Abstract
Leucine-rich repeat kinase 2 (LRRK2) has been associated with Parkinson’s disease (PD) and other disorders. However, its normal physiological functions and pathogenic properties remain elusive. Here we show that LRRK2 regulates the anterograde ER–Golgi transport through anchoring Sec16A at the endoplasmic reticulum exit sites (ERES). LRRK2 interacted and co-localized with Sec16A, a key protein in the formation of ERES. Lrrk2 depletion caused a dispersion of Sec16A from ERES and impaired ER export. In neurons, LRRK2 and Sec16A showed extensive co-localization at the dendritic ERES (dERES) that locally regulate the transport of proteins to the dendritic spines. A loss of Lrrk2 affected the association of Sec16A with dERES and impaired the activity-dependent targeting of glutamate receptors onto the cell/synapse surface. Furthermore, the PD-related LRRK2 R1441C missense mutation in the GTPase domain interfered with the interaction of LRRK2 with Sec16A and also affected ER–Golgi transport, while LRRK2 kinase activity was not required for these functions. Therefore, our findings reveal a new physiological function of LRRK2 in ER–Golgi transport, suggesting ERES dysfunction may contribute to the pathogenesis of PD.
Keywords: dendritic ERES (dERES), ER exit sites (ERES), ER–Golgi transport, Leucine-rich repeat kinase 2 (LRRK2), Sec16A
See also: B Lu (October 2014)
Introduction
Missense mutations in LRRK2 have been initially linked to a late-onset autosomal dominant form of familial Parkinson’s disease (PD) (Paisan-Ruiz et al, 2004; Zimprich et al, 2004). The genome-wide association studies have further revealed that LRRK2 gene loci are also associated with sporadic PD, Crohn’s disease, and leprosy (Barrett et al, 2008; Satake et al, 2009; Simon-Sanchez et al, 2009; Zhang et al, 2009). LRRK2 encodes a large multi-domain protein, containing one Ras of complex GTPase (ROC) domain, one serine/threonine kinase domain, and multiple protein–protein interaction domains (Mata et al, 2006). LRRK2 has been shown to regulate a variety of cellular events, such as microtubule and actin cytoskeleton assembly, protein translation, microRNA maturation, nuclear factor of activated T-cells (NFAT) signaling, endocytosis, neuronal morphogenesis, and autophagy/lysosome-mediated protein degradation (Jaleel et al, 2007; Gandhi et al, 2008; Imai et al, 2008; Plowey et al, 2008; Lin et al, 2009; Parisiadou et al, 2009, 2014; Liu et al, 2011; Matta et al, 2012; Orenstein et al, 2013). In addition, our previous study in transgenic mice demonstrates that overexpression of LRRK2 causes Golgi fragmentation in neurons (Lin et al, 2009). PD-related-α-synuclein has also been shown to block ER–Golgi transport by inhibiting the docking and fusion of coat protein complex II (COPII) vesicles with Golgi apparatus (Cooper et al, 2006; Gitler et al, 2008). However, how LRRK2 regulates ER–Golgi trafficking is unclear.
ER–Golgi export is initiated by formation and secretion of COPII vesicles at the specialized ER domains, namely ER exit sites (ERES) or transitional ER (tER) (Miller & Barlowe, 2010; Zanetti et al, 2012). ER-associated membrane protein Sec12 mediates the assembly of COPII vesicles at ERES by activating GTPase Sar1, which subsequently recruits inner layer heterodimer protein complex Sec23 and Sec24 (Sec23/24) as well as outer layer protein complex Sec13 and Sec31 (Sec13/31) for the capture of cargo proteins into nascent buds and formation of a structural cage around the budding vesicles, respectively (Zanetti et al, 2012). Additionally, Sec16A, a large hydrophilic peripheral membrane protein, has been identified recently as an essential component required for the formation of ERES and COPII vesicle trafficking (Espenshade et al, 1995; Connerly et al, 2005; Watson et al, 2006; Hughes et al, 2009). However, what acts at the upstream of Sec16A that regulates the recruitment of Sec16A onto ERES is poorly understood (Miller & Barlowe, 2010).
In this study, we identified Sec16A as an interacting protein of LRRK2 by mass spectrometry proteomic screening of LRRK2-associated proteins. Immunostaining further revealed the co-localization of LRRK2 and Sec16A at ERES in cell lines and primary cultures. LRRK2 and Sec16A also extensively co-localized at the dendritic ERES (dERES) in neurons. Although inhibition of Sec16A did not affect the expression and subcellular distribution of LRRK2, a loss of Lrrk2 significantly reduced the association of Sec16A with ERES and impaired the clustering of ERES and transport of COPII vesicles, suggesting that LRRK2 may play an important role in anchoring Sec16A at ERES and thereby regulating COPII vesicle formation and transport. In neurons, inhibition of Lrrk2 led to dissociation of Sec16A from dERES and impaired the activity-dependent transport of glutamate receptors onto cell surface. We further evaluated the impact of different LRRK2 functional domains and PD-related missense mutations on the ERES localization of Sec16A and COPII vesicle transport and found that a PD-related R1441C missense mutation in the ROC domain of LRRK2 interfered with the interaction of LRRK2 with Sec16A and impaired ER export. Together, these findings reveal a new physiological function of LRRK2 that acts upstream of Sec16A in the formation of ERES during ER–Golgi transport, implying an involvement of ERES dysfunction in the pathogenesis of PD.
Result
LRRK2 interacts and co-localizes with ERES protein Sec16A
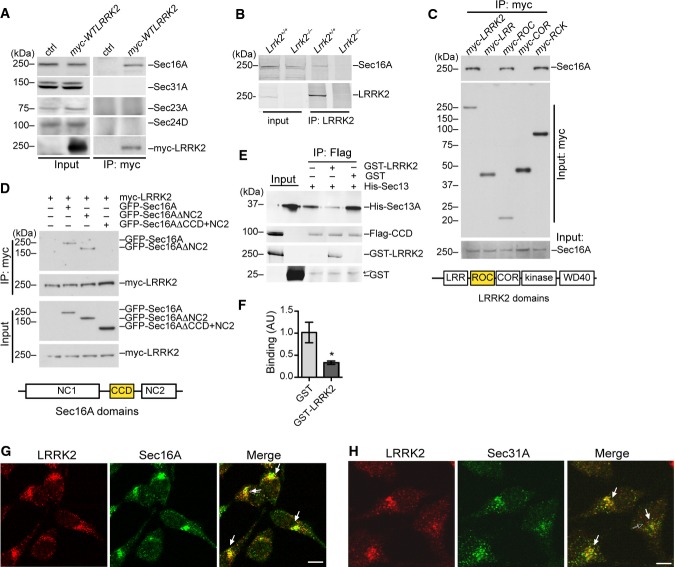
In an effort to identify LRRK2-interacting proteins, we performed mass spectrometric analyses of proteins that co-immunoprecipitated (co-IPed) with LRRK2 in HEK293T cells transiently transfected with amino-terminal myc-tagged human wild-type (WT) LRRK2 full-length cDNA (myc-WTLRRK2). We identified Sec16A, an ERES-associated protein, was selectively pulled down with LRRK2 (Supplementary Fig S1A). We further confirmed the interaction of endogenous Sec16A with myc-WTLRRK2 in transfected HEK293T cells by co-IP (Fig 1A). The specificity of a custom-made Sec16A antibody (HL7200) that recognizes both mouse and human Sec16A proteins was verified in HEK293T cells transfected with either a GFP-tagged human Sec16A full-length cDNA (GFP-Sec16A) or Sec16A siRNA oligos (Supplementary Fig S1B and C). Moreover, we demonstrate the association of LRRK2 with Sec16A in vivo by co-IP of endogenous LRRK2 and Sec16A proteins from the brain extract of Lrrk2 WT (Lrrk2+/+) but not knockout (Lrrk2−/−) mice using an anti-LRRK2 antibody N138 (Fig 1B). By contrast, other ERES and COPII vesicle-associated Sec proteins such as Sec31A, Sec23A, and Sec24D proteins did not co-IP with LRRK2 (Fig 1A), indicating a selective interaction between LRRK2 and Sec16A at the ERES. Further experiments revealed the ROC domain of LRRK2 and the central conserved domain (CCD) of Sec16A were responsible for the interaction between LRRK2 and Sec16A (Fig 1C and D). A previous study shows that Sec16A also binds to COPII component Sec13A through its CCD (Whittle & Schwartz, 2010). To investigate whether LRRK2 competes with Sec13A in binding with the CCD domain of Sec16A, we performed in vitro recombinant protein binding assays using FLAG-tagged Sec16A CCD (Flag-CCD) purified from HEK293 cells, and GST-tagged LRRK2 (GST-LRRK2) and His-tagged Sec13A (His-Sec13A) proteins prepared from E. coli. Consistent with the early IP data, we found GST-LRRK2 and Flag-CCD recombinant proteins co-IPed in this cell-free assay (Fig 1E). Furthermore, the presence of GST-LRRK2 but not the control GST proteins suppressed the binding of Sec13A with the CCD of Sec16A (Fig 1E and F). Together, these co-IP experiments identify Sec16A as a new LRRK2-interacting protein that binds to the ROC domain of LRRK2 through its CCD domain.
Figure 1. LRRK2 interacts and co-localizes with Sec16A at ERES.
A Protein extracts from HEK293T cells transiently transfected with myc-LRRK2 WT were immunoprecipitated (IPed) with an anti-myc antibody and probed with Sec16A, Sec31A, Sec23A, Sec24D, and myc antibodies. Two percent of total extracts were subjected to Western blot analyses as the input.
B Protein extracts from the cerebral cortex of Lrrk2+/+ and Lrrk2−/− mice were IPed with a LRRK2 antibody (N138) and probed with Sec16A antibody.
C Lysates prepared from HEK293T cells transiently transfected with various myc-tagged LRRK2 domain constructs (LRR, ROC, COR, and RCK domains) were subjected to immunoprecipitation with an anti-myc antibody and Western blot analyses with an anti-Sec16A antibody. The diagram outlines the structural domains of LRRK2 protein. LRR: leucine-rich repeat domain, ROC: Ras of complex proteins domain, COR: C-terminal of ROC domain, RCK: ROC, COR, and kinase domains.
D Lysates obtained from HEK293T cells co-transfected with myc-LRRK2 and GFP-tagged Sec16A full-length and C-terminal deletion constructs were subjected to immunoprecipitation with an anti-myc antibody, followed by anti-GFP immunoblotting. Sec16AΔNC2 and Sec16AΔCCD+NC2 constructs contain residues 1–1,589 and 1–931 of Sec16A, respectively. The diagram shows the structural domains of Sec16A protein. NC1: non-conserved domain 1, CCD: central conserved domain, NC2: non-conserved domain 2.
E, F Sec16A CCD and Sec13A recombinant proteins were co-IPed in the presence of LRRK2 or GST recombinant proteins (E). An arrow points to the non-specific band. The bar graph (F) depicts the binding affinity between Sec16A CCD and Sec13A competed with GST or LRRK2. The data are presented as means ± SEM. N = 4 independent experiments. P < 0.05.
G HeLa cells were double-labeled with LRRK2 (red) and Sec16A (green) antibodies. Arrows point out the juxtanuclear localization of LRRK2 and Sec16A. Scale bar: 10 μm.
H HeLa cells were co-stained with LRRK2 (red) and Sec31A (green) antibodies. Arrows point out the juxtanuclear localization of LRRK2 and Sec31A. Scale bar: 10 μm.
Source data are available online for this figure.
