Peptidoglycans | NF-κB, MAPK |
| NLRP1 | Cytosol | Type III secretion system | ASC, Caspase-1 |
| NLRP3 | Cytosol | MDP | ASC, Caspase-1 |
| NLRC4 | Cytosol | mRNA/flagellin | ASC, Caspase-1 |
| RNA pol III | Cytosol | dsRNA | MAVS, TBK1, IRF3 |
| DAI | Cytosol | AT-rich B-DNA | TBK1, IRF3, NF-κB |
| LRRFIP1 | Cytosol | dsRNA, dsDNA | β-catenin, IRF3 |
| IFI16/p204 | Cytosol | dsDNA | STING, TBK1, IRF3 |
| AIM2 | Cytosol | dsDNA | ASC, Caspase-1 |
| RIG-I | Cytosol | 3p-RNA | MAVS, IRF3, IRF7, NF-κB, ASC, Caspase-1 |
| MDA5 | Cytosol | Double-stranded RNA | MAVS, IRF3, IRF7, NF-κB |
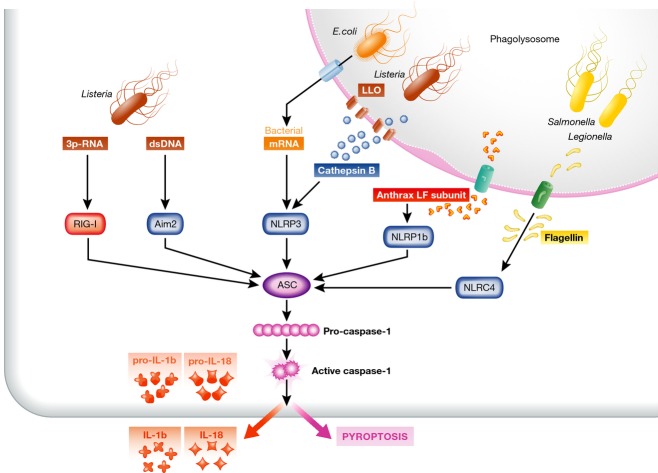
Besides induction of type I IFN and inflammatory cytokines, activation of cytosolic PRRs can lead to inflammasome activation that is characterized by ASC-mediated proteolytic cleavage of pro-IL-1β and pro-IL-18 to biologically active pro-inflammatory IL-1β and IL-18. Inflammasomes are multicomponent protein complexes that assemble as a platform upon recognition of non-self nucleic acids in the cytosol or upon recognition of certain danger signals (Fig 2 and Table 1) (Martinon & Tschopp, 2007; Latz et al, 2013). The outcome of inflammasome activation is the cleavage and release of the biologically active forms of IL-1β, IL-18, and IL-33. Also, inflammasome-induced activation of caspase-1 catalyzes the proteolytic activation of caspase-7, rather than caspase-3, in the cytosol, triggering pyroptotic cell death and consequently local inflammation. This inflammasome-induced pyroptotic cell death contributes to the elimination of invading pathogens (Miao et al, 2010; Latz et al, 2013). While caspase-1 processing is required for the release of IL-1β and IL-18, it is dispensable for pyroptosis, suggesting that caspase-1 can also be activated in a non-proteolytic way to trigger cell death (Zitvogel et al, 2012). Caspase-11 activation can lead to inflammasome-independent generation of IL-1β by degradation of TRPC1 (Py et al, 2014), a family member of TRP receptors that serve as sensors for various physiological cell functions (Clapham, 2003). Activation of NOD-like receptors (NLRs), Aim2, and RIG-I all trigger inflammasome activation upon sensing of bacterial flagellin, DNA, and RNA, respectively (Fig 2). Cytosolic presence of cathepsin B or ATP leakage, that indicates vesicle damage induced by phagocytosed bacteria, activates NLRP3 and NRLP1 inflammasomes (Duncan et al, 2009; Meixenberger et al, 2010; Ali et al, 2011).
Figure 2. Inflammasome activation in response to bacterial infection.
Inflammasome activation is achieved by different PRRs and through sensing of cell stress or presence of particular bacterial products. Detection of small size nucleic acids secreted from Listeria triggers by AIM2 and RIG-I leads to inflammasome activation. Pathogenic Escherichia coli and Listeria monocytogenes are sensed via NLRP3 that detects bacterial mRNA and membrane damage—cathepsin B—in the cytosol of infected cells. NLRP1b detects the lethal toxin (LT) of the Bacillus anthracis that reaches the cytosol of the host cell through the bacterial toxin protective antigen (PA). Furthermore, intracellular Salmonella typhimurium, Shigella flexneri, and Legionella pneumophila are recognized through NLRC4 inflammasome that detects the monomeric flagellin that is secreted through bacterial type III and IV secretion systems into the cytosol. Inflammasome-mediated caspase-1 activation leads to the release of IL-1β and IL-18 as well as to pyroptotic cell death that both serve to restrict further pathogen expansion.
