Abstract
Hematopoietic stem cells (HSCs) require multiple molecular inputs for proper specification, including activity of the Notch signaling pathway. A requirement for the Notch1 and dispensability of the Notch2 receptor has been demonstrated in mice, but the role of the remaining Notch receptors has not been investigated. Here, we demonstrate that three of the four Notch receptors are independently required for the specification of HSCs in the zebrafish. The orthologues of the murine Notch1 receptor, Notch1a and Notch1b, are each required intrinsically to fate HSCs, just prior to their emergence from aortic hemogenic endothelium. By contrast, the Notch3 receptor is required earlier within the developing somite to regulate HSC emergence in a non-cell-autonomous manner. Epistatic analyses demonstrate that Notch3 function lies downstream of Wnt16, which is required for HSC specification through its regulation of two Notch ligands, dlc and dld. Collectively, these findings demonstrate for the first time that multiple Notch signaling inputs are required to specify HSCs and that Notch3 performs a novel role within the somite to regulate the neighboring precursors of hemogenic endothelium.
Keywords: hematopoietic stem cell, hemogenic endothelium, Notch, somite
Introduction
The developmental ontogeny of the hematopoietic system is complex and proceeds through four ordered, temporal waves during vertebrate development. The first two generate primitive erythroid and myeloid cells, which arise through the direct specification of mesoderm to rapidly generate cells capable of transporting oxygen throughout the developing embryo and providing immunity, respectively (Davidson & Zon, 2004; Tober et al, 2007; Le Guyader et al, 2008; Orkin & Zon, 2008). Next, these primitive waves are followed by specification of two definitive waves, the first giving rise to erythromyeloid progenitors (EMPs), transient precursors that give rise to cells of the erythroid and myeloid pathways (Palis et al, 1999; Bertrand et al, 2007), and the second to hematopoietic stem cells (HSCs), which have both the ability to self-renew and differentiate into the complete repertoire of mature blood cells for an organism’s lifespan. HSC specification is spatially conserved across vertebrate species and involves the transdifferentiation of hemogenic endothelium in the ventral wall of the dorsal aorta (DA) (de Bruijn et al, 2000; Zovein et al, 2008; Bertrand et al, 2010a; Boisset et al, 2010; Kissa & Herbomel, 2010). Numerous key studies have demonstrated that HSC specification requires specific molecular inputs from a number of signaling pathways, including Notch.
Notch signaling is a conserved cell-to-cell signaling pathway responsible for a multitude of critical cell-fate decisions during the lifespan of metazoan organisms (Lai, 2004; Kopan & Ilagan, 2009). In mammals and zebrafish, Notch signaling occurs through the interaction of many proteins. First, one of four transmembrane Notch receptors (Notch1, Notch2, Notch3, and Notch4 in mice; Notch1a, Notch1b, Notch2, and Notch3 in zebrafish) on a signal-receiving cell binds to a Notch ligand, termed Jagged and Delta, on a signal-emitting cell (Rebay et al, 1991). Ligand-dependent activation of Notch signaling requires cleavage of the Notch receptor, first by members of ADAM TACE metalloproteases at the S2 site (Brou et al, 2000; Bozkulak & Weinmaster, 2009), then by γ-secretase at the S3 site to release a Notch intracellular domain (NICD), which translocates to the nucleus (Mumm et al, 2000) to modulate transcription of Notch target genes (Kopan & Ilagan, 2009). The specification, lineage commitment, and maintenance of many tissues require the precise regulation of Notch signaling.
Notch signaling is especially important for the initial specification of the adult hematopoietic system during embryogenesis. While Notch signaling is dispensable for the generation of transient, embryonic blood cells (Bertrand et al, 2010b), it is absolutely required for the generation of HSCs across vertebrate phyla (Krebs et al, 2000; Kumano et al, 2003; Hadland et al, 2004; Burns et al, 2005; Robert-Moreno et al, 2005). Several Notch pathway mutants that fail to specify the DA have defects in HSCs (Krebs et al, 2000, 2004; Lawson et al, 2001; Duarte et al, 2004), suggesting that the DA is a morphogenetic prerequisite to HSCs. However, several studies have demonstrated that HSC specification can be rescued even in the context of impaired DA formation (Burns et al, 2005; Ren et al, 2010), confounding a clear necessity for a properly formed DA in subsequent HSC formation. Furthermore, mutants in the Notch ligand Jagged1 are deficient in HSCs but have normal arterial formation, suggesting that HSC formation has unique Notch requirements distinct from those required for arterial fate (Robert-Moreno et al, 2008). Previous work from our laboratory demonstrated that Wnt16 regulates the somitic expression of two Notch ligands, dlc and dld. While the somitic expression of dlc and dld was dispensable for DA specification, it was required for the formation of the sclerotome compartment of the somite and subsequent HSC specification (Clements et al, 2011). We reasoned that if Notch signaling performs differential functions in the somites versus the endothelium, then this specificity might be achieved through the discrete use of specific Notch receptors during these different processes.
In this report, we investigated which of the four Notch receptors are required for HSC specification, and when and where each of these requirements is needed. We have determined that Notch1a and Notch1b are autonomously required in the precursors of hemogenic endothelium, whereas Notch3 is dispensable in the endothelium and instead required in the somites to indirectly specify the HSC program. Furthermore, we demonstrate that this novel Notch requirement functions within the Wnt16/Notch signaling pathway that we previously showed is necessary to specify the sclerotome and HSCs.
Results
Notch3 is required for HSC specification
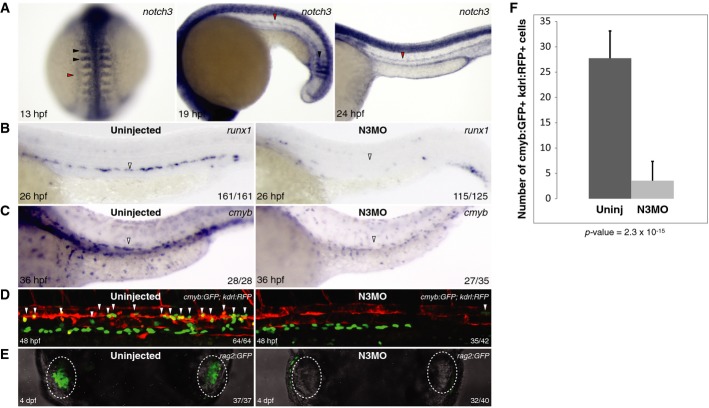
Previous studies have investigated the role of two of the four murine Notch receptors in HSC specification. Notch1 is required cell-autonomously for HSC specification, while Notch2 is dispensable (Kumano et al, 2003; Hadland et al, 2004). However, the role of Notch3 or Notch4 has not previously been explored. First, we characterized the expression pattern of notch3 in zebrafish embryos at 13, 19, and 24 h post-fertilization (hpf) and identified expression in HSC-related tissues. Notch3 was expressed widely throughout posterior lateral mesoderm (PLM) and somites at 13 hpf (Fig 1A). At 19 hpf, notch3 expression was reduced in mature somites but maintained in the 3–5 youngest somites and nascent endothelium. At 24 hpf, notch3 was largely restricted to the endothelium. To examine whether Notch3 was required for HSC specification, we knocked down its expression with a notch3 splice-blocking morpholino (Ma & Jiang, 2007). Whole-mount in situ hybridization (WISH) analysis of runx1 and cmyb, two early markers of hematopoietic stem and progenitor cells (HSPC) in the DA, were greatly reduced in notch3 morphants compared to uninjected control embryos (Fig 1B and C). Consistent with these WISH results, confocal imaging of cmyb:GFP; kdrl:RFP embryos indicated that cmyb+kdrl+ double-positive HSPCs (Bertrand et al, 2010a) that normally emerge from hemogenic endothelium were absent at 48 hpf in notch3 morphants (Fig 1D). Furthermore, rag2:GFP+ T lymphocytes, which are dependent upon upstream HSC precursors, were completely absent at 4 dpf in notch3 morphants (Fig 1E). Quantification of the number of cmyb+kdrl+ double-positive HSPCs demonstrated that differences between uninjected and notch3 morphants are statistically significant (Fig 1F). Together, these results indicate that Notch3 is required for the specification of HSCs.