Next, to determine whether LRRK2 and Sec16A co-localize at ERES, we performed co-immunostaining of endogenous LRRK2 and Sec16A in HeLa cells. An extensive overlap of LRRK2 and Sec16A immunostaining was found in the juxtanuclear region of HeLa cells with a few being scattered around the cytosol and cell periphery (Fig 1G). In addition, co-immunostaining experiments also revealed a substantial co-localization of LRRK2 with Sec31A, a well-characterized ERES and COPII coat protein near the nucleus of HeLa cells (Fig 1H). These results together demonstrate that endogenous LRRK2 co-localizes with Sec16A at the ERES.
Two different LRRK2 antibodies, OC83A and N138 that recognize both human and mouse LRRK2 protein (Cho et al, 2013), were used to determine the subcellular localization of endogenous human LRRK2 in HeLa cells (Supplementary Fig S1D and E). In addition, LRRK2 also showed partial co-staining with ER and Golgi markers calnexin and syntaxin 6 (Syn6), respectively (Supplementary Fig S1F). Using an additional rabbit polyclonal antibody (4EC9E) against endogenous mouse LRRK2 (Parisiadou et al, 2009), we observed a similar juxtanuclear distribution of LRRK2 in Lrrk2+/+ mouse fibroblast cells but not in Lrrk2−/− cells (Supplementary Fig S1G).
To further confirm the localization of LRRK2 protein at ERES, we also performed subcellular fractionation of HEK293T cells. LRRK2 protein was present in the cytosol, microsome, and mitochondria fractions, but was more abundant in the microsome fraction that was enriched with ERES and COPII proteins Sec31A and Sec23A (Supplementary Fig S1H). Moreover, fast protein liquid chromatography (FPLC) experiments revealed that the majority of LRRK2 protein was co-eluted with Sec16A with a corresponding molecular weight around 700 kDa (Supplementary Fig S1I). Taken together, these data demonstrate that LRRK2 may form high molecular complex with Sec16A at the ERES.
LRRK2 regulates the juxtanuclear ERES localization of Sec16A
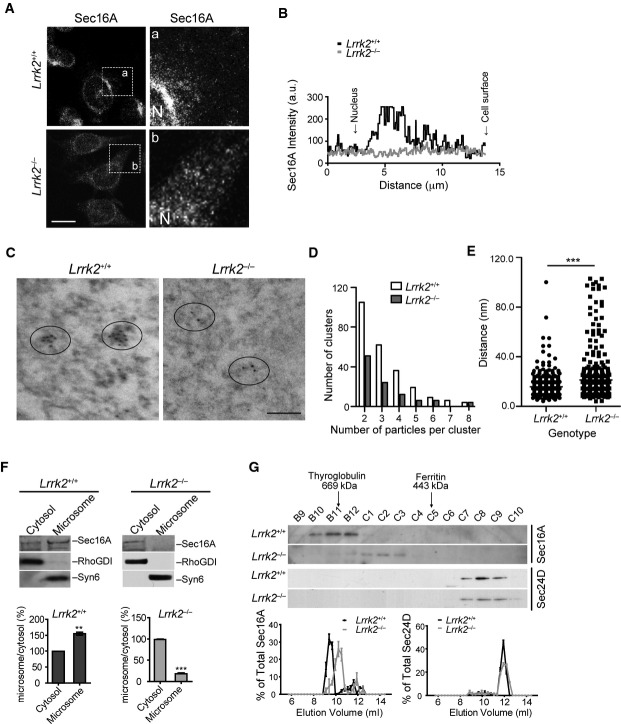
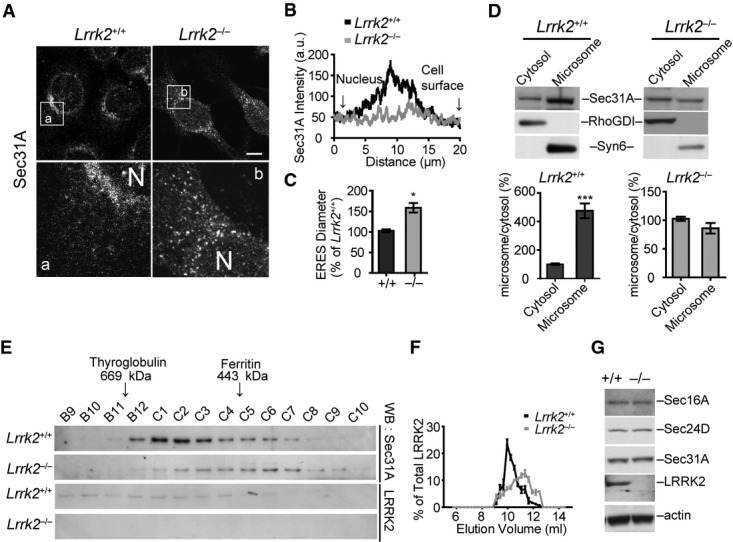
Sec16A is essential for ERES formation and COPII vesicle trafficking (Yorimitsu & Sato, 2012). To investigate whether LRRK2 regulates the function of Sec16A at ERES, we examined the subcellular distribution of Sec16A in Lrrk2+/+ and Lrrk2−/− immortalized mouse fibroblast cell lines obtained from newborn Lrrk2+/+ and Lrrk2−/− pups (Law et al, 2014). A substantial reduction of juxtanuclear distribution of Sec16A was observed in the Lrrk2−/− fibroblast cell lines and primary fibroblasts compared to the controls (Fig 2A and B; Supplementary Fig S2A). This alteration of Sec16A staining in Lrrk2−/− cells was further confirmed in Lrrk2+/+ and Lrrk2−/− fibroblast cell lines transfected with GFP-Sec16A (Supplementary Fig S2B). We further performed immnuno-electron microscopy (EM) of Sec16A on Lrrk2+/+ and Lrrk2−/− fibroblasts. We counted the number of Sec16A-positive clusters with two or more gold particles within 100 nm radius and measured the shortest distance between two particles. We found fewer clusters but longer spacing between the Sec16A-positive particles in Lrrk2−/− cells (Fig 2C–E). These immune-EM data are consistent with the observations with light microscopy that Sec16A staining was more spreading in Lrrk2−/− cells. On the other hand, either knockdown or overexpression of Sec16A did not affect the subcellular localization of LRRK2 (Supplementary Fig S2C and D). Taken together, these data indicate that LRRK2 may act upstream of Sec16A in maintaining the juxtanuclear ERES localization of Sec16A in cells.
Figure 2. The subcellular distribution of Sec16A is altered in LRRK2-deficient cells.
A, B Immunostaining of Sec16A in immortalized fibroblasts derived from Lrrk2+/+ and Lrrk2−/− mice (A). Images in a/b are of a high magnification from the boxed areas. N marks the location of nucleus. The line graph shows the distribution of Sec16A signal intensity from the edge of nucleus to the cell periphery. Scale bar: 10 μm.
C–E Immuno-EM shows Sec16A distribution in Lrrk2+/+ and Lrrk2−/− fibroblast cell lines (C). Ellipses encircle Sec16A-positive particles. Bar graph shows the distribution of Sec16A-positive clusters with two or more particles in Lrrk2+/+ and Lrrk2−/− cells (D). P < 0.05. Scatter plot depicts the shortest distance between two adjacent Sec16A-positive particles in Lrrk2+/+ and Lrrk2−/− cells (E). ***P < 0.001.
F Western blot analyses show Sec16A in the cytosol and membrane fractions of 1-month-old Lrrk2+/+ and Lrrk2−/− mouse brains. Rho GDP dissociated inhibitor (RhoGDI) and syntaxin 6 (Syn6) were used as the markers for the cytosol and membrane fractions, respectively. Bar graphs show the ratio of Sec16A in membrane versus cytosol fractions. Data are presented as means ± SEM for three independent experiments. **P < 0.01, ***P < 0.001.
G Fast protein liquid chromatography (FPLC) analyses of Sec16A and Sec24D in 1-month-old Lrrk2+/+ and Lrrk2−/− mouse brain extracts. Line graphs show the distributions of Sec16A and Sec24D in each elute versus the sum of total elutes.
Source data are available online for this figure.
We next performed differential fractionation to determine whether LRRK2 is important for the microsome association of Sec16A. Cytosolic and microsome fractions were obtained from Lrrk2+/+ and Lrrk2−/− mouse brain extracts and then assessed by Western blot analyses, in which RHO protein GDP dissociation inhibitor (RhoGDI) and Syn6 were used as the markers for the cytosol and microsome fractions, respectively (Fukumoto et al, 1990; Bock et al, 1997). Sec16A protein mainly resided in the microsome fraction of Lrrk2+/+ mouse brain extracts, whereas in Lrrk2−/− mouse brain extracts, more Sec16A protein presented into the cytosol fraction (Fig 2F). These results suggest that LRRK2 plays an important role in anchoring Sec16A at ERES.
We also performed FPLC analyses of brain extracts prepared from 1-month-old Lrrk2+/+ and Lrrk2−/− mice to investigate whether the molecular weight of Sec16A-containing protein complexes is shifted under the Lrrk2 knockout condition. Indeed, a significant redistribution of Sec16A into lower molecular weight protein complex was observed in Lrrk2−/− mouse brain samples (Fig 2G). As a control, the molecular weight of COPII vesicle inner layer protein Sec24D-containing protein complex remained unchanged in Lrrk2−/− samples (Fig 2G). Together these data provide evidence that LRRK2 is important in selectively interacting with Sec16A to form high molecular weight protein complex at the ERES.
Loss of LRRK2 impairs the organization of ERES and COPII vesicle formation
To further investigate the potential function of LRRK2 in ERES formation and COPII vesicle transport, we examined the subcellular distribution of Sec31A in Lrrk2−/− fibroblasts. In control Lrrk2+/+ cells, Sec31A displayed typical ERES localization concentrated near the nucleus, while less juxtanuclear enrichment of Sec31A staining was also observed in Lrrk2−/− fibroblasts (Fig 3A and B). In addition, the diameter of Sec31A-positive COPII vesicles in Lrrk2−/− cells appeared larger compared with the control cells (Fig 3C). The similarity between the alteration of subcellular distribution of ERES and COPII proteins Sec16A and Sec31A in Lrrk2−/− cells further demonstrates the involvement of LRRK2 in maintaining the normal structure and function of ERES and COPII vesicles.
Figure 3. The subcellular distribution of COPII protein Sec31A is altered in Lrrk2-deficient cells.
A–C Immunostaining of Sec31A in Lrrk2+/+ and Lrrk2−/− primary fibroblasts (A). Bottom panels show the enlargement of the boxed areas in the top panels. N indicates the location of the nucleus. Scale bar: 10 μm. Line graph shows the distribution of Sec31A signal from the edge of nucleus to the cell periphery (B). Bar graph depicts the diameter of Sec31A-positive puncta in Lrrk2+/+ and Lrrk2−/− cells (C). Data are presented as means ± SEM. 10–15 cells were analyzed for each genotype. *P < 0.05.
D Western blot analyses of Sec31A in the cytosol and microsome fractions of Lrrk2+/+ and Lrrk2−/− mouse brains. RhoGDI and Syn6 were used as the marker for the cytosol and microsome fractions, respectively. Bar graphs show the ratio of Sec31A in the microsome fraction versus the cytosol fraction. Data are presented as means ± SEM for three independent experiments. ***P < 0.001.
E, F FPLC analysis of Sec31A in Lrrk2+/+ and Lrrk2−/− mouse brain extracts (E). Line graph shows the distributions of Sec31A in each elute versus the sum of total elute (F).