Pathogenic bacteria induce inflammasome activation via several pathways. Gram-positive bacteria like Listeria can trigger the NLRs-induced inflammasome activation by secreting pore-forming proteins leading to leakage of phagolysosomal constituents such as cathepsins into the cytosol that are then recognized by NLRP3 (Meixenberger et al, 2010). Gram-negative bacteria such as Salmonella (Mariathasan et al, 2004) or Shigella (Suzuki et al, 2007) use type III secretion systems for cytosolic delivery of flagellin that is detected by NLRC4 (Franchi et al, 2009). Furthermore, the DNA-recognizing PRR Aim2 is reported to sense DNA of several intracellular bacteria, such as Listeria monocytogenes (Fernandes-Alnemri et al, 2009) and Legionella pneumonia (Ge et al, 2012). Similar to NLRP3, binding of non-self DNA to Aim2 will trigger the supramolecular assembly of ASC-dimers and the subsequent release of the pro-inflammatory cytokines IL-1β and IL-18 together with induction of pyroptosis. Besides this, a non-canonical pathway of inflammasome activation exists, where type III and type IV secretion systems of Gram-negative bacteria like Legionella pneumophila and Yersinia pseudotuberculosis induce caspase-11-dependent release of IL-1β and cell death through the NLRP3/ASC inflammasome (Casson et al, 2013). Certain Gram-negative bacteria trigger TRIF-mediated NLRP3 inflammasome activation that in turn activates caspase-11 to synergize with the assembled NLRP3 inflammasome to regulate caspase-1 activation, caspase-1-independent IL-1β release, and cell death (Rathinam et al, 2012). More recently, it was shown that particular serotypes of LPS from Escherichia coli or Salmonella typhimurium, upon delivery into the cytosol of macrophages, can activate caspase-11, release of IL-1β, and induce pyroptotic cell death (Kayagaki et al, 2013). Thus, inflammasome activation in response to bacterial infection is achieved via various PRRs.
Sensing the threat of bacterial infection to scale anti-bacterial immunity
Myeloid cells of the immune system serve a dual function in early containment and direct elimination of invading bacteria as well as in the sensing and scaling the threat of bacterial infection to mount appropriate immune responses against pathogenic bacteria. Detection of PAMPs such as bacterial cell wall constituents or CpG-rich bacterial DNA by TLRs does not allow the macrophage to discriminate between bacterial debris and the presence of viable and virulent bacteria. Breaching of plasma membranes or spillage of proteolytic enzymes from damaged phagolysosomes, however, indicates not only bacterial viability but also virulence. Nonpathogenic bacteria typically do not enter the cytosol, and they do not deliver virulence factors into the cytosol of host cells. Rapid and robust induction of immunity is necessary to fight highly virulent bacteria, whereas elimination of bacterial degradation products does not require the same strength of response. Moreover, detecting the threat of a bacterial infection occurs in the context of bacterial strategies to interfere with the host’s immune response revealing a co-evolutionary race and continuous ‘hide and seek’ interaction between infecting bacteria and the host’s immune system. For scaling, the necessary extent and duration of anti-bacterial immunity myeloid cells use the complex information they receive from the different PRRs in distinct subcellular compartments. While membrane-anchored TLRs recognize bacterial cell wall constituents and nucleic acids from degraded bacteria, only cytosolic PRRs recognize bacterial nucleic acids released from viable bacteria (Sander et al, 2011; Abdullah et al, 2012; Rathinam et al, 2012). Of note, DNA- and RNA-sensing receptors recognize bacterial nucleic acids independent of the particular bacterial species. This suggests that detection of infection with pathogenic intracellular bacteria may follow a ‘red button’ principle to initiate and scale the extent of anti-bacterial defense that is based on cytosolic alarm signals rather than the specific detection of particular pathogenic bacterial species.
Intracellular bacteria such as Mycobacterium tuberculosis, Listeria monocytogenes, and Legionella pneumophila persist in host cells by preventing phagosomal maturation, by resisting microbicidal effector functions of the host cell or by escape from phagolysosomes into the cytosol (Diacovich & Gorvel, 2010). Escape from the phagolysosomal compartment is clearly an indication of bacterial virulence and is caused by virulence factors, for example listeriolysin (LLO) in case of Listeria monocytogenes. However, bacteria have evolved various immune escape mechanisms by interfering with recognition through PRRs or modulating signaling downstream of receptor activation. Certain bacteria impair MyD88-mediated signaling downstream of TLRs and thereby compromise induction of innate immune responses (Johannessen et al, 2013). Some bactericidal mechanisms induced by the activation of the PRRs trigger induction of virulence factors by the intracellular pathogens Listeria, Shigella, and Mycobacterium and therefore may enhance bacterial pathogenicity (Arpaia & Barton, 2013).
Once in the cytosol, bacteria like Listeria monocytogenes, Shigella flexneri, and Burkholderia pseudomallei can escape detection through surface and phagosomal/endosomal PRRs by rapid migration and by infecting a neighboring cell, thus avoiding the extracellular environment and detection by PRRs at the cell surface or phagosomal compartments. Following LLO-mediated escape from the phagosome, Listeria monocytogenes within the cytosol employs ActA for uni-directional propelling via actin-polymerization, which is required for continuous evasion from killing (Chakraborty et al, 1995; Skoble et al, 2000). Salmonella spp. employs other escape mechanisms and down-regulates expression of flagellin and the secretion system T3SS. Thus, Salmonella avoid detection of flagellin by TLR5 at the cell membrane (Hersh et al, 1999; Andersen-Nissen et al, 2005) and by NLRC4 within the cytosol (Perez-Lopez et al, 2013). Several other pathogenic bacteria also evade inflammasome activation (Cunha & Zamboni, 2013). Since the inflammasome is crucial to mount effective innate immune responses against intracellular bacterial pathogens like Salmonella typhimurium, such evasion of cytosolic immune sensing may impair protective immunity. Furthermore, Mycobacterium tuberculosis prevents activation of caspase-1 triggered by the administration of NLRP3 agonists and by inactivating small GTPases (Mishra et al, 2013; Sun et al, 2013), suggesting that bacteria actively repress detection by PRRs. Within the gastrointestinal tract, distinction between pathogenic and commensal bacteria is achieved through NLRC4/inflammasome-driven production of IL-1β that promotes host intestinal defense (Franchi et al, 2012). These findings strengthen the notions that inflammasome activation is important for appropriate immune responses against pathogenic bacteria and that cytosolic pattern recognition is involved in distinction between pathogenic and commensal bacteria.