Figure 1. Notch3 is required for HSPC specification.
A WISH of notch3 viewed dorsally at 13 hpf (left) and laterally in the trunk at 19 (middle) and 24 hpf (right). Black arrowheads denote somitic expression; red arrowheads denote PLM expression at 13 hpf and endothelial expression at 19 and 24 hpf.
B, C WISH of the HSPC marker runx1 at 26 hpf (B) and cmyb at 36 hpf (C) on uninjected and notch3 morphants. Arrowheads indicate HSPCs in the DA.
D, E Confocal fluorescence microscopy images of transgene reporter expression in cmyb:GFP; kdrl:RFP trunk region at 48 hpf (D) and rag2:GFP at 4 dpf (E) transgenics uninjected or with notch3 morpholino injected. Arrowheads in cmyb:GFP; kdrl:RFP embryos indicate double-positive HSPCs, and dotted lines in rag2:GFP embryos outline the thymic lobes where GFP+ lymphoid cells should reside.
F Enumeration of cmyb:GFP+; kdrl:RFP+ cells in the floor of the DA at 48 hpf. Bars represent mean ± SEM of double-positive cells for uninjected (n = 12) and notch3 morphants (n = 20). P = 2.3 × 10−15.
To investigate whether the reduction of HSPCs in notch3 morphants was caused by defects in vasculature formation, we examined several markers of endothelium and DA specification by WISH (Fig 2). Kdrl expression was reduced in intersomitic vessels and moderately upregulated in trunk endothelium, whereas aortic expression of efnb2a and dlc was unaffected in notch3 morphants, indicating that DA formation occurs normally and is not likely an explanation for reduced HSPC number (Fig 2C–E). As our previous work suggested that somite and sclerotome formation is linked to HSPC formation (Clements et al, 2011), we investigated the expression of the somite marker myod and the sclerotome-specific markers foxc1b and twist1b in notch3 morphants. The somites in notch3 morphants were specified but exhibited moderate upregulation of myod expression in ventral domains (Fig 2F). In contrast, sclerotomal expression of foxc1b and twist1b was greatly reduced in notch3 morphants compared to uninjected embryos (Fig 2G and H). We confirmed that the loss of HSPCs and sclerotome observed in notch3 morphants is specifically due to loss of function of the notch3 gene and not due to off-target effects from morpholino injection as evidenced by similar defects in notch3fh332 mutants (Quillien et al, 2014) (Supplementary Fig S1). Earlier examination of notch3 morphants and notch3fh332 mutants at 17 hpf showed reduction of foxc1b, twist1a, and twist1b expression, indicating that sclerotome specification is impaired (Supplementary Fig S2). These results indicate that Notch3 is essential for sclerotome and HSPC specification, but is largely dispensable for DA formation.
Figure 2. Notch3 is dispensable for DA, but required for sclerotome.
A Brightfield image of a 26 hpf zebrafish.
B Cartoon cross section of the embryonic trunk marking somites in light blue, sclerotome in purple, venous endothelium in yellow, and aortic endothelium in orange.
C–H WISH of uninjected and notch3 morphants at 26 hpf for the endothelial marker kdrl (C), dorsal aorta markers efnb2a (D) and dlc (E), the somite marker myod (F), and sclerotome markers foxc1b (G) and twist1b (H). Magnified panels are shown for somitic and sclerotomal markers in lower left corner. Arrowheads indicate tissue-specific gene expression.
We next wished to determine possible roles for the remaining Notch receptors Notch1a, Notch1b, and Notch2 in HSC specification. The expression pattern of notch1a and notch1b was similar to that of notch3, whereas notch2 was exclusively observed in the somites at these developmental stages (Supplementary Fig S3A–C). We utilized a splice-blocking morpholino for notch1a (Ma & Jiang, 2007) and designed splice-blocking morpholinos for notch1b and notch2 (Supplementary Fig S3D and E). Loss of function of notch1a and notch1b, but not notch2, resulted in loss of runx1 expression in the DA (Supplementary Fig S4A), consistent with the requirement for Notch1 but not Notch2 in murine HSC specification (Kumano et al, 2003). Notch1 mutant mice have vascular defects including a failure to specify DA (Krebs et al, 2000). In agreement with these findings, we observed variable loss of intersomitic vessels and defective aortic efnb2a and dlc expression in notch1a morphants, whereas notch1b morphants had only mild defects in dlc (Supplementary Fig S4B–D). Notch2 morphants displayed loss of intersomitic kdrl and dlc expression but maintained trunk endothelium and aortic markers. Notch1a, notch1b, and notch2 morphants showed normal myod+ somites and foxc1b+/twist1b+ sclerotome (despite affected somite boundaries in notch1a morphants), suggesting that formation of somites does not require these Notch receptors (Supplementary Fig S4E–G). We confirmed that the tissue-specific defects observed in notch1a and notch2 morphants are not due to off-target morpholino effects as evidenced by similar aortic defects in runx1 and efnb2a expression in notch1ab420 mutants (Gray et al, 2001) and loss of intersomitic vessel expression of dlc but normal aortic expression of runx1 in notch2el517 mutants (Supplementary Fig S5A–D). Although singular loss of notch1b or notch3 does not result in a loss of aortic efnb2a, a recent study demonstrated that both receptors are required synergistically for aorta specification (Quillien et al, 2014). Injection of notch1b morpholino #2 from Quillien et al resulted in the reduction of HSPCs but did not affect efnb2a, validating our observed notch1b morphant phenotype. Furthermore, coinjection of notch1b morpholino #2 and our notch3 morpholino resulted in the loss of aortic efnb2a, supporting the previous finding that loss of notch3 alone is tolerated by the aortic program in the presence of functional notch1b (Supplementary Fig S5E and F). Consistent with the endothelial-specific effects observed in notch1a and notch1b morphants, we found coexpression of notch1a/notch1b within and around efnb2a and runx1 expression domains in wild-type embryos (Supplementary Fig S6A–D). These data suggest that the role of the paralogous notch1a and notch1b genes in zebrafish is functionally conserved to that described for the murine Notch1 gene.