G Western blots show Sec16A, Sec24D, Sec31A, LRRK2 expression in the whole-brain extracts of 1-month-old Lrrk2+/+ and Lrrk2−/− mice. The expression of actin was used as the loading control.
Source data are available online for this figure.
Similar to Sec16A protein, less Sec31A was detected in the microsome fraction of Lrrk2−/− mouse brain extracts (Fig 3D). FPLC experiments also revealed a significant shift of Sec31A-containing protein complex to lower molecular weights in Lrrk2−/− mouse brain extracts (Fig 3E and F). Meanwhile, the overall expression levels of Sec16A and Sec31A were comparable in Lrrk2+/+ and Lrrk2−/− mouse brain extracts (Fig 3G). These results together establish an important function of LRRK2 in anchoring ERES and COPII vesicle-associated protein complex at the ERES.
Loss of Lrrk2 impairs ERES function and COPII vesicle transport
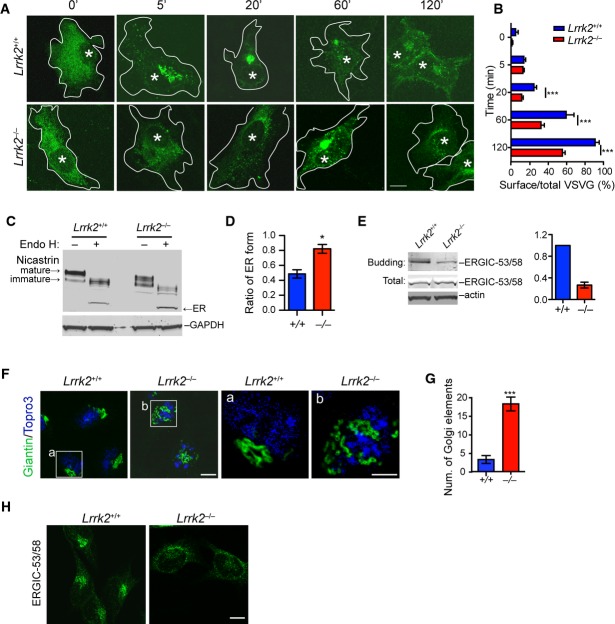
To investigate whether LRRK2 regulates COPII vesicle transport, we examined the kinetics of export and subsequent transport of vesicular stomatitis virus G protein tagged with green fluorescent protein (VSVG-GFP), a well-characterized ER–Golgi export reporter (Presley et al, 1997), in Lrrk2+/+ and Lrrk2−/− fibroblast cells. In control Lrrk2+/+ fibroblasts, VSVG-GFP became clearly present at the ERES 5 min after permissive temperature, reached the Golgi complex by 20 min, and mostly arrived at the cell surface by 120 min (Fig 4A and B). By contrast, in Lrrk2−/− cells, VSVG-GFP exhibited significant delay in transporting from ER to cell surface (Fig 4A and B). It has been shown previously that PD-related mutant α-synuclein impairs the transport of nicastrin from ER to Golgi (Chung et al, 2013). We then compared the ER–Golgi transport of endogenous protein nicastrin in Lrrk2+/+ and Lrrk2−/− fibroblast cells using an Endo H assay. We treated Lrrk2+/+ and Lrrk2−/− cell extracts with or without Endo H and then examined the expression of various forms of nicastrin by Western blot. We found more ER form of nicastrin in the Lrrk2−/− cell extracts (Fig 4C and D). In addition, we performed in vitro budding assay using microsomes purified from Lrrk2+/+ and Lrrk2−/− mouse fibroblasts. Using ER–Golgi intermediate compartment (ERGIC) marker protein ERGIC-53/58 as the readout, we found the budding reaction was attenuated in Lrrk2−/− cells as compared to the controls (Fig 4E). As controls, the total level of ERGIC-53/58 was comparable between Lrrk2+/+ and Lrrk2−/− cell lysates (Fig 4E). Our data suggest that LRRK2 may facilitate ERES-mediated COPII transport via maintaining the normal organization of ERES through association with ERES residential protein Sec16A.
Figure 4. COPII transportation is impaired in LRRK2-deficient cells.
A, B Tracing of VSVG-GFP in transfected Lrrk2+/+ and Lrrk2−/− fibroblasts at different time points (A). Asterisks indicate the nuclei. Bar graph shows the ratio of surface VSVG-GFP versus the total VSVG-GFP in cells at different time points (B). Data are presented as means ± SEM. Of 15 cells were analyzed for each genotype per time point. ***P < 0.001. Scale bar: 10 μm.
C, D Western blot shows the ER form of nicastrin in Lrrk2+/+ and Lrrk2−/− fibroblasts after treated with Endo H (C). Bar graph depicts the ratio of ER to non-ER forms of nicastrin in Endo H-treated cells (D). Data are presented as means ± SEM. N = 5 independent experiments. *P < 0.05.
E In vitro budding assay shows the levels of ERGIC-53/58 derived from Lrrk2+/+ and Lrrk2−/− microsomes purified from mouse fibroblasts. The expression of ERGIC-53/58 in total cell lysates was also examined by Western blot. Actin was used as the loading control. Bar graph depicts the ratio of budding ERGIC-53/58 in Lrrk2−/− microsomes normalized against Lrrk2+/+ controls from three independent experiments.
F, G Representative images show Golgi marker giantin (green) staining in Lrrk2+/+ and Lrrk2−/− fibroblasts (F). Nucleus was labeled with Topro3. Scale bar: 10 μm. Bar graph depicts the number of discrete Golgi elements in each genotype type (G). Data are presented as means ± SEM. Of 25 cells were analyzed for each genotype. ***P < 0.001.
H Immunostaining of ERGIC-53/58 in fibroblasts derived from Lrrk2+/+ and Lrrk2−/− mice. Scale bar: 10 μm.
Source data are available online for this figure.
A previous report also shows that Sec16A-deficient cells display fragmented and dispersed Golgi structure due to the dysfunction of ERES-mediated COPII vesicle transport (Watson et al, 2006). In accordance with this early finding, Golgi apparatus, which formed a compact ribbon structure near the nucleus of Lrrk2+/+ fibroblasts visualized by Golgi marker giantin staining, became scattered more widely around the nucleus of Lrrk2−/− cells (Fig 4F and G). A similar scattering of ERGIC revealed by ERGIC-53/58 staining was also found in Lrrk2−/− fibroblast cells (Fig 4H). Taken together, these observations further support the involvement of LRRK2 in ERES-mediated COPII vesicle transport.
Impaired ERES organization and COPII vesicle transport in Lrrk2 R1441C knock-in cells
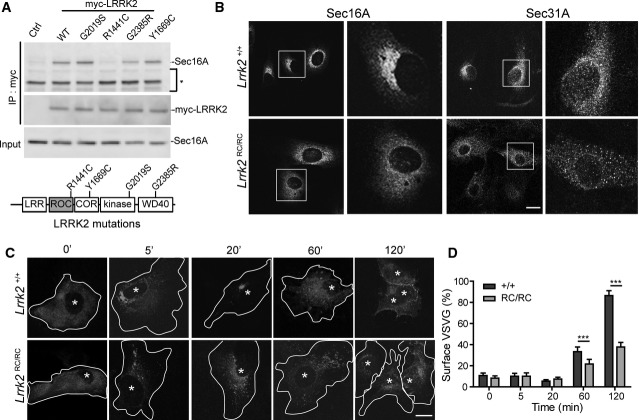
To investigate the effects of PD-associated familial mutations on the interaction between LRRK2 and Sec16A, we conducted co-IP analyses in HEK293T cells transiently transfected with myc-tagged WT and PD-related G2019S, R1441C, G2385R, and Y1669C LRRK2 expression constructs. Interestingly, the R1441C missense mutation in the ROC domain of LRRK2 impaired the interaction with Sec16A, while other PD-related LRRK2 mutations showed comparable interaction with Sec16A as the WT protein (Fig 5A). Furthermore, a GTPase-dead missense mutation at the ROC domain of LRRK2 (K1347A) also compromised the interaction of LRRK2 with Sec16A, whereas a LRRK2 kinase-dead missense mutation D1994N had no impact on the interaction with LRRK2 (Supplementary Fig S3A). To further investigate whether R1441C LRRK2 compromises the function of Sec16A in ER–Golgi transport, we examined the subcellular distribution of Sec16A and Sec31A proteins in primary fibroblast cells derived from R1441C Lrrk2 homozygous knock-in (RC/RC) mice (Tong et al, 2009). Similar to what happened in Lrrk2−/− fibroblast cells, more diffused cytosolic staining of Sec16A and Sec31A proteins was observed in Lrrk2RC/RC cells (Fig 5B). FPLC experiments further revealed the reduced Sec16A-containing high molecular weight protein complex in Lrrk2RC/RC mouse brain extracts (Supplementary Fig S3B). Moreover, we found that the delivery of VSVG-GFP from ER to the cell surface was significantly delayed in Lrrk2RC/RC cells (Fig 5C and D). Meanwhile, no significant alteration of LRRK2 subcellular distribution was observed in Lrrk2RC/RC cells as compared to control Lrrk2+/+ cells (Supplementary Fig S3C). In addition, the overall expression of Sec16A was comparable between the control (+/+) and Lrrk2RC/RC mouse brains (Supplementary Fig S3D).
Figure 5. Sec16A subcellular distribution and COPII vesicle transport are affected in Lrrk2 R1441C knock-in fibroblasts.
A Co-immunoprecipitation of Sec16A and LRRK2 with an anti-myc antibody in HEK293T cells transiently transfected with myc-tagged WT and PD-related G2019S, R1441C, G2385R, and Y1699C mutant LRRK2 constructs. The non-transfected cells served as the negative control (Ctrl). The non-specific bands were marked by asterisk.
B Representative images show Sec16A and Sec31A staining in Lrrk2+/+ and Lrrk2RC/RC fibroblasts. Scale bar: 10 μm.
C, D Tracing of VSVG-GFP in transfected Lrrk2+/+ and Lrrk2RC/RC fibroblasts at multiple time points (C). Asterisks indicate the nuclei. Bar graph shows the ratio of surface VSVG-GFP versus the total VSVG-GFP in cells at different time points (D). Data are presented as means ± SEM. 15 cells were analyzed for each genotype and time point. ***P < 0.001.
Source data are available online for this figure.
In support of the notion that loss of Lrrk2 leads to the subcellular redistribution of Sec16A, reintroduction of myc-WTLRRK2 by transient transfection restored the juxtanuclear ERES localization of endogenous Sec16A in Lrrk2−/− primary fibroblasts (Supplementary Fig S3E). Similarly, transfection of myc-WTLRRK2 also improved the association of endogenous Sec16A onto the ERES of Lrrk2RC/RC primary fibroblasts (Supplementary Fig S3F). By contrast, both myc-R1441C and myc-K1347A LRRK2 failed to recruit Sec16A at the ERES in Lrrk2−/− cells (Supplementary Fig S3G and H). On the other hand, a kinase-dead D1994N and PD-related G2019S, G2385R, and Y1669C missense mutations were able restore the juxtanuclear localization of Sec16A in Lrrk2−/− cells (Supplementary Fig S3I–L). It is noticeable that the level of Sec16A staining was stronger in cells transfected with LRRK2 (Supplementary Fig S3E, F, I–L). The stronger Sec16A staining may reflect more Sec16A proteins were associated with ERES and membrane structures in these cells since most cytosolic proteins would lose after methanol fixation. Together, these data demonstrate the ROC domain of LRRK2 is important in anchoring Sec16A at ERES organization, suggesting that LRRK2 R1441C missense mutation may impair the ER–Golgi secretory pathway in the pathogenesis of PD.