Intracellular bacteria can also modify cellular functions. For example, while still being in phagosomal compartments, Salmonella inject pro-apoptotic molecules via their type III secretion system into infected macrophages and thereby cause further dissemination of bacterial infections by release of bacteria from dying macrophages. Interestingly, human and mouse NLRC4 recognize bacterial type III secretion needle protein that initiates rapid inflammasome activation upon development of bacterial virulence (Zhao et al, 2011; Yang et al, 2013). Mycobacteria that persist in phagosomal compartments in macrophages regulate cellular metabolism to their advantage to support intracellular survival such as induction of cholesterol accumulation (Mattos et al, 2014) and regulation of autophagy. Pharmacologic re-activation of autophagy forms the basis for effective anti-mycobacterial drug action (Kim et al, 2012). Furthermore, disruption of glycolipid synthesis results in an increase in the release of the pro-inflammatory cytokines from infected cells and infection with M. tuberculosis overproducing glycolipids inhibited the release of these pro-inflammatory mediators (Reed et al, 2004). Therefore, differences in mycobacterial lipid metabolism may modify the host’s immune response.
Intracellular Gram-positive bacteria like Listeria employ another strategy and confound the host’s transcriptional response by the secretion of small RNAs. Upon infection of the host cell, Listeria shows dramatic changes in its gene expression including generation of non-coding RNAs (Toledo-Arana et al, 2009). However, only few details are so far known how RNA or DNA secreted by cytosolic bacteria may affect anti-bacterial immune response (Caldelari et al, 2013). In support of this reasoning, Listeria lacking the secA2 secretion system, that is required for secretion of bacterial proteins and nucleic acids into the cytosol (Lenz & Portnoy, 2002), are more rapidly eliminated by macrophages (Lenz et al, 2003; Muraille et al, 2007; Abdullah et al, 2012).
Detection of secreted nucleic acids through PRRs such as RIG-I and cGAS/STING likely represents the Achilles heel of bacteria that have reached the cytosol (Abdullah et al, 2012). Since it is likely that substantial amounts of bacterial nucleic acids are required to influence the host cell transcriptome, the sensitive and rapid detection of those nucleic acids by cytosolic RIG-I and cGAS/STING may be particularly important to prevent deterioration of cellular defense function during cytosolic bacterial infection. Thus, cytosolic immune sensing of secreted nucleic acids provides a mean to differentiate between dead bacteria and bacterial debris and thereby allows macrophages to rapidly initiate appropriate defense mechanisms before bacterial escape mechanisms take action.
For virulent bacteria that do not gain excess to the cytosolic compartment, other mechanisms operate to achieve a commensurate response to infection. It was recently found that low concentrations of LPS drive a TLR4-dependent pro-inflammatory response that is characterized by release of cytokines and alerts the host to the presence of bacterial infection (Kawai & Akira, 2010). Higher quantities of LPS, however, can reach the cytosol and trigger inflammasome activation, IL-1β production, and pyroptotic cell death (Kayagaki et al, 2013). This distinct response pattern, which depends on the concentration of LPS rather than detection of qualitatively different molecular patterns, adds further complexity to shape the response and magnitude of innate immune responses against bacteria.
Scaling up anti-bacterial immunity and immune sensing
Pro-inflammatory mediators released from macrophages as a consequence of PRR activation initiate a local inflammatory response and through induction of chemokines recruit further immune cells to the site of infection. Various immune cell populations such as neutrophils, inflammatory monocytes, natural killer (NK) cells, and dendritic cells (DCs) are involved in local defense against bacterial infection. Early after infection, neutrophils migrate toward the site of infection, attracted by IL-6 and IL-8 and chemokines that are secreted by infected cells. Neutrophils amplify this inflammatory response by secreting further inflammation-inducing mediators and chemokines for the recruitment of inflammatory monocytes and DCs. Such recruitment of immune cell populations serves the purpose to rapidly contain and eliminate invading bacteria at the site of initial infection and to allow for antigen sampling by DCs and monocytes/macrophages leading to induction of sterile clearance and protective immune memory (Schnare et al, 2001; Iwasaki & Medzhitov, 2010).
Macrophages located within liver sinusoids are most prominent in removing bacteria circulating in the blood through a concerted action involving granulocytes trapping bacteria in NETs and platelets enhancing macrophage binding and clearance of bacteria (Lee et al, 2010; Wong et al, 2013). Macrophage activation by PRRs further orchestrates a complex anti-bacterial immune response. The triggering of surface-bound TLRs by live bacteria or bacterial degradation products leads to expression of cytokines such as TNF, IL-12, and IL-18 that in turn cause local NK-cell activation and IFN-γ production. This stimulates bactericidal activity in macrophages to eliminate phagocytosed bacteria by increasing phagolysosomal fusion. Macrophages and granulocytes produce ROS early after uptake of bacteria, which prevents bacterial escape into the cytosol. The killing of ingested bacteria through cytokine-mediated enhancement of bactericidal activity in phagocytosing cells such as macrophages and granulocytes constitutes a positive feed-forward loop to gain rapid control over infecting bacteria (Lee et al, 2010; Wong et al, 2013).
If bacteria are not readily eliminated by these measures, a further layer of immune defense is activated. Damage to the phagosome causes leakage of cathepsins into the cytosol triggering NLRP3 activation. Direct cytosolic recognition of bacterial constituents such as flagellin by NLRC4, anthrax subunit proteins by NLRP1b, or bacterial nucleic acids by RIG-I or Aim2 leads either to production of type I IFN or to inflammasome activation (Fig 3). Caspase activation further augments bacterial killing within the phagolysosome. Acidification of phagosomes containing Gram-positive bacteria is regulated by the NLRP3 inflammasome and caspase-1. Active caspase-1 accumulates on phagosomes and acts locally to control the pH by modulating buffering by the NADPH oxidase NOX2 (Sokolovska et al, 2013). Gene expression induced by IFN-γ upregulates phagosomal defense mechanisms that limit the bacterial escape from phagosomes (Myers et al, 2003; Lindgren et al, 2004). Inflammasome activation contributes to elimination of intracellular bacteria and at the same time further enhances recruitment of immune cells and induction of sterile clearance as well as protective antigen-specific immunity. These positive feedback loops initiated at the different subcellular locations by different PRRs converge on cytosolic immune sensing and inflammasome activation to reinforce anti-bacterial immunity and facilitate rapid control of bacterial spread.