Notch3 is required non-cell-autonomously for HSC specification
By temporal induction of an NICD transgene, we previously showed that a Notch signal downstream of wnt16 in the somite was required for HSC specification during a brief permissive window beginning at 14 hpf. This finding helped us determine that this requirement was non-cell-autonomous, since the earliest Notch signaling events in HSC precursors were not detectable until 20–22 hpf (Clements et al, 2011). We therefore investigated whether similar induction of NICD could rescue HSCs in notch3 morphants. To perform these experiments, we utilized hsp70:gal4; UAS:NICD-myc double-transgenic animals to temporally control Notch signaling. Expression of NICD-Myc protein was detected by whole-mount immunofluorescence within an hour and up to 24 h after induction as previously reported (Clements et al, 2011). Induction of NICD in uninjected embryos did not affect the number of runx1+ HSPCs (unpublished observations). Early induction of NICD at 14 hpf rescued runx1+ HSPCs in notch3 morphants, while late induction at 20 hpf did not (Fig 3A–C), suggesting that Notch3 may mediate the early, non-cell-autonomous HSC specification requirement. In contrast, notch1a and notch1b morphants were robustly rescued by 14 hpf induction and, more importantly, by 20 hpf induction (Supplementary Fig S7A–D).
Figure 3. Specific temporal activation of Notch signaling is sufficient to rescue HSPCs in notch3 morphants.
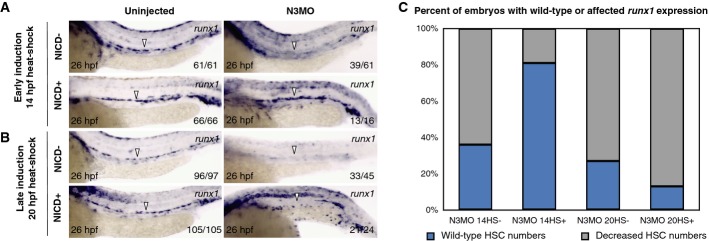
A, B WISH for runx1 in 26 hpf hsp70:gal4; UAS:NICD-myc uninjected or notch3 morphant transgenic embryos with heat-shock induction at 14 hpf (A) or 20 hpf (B), with or without enforced NICD expression. Arrowheads indicate the presence or absence of HSPCs at the midline.
C Quantitation of results recording percentages of embryos displaying normal or decreased numbers of runx1+ HSPCs at 26 hpf in notch3 morphants with heat-shock induction conditions.
The observation that notch3 morphants had different temporal requirements for NICD-mediated HSC rescue when compared to notch1a and notch1b morphants suggests that there are at least two temporal windows in which Notch signaling is important for HSC specification. We predicted that global Notch inhibition that spanned 14 hpf would phenocopy loss of notch3, while inhibition at 20 hpf would phenocopy loss of either notch1a or notch1b. To pharmacologically inhibit Notch signaling, we utilized the γ-secretase inhibitor DBZ, treating embryos during 6–15 hpf or 15–26 hpf developmental time windows and subsequently assaying for tissue-specific effects. We observed a reduction in runx1 intensity in the DA of embryos treated during either drug treatment window, indicating that Notch signaling was required for HSPC specification during both windows (Fig 4A). We observed sclerotome malformation when embryos were treated between 6 and 15 hpf that phenocopied notch3 morphants, but no effect when embryos were treated between 15 and 26 hpf (compare Figs 4B and 2E). In contrast, the DA transcripts efnb2a and dlc were severely reduced during the 15–26 hpf window resembling notch1a and notch1b morphants (compare Fig 4C and D to Supplementary Fig S4C and D), but unaffected by drug treatment during 6–15 hpf. Interestingly, the loss of efnb2a and dlc caused by DBZ treatment from 15 to 26 hpf was more dramatic than that observed in notch1a or notch1b morphants, suggesting that each may have non-redundant requirements or that remaining Notch receptors, likely Notch3 (Supplementary Fig S5F), may partially compensate for the loss of either Notch1a or Notch1b during DA specification. These results indicate that Notch signaling performs transient and non-redundant roles during somitogenesis compared to DA formation that are both essential for HSC production.
Figure 4. Notch signaling is required during two distinct time windows for specification of sclerotome and dorsal aorta.
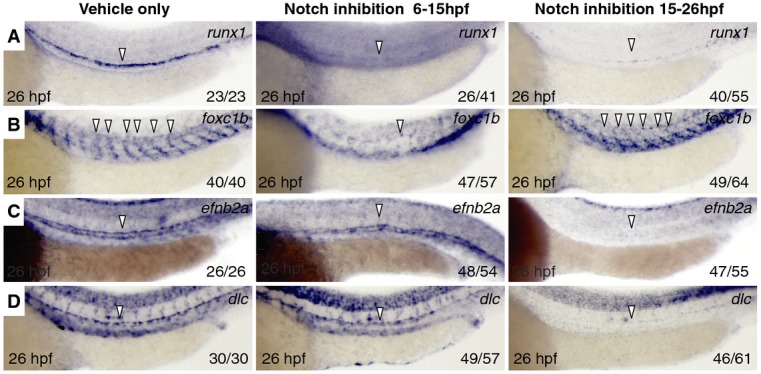
A–D WISH for runx1 (A), foxc1b (B), efnb2a (C), and dlc (D) in 26 hpf embryos treated with DMSO vehicle (left), 4 μM γ-secretase Notch inhibitor DBZ at 6–15 hpf (middle), or 15–26 hpf (right). Arrowheads indicate tissue-specific expression.