LRRK2 is co-localized with Sec16A at somatic and dendritic ERES in hippocampal neurons
We next investigated whether LRRK2 is co-localized with Sec16A at ERES in neurons. Co-immunostaining of LRRK2 and Sec16A in cultured Lrrk2+/+ mouse hippocampal neurons revealed discrete LRRK2-positive signals scattered in the soma, of which around 20% of LRRK2-labeled puncta showed co-staining with Sec16A (Fig 6A). In addition to somatic ERES, neurons also possess dendritic ERES (dERES), which contain typical components of COPII vesicles, such as Sec23/Sec24, Sec13/Sec31, and GTPase Sar1 (Aridor et al, 2004). The dERES is critical in the assembly and export of dendritic proteins (Horton & Ehlers, 2004). Interestingly, we observed a substantial overlap of LRRK2 and Sec16A staining as small puncta along the dendrites of cultured Lrrk2+/+ mouse hippocampal neurons (Fig 6B). Similarly, we found LRRK2 was co-localized with Sec31A at the small puncta in dendrites (Fig 6C). These observations demonstrate the presence of LRRK2 and Sec16A at the ERES in neurons, especially at the dERES.
Figure 6. LRRK2 co-localizes with Sec16A proteins at the dERES of cultured hippocampal neurons.
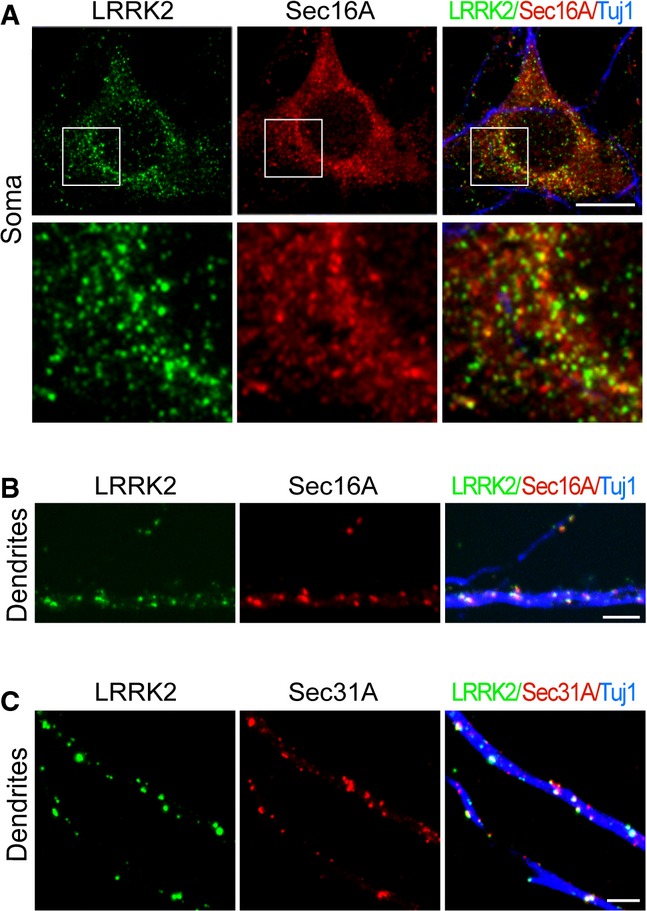
A, B Representative images show LRRK2 (green) and Sec16A (red) staining in the soma (A) and dendrites (B) of Lrrk2+/+ hippocampal neurons at 14DIV. Neurons were marked by staining with an antibody (Tuj1) against βIII-tubulin (blue). Scale bar: 10 μm.
C Representative images show of LRRK2 (green) and Sec31A (red) in dendrites of hippocampal neurons at 14DIV. Neurons were visualized by staining with an antibody (Tuj1) against βIII-tubulin (blue). Scale bar: 10 μm.
Sec16A is detached from the dERES in dendrites of Lrrk2−/− neurons
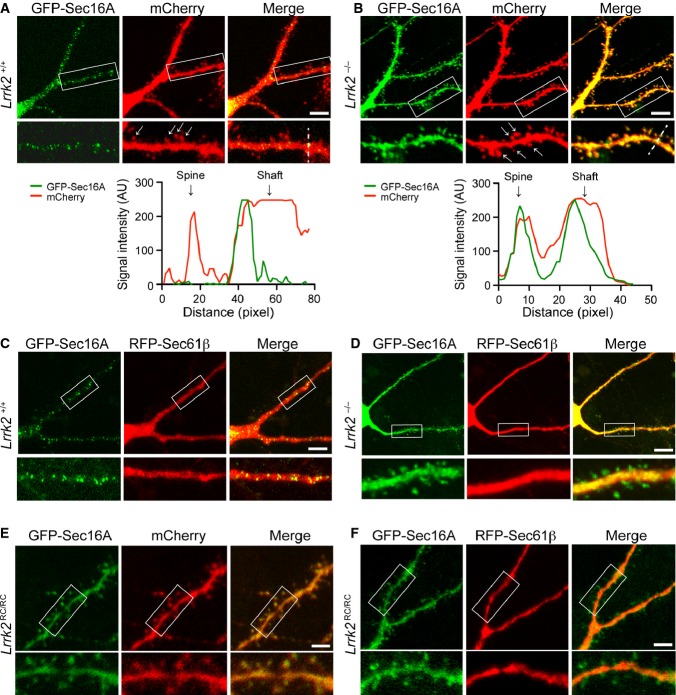
We showed earlier that LRRK2 plays an important role in anchoring Sec16A onto ERES in mouse fibroblasts (Fig 2). To determine whether LRRK2 is also responsible for anchoring Sec16A at dERES in neurons, we compared the subcellular localization of Sec16A in dendrites of Lrrk2+/+ and Lrrk2−/− hippocampal neurons after 21 days in culture (DIV21). Neurons were co-transfected with GFP-Sec16A and membrane-bound fluorescent protein mCherry. The expression of mCherry helps to illuminate the contour of dendritic shaft and spines (Zhong et al, 2009). In Lrrk2+/+ neurons, the Sec16A-positive punctate staining was exclusively distributed along the dendritic shaft, and no Sec16A staining was detected within dendritic spines (Fig 7A). By contrast, in Lrrk2−/− neurons, the Sec16A staining displayed an evenly distribution pattern in both dendritic shaft and spines and showed extensive overlap with mCherry (Fig 7B). These data suggest that LRRK2 may play an important role in retaining Sec16A at dERES, whereas loss of Lrrk2 impairs the association of Sec16A with dERES.
Figure 7. Alteration of Sec16A subcellular distribution in dendrites of Lrrk2−/− and Lrrk2RC/RC hippocampal neurons.
A, B Representative images show subcellular localization of Sec16A in dendrites of Lrrk2+/+ (A) and Lrrk2−/− (B) hippocampal neurons (21DIV) co-transfected with GFP-Sec16A and mCherry. The bottom panels show enlargement of the boxed areas in the top panels. Arrows point to the dendritic spines. Scale bar: 10 μm. Line graphs show the distribution of signal intensities of GFP-Sec16A and mCherry along the dash line crossing both dendritic spines and shaft.
C, D Representative images show subcellular localization of Sec16A in dendrites of Lrrk2+/+ (C) and Lrrk2−/− (D) hippocampal neurons (21DIV) co-transfected with GFP-Sec16A and RFP-Sec61β. The bottom panels show enlargement of the boxed areas in the top panels. Scale bar: 10 μm.
E, F Images show the subcellular localization of Sec16A in dendrites of Lrrk2RC/RC hippocampal neurons (21DIV) co-transfected with GFP-Sec16A and mCherry (E), as well as GFP-Sec16A and RFP-Sec61β(F). The bottom panels show enlargement of the boxed areas in the top panels. Scale bar: 10 μm.
To further investigate whether Sec16A remains associated with ER in Lrrk2−/− neurons, we co-transfected cultured Lrrk2+/+ and Lrrk2−/− hippocampal neurons with red fluorescent protein (RFP)-tagged ER tubular protein Sec61β and GFP-Sec16A. In Lrrk2+/+ neurons, GFP-Sec16A-associated small puncta decorated along the Sec61β-positive ER tubules located mainly within the dendritic shaft (Fig 7C). In Lrrk2−/− neurons, while Sec61β signals remained strictly within the dendritic shaft, Sec16A staining was presented in both the dendritic shaft and spines, indicating that a pool of Sec16A within the dendritic spines was detached from the ER and stays in the cytosol (Fig 7D). These results suggest that loss of Lrrk2 may not affect the overall dendritic ER structure in neurons, but may fail to maintain Sec16A at the dERES.
We next examined the impact of LRRK2 R1441C mutation on the distribution of Sec16A in dendrites of cultured Lrrk2RC/RC hippocampal neurons transfected with GFP-Sec16A, mCherry, and RFP-Sec61β. Similar to its distribution pattern in Lrrk2−/− neurons, Sec16A signal was found in both the dendritic shaft and dendritic spine of Lrrk2RC/RC neurons, while the distribution of ER tubular protein Sec61β remained exclusively along the dendritic shaft (Fig 7E and F). These observations indicate that PD-related R1441C mutation may also impair the anchoring of Sec16A by LRRK2 at the dERES in neurons.
Loss of LRRK2 impairs the activity-dependent targeting of NMDA receptors to cell surface
N-methyl-D-aspartate (NMDA)-type glutamate receptors (NMDARs) are critical for synaptic transmission and plasticity (Malenka & Nicoll, 1993). Increasing evidence suggests that NMDARs are transported to the synaptic surface mainly through the dERES-dependent secretory pathway (Aridor et al, 2004; Jeyifous et al, 2009). In addition, the cell surface targeting of NMDRs is regulated by synaptic and neuronal activities, in which chronic inhibition of neuron activities leads to increase cell surface targeting of NMDARs (Rao & Craig, 1997; Crump et al, 2001; Mu et al, 2003). Consistent with these earlier findings, we found that tetrodotoxin (TTX) treatment, which suppresses the neuronal activities, led to a significant increase of cell surface localization of NMDAR subunits NR1, NR2A, and NR2B in cultured neurons (Fig 8A–D). However, the extent of TTX-induced cell surface targeting of NMDARs was significantly decreased in Lrrk2−/− neurons compared to the Lrrk2+/+ ones (Fig 8A–D). By contrast, the surface targeting of α-amino-3-hydroxyl-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors (AMPARs), GluR1 and GluR2, was comparable between TTX-treated Lrrk2+/+ and Lrrk2−/− neurons (Supplementary Fig S4A and B). Meanwhile, bicuculline treatment, which increases neuronal activity, showed no significant impact on NR1, NR2A, and NR2B surface targeting in both Lrrk2+/+ and Lrrk2−/− neurons (Fig 8A–D). We further examined the cell surface targeting of NR2B in Lrrk2RC/RC neurons treated with vehicle or TTX and observed a significant reduction of TTX-induced NR2B translocation at cell surface in Lrrk2RC/RC neurons compared to Lrrk2+/+ neurons (Fig 8E, Supplementary Fig S4C). These data suggest that TTX-induced cell surface targeting of NMDARs is selectively compromised in Lrrk2−/− and Lrrk2RC/RC neurons.
Figure 8. Impaired activity-dependent NMDA receptor trafficking in Lrrk2−/− and Lrrk2RC/RC neurons.