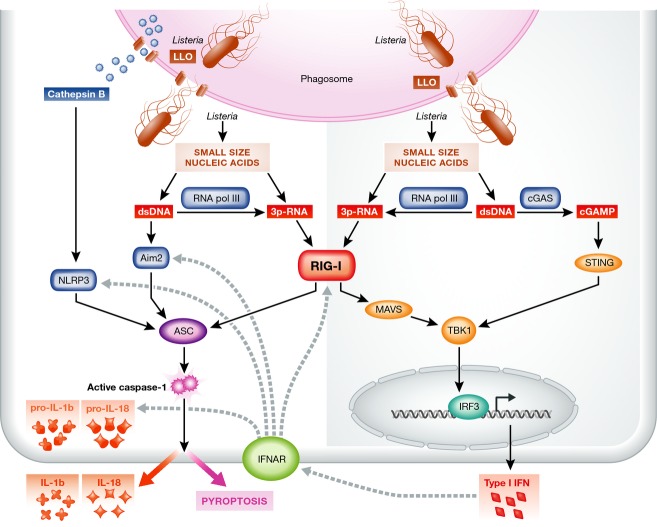
Figure 3. Prominent role of RIG-I in orchestrating immunity against viable and virulent intracellular Listeria.
Schematic representation of the different PRRs and amplification mechanisms that operate in detection of intracellular infection with viable and virulent Listeria monocytogenes. Active secretion of small size nucleic acids through SecA2 from cytosolic Listeria is detected by DNA-sensing Aim2 as well as RNA-sensing RIG-I. RIG-I signaling is unique since it leads to concomitant activation of type I IFN induction through MAVS and inflammasome activation through ASC. This ensures that independent of cell surface or endosomal PRR activation in case of bacterial escape by direct cell-to-cell infection, the levels of cytosolic PRRs are increased, and through NF-κB activation, the expression of the pro-forms of IL-1β and IL-18 is augmented. The combination of type I IFN induction and inflammasome activation is important for rapid generation of protective anti-bacterial immunity.
During infection, the capacity for detection of bacterial infection by PRRs is increased by several mechanisms. The initial expression of TNF, type I IFNs, or IFN-γ leads to augmented transcription of genes coding for PRRs, thus increasing the expression levels of those receptors and also of pro-IL-1β or pro-IL-18 (Coers et al, 2007; Henry et al, 2007; Mancuso et al, 2007). In particular, the enhanced interferon-mediated expression of guanylate binding proteins (GBP) facilitates improved cytosolic sensing of LPS (Kim et al, 2011; Pilla et al, 2014). Furthermore, interferon-induced GBPs mediate inflammasome assembly and thereby promote the development of effector functions downstream of inflammasome activation (Shenoy et al, 2012). Two other important mechanisms further amplify detection of microbial DNA in the cytosol. First, RNA polymerase III detects double-stranded DNA and transcribes it into RNA-ligands, thereby allowing their detection by RIG-I (Ablasser et al, 2009). Second, DNA recognition by the nucleotidyl transferase cGAS leads to generation of the second messengers cyclic GMP-AMP that are recognized by STING leading to downstream IRF3 activation and enhanced IFN induction (Xiao & Fitzgerald, 2013). These amplifying mechanisms enhance nucleic acid sensing and ensure that even small concentrations of microbial nucleic acids detected within the cytosol are sufficient to initiate a robust and protective anti-bacterial immune response.
Dynamics of anti-bacterial immunity adapted to the threat of infecting bacteria
Detection of intracellular bacterial infection occurs in the context of bacterial replication and bacteria-mediated repression of PRR activation and therefore requires rapid yet sensitive and specific detection via PRRs. If intracellular bacteria have evaded phagolysosomal destruction and escaped into the cytosol, the detection of those invading bacteria needs to occur rapidly. Cytosolic sensing of bacterial nucleic acids has been found to be a key event in detection of intracellular viable bacteria (Sander et al, 2011; Abdullah et al, 2012). Since nucleic acids derived from dead bacteria do not trigger cytosolic immune sensing (Abdullah et al, 2012), the active release of bacterial nucleic acids in the cytosol during infection with Gram-positive or Gram-negative intracellular bacteria has led to the term ‘vita-PAMP’ that indicates both, viability and virulence of invading intracellular bacteria (Sander et al, 2011).
Activation of most cytosolic PRRs leads either to type I IFN production or to inflammasome activation. RIG-I does not show this functional dichotomy but rather has the ambivalent function to elicit at the same time both type I IFN production and inflammasome activation (Poeck et al, 2010; Pothlichet et al, 2013) (Fig 3). Even in the presence of bacterial escape at the cell surface or phagolysosomal compartments, this concomitant induction of type I IFN and inflammasome activation serves to increase detection in neighboring cells by the mechanisms described above. At the same time, it will contain intracellular infection through inflammasome-mediated pyroptosis that also leads to chemokine-dependent recruitment of further immune cells (Miao et al, 2010). A similar ambivalent response pattern has been observed during infection with pathogenic Gram-negative bacteria, where activation of TRIF signaling stimulates both, type IFN induction and non-canonical inflammasome activation via caspase-11 (Rathinam et al, 2012). However, caspase-11 activation in the absence of caspase-1, that is lack of proteolytic cleavage of pro-IL-1β and pro-IL-18, is not sufficient to control infection and is even detrimental to the outcome of Salmonella infection (Broz et al, 2012). Thus, the simultaneous induction of type I IFN and inflammasome activation leading to the release of IL-1β and IL-18 appears to provide an advantage for immune protection against intracellular bacteria. Along this line, lack of type I IFN signaling in infected cells leads to reduced immunity against Gram-negative intracellular bacteria (Rathinam et al, 2012). For infection with intracellular Gram-positive bacteria, the situation is similar. Clearance of infection with Francisella tularensis is impaired in the absence of type I IFN signaling (Fernandes-Alnemri et al, 2010), and type I interferon receptor (IFNAR) signaling is required for inflammasome activation during Listeria infection (Henry et al, 2007). Furthermore, there is an unequivocal role for type I IFN in recruitment of immune cells to the site of infection with Listeria (Jia et al, 2009).