To determine whether spatially restricted expression of NICD was sufficient to rescue HSPCs in notch3 morphants, we utilized tissue-specific drivers of Gal4. To drive NICD within the HSC lineage, we utilized a kdrl:miniGAL4 transgenic line whereby expression is targeted to the vasculature, including hemogenic endothelial cells (Supplementary Fig S8A). Since notch3 is expressed in the sclerotome (Supplementary Fig S8B), and since notch3 morphants displayed defects in both sclerotome and HSPCs, we asked whether enforced somitic expression of NICD could rescue HSPCs in notch3 morphants. To perform these experiments, we utilized the phldb1:gal4-mCherry transgenic line (Distel et al, 2009), which drives robust expression specifically in the somite (Supplementary Fig S8C–G). Double-transgenic α-actin:GFP; phldb1:gal4-mCherry embryos showed high levels of mCherry in all GFP+ cells by fluorescence-activated cell sorting (FACS), indicating phldb1:gal4-mCherry is expressed widely in somitic tissues (Supplementary Fig S8G). Enforced expression of NICD in the somite rescued runx1+ HSPCs in notch3 morphants with significantly greater frequency than endothelial-driven NICD did in our analyses (Fig 5A–C). In contrast, HSPCs in notch1a and notch1b morphants were rescued with the vascular-specific kdrl:gal4 NICD driver but not with the somite-specific phldb1:gal4-mCherry driver in our analyses (Supplementary Fig S9A–D). These findings indicate that notch1a and notch1b are required for activation of Notch signaling within the endothelium, but not the somites, to specify HSCs. We next asked whether the induction of NICD sufficient to rescue HSCs in notch3 or notch1a morphants could also rescue the defects observed in the sclerotome and DA, respectively. NICD induction globally at 14 hpf or somitically using the phldb1:gal4-mCherry driver in notch3 morphants also restored expression of twist1b in the sclerotome (Supplementary Fig S10A and B). Similarly, global NICD induction at 20 hpf or in vascular cells using the kdrl:miniGAL4 driver restored expression of efnb2a in the DA (Supplementary Fig S10C and D). Together, these results demonstrate that Notch3 is required in the somite at 14 hpf to specify sclerotome and HSPCs and that Notch signaling is then needed again for HSPC fate via subsequent function of Notch1a and Notch1b in the vasculature at 20 hpf.
Figure 5. Specific spatial activation of Notch signaling is sufficient to rescue HSPCs in notch3 morphants.
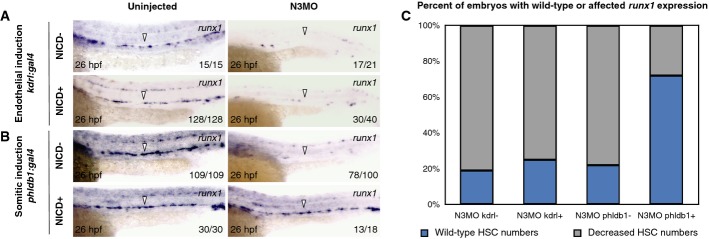
A, B WISH for runx1 in 26 hpf kdrl:gal4 (A) or phldb1:gal4-mcherry (B) crossed to UAS:NICD-myc transgenic embryos either uninjected or injected with notch3 morpholino, with or without enforced NICD expression. Arrowheads indicate the presence or absence of HSPCs at the midline.
C Quantitation of results recording percentages of embryos displaying normal or decreased numbers of runx1+ HSPCs at 26 hpf in notch3 morphants with tissue-specific induction conditions.
Notch3 is required downstream of somitic dlc, dld, and wnt16 for HSC specification
Because wnt16, dlc, dld (Clements et al, 2011), and notch3 are each important for sclerotome formation, and because loss of wnt16 or notch3 can be rescued only by early NICD induction, we investigated the functional relationship between dlc, dld, and notch3 in HSC specification. We tested whether there was synergy between dlc or dld and notch3 by combinatorial low-dose knockdown experiments and assessed the severity of affected phenotypes. Heterozygotes from the dlc mutant, beamter (Julich et al, 2005), low-dose (5 ng) dld morphants, or low-dose (5 ng) notch3 morphants each showed a partial loss of runx1, twist1b, and foxc1b expression compared to controls (Fig 6A–D). Combinatorial knockdown of dlc/notch3 or dld/notch3, however, had more severe effects on HSPCs and sclerotome than any of the single knockdown controls, suggesting that dlc and dld interact with notch3 (Fig 6E and F). To test whether notch3 was genetically upstream of dlc and dld expression in somites, we examined whether notch3 morphants had defects in somitic dlc or dld expression. We observed no significant reduction in dlc or dld in notch3 morphants compared to uninjected embryos, suggesting that notch3 is not required to induce dlc or dld expression (Supplementary Fig S11A and B). In addition, HSPC formation in notch3 morphants was not rescued by combined dlc/dld mRNA injection, a strategy that was able to rescue HSPCs following loss of wnt16 (Fig 7A–D) (Clements et al, 2011). Collectively, these findings suggest that Notch3 function lies downstream of DeltaC/DeltaD function. This hypothesis is supported by the finding that loss of notch3 inhibited the rescue of HSPCs by dlc/dld mRNA in wnt16 morphants (Fig 7E). These results suggest that Notch3 is necessary to receive signals from DeltaC and/or DeltaD in the sclerotome to relay further signals to the precursors of HSCs that are required for their proper specification (Supplementary Fig S12).
Figure 6. Notch3 cooperates synergistically with dlc and dld to specify HSPCs.
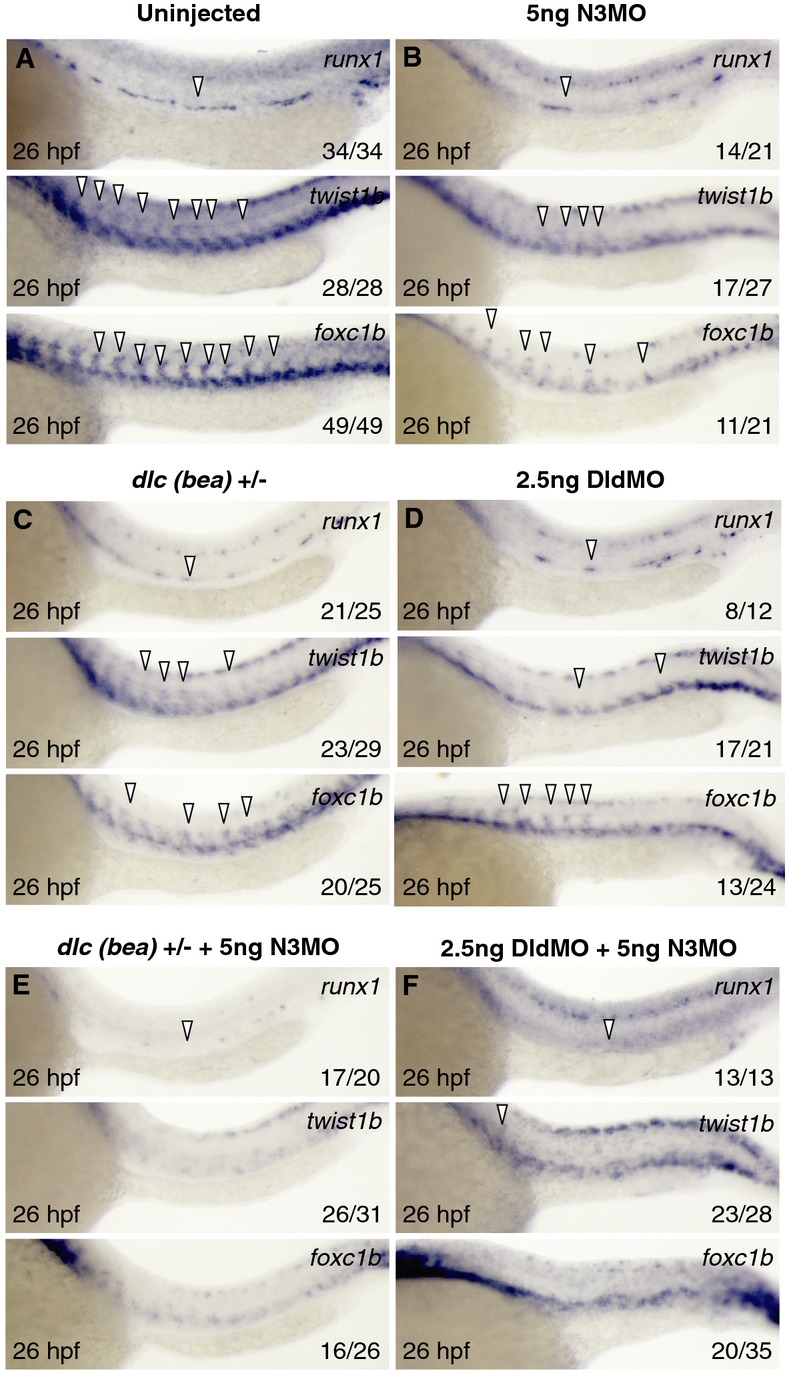
A–F WISH for runx1, twist1b, and foxc1b at 26 hpf in uninjected (A), low-dose knockdown of notch3 (B), heterozygotes for dlc mutant bea (C), low-dose knockdown of dld (D), bea heterozygotes with low-dose knockdown of notch3 (E), and combinatorial low-dose knockdown of dld and notch3 (F). Arrowheads indicate tissue-specific expression.