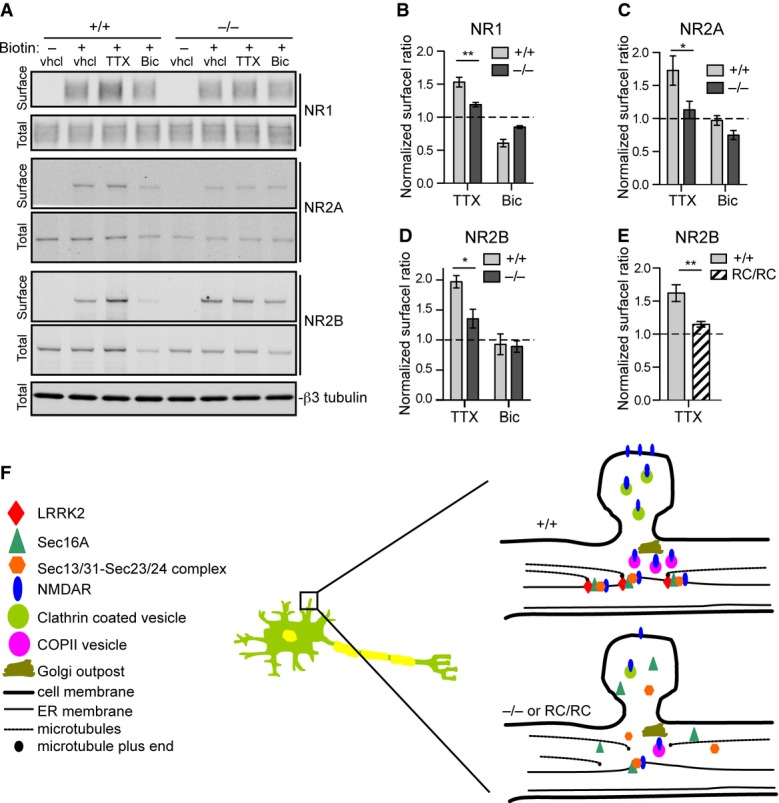
A Western blots show the levels of biotinylated cell surface and total NMDAR subunits NR1, NR2A, and NR2B in cultured Lrrk2+/+ and Lrrk2−/− cortical neurons (14DIV) treated with vehicle (vhcl), tetrodotoxin (TTX), and bicuculline (Bic), respectively. βIII-tubulin was used as the loading control for the lysate.
B–D Bar graphs show the ratio of NR1 (B), NR2A (C), and NR2B (D) at cell surface versus total proteins. The data from TTX and Bic-treated samples were normalized with vehicle-treated samples. Three and six independent experiments were performed on Lrrk2+/+ and Lrrk2−/− neurons, respectively. Two-way ANOVA shows significant interaction between genotype and TTX treatment for NR1 (F = 17.49, P = 0.0013), NR2A (F = 9.07, P = 0.027), and NR2B (F = 6.68, P = 0.022). *P < 0.05. **P < 0.01.
E Bar graph shows the ratio of NR2B at cell surface versus total proteins from vehicle and TTX-treated cultured Lrrk2+/+ and Lrrk2RC/RC cortical neurons (14DIV). The data from TTX-treated samples were normalized with vehicle-treated samples. Three independent experiments were performed on Lrrk2+/+ and Lrrk2RC/RC neurons. Two-way ANOVA shows significant interaction between genotype and TTX treatment for NR2B (F = 12.37, P = 0.0079). **P < 0.01.
F Schematic diagram illustrates a working model on how activity-dependent surface trafficking of NMDARs is impaired by redistribution of Sec16A in Lrrk2−/− and Lrrk2RC/RC neurons. LRRK2 may anchor the dERES near the dendritic spines through interaction with the dynamic ends of microtubules that are enriched at the base of dendritic spines (Jaworski et al, 2009), and facilitates the cargo transport from dERES to the dendritic spines in response to strong neuron activation.
Source data are available online for this figure.
Our findings from neurons further support the notion that LRRK2 is important in anchoring Sec16A at ERES. Interestingly, the dynamic ends of microtubules also enrich at the base and stem of dendritic spines, while LRRK2 prefers to bind with the dynamic end of microtubules (Jaworski et al, 2009; Law et al, 2014). LRRK2 may anchor the dERES near the dendritic spines through interaction with the dynamic ends of microtubules and facilitates the cargo transport from dERES to the dendritic spines in response to strong neuron activation, whereas the impairment of surface targeting of NMDARs in TTX-treated Lrrk2−/− and Lrrk2RC/RC neurons may reflect a dysfunction of dERES-mediated protein transport induced by the redistribution of Sec16A from dERES (Fig 8F).
Discussion
There is increasing evidence that LRRK2 participates retrograde vesicle transport in endosomes and lysosomes through interacting with Rab5, Rab7, Rab7L, and other proteins (Heo et al, 2010; Dodson et al, 2012; MacLeod et al, 2013; Beilina et al, 2014). Our study is the first to demonstrate the involvement of LRRK2 in the ER–Golgi anterograde vesicle transport. LRRK2 may play important roles in both anterograde and retrograde secretory pathways. In the present study, we identified LRRK2 as an upstream regulator of Sec16A in regulating ERES formation and COPII vesicle transport. A loss of Lrrk2 altered both the association of Sec16A with ERES and the clustering of ERES near the nucleus. The change in ERES organization may contribute to the impairment of cargo transport from ER to Golgi observed in Lrrk2−/− cells. Interestingly, PD-related LRRK2 R1441C missense mutation that comprises the interaction of LRRK2 with Sec16A also causes a similar impairment in ERES formation and function. PD-related α-synuclein has been shown previously to block ER–Golgi transport by inhibiting the docking and fusion of COPII vesicles with Golgi (Cooper et al, 2006; Gitler et al, 2008). In addition, our early study demonstrates that co-expression of both PD-related mutant α-synuclein and LRRK2 exacerbates Golgi fragmentation in neurons of transgenic mice (Lin et al, 2009). These findings indicate that both PD-related α-synuclein and LRRK2 are involved in early ER export, of which LRRK2 regulates the initial formation of ERES and COPII vesicles through interaction with ERES protein Sec16A, whereas α-synuclein works on the later stages of COPII vesicle fusion with cis-Golgi. Our study also suggests a potential loss-of-function mechanism of the LRRK2 R1441C mutation in the pathogenesis of PD.
LRRK2 co-localizes with Sec16A at ERES
Previous studies indicate the presence of LRRK2 in cytosol and various membrane structures, such as synaptic vesicles, multivesicular bodies, lysosomes, Golgi apparatus, plasma membrane, mitochondria, and ER (Biskup et al, 2006; Hatano et al, 2007; Alegre-Abarrategui et al, 2009; Mandemakers et al, 2012; Matta et al, 2012). However, it is worth pointing out that many of early studies were performed in heterologous overexpression systems, which may not faithfully recapitulate the subcellular localization of endogenous LRRK2 protein. In the present study, we found endogenous LRRK2 is co-localized with endogenous Sec16A and Sec31A at ERES in HeLa cells, primary and immortalized fibroblasts, and neurons. We further confirmed these observations using Lrrk2−/− cells as the negative controls. Besides immunocytochemistry, we also performed a number of complementary biochemical assays that further establish the physical association of LRRK2 with Sec16A in the same protein complex. A structural study shows that Sec16A binds to Sec13A to form edge elements similar to those formed by Sec13-31, suggesting the interaction of Sec13-16 with Sec23-24 at the ERES may set up the platform for the eventual COPII cage polymerization of Sec13-31 and Sec23-24 proteins (Whittle & Schwartz, 2010). A competition of LRRK2 with Sec13A in binding with the CCD of Sec16A raises an interesting perspective that LRRK2 may involve in COPII vesicle formation at the ERES, although the detailed mechanism remains to be determined. Together, these findings demonstrate that LRRK2 interacts and co-localizes with Sec16A at ERES, suggesting that LRRK2 may play an important role in regulating ERES formation and COPII vesicle transport.
LRRK2 acts upstream of Sec16A in the formation of ERES
Sec16A serves as a molecular scaffold required for the formation and function of ERES in ER export (Miller & Barlowe, 2010). However, less is known about what acts upstream of Sec16A that recruits Sec16A onto ERES at the first place. While the loss or overexpression of Sec16A has no effect on the juxtanuclear distribution of LRRK2 in cells, the depletion of LRRK2 leads to dispersion of Sec16A into cytosol, suggesting that LRRK2 may function as a scaffolding protein at the ER surface prior to the recruitment of Sec16A in ERES formation.
A previous study shows that extracellularly regulated kinase 7 (ERK7) regulates the level of Sec16A at ERES upon serum or amino acid starvation, in which ERK7 phosphorylates the C-terminus of Sec16A that leads to the dispersion of Sec16A into cytosol and inhibits protein secretion (Zacharogianni et al, 2011). Unlike ERK7, LRRK2 binds to the middle CCD domain of Sec16A and recruits Sec16A onto ERES under the normal cell culture condition. In addition, the kinase activity of LRRK2 seems not required in regulating the association of Sec16A with ERES, since the introduction of LRRK2 kinase-dead mutation (D1994N) can also restore the juxtanuclear localization of Sec16A in Lrrk2−/− cells. Besides Sec16A, there also exists Sec16B, a shorter isoform of Sec16A in the mammalian cells (Bhattacharyya & Glick, 2007). However, Sec16B may have a different role in ER export compared to Sec16A, since Sec16B cannot compensate for the loss of Sec16A in cells (Budnik et al, 2011). In line with this view, LRRK2 shows no interaction with Sec16B (Supplementary Fig S5), suggesting that LRRK2 may selectively form molecular complex with Sec16A in the formation of ERES in cells. Taken together, our findings establish LRRK2 as a key upstream scaffold protein at ER that may selectively anchor Sec16A during the formation of ERES under the normal cellular condition.
LRRK2 regulates microtubule and actin dynamics especially in developing neurons (Parisiadou & Cai, 2010). Microtubules are essential in ER-to-Golgi transport (Presley et al, 1997). There is substantial evidence demonstrating that microtubules associate with ERGIC and mediate the vesicle transport via dynein and kinesin motor proteins from ERGIC to Golgi (Appenzeller-Herzog & Hauri, 2006). However, whether microtubules directly contribute to the vesicle transport from ERES to ERGIC is less clear (Zanetti et al, 2012). On one hand, depolymerization of microtubules seems not affect the transport of COPII cargos from ERES to ERGIC, suggesting a microtubule-independent mechanism of ERES export to ERGIC (Presley et al, 1997; Scales et al, 1997; Hammond & Glick, 2000). On the other hand, Watson and colleagues show that ERES co-localizes with microtubules and can rapidly associate with the newly polymerized microtubules (Watson et al, 2005). Dynactin p150glued, a microtubule plus end binding protein, mediates the association of ERES with the dynamic end of microtubules through interaction with COPII protein Sec23A at ERES (Watson et al, 2005). Interestingly, a recent study also demonstrates that LRRK2 preferentially binds to the dynamic microtubules that undergo rapid polymerization and depolymerization (Law et al, 2014). LRRK2 may play a similar role as p150glued in association with both ERES protein Sec16A and microtubule network. Like p150glued, the binding of LRRK2 with the dynamic microtubules may direct the clustering of Sec16A and other ERES proteins at the vicinity of nucleus. To test this hypothesis, we treated Lrrk2+/+ and Lrrk2−/− fibroblasts with microtubule-depolymerizing agent nocodazole and microtubule-polymerizing agent taxol. Nocodazole treatment caused dispersion of Sec16A staining in Lrrk2+/+ cells, a phenomenon similar to non-treated Lrrk2−/− fibroblasts (Supplementary Fig S6A). In parallel, nocodazole treatment led to further dispersion of Sec16A staining in Lrrk2−/− fibroblasts, which showed a rather homogenous staining pattern in Lrrk2−/− cells (Supplementary Fig S6B). By contrast, taxol treatment did not significantly alter the subcellular distribution of Sec16A in Lrrk2+/+ fibroblasts, but appeared to increase the clustering of Sec16A staining near the nucleus of Lrrk2−/− cells (Supplementary Fig S6). However, the treatment of taxol did not fully restore the juxtanuclear clustering of Sec16A in Lrrk2−/− cells compared to the non-treated Lrrk2+/+ fibroblasts (Supplementary Fig S6). It is worth to point out that a lack of Lrrk2 did not cause overt disruption of microtubule network compared cells treated with nocodazole (Supplementary Fig S6B). Nonetheless, taxol may still help to stabilize and align microtubules in the absence of Lrrk2, which may allow the stable microtubules concentrate near the nucleus (Supplementary Fig S6B). ERES has been shown to attach to the stable microtubules (Mizuno & Singer, 1994). We suspect that one function of LRRK2 is to direct the attachment of ERES protein complex to the dynamic ends of microtubules, which may contribute to the clustering of ERES near the nucleus. In addition, LRRK2 is also important in anchoring Sec16A at ERES. A loss of Lrrk2 led to more Sec16A into cytosol and less attached to the microsomes. Our findings raise an interesting perspective that LRRK2 may play two roles in organizing ERES. The first one is to bind with Sec16A and facilitate the attachment of Sec16A with ERES. The second one is by binding with the dynamic end of microtubules, LRRK2 not only stabilizes the microtubules but also establishes the polarity of ERES clustering in the vicinity of nucleus. Together, our study suggests that LRRK2 plays an important role in clustering ERES proteins near the nucleus through association with both Sec16A and the dynamic ends of microtubules.