Beyond the beneficial effect of type I IFN in innate immune responses against infection with intracellular bacteria, it plays also an important role in the induction of protective specific immunity. Animals devoid of lymphocytes and lacking adaptive immunity succumb to Listeria at day 10 after infection (Ladel et al, 1994). While the different immune cell populations contributing to Listeria-specific immunity have been well characterized (Pamer, 2004), little is known about the molecular mechanisms contributing to induction of protective bacteria-specific immunity. Inflammasome activation through release of IL-1β and IL-18 leads to stimulation and generation of T helper 17 cells or T helper 1 cells, respectively (Dostert et al, 2013), that are instrumental for infection control. Furthermore, inflammasome signals amplify innate bactericidal capacities of T helper 1 cells (O’Donnell et al, 2014). These reports revealed a direct activity of inflammasome-induced signals to amplify the anti-bacterial activity of T cells.
Although inflammasome activation and induction of innate immune responses are instrumental in the functional maturation of antigen-presenting cells, we do not know the exact contribution of inflammasome activation to the immediate anti-bacterial CD8 T-cell response. Cross-priming is a critical event in the induction of protective CD8 T-cell immunity against intracellular infection (Kurts et al, 2010) and against Listeria infection in particular (Jung et al, 2002). Type I IFN has been shown to increase cross-priming of CD8 T cells by antigen-presenting DCs (Le Bon et al, 2003). IL-1R-signaling in DCs can replace activation of PRRs promoting CD8 T-cell immunity (Pang et al, 2013). Consequently, in the absence of IFNAR-signaling, a loss of memory CD8 T-cell formation has been observed (Xiao et al, 2009). Importantly, a reduction in cytosolic detection of Listeria mutants lacking the secretory protein SecA2 leads to reduced protection through CD8 T cells upon re-infection with wild-type Listeria (Muraille et al, 2007; Rahmoun et al, 2011). Furthermore, IL-18 generated during inflammasome activation regulates non-cognate effector function by memory CD8 T cells, thus resulting in a broad enhancement of protective immunity (Kastenmuller et al, 2012; Kupz et al, 2012). Together with the recognized role of inflammasome activation during vaccination (Eisenbarth & Flavell, 2009), these results indicate an essential role of inflammasome activation in protective T-cell immunity beyond the relevance for innate immunity.
Notwithstanding this role of inflammasome activation in anti-bacterial defense, inflammasome-mediated induction of pyroptotic cell death and the ensuing inflammatory reaction can cause immunopathology in infected tissues (Cohen & Prince, 2013). While strong inflammatory responses during acute infection help to control local bacterial infection, longer lasting infection through the amplification loops discussed above can promote deleterious inflammation and tissue damage. Well-known immune regulatory mediators such as IL-10 or nitric oxide are released from activated immune cells and limit immunopathology (Bogdan, 2001; Saraiva & O’Garra, 2010; Nairz et al, 2013; Teixeira-Coelho et al, 2014). However, type I IFN beyond its beneficial effect during the initial phase of infection has further effects (Decker et al, 2005). Importantly, type I IFN can control inflammasome activation and subsequently release of IL-1β and IL-18 (Guarda et al, 2011). Further regulatory functions of type I IFN include augmented IL-10 expression and enhanced sensitivity of T cells or macrophages to undergo apoptosis (Guarda et al, 2011; Lee et al, 2012; Robinson et al, 2012).
This indicates that type I IFN acts differently during two distinct phases of infection: during acute infection, type I IFN enhances bacterial detection by PRRs and amplifies innate immune responses. During protracted or chronic infection associated with continuous and high-level expression, type I IFN rather regulates inflammasome activation. Since type I IFN is induced upon infection with various bacteria such as Listeria monocytogenes (Woodward et al, 2010; Abdullah et al, 2012), Salmonella typhimurium or group B streptococci (Parker et al, 2011), Francisella (Henry et al, 2007), and Mycobacteria (Stanley et al, 2007) it is possible that type I IFN may affect bacterial clearance via these regulatory properties. Indeed, there is evidence that Listeria infection is more rapidly cleared in the absence of signaling through the INFAR (Archer et al, 2014). Furthermore, chronic mycobacterial infection in humans is associated with high local IFN levels in infected tissues that impair protective immunity (Stanley et al, 2007; Teles et al, 2013). It is unclear, however, whether timing of exposure to, local levels of type I IFN, or the combination of both determine the induction or regulation of innate and protective immunity.