Figure 7. Wnt16, dlc/dld, and notch3 function in a linear pathway to specify HSPCs.
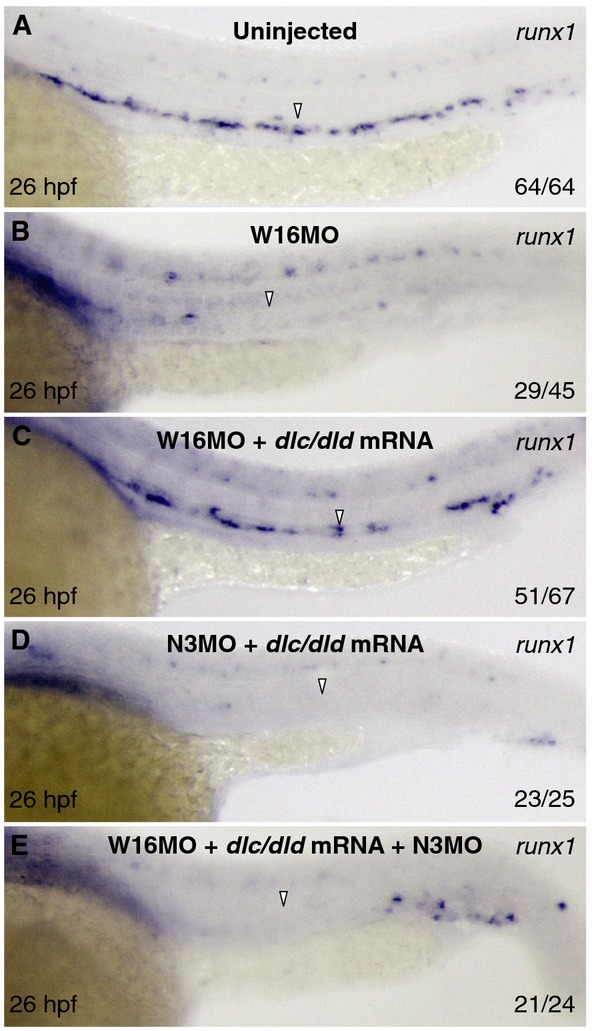
A–E Expression of runx1 at 26 hpf in uninjected (A) or injected with wnt16MO (B), wnt16MO with 50 pg dlc/dld mRNA (C), notch3MO with 50 pg dlc/dld mRNA (D), and wnt16MO/notch3MO with 50 pg dlc/dld mRNA (E). Arrowheads indicate the presence or absence of HSPCs.
Discussion
Previous studies have demonstrated that Notch signaling is required for HSC specification (Kumano et al, 2003; Hadland et al, 2004; Burns et al, 2005; Robert-Moreno et al, 2005, 2008; Yoon et al, 2008; Bertrand et al, 2010b). Of the four murine Notch receptors, Notch1 is required cell-autonomously, while Notch2 is dispensable (Kumano et al, 2003; Hadland et al, 2004); the roles of the remaining receptors have not been addressed. Here, we demonstrate that Notch3 is required to activate Notch signaling in the somite by 14 hpf and that this activation is required for HSC specification.
Despite the fact that Notch receptors are widely conserved across vertebrate species, there are evolutionary differences in the Notch receptor genes of mammals and zebrafish. One of the most notable is the presence of two Notch1-related homologues, notch1a and notch1b in zebrafish. Notch1 has high amino acid identity to both Notch1a and Notch1b, and phylogenetic reconstruction analyses have suggested that notch1a and notch1b arose from a gene duplication event that occurred early during teleost evolution (Westin & Lardelli, 1997; Kortschak et al, 2001). Despite Notch1 and Notch2 genes sharing high amino acid similarity, Notch2 is dispensable for HSC specification in the mouse (Kumano et al, 2003). We show that Notch2 is also dispensable in zebrafish for HSC generation, indicating that the individual roles of Notch receptors may be conserved across vertebrates. Supporting this hypothesis, our experiments show that the combinatorial actions of Notch1a and Notch1b in zebrafish functionally phenocopy the activity of Notch1 in other vertebrates.
Several lines of evidence indicate that Notch1a and Notch1b have distinct but overlapping roles in hemogenic endothelium. We show that notch1a and notch1b are both expressed in endothelium by FISH, but in other tissues, each receptor’s expression pattern is more distinct but additively resemble the wider expression pattern of murine Notch1 (Westin & Lardelli, 1997). We show that notch1a morphants have reduced HSCs and reduced aortic efnb2a and dlc expression, while notch1b morphants had reduced numbers of HSCs and reduced aortic dlc levels. Additively, these phenotypes resemble Notch1 mutant mice (Krebs et al, 2004), which suggests that the zebrafish Notch1b receptor has evolved to be more HSC specific. We also demonstrate that Notch1a and Notch1b have a functional role in endothelial cells; when NICD was specifically expressed in kdrl+ endothelium, runx1+ HSPC formation was rescued in both notch1a and notch1b morphants. In addition, aortic efnb2a expression was restored in notch1a morphants. The function of Notch1a and Notch1b receptors is required beginning at approximately 20 hpf, the time at which HSC precursors first experience Notch signaling (Clements et al, 2011). This timing is consistent with our findings that global pharmacological inhibition of Notch between 15 and 26 hpf specifically blocked DA and HSPC specification. Our data demonstrate that like Notch1, Notch1a and Notch1b both perform a cell-autonomous role in HSC specification.