LRRK2 regulates COPII vesicle trafficking
ERES facilitate the generation of COPII vesicles containing cargo proteins, which then form larger vesicular tubular transport carriers by vesicle fusion that travel along microtubules to merge with Golgi apparatus for the completion of cargo protein transport (Miller & Barlowe, 2010). Since Sec16A is essential in the formation of ERES and COPII vesicle secretion (Watson et al, 2006; Hughes et al, 2009), we suspect that LRRK2 may regulate the COPII vesicle transport mainly through anchoring Sec16A at ERES. The dispersion of juxtanuclear ERES cluster in Lrrk2−/− cells may thereby compromise the effectiveness of COPII vesicle formation at ERES, a phenomenon that has been extensively described in the Sec16A-deficient cells (Connerly et al, 2005; Watson et al, 2006). Consistently, we present multiple lines of evidence demonstrating that Lrrk2 depletion impairs the proper formation of ERES at the initial stage of COPII vesicle trafficking, including the increase of cytosolic distribution of Sec16A and Sec31A and the alteration of Sec16A and Sec31A protein complex formation in Lrrk2−/− cells. Therefore, our findings establish a new function of LRRK2 that regulates COPII vesicle transport through anchoring of Sec16A at ERES clustered near the nucleus of cells.
The impact of PD-related LRRK2 R1441C mutation on Sec16A ERES localization and COPII vesicle transport
Sec16A interacts with the ROC domain of LRRK2 that displays GTPase activities (Li et al, 2007). The PD-related R1441C missense mutation locates in the ROC domain of LRRK2 that disrupts the binding of LRRK2 with Sec16A. As a result, homozygous Lrrk2 R1441C knock-in cells show a similar dispersion of Sec16A into cytosolic space and impairment of COPII vesicle transport as happened in Lrrk2−/− cells. The LRRK2 R1441C missense mutation also impairs the GTPase activities of LRRK2 (Guo et al, 2007; Li et al, 2007). However, whether the LRRK2’s GTPase activity regulates the ERES localization and function of Sec16A remains unclear. The K1347A missense mutation that abolishes the GTPase function of LRRK2 also disrupts the interaction of LRRK2 with Sec16A and fails to anchor Sec16A at the ERES. Therefore, it remains interesting to identify LRRK2 mutations that lack the GTPase activity but preserve the association with Sec16A to evaluate the contribution of GTPase activity of LRRK2 to ERES formation and function.
The ROC domain of LRRK2 shares homology with the GTPase domain of Sar1 (data not shown). The activation of Sar1 GTPase activities results in the assembly of COPII vesicle coat proteins at the ERES membranes (Watson et al, 2006). A previous report also shows that Sec16A interacts with Sar1 and may indirectly regulate the Sar1 activity (Yorimitsu & Sato, 2012). Although the relationship between Sec16A and Sar1 remains to be clarified Sec16A may likely act upstream of Sar1 in COPII vesicle transport at ERES (Yorimitsu & Sato, 2012). In addition to the GTPase domain, LRRK2 also possesses a kinase domain (West et al, 2005). The PD-related G2019S missense mutation increases the kinase activity of LRRK2, while the D1994N mutation diminishes the kinase activity of LRRK2 (Smith et al, 2006). We found the D1994N mutation can restore the juxtanuclear ERES localization of Sec16A in Lrrk2−/− cells, suggesting the kinase activity of LRRK2 may not directly regulate the association of Sec16A with LRRK2 at ERES. These observations also suggest that different PD-related LRRK2 mutations perhaps cause the disease through different pathogenic pathways. Noticeably, PD patients carrying LRRK2 mutations also display pleomorphic pathological phenotypes (Bonifati, 2006; Whaley et al, 2006), implicating a diverse molecular and cellular pathogenic pathways induced by different LRRK2 mutations.
LRRK2 regulates the formation and function of dERES in neurons
In neurons, there exist two secretory pathways for transport of proteins to dendritic spines (Ramirez & Couve, 2011). The canonical pathway includes the formation of secretory vesicles at the centralized ERES and Golgi compartment in the soma and then the movement of secretory vesicles along the microtubule network to deliver the proteins to the dendritic spines (Kennedy & Ehlers, 2006). Alternatively, dendritic proteins can be transported locally from dERES to Golgi outpost in dendrites (Horton & Ehlers, 2004). Accumulating evidence demonstrates the significance of local dendritic protein transport that employs the same COPII vesicle trafficking machinery transporting proteins from dERES to Golgi outpost and eventually to dendritic surface (Aridor et al, 2004; Horton & Ehlers, 2004; Ramirez & Couve, 2011). The dERES controls the assembly and export of dendritic proteins, particularly the NMDARs (Jeyifous et al, 2009). In agreement with these early studies, we found that loss of Lrrk2 causes dispersion of Sec16A from dERES and impairs the activity-dependent trafficking of NMDARs to the cytoplasmic membrane in Lrrk2−/− hippocampal neurons. We therefore propose that LRRK2 may regulate the activity-dependent targeting of NMDARs to cell/dendritic spine surface by anchoring Sec16A at dERES. The PD-related LRRK2 R1441C mutation also disrupts the association of Sec16A with dERES in neurons. Correlatively, the R1441C missense mutation impairs the activity-dependent trafficking of NMDRs in neurons. Dysfunction of neurotransmission underlies the neuronal basis of PD and many other neurological disorders (Shen, 2010). This newly defined function of LRRK2 that locally regulates the dendritic protein transport at dERES may not only provide new molecular clues on how dERES are organized and regulated in neurons, but also imply the involvement of dERES impairment as a new pathogenic mechanism in PD.
Materials and Methods
LRRK2 interaction partner identification by mass spectrometry
Myc-LRRK2 overexpressed in HEK293T cells was immunoprecipitated using an anti-myc antibody. LRRK2-containing complexes were resolved on SDS–PAGE and stained with colloidal Coomassie Blue. Bands were excised, in-gel trypsin digested, and analyzed with LC-MS/MS mass spectrometry.
Antibodies
To observe the human LRRK2 proteins in HeLa cells by immunocytochemistry, the rabbit polyclonal LRRK2 antibody (OC83A) generated via immunization with a histidine-tagged recombinant protein with a sequence corresponding to human LRRK2 residues 982 to 1,280 was used (Cho et al, 2013). For the immunocytochemistry of mouse LRRK2, two different LRRK2 antibodies were used: anti-LRRK2 monoclonal antibody produced from mouse (N138, purchased from NeuroMab) and the rabbit polyclonal LRRK2 antibodies (4EC9E and 4C84E) described previously (Parisiadou et al, 2009). For the Western blot analysis, anti-LRRK2 monoclonal antibody produced from rabbit (MJFF-2; c41-2, obtained from the Michael J. Fox Foundation) as well as 4C84E polyclonal LRRK2 antibody was used. To detect mouse and human Sec16A proteins, a custom antibody raised against amino acids DEKKNQWVNLNEPEEEKK in guinea pig by a commercial service (Covance, Denver, PA, USA). Antibodies were from the following sources: Sec31A, Sec23A, Tom20, laminB, and Sec24D (Santa Cruz Biotechnology, Inc.); Syntaxin 6, ERGIC-53/58, β-tubulin III, and MAP2 (Sigma-Aldrich); human Sec31A, Giantin, GM130, and NR2B (BD Transduction Laboratories); human Sec23A (Abcam); RhoGDI (Cell Signaling Technology); NR1, NR2A, and NR2B (BD Biosciences); GluR1 (Calbiochem); GluR2/3 (BD Pharmingen); GFP and Myc (Roche).
Cell culture (cell line, primary neuron, primary fibroblast)
HEK293T, HeLa, Lrrk2+/+, and Lrrk2−/− immortalized mouse fibroblast cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM high glucose) containing 10% fetal bovine serum (FBS) in 5% CO2 incubator at 37°C. Mouse primary hippocampal or cortical neuron cultures were prepared from hippocampus or cortex of newborn (postnatal day 0) pups, as described previously (Parisiadou et al, 2009). Briefly, tissues were dissociated by papain (Worthington), and then, the cells were plated in poly-D-lysine pre-coated wells in Basal Eagle Medium (Sigma) containing B27, N2, 1 μM L-glutamine, and penicillin/streptomycin (all from Invitrogen). The medium was changed every 2 days. Mouse skin primary fibroblast cells were derived from the dorsal skin of postnatal day 0 (P0) pups. Briefly, the skins were minced and suspended in HEPES-buffered DMEM supplemented with 0.25% trypsin and 0.01% DNase I and incubated at 37°C for 20 min. The tissues were then transferred to DMEM supplemented with 10% fetal bovine serum and dissociated by repeated trituration. The dispersed cells were plated in one well of a 24-well plate. Cells were subsequently immortalized with large T antigen (Cai et al, 2001). Primary cells at passage below seven were used for the experiments.
Plasmids
GFP-Sec16A (Bhattacharyya & Glick, 2007) (Addgene plasmid #15776) and EGFP-VSVG (Presley et al, 1997) (Addgene plasmid #11912) plasmids were obtained from Addgene (Cambridge, MA). GFP-Sec16B and GFP-Sec31A plasmids were purchased from Origene (Rockville, MD). RFP-Sec61β construct was obtained from Dr. Craig Blackstone of NINDS (Bethesda, MD), myc-tagged LRRK2 constructs were obtained from Dr. Mark Cookson of NIA (Bethesda, MD), and mCherry expression vector was obtained from Dr. Haining Zhong of Vollum Institute (Portland, OR).
Transfection and gene silencing
All plasmid transfections were performed using Fugene HD (Roche). SiRNA for human LRRK2 was obtained from Santa Cruz Biotechnology. SiRNA duplexes were transfected into the HeLa cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. SiRNA duplexes for Sec16A were of the ON-TARGETplus SMARTpool type obtained from Thermo Fisher Scientific. For efficient knockdown, the cells were transfected twice with a 24-h interval. All experiments were performed 72 h after the first siRNA transfection.