Concluding remarks
Taken together, multiple PRRs in different cellular compartments cooperate to create a dense network of surveillance in order to achieve most sensitive detection of infection with intracellular bacteria and to rapidly mount commensurate protective immunity. Distinction of dead from viable and virulent intracellular bacteria results from immune sensing of PAMPs and in particular recognition of functional properties of viable and pathogenic bacteria, that is secreted bacterial nucleic acids. This extends the existing concepts that PRRs distinguish foreign from self, and dangerous from harmless supporting that PRRs closely cooperate to generate decisive information concerning viability and virulence of intracellular bacteria that allows a commensurate immune response to infection. The combination of type I IFN expression and inflammasome activation early after infection fosters development of potent innate immune responses and protective T-cell immunity that are required to rapidly contain infection with pathogenic intracellular bacteria. However, type I IFN also regulates innate and adaptive immune responses which suggests that induction and control of anti-bacterial immune responses are continuously balanced right from the beginning of infection and that the timing of type I IFN induction and its concentration within the microenvironment are key factors determining infection control or persistence. Moreover, the mechanistic principles governing successful immunity against bacterial infection may be employed to develop novel immune therapies aiming at defeating persistent infection with intracellular bacteria without causing severe tissue damage by excessive immunity.
Acknowledgments
Funding by the Deutsche Forschungsgemeinschaft (SFBs 704, 670, TR57, TR36 and Excellence Cluster ImmunoSensation) and by the BMBF (PeTrA).
Glossary
- Aim2
(absent in melanoma 2) cytosolic DNA sensor
- ASC
(Apoptosis-associated speck-like protein containing a caspase-recruitment domain) signaling adapter molecule for canonical inflammasome activation
- DAMP
danger-associated molecular pattern
- DC
dendritic cell
- IFNAR
type I interferon receptor
- IFN
interferon
- IRF3
interferon responsive factor 3
- MAVS
(mitochondrial antiviral signaling protein) cytosolic signaling adapter molecule downstream of RIG-I or MDA5
- MDA5
(melanoma differentiation associated antigen 5) cytosolic RNA sensor
- MyD88
(myeloid differentiation primary response gene 88) signaling adapter molecule downstream of TLRs mediating NF-κB activation
- NLRP3
(NOD-like receptor family, pyrin domain containing 3)inflammasome component
- NOD
(nucleotide-binding oligomerization domain) cytosolic pattern recognition receptor
- PAMP
pathogen-associated molecular pattern
- PRR
pattern recognition receptor
- RIG-I
(retinoic acid inducible gene I) cytosolic RNA sensor
- SecA2
auxiliary protein secretion system in bacteria
- STING
(stimulator of interferon genes) cytosolic DNA sensor and signaling adapter molecule
- TBK1
(tank binding kinase) signaling adapter molecule
- TLR
Toll-like receptor
- TRIF
(TIR-domain-containing adapter-inducing interferon-β) signaling molecule downstream of TLR4 and TLR3
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, Civril F, Hopfner KP, Kurts C, Ruland J, Hartmann G, Chakraborty T, Knolle PA. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Ali SR, Timmer AM, Bilgrami S, Park EJ, Eckmann L, Nizet V, Karin M. Anthrax toxin induces macrophage death by p38 MAPK inhibition but leads to inflammasome activation via ATP leakage. Immunity. 2011;35:34–44. doi: 10.1016/j.immuni.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer KA, Durack J, Portnoy DA. STING-dependent type I IFN production inhibits cell-mediated immunity to Listeria monocytogenes. PLoS Pathog. 2014;10:e1003861. doi: 10.1371/journal.ppat.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Barton GM. The impact of Toll-like receptors on bacterial virulence strategies. Curr Opin Microbiol. 2013;16:17–22. doi: 10.1016/j.mib.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, Keilty JJ, Gosselin ML, Robison KE, Wong GH, Glucksmann MA, DiStefano PS. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J Biol Chem. 1999;274:12955–12958. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol. 2012;12:215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari I, Chao Y, Romby P, Vogel J. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb Perspect Med. 2013;3:a010298. doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson CN, Copenhaver AM, Zwack EE, Nguyen HT, Strowig T, Javdan B, Bradley WP, Fung TC, Flavell RA, Brodsky IE, Shin S. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog. 2013;9:e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T, Ebel F, Domann E, Niebuhr K, Gerstel B, Pistor S, Temm-Grove CJ, Jockusch BM, Reinhard M, Walter U. A focal adhesion factor directly linking intracellularly motile Listeria monocytogenes and Listeria ivanovii to the actin-based cytoskeleton of mammalian cells. EMBO J. 1995;14:1314–1321. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- Coers J, Vance RE, Fontana MF, Dietrich WF. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 2007;9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- Cohen TS, Prince AS. Activation of inflammasome signaling mediates pathology of acute P. aeruginosa pneumonia. J Clin Invest. 2013;123:1630–1637. doi: 10.1172/JCI66142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha LD, Zamboni DS. Subversion of inflammasome activation and pyroptosis by pathogenic bacteria. Front Cell Infect Microbiol. 2013;3:76. doi: 10.3389/fcimb.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- Diacovich L, Gorvel JP. Bacterial manipulation of innate immunity to promote infection. Nat Rev Microbiol. 2010;8:117–128. doi: 10.1038/nrmicro2295. [DOI] [PubMed] [Google Scholar]