In contrast to Notch1, the role of Notch3 in HSC specification is poorly understood. We investigated whether Notch3 acts cell-autonomously or non-cell-autonomously to specify HSPCs by specific temporal and spatial induction of NICD in notch3 morphants. In contrast to the rescue of HSCs by induction of NICD at 20 hpf in notch1a and notch1b morphants, notch3 morphants could only be rescued by earlier NICD induction at 14 hpf. This suggests that Notch3 is required during a brief permissive window before HSC precursors experience Notch signaling directly. Confirming the requirement for Notch signaling during this permissive window, pharmacological inhibition of Notch signaling during 6–15 hpf showed a strong reduction in HSPC numbers. Our results using tissue-specific drivers of NICD in notch3 morphants showed that there were also precise spatial requirements for HSC rescue. Enforced expression of NICD in the somites was sufficient to rescue HSCs in notch3 morphants, whereas endothelial-specific expression was not, despite the fact that notch3 is expressed in the DA (Lawson et al, 2002). These results indicate that Notch3 activation is required in the somites, but not in endothelium, to specify HSCs. The temporal and tissue-specific rescue of HSPCs by NICD in notch3 morphants was also accompanied by the rescue of sclerotome-specific transcripts. These data suggest that the molecular requirements of sclerotome formation are closely linked to HSC specification. Collectively, our data demonstrate that Notch3 is required non-cell-autonomously in the somites to specify HSPCs.
We established that there is a genetic relationship between dlc/dld and notch3 during HSPC specification by combinatorial low-dose knockdown of these genes. Furthermore, the partial knockdown of dlc or dld was synergistic with a low-dose knockdown of notch3, indicating that each of these genes are involved in a linear genetic pathway. In this pathway, wnt16 is genetically upstream of somitic dlc/dld but is dispensible for somitic notch3 (Clements et al, 2011). However, unlike wnt16, notch3 is not genetically upstream of dlc/dld as evidenced by maintenance of dlc/dld in the somites of notch3 morphants. This is bolstered by the finding that reduced numbers of HSCs in notch3 morphants could not be rescued with coinjection of dlc/dld mRNA. Additionally, HSPC rescue in wnt16 morphants coinjected with dlc/dld mRNA was inhibited by the coinjection of the notch3 morpholino. The simplest explanation is that Dlc and/or Dld directly activates Notch3. Why both ligands might be required to activate Notch3 is unclear. The roles of mammalian homologues Dll3 and Dll1 appear to be conserved with DeltaC and DeltaD, respectively, as evidenced by their conserved expression pattern in somitic tissues and loss of sclerotome and/or vertebral malformations in Dll3 and Dll1 loss of function animals (Hrabe de Angelis et al, 1997; Takahashi et al, 2003; Chapman et al, 2010), though a role in HSC specification has not been described. No studies to our knowledge have demonstrated that Notch3 is an obligate receptor for Dll3 or Dll1. On the contrary, mammalian cell culture experiments have demonstrated that Notch1 and Notch3 can bind a range of Delta and Jagged ligands, suggesting that binding between receptors and ligands is promiscuous (Shimizu et al, 2000). Direct binding has been reported between Dll3 and Notch1; however, this interaction is inhibitory and occurs in cis, suggesting that one of the functions of Dll3 is to suppress Notch1 signaling cell-autonomously (Ladi et al, 2005; Chapman et al, 2010). Intriguingly, Dll3 and Dll1 display non-redundant and even counteracting functions in somitogenesis (Takahashi et al, 2003; Ladi et al, 2005; Geffers et al, 2007). Studies in zebrafish may offer an explanation, since both DeltaC and DeltaD within the presomitic mesoderm (PSM) can form heterodimers. Both ligands were observed to be endocytosed together from the plasma membrane, suggesting that both ligands may be required together to effectively activate Notch signaling (Wright et al, 2011). This hypothesis could explain our previous results demonstrating that both dlc and dld are combinatorially required to restore HSC formation in wnt16 morphants (Clements et al, 2011). While the lack of a specific Notch3 antibody in zebrafish precludes testing whether or not Notch3 directly interacts with Dlc/Dld by biochemical or histological approaches, our results demonstrate that wnt16-induced dlc/dld requires the presence of notch3 to promote HSC emergence.
Notch3 may be required for a specific morphogenetic process and/or activation of another signaling cascade required by HSCs. We have shown that wnt16, dlc, dld, and notch3 are all required for sclerotome and HSPC formation, but dispensable for DA specification. Notch3 function may be required to specify the sclerotome, which in turn is required to specify HSCs through provision of a relay signal to neighboring PLM cells. Another potential mechanism to explain the link between sclerotome and HSC specification is that sclerotome may give rise to vascular smooth muscle precursors that support the endothelium, as it does in chick and mouse (Pouget et al, 2008; Wasteson et al, 2008). This is an attractive hypothesis, as Notch signaling is required for the proper emigration of vascular smooth muscle precursors from the somite to the dorsal aorta in the chick embryo (Sato et al, 2008). Recent studies performed in embryonic stem cells have confirmed that VSMCs do not directly give rise to hemogenic endothelium, indicating that if VSMCs have a role in HSC specification, it is indirect (Stefanska et al, 2014). It is currently unknown if the sclerotome is specifically required for HSC specification, but previous studies have established that somites are, through their production of VEGF, required for HSC formation (Ciau-Uitz et al, 2010; Leung et al, 2013). Our data elucidate an additional molecular pathway in which the somites are essential for the establishment of HSC fate.
Our study elucidates a previously unappreciated role for notch3 in the somites that is required for HSC specification, and that the Notch1 orthologues notch1a and notch1b are both required cell-autonomously in hemogenic endothelium for HSC formation. These data should prove important for future studies focused on the identification of unique targets downstream of each required Notch receptor essential for HSC specification.