Immunoprecipitation
Brain tissues from Lrrk2+/+ and Lrrk2−/− mice were homogenized in 50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 50 mM NaF, 10 mM glycerolphosphate, 2 mM EGTA, 2 mM EDTA, 1% NP-40 supplemented with protease inhibitor mixture (Roche, Applied Science, IN), and phosphatase inhibitor cocktail (Pierce, Rockford, IL). About 500 μg extracts were used for immunoprecipitation and diluted to a final volume of 1,000 μl with cold IP buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 50 mM NaF, 10 mM glycerolphosphate, 2 mM EGTA, 2 mM EDTA, 1% NP-40 protease inhibitor mixture, and phosphatase inhibitor cocktail). The lysates were incubated with a rabbit monoclonal anti-LRRK2 primary antibody (Epitomics, CA) overnight, and the antibody-bound protein complexes were collected with agarose protein A (Pierce Rockford, IL, USA) for 1 h at 4°C. To precipitate overexpressed myc-tagged LRRK2 proteins, anti-c-myc agarose (50 μl beads/1 ml of lysates, Sigma) was incubated for 4 h at 4°C. The beads were washed five times with the IP buffer, and the immune complexes were eluted with SDS buffer.
In vitro recombinant protein binding assay
Amplified PCR fragments for the CCD domain (900-1621aa) of Sec16A were cloned into a 3 × FLAG-CMV-10 vector (Sigma). The construct of Sec16A CCD domain was verified by sequencing and was transfected into HEK293 cells. After 48-h incubation, the cells were collected and lysed using RIPA buffer (Sigma) including protease inhibitor cocktail (Thermo). The protein lysate was then prepared after centrifugation at 13,000 × g for 10 min at 4°C. The immunobeads were prepared by coupling 2 μl rabbit anti-FLAG antibody (Sigma) to 10 μl of protein G dynabeads (Invitrogen) for 1 h at RT on a rotator in 300 μl RIPA buffer. The antibody-coupled beads were subsequently incubated in 1 ml of protein lysate for 1 h at 4°C. The protein complex-coupled beads were washed first using RIPA buffer once and then using IP buffer (20 mM Tris–HCl, pH 7.3; 150 mM NaCl; 2 mM EDTA; 1% NP-40) twice. The complex was then incubated separately with 0.3 μM His-tagged Sec13A (Fitzgerald industry) alone, 0.3 μM His-tagged Sec13A with 0.1 μM GST-tagged LRRK2 (Life Technologies), and 0.3 μM His-tagged Sec13A with 0.1 μM GST (Life Technologies) in 300 μl IP buffer for 3 h at 4°C. The complex was then washed using IP buffer three times. The proteins retained on the beads were analyzed by Novex gel (Invitrogen) and immunoblotting using mouse anti-FLAG antibody (Sigma), rabbit anti-Sec13A (Thermo), and goat anti-GST (Life Technologies).
Immunocytochemistry
Cells grown on coverslips (Thermo Fisher Scientific) were washed two times with PBS. The cells were then fixed with −20°C methanol for 7 min on ice. The fixed cells were washed three times at 5-min intervals using PBS. The cells were then incubated with blocking buffer (10% normal serum in PBS) for 1 h. After PBS washing, the cells were labeled with appropriate primary antibodies diluted in blocking buffer for 3 h at room temperature. The coverslips were then washed five times with PBS at 5-min intervals. Secondary antibodies were diluted appropriately in blocking buffer and incubated at room temperature for 1 h. After washing five times with PBS, coverslips were mounted on microscopic slide.
Microscope image acquisition
Fluorescence images were captured using a laser scanning confocal microscope (LSM 510; Zeiss, Thornwood, NJ). Images were scanned using a 63× 1.4 numerical aperture (NA) oil objective or a 100× 1.4 NA oil objective lens. A zoom factor of 2 was used to obtain maximum resolution of 100× images. Some images were acquired in z-series stack scans at 1-μm intervals from individual fields to determine the protein–protein co-localization. Maximum intensity projections were used to represent confocal stacks.
Transmission electron microscopy (TEM)
Samples were fixed in 4% paraformaldehyde, 0.1% glutaraldehyde, 3 mM MgCl2, and 0.1 M sodium cacodylate buffer, pH 7.2 for at least 1 h at 4°C. After buffer rinse, samples were postfixed in 0.5% osmium tetroxide, 0.8% potassium ferrocyanide in buffer (0.5 h) on ice in the dark. Following a 0.1 M maleate buffer rinse, plates were stained with 2% uranyl acetate in 0.1 M maleate buffer (0.22 μm filtered, 1 h, dark), dehydrated in a graded series of ethanol and embedded in Eponate 12 (Ted Pella) resin. Samples were polymerized at 60°C overnight. Thin sections, 60–90 nm, were cut with a diamond knife on the Reichert-Jung Ultracut E ultramicrotome and picked up with formvar-coated 200-mesh nickel grids. Grids were etched with 5% SMP (sodium meta-periodate), followed by reduction in 50 mM NH4Cl, blocked in 10% donkey serum and primary antibody overnight. Primary antibody was amplified with 12 nm gold followed by 2% glutaraldehyde in 0.1 M sodium cacodylate and stained with 2% uranyl acetate and observed with a Philips CM120 at 80 kV. Images were captured with an AMT CCD (1K × 1K) camera.
Preparation of cell fractions
Harvested cells or mouse brain tissues were homogenized using a Teflon Pestle (Thomas Scientific) in 20 mM HEPES-KOH, pH 7.6, 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, 0.5 mM PMSF, and 2 mg/ml BSA, and centrifuged at 800 g at 4°C for 10 min to isolate the postnuclear supernatant. Mitochondria were pelleted by centrifugation at 10,000 g at 4°C for 20 min. The supernatant fraction was centrifuged again for 30 min at 100,000 g to isolate cytosolic and microsome protein fractions as described before (Jin et al, 2010).
Endo H assay
Fibroblasts from Lrrk2+/+ and Lrrk2−/− mice were cultured and lysed using RIPA buffer in the presence of protease inhibitor cocktail (Roche). Supernatant was collected after centrifugation at 12,000 × g, 4°C for 20 min. Protein concentration was determined using BCA assay (Pierce). Endoglycosidase (Endo) H (New England Biolabs) digestion was performed based on the manufacturer’s instruction. Briefly, 30 μg of samples from Lrrk2+/+ and Lrrk2−/− fibroblasts were assembled in 9 μl reaction volume with 1 μl of denaturing buffer and boiled for 10 min at 100°C. Then, 2 μl of G5 buffer, 1 μl of Endo H, and 7 μl of H2O were added to the denatured reaction and incubated in 37°C for 2 h. Rabbit anti-nicastrin antibody (Cell Signaling) was used for Western blot 1:1,000.
In vitro budding assay
Budding reactions were carried out as described using microsomes purified Lrrk2+/+ and Lrrk2−/− mouse fibroblasts. Equal amount of proteins from Lrrk2+/+ and Lrrk2−/− microsomes was used in each assay. The budding reaction was assembled using rat liver cytosol at 4 mg/ml. An ATP-regenerating system was added resulting in final concentrations of 1 mM ATP, 40 mM creatine phosphate, 0.2 mg/ml creatine phosphokinase, and 0.1 mM GTP in buffer [20 mM HEPES-KOH (pH 7.2), 110 mM pot acetate, 2 mM Mg-acetate-KHM]. Budding reactions were assembled on ice and incubated for 30 min at 30°C, then put on ice. The donor membranes were removed by a 20-min 12,000 × g centrifugation, and the supernatant fraction was further centrifuged for 30 min at 55,000 rpm at 4°C using a TLA100 rotor in a Beckman Optima TLX ultracentrifuge to sediment the vesicle products.
Gel filtration
HEK293T cells were collected in PBS containing protease inhibitor cocktail (Roche) and lysed by freeze–thaw cycles in liquid nitrogen. For the mouse brain extracts, 0.1% Triton X-100 was added in PBS. Clear supernatant was isolated by centrifugation and then passed through 0.45-μm filters (Nanosep MF, Pall Life Sciences). 200 μl samples were injected into the column, and each fraction was collected. BioAssist G4SWXL column (7.8 mm × 30.0 cm, Tosoh Bioscience) with PBS as the mobile phase (0.8 ml/min, 4°C) was used for the size-exclusion chromatography. Aliquots were analyzed by SDS–PAGE (4-20% Tris-glycine gel) in MOPS buffer (Invitrogen) followed by Western blot analysis for Sec proteins. Thyroglobulin (660 kDa), ferritin (440 kDa), and IgM (∼1 mDa) were used as standards (Rudenko et al, 2012). Distribution of Sec protein in each fraction was presented by measuring the integrated absorbance of Sec protein band corrected to the total amount of immunoreactivity in all fractions.
VSVG transport assay
Fibroblast cells were transfected with VSVG-GFP plasmid using Fugene HD (Roche). Immediately after transfection, cells were incubated at 40°C to accumulate the temperature-sensitive forms of VSVG-GFP protein in ER. After 24 h of incubation at 40°C, the cells were transferred to the permissive temperature at 32°C for various times before being fixed with −20°C methanol for immunofluorescence microscopy. MetaMorph software was used to quantify surface VSVG.
Transfection of primary neurons
Primary hippocampal or striatal neurons were transfected between P15 and P21DIV using the calcium phosphate method. Briefly, neuronal cultures were incubated with the DNA–calcium phosphate precipitate for approximately 1.5 h. The precipitate was then dissolved by the incubation of the cells in a medium that had been pre-equilibrated in a 10% CO2 incubator. The cells were in turn transferred to their original conditioned medium and fixed at about 24–48 h after transfection.
Biotinylation of neuronal surface proteins
Cortical neurons were cultured in Biocoat Poly-D-Lysine Cellware 6-well plate (BD Biosciences) and chronically treated with vehicle, 2 μM tetrodotoxin (TTX, Sigma) or 40 μM Bicuculline (Sigma) on 10DIV. After 48-h treatment, neurons were washed with PBS containing 0.1 mM CaCl2 and 1 mM MgCl2 (PBS/CM), incubated with 1 ml biotin solution (0.5 mg/ml Sulfo-NHS-SS-Biotin in cold PBS/CM, Thermo Fisher Scientific) for 20 min at 4°C, and then washed subsequently with PBS/CM, 0.1 M glycine solution, and TBS (25 mM Tris–HCl pH 7.4, 137 mM NaCl) buffer. Cells were harvested in 250 μl RIPA buffer (Sigma) supplemented with protease inhibitor and phosphatase inhibitor cocktail, lysed on ice for 30 min, and centrifuged at 16,000 g for 15 min at 4°C. Resulting supernatant containing equal amount of total protein was incubated with 50 μl neutral-avidin agarose (Pierce) at 4°C for 2 h with gently rotating. After washing in RIPA buffer for five times, the biotin-labeled surface protein was eluted with SDS sample buffer by heating at 70°C for 10 min. Total proteins and isolated biotinylated surface proteins were analyzed by immunoblotting.
Statistical analysis
All experiments were performed at least three times independently for each condition and presented as the mean ± standard error of the mean (SEM). Statistical significance was determined using Student’s t-test followed by a Mann–Whitney test, one-way ANOVA with Tukey–Kramer multiple comparison test, or two-way ANOVA with Bonferroni post-tests. Calculations were performed using GraphPad Prism5 software (GraphPad Software, Inc., La Jolla, CA).