- Dostert C, Ludigs K, Guarda G. Innate and adaptive effects of inflammasomes on T cell responses. Curr Opin Immunol. 2013;25:359–365. doi: 10.1016/j.coi.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gao X, Huang MT, O’Connor BP, Thomas CE, Willingham SB, Bergstralh DT, Jarvis GA, Sparling PF, Ting JP. Neisseria gonorrhoeae activates the proteinase cathepsin B to mediate the signaling activities of the NLRP3 and ASC-containing inflammasome. J Immunol. 2009;182:6460–6469. doi: 10.4049/jimmunol.0802696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC, Flavell RA. Innate instruction of adaptive immunity revisited: the inflammasome. EMBO Mol Med. 2009;1:92–98. doi: 10.1002/emmm.200900014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, Eisenlohr L, Landel CP, Alnemri ES. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, Nunez G. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Jeong WI, Tian Z. Liver: an organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- Ge J, Gong YN, Xu Y, Shao F. Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc Natl Acad Sci USA. 2012;109:6193–6198. doi: 10.1073/pnas.1117490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003a;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003b;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, Farlik M, Decker T, Du Pasquier RA, Romero P, Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, Thomas BJ, Malosse C, Gantier MP, Casillas LN, Votta BJ, Bertin J, Boneca IG, Sasakawa C, Philpott DJ, Ferrero RL, Kaparakis-Liaskos M. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe. 2014;15:623–635. doi: 10.1016/j.chom.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia T, Leiner I, Dorothee G, Brandl K, Pamer EG. MyD88 and Type I interferon receptor-mediated chemokine induction and monocyte recruitment during Listeria monocytogenes infection. J Immunol. 2009;183:1271–1278. doi: 10.4049/jimmunol.0900460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M, Askarian F, Sangvik M, Sollid JE. Bacterial interference with canonical NFkappaB signalling. Microbiology. 2013;159:2001–2013. doi: 10.1099/mic.0.069369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Unutmaz D, Wong P, Sano G, De lSK, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, Pamer EG, Littman DR, Lang RA. In vivo depletion of CD11c(+) dendritic cells abrogates priming of CD8(+) T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmuller W, Torabi-Parizi P, Subramanian N, Lammermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell. 2012;150:1235–1248. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, Forsberg LS, Carlson RW, Dixit VM. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Lee HM, Shin DM, Kim W, Yuk JM, Jin HS, Lee SH, Cha GH, Kim JM, Lee ZW, Shin SJ, Yoo H, Park YK, Park JB, Chung J, Yoshimori T, Jo EK. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11:457–468. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Knolle PA, Loser E, Protzer U, Duchmann R, Schmitt E, zum BK, Rose-John S, Gerken G. Regulation of endotoxin-induced IL-6 production in liver sinusoidal endothelial cells and Kupffer cells by IL-10. Clin Exp Immunol. 1997;107:555–561. doi: 10.1046/j.1365-2249.1997.d01-959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupz A, Guarda G, Gebhardt T, Sander LE, Short KR, Diavatopoulos DA, Wijburg OL, Cao H, Waithman JC, Chen W, Fernandez-Ruiz D, Whitney PG, Heath WR, Curtiss R, 3rd, Tschopp J, Strugnell RA, Bedoui S. NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8(+) T cells. Nat Immunol. 2012;13:162–169. doi: 10.1038/ni.2195. [DOI] [PubMed] [Google Scholar]
- Kurts C, Robinson BW, Knolle PA. Cross-priming in health and disease. Nat Rev Immunol. 2010;10:403–414. doi: 10.1038/nri2780. [DOI] [PubMed] [Google Scholar]
- Ladel CH, Flesch IE, Arnoldi J, Kaufmann SH. Studies with MHC-deficient knock-out mice reveal impact of both MHC I- and MHC II-dependent T cell responses on Listeria monocytogenes infection. J Immunol. 1994;153:3116–3122. [PubMed] [Google Scholar]
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D, Borrow P, Tough DF. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat Immunol. 2003;4:1009–1015. doi: 10.1038/ni978. [DOI] [PubMed] [Google Scholar]
- Lee SE, Li X, Kim JC, Lee J, Gonzalez-Navajas JM, Hong SH, Park IK, Rhee JH, Raz E. Type I interferons maintain Foxp3 expression and T-regulatory cell functions under inflammatory conditions in mice. Gastroenterology. 2012;143:145–154. doi: 10.1053/j.gastro.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, Moriarty TJ, Wong CH, Zhou H, Strieter RM, van Rooijen N, Chaconas G, Kubes P. An intravascular immune response to Borrelia burgdorferi involves Kupffer cells and iNKT cells. Nat Immunol. 2010;11:295–302. doi: 10.1038/ni.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz LL, Mohammadi S, Geissler A, Portnoy DA. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc Natl Acad Sci USA. 2003;100:12432–12437. doi: 10.1073/pnas.2133653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz LL, Portnoy DA. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol Microbiol. 2002;45:1043–1056. doi: 10.1046/j.1365-2958.2002.03072.x. [DOI] [PubMed] [Google Scholar]
- Lindgren H, Golovliov I, Baranov V, Ernst RK, Telepnev M, Sjostedt A. Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol. 2004;53:953–958. doi: 10.1099/jmm.0.45685-0. [DOI] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R, Tomasello F, Gambuzza M, Macri G, Ruggeri A, Leanderson T, Teti G. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007;14:10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- Mattos KA, Oliveira VC, Berredo-Pinho M, Amaral JJ, Antunes LC, Melo RC, Acosta CC, Moura DF, Olmo R, Han J, Rosa PS, Almeida PE, Finlay BB, Borchers CH, Sarno EN, Bozza PT, Atella GC, Pessolani MC. Mycobacterium leprae intracellular survival relies on cholesterol accumulation in infected macrophages: a potential target for new drugs for leprosy treatment. Cell Microbiol. 2014;16:797–815. doi: 10.1111/cmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixenberger K, Pache F, Eitel J, Schmeck B, Hippenstiel S, Slevogt H, N’Guessan P, Witzenrath M, Netea MG, Chakraborty T, Suttorp N, Opitz B. Listeria monocytogenes-infected human peripheral blood mononuclear cells produce IL-1beta, depending on listeriolysin O and NLRP3. J Immunol. 2010;184:922–930. doi: 10.4049/jimmunol.0901346. [DOI] [PubMed] [Google Scholar]
- Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BB, Rathinam VA, Martens GW, Martinot AJ, Kornfeld H, Fitzgerald KA, Sassetti CM. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nat Immunol. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraille E, Narni-Mancinelli E, Gounon P, Bassand D, Glaichenhaus N, Lenz LL, Lauvau G. Cytosolic expression of SecA2 is a prerequisite for long-term protective immunity. Cell Microbiol. 2007;9:1445–1454. doi: 10.1111/j.1462-5822.2007.00883.x. [DOI] [PubMed] [Google Scholar]
- Myers JT, Tsang AW, Swanson JA. Localized reactive oxygen and nitrogen intermediates inhibit escape of Listeria monocytogenes from vacuoles in activated macrophages. J Immunol. 2003;171:5447–5453. doi: 10.4049/jimmunol.171.10.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz M, Schleicher U, Schroll A, Sonnweber T, Theurl I, Ludwiczek S, Talasz H, Brandacher G, Moser PL, Muckenthaler MU, Fang FC, Bogdan C, Weiss G. Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J Exp Med. 2013;210:855–873. doi: 10.1084/jem.20121946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell H, Pham OH, Li LX, Atif SM, Lee SJ, Ravesloot MM, Stolfi JL, Nuccio SP, Broz P, Monack DM, Baumler AJ, McSorley SJ. Toll-like receptor and inflammasome signals converge to amplify the innate bactericidal capacity of T helper 1 cells. Immunity. 2014;40:213–224. doi: 10.1016/j.immuni.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- Pang IK, Ichinohe T, Iwasaki A. IL-1R signaling in dendritic cells replaces pattern-recognition receptors in promoting CD8(+) T cell responses to influenza A virus. Nat Immunol. 2013;14:246–253. doi: 10.1038/ni.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Martin FJ, Soong G, Harfenist BS, Aguilar JL, Ratner AJ, Fitzgerald KA, Schindler C, Prince A. Streptococcus pneumoniae DNA initiates type I interferon signaling in the respiratory tract. mBio. 2011;2:e00016–00011. doi: 10.1128/mBio.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lopez A, Rosales-Reyes R, Alpuche-Aranda CM, Ortiz-Navarrete V. Salmonella downregulates Nod-like receptor family CARD domain containing protein 4 expression to promote its survival in B cells by preventing inflammasome activation and cell death. J Immunol. 2013;190:1201–1209. doi: 10.4049/jimmunol.1200415. [DOI] [PubMed] [Google Scholar]
- Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA, Coers J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci USA. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, Kato H, Akira S, Inoue S, Endres S, Peschel C, Hartmann G, Hornung V, Ruland J. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Pothlichet J, Meunier I, Davis BK, Ting JP, Skamene E, von Messling V, Vidal SM. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog. 2013;9:e1003256. doi: 10.1371/journal.ppat.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py BF, Jin M, Desai BN, Penumaka A, Zhu H, Kober M, Dietrich A, Lipinski MM, Henry T, Clapham DE, Yuan J. Caspase-11 controls interleukin-1beta release through degradation of TRPC1. Cell Rep. 2014;6:1122–1128. doi: 10.1016/j.celrep.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmoun M, Gros M, Campisi L, Bassand D, Lazzari A, Massiera C, Narni-Mancinelli E, Gounon P, Lauvau G. Priming of protective anti-Listeria monocytogenes memory CD8+ T cells requires a functional SecA2 secretion system. Infect Immun. 2011;79:2396–2403. doi: 10.1128/IAI.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, Fitzgerald KA. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE., 3rd A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol. 2012;13:954–962. doi: 10.1038/ni.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, Komatsu R, Miura N, Adachi Y, Ohno N, Shibuya K, Yamamoto N, Kawakami K, Yamasaki S, Saito T, Akira S, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. Toll-like receptors control activation of adaptive immune responses. Nat Immunol. 2001;2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, MacMicking JD. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J Cell Biol. 2000;150:527–538. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolovska A, Becker CE, Ip WK, Rathinam VA, Brudner M, Paquette N, Tanne A, Vanaja SK, Moore KJ, Fitzgerald KA, Lacy-Hulbert A, Stuart LM. Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function. Nat Immunol. 2013;14:543–553. doi: 10.1038/ni.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- Sun J, Singh V, Lau A, Stokes RW, Obregon-Henao A, Orme IM, Wong D, Av-Gay Y, Hmama Z. Mycobacterium tuberculosis nucleoside diphosphate kinase inactivates small GTPases leading to evasion of innate immunity. PLoS Pathog. 2013;9:e1003499. doi: 10.1371/journal.ppat.1003499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Inohara N, Sasakawa C, Nunez G. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, Brown GD. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira-Coelho M, Guedes J, Ferreirinha P, Howes A, Pedrosa J, Rodrigues F, Lai WS, Blackshear PJ, O’Garra A, Castro AG, Saraiva M. Differential post-transcriptional regulation of IL-10 by TLR2 and TLR4-activated macrophages. Eur J Immunol. 2014;44:856–866. doi: 10.1002/eji.201343734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RM, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, Komisopoulou E, Kelly-Scumpia K, Chun R, Iyer SS, Sarno EN, Rea TH, Hewison M, Adams JS, Popper SJ, Relman DA, Stenger S, Bloom BR, Cheng G, Modlin RL. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 2013;339:1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Memet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- Wong CH, Jenne CN, Petri B, Chrobok NL, Kubes P. Nucleation of platelets with blood-borne pathogens on Kupffer cells precedes other innate immunity and contributes to bacterial clearance. Nat Immunol. 2013;14:785–792. doi: 10.1038/ni.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Mol Cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Casey KA, Jameson SC, Curtsinger JM, Mescher MF. Programming for CD8 T cell memory development requires IL-12 or type I IFN. J Immunol. 2009;182:2786–2794. doi: 10.4049/jimmunol.0803484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci USA. 2013;110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Kepp O, Galluzzi L, Kroemer G. Inflammasomes in carcinogenesis and anticancer immune responses. Nat Immunol. 2012;13:343–351. doi: 10.1038/ni.2224. [DOI] [PubMed] [Google Scholar]