Materials and Methods
Zebrafish husbandry
Zebrafish strains AB*, Tg(UAS:myc-Notch1a-intra)kca3 (Scheer & Campos-Ortega, 1999), Tg(hsp70 l:gal4)1.5kca4 (Scheer & Campos-Ortega, 1999), Tg(actc1b:GFP)zf13 (Higashijima et al, 1997), Tg(-80.0myf5:EGFP)zf37 (Chen et al, 2007), Tg(rag2:EGFP)zdf8 (Langenau et al, 2003), Tg(phldb1:gal4-mCherry) (Distel et al, 2009), Tg(kdrl:EGFP)la116 (Choi et al, 2007), Tg(cmyb:EGFP)zf169 (North et al, 2007), dlctit446/tit446 (from Tübingen 2000 screen), notch3fh332 (Quillien et al, 2014), and notch1ab420 (Gray et al, 2001) were maintained, injected, and staged as described (Westerfield, 2004) and in accordance with IACUC guidelines. Tg(kdrl:miniGAL4) was generated by cloning a 6-kb genomic fragment immediately upstream of the transcription start site from a plasmid carrying kdrl:R-CFP (Cross et al, 2003) and inserted into pCR8 (Invitrogen). The resulting plasmid was recombined into a Tol2 transgenesis vector pColdHeart-Gtwy-miniGAL4 (Campbell et al, 2007) and coinjected with Tol2 mRNA into 1-cell-stage embryos. A stable transgenic line with a single insertion was established. The notch2el517 mutant was generated using TALENS targeting exon 4 of the notch2 gene using the following target sequences: left TALEN: 5′-TTGTGTGAACACCATAGGCT-3′; right TALEN: 5′-TCCGGTGAAGCCAGGTTGGC-3′. TALEN RNAs were synthesized using the mMessage mMachine T7 Ultra kit (Ambion) and injected into 1-cell embryos at 100 ng/μl. Founders were identified among the injected animals by PCR followed by digestion with ClaI. The following primers were used for genotyping: notch2-F: 5′-GAGCAAGAGGACGCATGTCT-3′; notch2-R: 5′-GCTGCGGTAAAATCCCATTA-3′. Stable mutant alleles were isolated in the F1 generation. The notch2el517 allele contains a 104-bp deletion that causes a frameshift and premature stop codon. Heat shocks were performed at the times indicated for 45 min at 37°C as previously described (Burns et al, 2005).
Microinjection of morpholinos, RT–PCR, and mRNA
The following morpholino antisense oligonucleotides were synthesized by Gene Tools, LLC and suspended as 25 mg/ml stocks in DEPC ddH2O and diluted to injection strengths: 5 ng wnt16-MO, 5 ng dld-MO2 (Clements et al, 2011), 10 ng notch1a-sp MO1, 10 ng notch3-sp MO (Ma & Jiang, 2007), 10 ng notch1bMO GTCGAGAATCTTATCACTTACTTGC, 10 ng notch2MO TTCGAATGTGAAAGTCTTACCTGCA, 2.5 ng notch1bMO2 (Quillien et al, 2014). For RT–PCR, RNA was isolated from groups of 30 uninjected or morpholino-injected embryos at 26 hpf, and cDNA was prepared as previously described (Clements et al, 2009). PCR on cDNA was amplified with notch1b-sp-F TGCATCTTTTCTTCGTGAAAC, notch1b-sp-R GGATTGGAAGCAAGGGTTG, notch2-sp-F CAAAATATGGGCCAATTACCC, notch2-sp-R GACAGACATGCGTCCTCTTGC, b-actin-sp-F AAGATCAAGATCATTGCC, and b-actin-sp-R TTGTCGTTTGAAGTTTCTC with Taq polymerase (Invitrogen, Philadelphia, PA) as previously described (Clements et al, 2009). Full-length dlc and dld mRNA was synthesized as described (Clements et al, 2011). Injections were performed as described previously (Clements et al, 2009). Genotyping of notch3fh332 after phenotypic analysis was performed as described previously (Quillien et al, 2014). Genotyping by PCR of notch2el517 animals was performed with notch2-F GAGCAAGAGGACGCATGTCT and notch2-R GCTGCGGTAAAATCCCATTA.
WISH, immunofluorescence, and microscopy
Single enzymatic and double-fluorescence whole-mount in situs were performed as previously described (Clements et al, 2011). Antisense RNA probes for the following genes were prepared using probes containing digoxigenin or fluorescein-labeled UTP: runx1, kdrl, efnb2a, dlc, myod, foxc1b, twist1a, twist1b, notch1a, notch1b, and notch3 as previously described (Clements et al, 2011). Whole-mount immunofluorescence was performed using anti-Myc monoclonal 9E10 antibodies at 1:200 (Covance) and Dylight488 AffiniPure donkey anti-mouse IgG secondary antibodies (Jackson Immunoresearch Laboratories) at 1:100 as described previously (Clements et al, 2011). Fluorescence images of transgenic embryos and embryo samples were imaged using confocal microscopy (Leica, SP5) and processed using Volocity software (Perkin-Elmer) as previously described (Bertrand et al, 2010a).
Pharmacological inhibition of Notch signaling
Dibenzazepine (DBZ) γ-secretase inhibitor (Calbiochem) was dissolved in DMSO at a concentration of 2 mM. Zebrafish embryos were incubated in 3 ml of 4 μM DBZ solution in the dark from 6 to 15 or 15 to 26 hpf followed by fixation with 4% PFA.
Fluorescence-activated cell sorting
Kdrl:GFP; phldb1:gal4-mCherry, and α-actin:GFP; phldb1:gal4-mCherry embryos were collected at 17 hpf and processed for FACS as previously described (Bertrand et al, 2007).
Acknowledgments
The authors wish to thank D. Langenau for providing somite-specific transgenic lines; B. Weijts, Y. Lee, C. Hochmuth for providing critical evaluation of the manuscript; K. Ong for technical assistance; and NIH CMG T32 Training Grant (AK), Innovative Science Award #12PILT12860010 from the American Heart Association and R01-DK074482 from the National Institutes of Health (DT) for support.