Acknowledgments
This work was supported in part by The Intramural Research Programs of National Institute on Aging (AG000944) and National Institute of Allergy and Infectious Diseases. We thank Dr. Michael Lenardo of NIAID for his support in identification of LRRK2-interacting proteins, Dr. Eric Anderson of Proteomics and Mass Spectrometry Facility, National Institute of Diabetes and Digestive and Kidney Diseases for Mass Spectrometry analysis, Dr. Haining Zhong for providing mCherry expression vector, Dr. Craig Blackstone of National Institute of Neurological Disorders and Stroke (NINDS) for providing RFP-Sec61β plasmid, and Drs. Peng Cheng and Randy Sheckman for their input on in vitro budding assay.
Author contributions
HJC, ZL, and HC conceived the project, designed the experiments, and wrote the manuscript. HJC produced the majority biochemical and cellular biology data and analyzed the data in Figs 1–5 and Supplementary Figs S1, S2, S3, and S5. CX generated cell biology data in Figs 6–7 and Supplementary Fig S6. JY produced biochemical data from cultured neurons in Fig 8 and Supplementary Fig S4. PR generated and analyzed data in Fig 4C–E with help of XC. XC and JW generated and analyzed data in Fig 1E and F. LP, SL, GL, and BM provided antibodies and mouse brain tissues. JD provided bioinformatics support.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Hauri HP. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- Aridor M, Guzik AK, Bielli A, Fish KN. Endoplasmic reticulum export site formation and function in dendrites. J Neurosci. 2004;24:3770–3776. doi: 10.1523/JNEUROSCI.4775-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilina A, Rudenko IN, Kaganovich A, Civiero L, Chau H, Kalia SK, Kalia LV, Lobbestael E, Chia R, Ndukwe K, Ding J, Nalls MA, Olszewski M, Hauser DN, Kumaran R, Lozano AM, Baekelandt V, Greene LE, Taymans JM, Greggio E, et al. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc Natl Acad Sci USA. 2014;111:2626–2631. doi: 10.1073/pnas.1318306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Glick BS. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol Biol Cell. 2007;18:839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, Faull RL, Emson PC, Torp R, Ottersen OP, Dawson TM, Dawson VL. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- Bock JB, Klumperman J, Davanger S, Scheller RH. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V. The pleomorphic pathology of inherited Parkinson’s disease: lessons from LRRK2. Curr Neurol Neurosci Rep. 2006;6:355–357. doi: 10.1007/s11910-996-0013-z. [DOI] [PubMed] [Google Scholar]
- Budnik A, Heesom KJ, Stephens DJ. Characterization of human Sec16B: indications of specialized, non-redundant functions. Sci Rep. 2011;1:77. doi: 10.1038/srep00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, Wong PC. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, Sun L, Ma B, Ding J, Vancraenenbroeck R, Lobbestael E, Baekelandt V, Taymans JM, He P, Troncoso JC, Shen Y, Cai H. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2013;22:608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, Mungenast AE, Muffat J, Mitalipova M, Pluth MD, Jui NT, Schule B, Lippard SJ, Tsai LH, Krainc D, Buchwald SL, et al. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342:983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerly PL, Esaki M, Montegna EA, Strongin DE, Levi S, Soderholm J, Glick BS. Sec16 is a determinant of transitional ER organization. Curr Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump FT, Dillman KS, Craig AM. cAMP-dependent protein kinase mediates activity-regulated synaptic targeting of NMDA receptors. J Neurosci. 2001;21:5079–5088. doi: 10.1523/JNEUROSCI.21-14-05079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson MW, Zhang T, Jiang C, Chen S, Guo M. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum Mol Genet. 2012;21:1350–1363. doi: 10.1093/hmg/ddr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, Barlowe C, Lindquist S. The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci USA. 2008;105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson’s disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond AT, Glick BS. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol Biol Cell. 2000;11:3013–3030. doi: 10.1091/mbc.11.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T, Kubo S, Imai S, Maeda M, Ishikawa K, Mizuno Y, Hattori N. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum Mol Genet. 2007;16:678–690. doi: 10.1093/hmg/ddm013. [DOI] [PubMed] [Google Scholar]
- Heo HY, Kim KS, Seol W. Coordinate regulation of neurite outgrowth by LRRK2 and its interactor, Rab5. Exp Neurobiol. 2010;19:97–105. doi: 10.5607/en.2010.19.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Secretory trafficking in neuronal dendrites. Nat Cell Biol. 2004;6:585–591. doi: 10.1038/ncb0704-585. [DOI] [PubMed] [Google Scholar]
- Hughes H, Budnik A, Schmidt K, Palmer KJ, Mantell J, Noakes C, Johnson A, Carter DA, Verkade P, Watson P, Stephens DJ. Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER. J Cell Sci. 2009;122:2924–2934. doi: 10.1242/jcs.044032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson’s disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, Krugers H, Defilippi P, Akhmanova A, Hoogenraad CC. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Jeyifous O, Waites CL, Specht CG, Fujisawa S, Schubert M, Lin EI, Marshall J, Aoki C, de Silva T, Montgomery JM, Garner CC, Green WN. SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat Neurosci. 2009;12:1011–1019. doi: 10.1038/nn.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Ehlers MD. Organelles and trafficking machinery for postsynaptic plasticity. Annu Rev Neurosci. 2006;29:325–362. doi: 10.1146/annurev.neuro.29.051605.112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law BM, Spain VA, Leinster VH, Chia R, Beilina A, Cho HJ, Taymans JM, Urban MK, Sancho RM, Ramirez MB, Biskup S, Baekelandt V, Cai H, Cookson MR, Berwick DC, Harvey K. A direct interaction between leucine-rich repeat kinase 2 and specific beta-tubulin isoforms regulates tubulin acetylation. J Biol Chem. 2014;289:895–908. doi: 10.1074/jbc.M113.507913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson’s disease R1441C/G mutants. J Neurochem. 2007;103:238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, Xie C, Long CX, Yang WJ, Ding J, Chen ZZ, Gallant PE, Tao-Cheng JH, Rudow G, Troncoso JC, Liu Z, Li Z, Cai H. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson’s-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lee J, Krummey S, Lu W, Cai H, Lenardo MJ. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat Immunol. 2011;12:1063–1070. doi: 10.1038/ni.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod DA, Rhinn H, Kuwahara T, Zolin A, Di Paolo G, McCabe BD, Marder KS, Honig LS, Clark LN, Small SA, Abeliovich A. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron. 2013;77:425–439. doi: 10.1016/j.neuron.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA. NMDA-receptor-dependent synaptic plasticity: multiple forms and mechanisms. Trends Neurosci. 1993;16:521–527. doi: 10.1016/0166-2236(93)90197-t. [DOI] [PubMed] [Google Scholar]
- Mandemakers W, Snellinx A, O’Neill MJ, de Strooper B. LRRK2 expression is enriched in the striosomal compartment of mouse striatum. Neurobiol Dis. 2012;48:582–593. doi: 10.1016/j.nbd.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Mata IF, Ross OA, Kachergus J, Huerta C, Ribacoba R, Moris G, Blazquez M, Guisasola LM, Salvador C, Martinez C, Farrer M, Alvarez V. LRRK2 mutations are a common cause of Parkinson’s disease in Spain. Eur J Neurol: The Off J Eur Feder Neurol Soc. 2006;13:391–394. doi: 10.1111/j.1468-1331.2006.01256.x. [DOI] [PubMed] [Google Scholar]
- Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, De Bock PJ, Morais VA, Vilain S, Haddad D, Delbroek L, Swerts J, Chavez-Gutierrez L, Esposito G, Daneels G, Karran E, Holt M, Gevaert K, Moechars DW, De Strooper B, et al. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- Miller EA, Barlowe C. Regulation of coat assembly–sorting things out at the ER. Curr Opin Cell Biol. 2010;22:447–453. doi: 10.1016/j.ceb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno M, Singer SJ. A possible role for stable microtubules in intracellular transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Sci. 1994;107(Pt 5):1321–1331. doi: 10.1242/jcs.107.5.1321. [DOI] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 2003;40:581–594. doi: 10.1016/s0896-6273(03)00676-7. [DOI] [PubMed] [Google Scholar]
- Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cortes E, Honig LS, Dauer W, Consiglio A, Raya A, Sulzer D, Cuervo AM. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16:394–406. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, Lobbestael E, Baekelandt V, Taymans JM, Sun L, Cai H. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisiadou L, Cai H. LRRK2 function on actin and microtubule dynamics in Parkinson disease. Commun Integr Biol. 2010;3:396–400. doi: 10.4161/cib.3.5.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisiadou L, Yu J, Sgobio C, Xie C, Liu G, Sun L, Gu XL, Lin X, Crowley NA, Lovinger DM, Cai H. LRRK2 regulates synaptogenesis and dopamine receptor activation through modulation of PKA activity. Nat Neurosci. 2014;17:367–376. doi: 10.1038/nn.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Ramirez OA, Couve A. The endoplasmic reticulum and protein trafficking in dendrites and axons. Trends Cell Biol. 2011;21:219–227. doi: 10.1016/j.tcb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Rao A, Craig AM. Activity regulates the synaptic localization of the NMDA receptor in hippocampal neurons. Neuron. 1997;19:801–812. doi: 10.1016/s0896-6273(00)80962-9. [DOI] [PubMed] [Google Scholar]
- Rudenko IN, Kaganovich A, Hauser DN, Beylina A, Chia R, Ding J, Maric D, Jaffe H, Cookson MR. The G2385R variant of leucine-rich repeat kinase 2 associated with Parkinson’s disease is a partial loss-of-function mutation. Biochem J. 2012;446:99–111. doi: 10.1042/BJ20120637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Pepperkok R, Kreis TE. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Shen J. Impaired neurotransmitter release in Alzheimer’s and Parkinson’s diseases. Neurodegener Dis. 2010;7:80–83. doi: 10.1159/000285511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Tong Y, Pisani A, Martella G, Karouani M, Yamaguchi H, Pothos EN, Shen J. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc Natl Acad Sci USA. 2009;106:14622–14627. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Forster R, Palmer KJ, Pepperkok R, Stephens DJ. Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat Cell Biol. 2005;7:48–55. doi: 10.1038/ncb1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci USA. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whaley NR, Uitti RJ, Dickson DW, Farrer MJ, Wszolek ZK. Clinical and pathologic features of families with LRRK2-associated Parkinson’s disease. J Neural Transm Suppl. 2006;70:221–229. doi: 10.1007/978-3-211-45295-0_34. [DOI] [PubMed] [Google Scholar]
- Whittle JR, Schwartz TU. Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biol. 2010;190:347–361. doi: 10.1083/jcb.201003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Sato K. Insights into structural and regulatory roles of Sec16 in COPII vesicle formation at ER exit sites. Mol Biol Cell. 2012;23:2930–2942. doi: 10.1091/mbc.E12-05-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharogianni M, Kondylis V, Tang Y, Farhan H, Xanthakis D, Fuchs F, Boutros M, Rabouille C. ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J. 2011;30:3684–3700. doi: 10.1038/emboj.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2012;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y, Cui Y, Yan XX, Yang HT, Yang RD, Chu TS, Zhang C, Zhang L, Han JW, Yu GQ, Quan C, Yu YX, Zhang Z, Shi BQ, Zhang LH, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- Zhong H, Sia GM, Sato TR, Gray NW, Mao T, Khuchua Z, Huganir RL, Svoboda K. Subcellular dynamics of type II PKA in neurons. Neuron. 2009;62:363–374. doi: 10.1016/j.neuron.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.