Author contributions
ADK, WKC, and DT designed all experiments. Whole-mount in situ hybridization, whole-mount immunofluorescence, and double-fluorescence in situ hybridization experiments were performed by ADK and CHM. Cell sorting experiments were performed by DLS and ADK. All other experiments were performed by ADK. DP and CM provided the Tg(kdrl:miniGAL4) reporter line. MD provided the Tg(phldb1:gal4-mCherry) reporter line. LAM and JGC provided the notch2el517 mutant. The manuscript was written by AK and edited by DLS, WK, and DT with critical input as described in Acknowledgements.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010a;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Cisson JL, Stachura DL, Traver D. Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood. 2010b;115:2777–2783. doi: 10.1182/blood-2009-09-244590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol. 2009;29:5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- de Bruijn MF, Speck NA, Peeters MC, Dzierzak E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 2000;19:2465–2474. doi: 10.1093/emboj/19.11.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Stringham SA, Timm A, Xiao T, Law MY, Baier H, Nonet ML, Chien CB. Slit1a inhibits retinal ganglion cell arborization and synaptogenesis via Robo2-dependent and -independent pathways. Neuron. 2007;55:231–245. doi: 10.1016/j.neuron.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Chapman G, Sparrow DB, Kremmer E, Dunwoodie SL. Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum Mol Genet. 2010;20:905–916. doi: 10.1093/hmg/ddq529. [DOI] [PubMed] [Google Scholar]
- Chen YH, Wang YH, Chang MY, Lin CY, Weng CW, Westerfield M, Tsai HJ. Multiple upstream modules regulate zebrafish myf5 expression. BMC Dev Biol. 2007;7:1. doi: 10.1186/1471-213X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN. FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol. 2007;304:735–744. doi: 10.1016/j.ydbio.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciau-Uitz A, Pinheiro P, Gupta R, Enver T, Patient R. Tel1/ETV6 specifies blood stem cells through the agency of VEGF signaling. Dev Cell. 2010;18:569–578. doi: 10.1016/j.devcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Clements WK, Ong KG, Traver D. Zebrafish wnt3 is expressed in developing neural tissue. Dev Dyn. 2009;238:1788–1795. doi: 10.1002/dvdy.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, Traver D. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature. 2011;474:220–224. doi: 10.1038/nature10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross LM, Cook MA, Lin S, Chen JN, Rubinstein AL. Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arterioscler Thromb Vasc Biol. 2003;23:911–912. doi: 10.1161/01.ATV.0000068685.72914.7E. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Zon LI. The “definitive” (and “primitive”) guide to zebrafish hematopoiesis. Oncogene. 2004;23:7233–7246. doi: 10.1038/sj.onc.1207943. [DOI] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Koster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci USA. 2009;106:13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A, Hirashima M, Benedito R, Trindade A, Diniz P, Bekman E, Costa L, Henrique D, Rossant J. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18:2474–2478. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffers I, Serth K, Chapman G, Jaekel R, Schuster-Gossler K, Cordes R, Sparrow DB, Kremmer E, Dunwoodie SL, Klein T, Gossler A. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J Cell Biol. 2007;178:465–476. doi: 10.1083/jcb.200702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Moens CB, Amacher SL, Eisen JS, Beattie CE. Zebrafish deadly seven functions in neurogenesis. Dev Biol. 2001;237:306–323. doi: 10.1006/dbio.2001.0381. [DOI] [PubMed] [Google Scholar]
- Hadland BK, Huppert SS, Kanungo J, Xue Y, Jiang R, Gridley T, Conlon RA, Cheng AM, Kopan R, Longmore GD. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood. 2004;104:3097–3105. doi: 10.1182/blood-2004-03-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M, McIntyre J, II, Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Julich D, Hwee Lim C, Round J, Nicolaije C, Schroeder J, Davies A, Geisler R, Lewis J, Jiang YJ, Holley SA. beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev Biol. 2005;286:391–404. doi: 10.1016/j.ydbio.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortschak RD, Tamme R, Lardelli M. Evolutionary analysis of vertebrate Notch genes. Dev Genes Evol. 2001;211:350–354. doi: 10.1007/s004270100159. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, Smith GH, Stark KL, Gridley T. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- Krebs LT, Shutter JR, Tanigaki K, Honjo T, Stark KL, Gridley T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K, Chiba S, Kunisato A, Sata M, Saito T, Nakagami-Yamaguchi E, Yamaguchi T, Masuda S, Shimizu K, Takahashi T, Ogawa S, Hamada Y, Hirai H. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170:983–992. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, Look AT. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, Weinstein BM. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–136. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, Mordelet E, Zapata A, Shinomiya H, Herbomel P. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111:132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- Leung A, Ciau-Uitz A, Pinheiro P, Monteiro R, Zuo J, Vyas P, Patient R, Porcher C. Uncoupling VEGFA functions in arteriogenesis and hematopoietic stem cell specification. Dev Cell. 2013;24:144–158. doi: 10.1016/j.devcel.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Jiang YJ. Jagged2a-notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS Genet. 2007;3:e18. doi: 10.1371/journal.pgen.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, Daley GQ, Orkin SH, Zon LI. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Pouget C, Pottin K, Jaffredo T. Sclerotomal origin of vascular smooth muscle cells and pericytes in the embryo. Dev Biol. 2008;315:437–447. doi: 10.1016/j.ydbio.2007.12.045. [DOI] [PubMed] [Google Scholar]
- Quillien A, Moore JC, Shin M, Siekmann AF, Smith T, Pan L, Moens CB, Parsons MJ, Lawson ND. Distinct Notch signaling outputs pattern the developing arterial system. Development. 2014;141:1544–1552. doi: 10.1242/dev.099986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Ren X, Gomez GA, Zhang B, Lin S. Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood. 2010;115:5338–5346. doi: 10.1182/blood-2009-09-244640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Moreno A, Espinosa L, de la Pompa JL, Bigas A. RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 2005;132:1117–1126. doi: 10.1242/dev.01660. [DOI] [PubMed] [Google Scholar]
- Robert-Moreno A, Guiu J, Ruiz-Herguido C, Lopez ME, Ingles-Esteve J, Riera L, Tipping A, Enver T, Dzierzak E, Gridley T, Espinosa L, Bigas A. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 2008;27:1886–1895. doi: 10.1038/emboj.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Watanabe T, Saito D, Takahashi T, Yoshida S, Kohyama J, Ohata E, Okano H, Takahashi Y. Notch mediates the segmental specification of angioblasts in somites and their directed migration toward the dorsal aorta in avian embryos. Dev Cell. 2008;14:890–901. doi: 10.1016/j.devcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Saito T, Kumano K, Hirai H. Physical interaction of Delta1, Jagged1, and Jagged2 with Notch1 and Notch3 receptors. Biochem Biophys Res Commun. 2000;276:385–389. doi: 10.1006/bbrc.2000.3469. [DOI] [PubMed] [Google Scholar]
- Stefanska M, Costa G, Lie ALM, Kouskoff V, Lacaud G. Smooth muscle cells largely develop independently of functional hemogenic endothelium. Stem Cell Res. 2014;12:222–232. doi: 10.1016/j.scr.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Inoue T, Gossler A, Saga Y. Feedback loops comprising Dll1, Dll3 and Mesp2, and differential involvement of Psen1 are essential for rostrocaudal patterning of somites. Development. 2003;130:4259–4268. doi: 10.1242/dev.00629. [DOI] [PubMed] [Google Scholar]
- Tober J, Koniski A, McGrath KE, Vemishetti R, Emerson R, de Mesy-Bentley KK, Waugh R, Palis J. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood. 2007;109:1433–1441. doi: 10.1182/blood-2006-06-031898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteson P, Johansson BR, Jukkola T, Breuer S, Akyurek LM, Partanen J, Lindahl P. Developmental origin of smooth muscle cells in the descending aorta in mice. Development. 2008;135:1823–1832. doi: 10.1242/dev.020958. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio Rerio) Eugene, OR: University Oren Press; 2004. [Google Scholar]
- Westin J, Lardelli M. Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Dev Genes Evol. 1997;207:51–63. doi: 10.1007/s004270050091. [DOI] [PubMed] [Google Scholar]
- Wright GJ, Giudicelli F, Soza-Ried C, Hanisch A, Ariza-McNaughton L, Lewis J. DeltaC and DeltaD interact as Notch ligands in the zebrafish segmentation clock. Development. 2011;138:2947–2956. doi: 10.1242/dev.066654. [DOI] [PubMed] [Google Scholar]
- Yoon MJ, Koo BK, Song R, Jeong HW, Shin J, Kim YW, Kong YY, Suh PG. Mind bomb-1 is essential for intraembryonic hematopoiesis in the aortic endothelium and the subaortic patches. Mol Cell Biol. 2008;28:4794–4804. doi: 10.1128/MCB.00436-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, Tallquist MD, Iruela-Arispe ML. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.